Abstract
Induction of antibodies that neutralize a broad range of human immunodeficiency virus type 1 (HIV-1) isolates is a major goal of vaccine development. To study natural examples of broad neutralization, we analyzed sera from 103 HIV-1-infected subjects. Among progressor patients, 20% of sera neutralized more than 75% of a panel of 20 diverse viral isolates. Little activity was observed in sera from long-term nonprogressors (elite controllers). Breadth of neutralization was correlated with viral load, but not with CD4 count, history of past antiretroviral use, age, gender, race/ethnicity, or route of exposure. Clustering analysis of sera by a novel method identified a statistically robust subgrouping of sera that demonstrated broad and potent neutralization activity.
Eliciting neutralizing antibodies (NAbs) against human immunodeficiency virus type 1 (HIV-1) is likely to be crucial for an optimally effective vaccine. To date, the antibodies elicited by vaccines have had weak activity against a limited spectrum of HIV-1 strains (10, 22, 33). However, many HIV-infected patients make NAbs, and a small fraction make extremely potent NAbs with broad cross-reactivity (3, 4, 9, 26, 29, 32). Understanding how a broadly reactive NAb response develops in some HIV-1-infected patients, and what viral epitopes are targeted, may provide important clues for vaccine design (18). The prevalence and clinical parameters associated with broadly reactive NAbs in serum have been the subject of much recent interest (11, 28, 29). We therefore examined the potency and breadth of neutralization in a large cohort of patients, compared breadth with clinical and demographic variables, and used clustering analysis to discern patterns in serum reactivity to diverse isolates.
In a previous study (9), we screened HIV-infected patient sera for neutralizing activity against a panel of five viral isolates, using a TZM-bl Env pseudovirus neutralization assay. We also established a more robust 20-viral-isolate panel that included 10 clade B, 5 clade A, and 5 clade C Env pseudoviruses (9, 16-18). In order to evaluate the prevalence of neutralization breadth in a more quantitative manner, we studied 103 patient sera against all 20 viruses. All patients participated in National Institutes of Health clinical protocols, were infected for at least 1 year, and were antiretroviral (ARV) naïve or had been off ARVs for at least 3 months at the time of sampling. All patients were presumed to be infected with clade B virus based on locations of current and former residences. Eighty-one of the patients were included in the previously published analysis (9). Twenty-five patients were long-term nonprogressors (LTNP; also called elite controllers) from the cohort described in references 23 and 24, who typically maintain a viral load (VL) of <50 RNA copies/ml and a stable CD4+ T-cell count without ARV therapy; this group had a median CD4+ T-cell count of 850 cells/μl and a median time since HIV diagnosis of 13.5 years. The other 78 patients had a median viral load of 4,931 RNA copies/ml, a median CD4+ T-cell count of 534 cells/μl, and a median of 12.5 years since diagnosis. This patient group includes both typical progressors and patients without CD4+ T-cell decline (referred to in prior reports as slow progressors). In our previous analysis (9), we found no differences in neutralization breadth between typical and slow progressors; therefore, for the purposes of this report, both patient groups are analyzed together and collectively referred to as progressors. Dates of diagnosis but not of seroconversion were available. We calculated both the 50% and 80% inhibitory doses (ID50 and ID80, respectively) for each isolate using the TZM-bl assay as described in reference 31.
Among progressor patients with readily detectible viremia, wide ranges of serum neutralization potency and breadth were observed (Fig. 1A). Using a cutoff ID50 of ≥100, we found that these sera neutralized a median of 10.5 (interquartile range [IQR], 5 to 14) out of 20 isolates. A total of 20% of these sera were broadly reactive, neutralizing at least 15 of 20 isolates on our panel. However, 50% of the sera neutralized 10 or fewer isolates, with several sera having very low activity despite years of untreated viremia. In contrast, sera from LTNP, with <50 copies of HIV RNA/ml plasma, had little neutralization activity, with a median of only 1 of 20 isolates neutralized with an ID50 of ≥100 (IQR, 0 to 2.5) (Fig. 1B). The range of neutralization activity in this group was similar to the lowest end of values for progressor patients. This observation concurs with previous data from our laboratory and others which show that, compared to patients with higher levels of viremia, LTNP make weak NAb responses, perhaps due to reduced antigenic stimulation of B cells (2, 9, 15, 20, 27). To ensure that we were measuring serum-mediated viral neutralization, we also incorporated ID80 values into the analysis. Using a cutoff of both an ID50 of ≥100 and ID80 of ≥15, sera of progressor patients neutralized a median of 9 (IQR, 2.8 to 11) isolates, with 15% of sera neutralizing >75% of viruses. In contrast, among LTNP the median was 0 (IQR, 0 to 1).
FIG. 1.
Neutralization of 20 isolates by patient sera. (A and B) ID50 value against each of 20 isolates in the TZM-bl assay is plotted for each patient. The red line indicates an ID50 of 100. Input dilution was 1:10; if no neutralization was observed, the ID50 is plotted as 5. (A) Progressors. (B) LTNP. (C) Viral load (RNA copies/ml plasma) versus geometric mean titer for each patient's serum. Red circles, LTNP; black circles, progressor patients.
Clinical and demographic data for all patients were compared to neutralization breadth. Two parameters were used to quantify breadth for each serum: the geometric mean ID50 against the 20 isolates and the number neutralized with both an ID50 of ≥100 and an ID80 of ≥15. The associations between breadth and clinical covariates were tested using nonparametric methods (Spearman's rho, Wilcoxon rank-sum, or Kruskal-Wallis test). A Bonferroni correction was used as follows to adjust for the multiple comparisons: reported P values are multiplied by 10 from the original P values and are considered significant if they are below 0.05. Viral load had a modest positive association with breadth, as measured both by geometric mean ID50 (P < 0.001; r = 0.68) (Fig. 1C) and by the number of isolates neutralized (P < 0.001; r = 0.63). When the analysis was restricted to progressor patients, this relationship still held (P = 0.008 and r = 0.37 for geometric mean ID50; P = 0.050 and r = 0.31 for number neutralized). CD4+ T-cell count showed a modest negative correlation with breadth by both measures (P = 0.025 and r = −0.29 for geometric mean ID50; P = 0.031 and r = −0.29 for number neutralized), but this relationship was not significant when LTNP were excluded from the analysis. Years since diagnosis, HLA class II alleles, risk group, history of using ARVs, race, ethnicity, gender, and age were not associated with breadth by either measure. Thus, the only strong predictor of breadth found in this cohort is viral load.
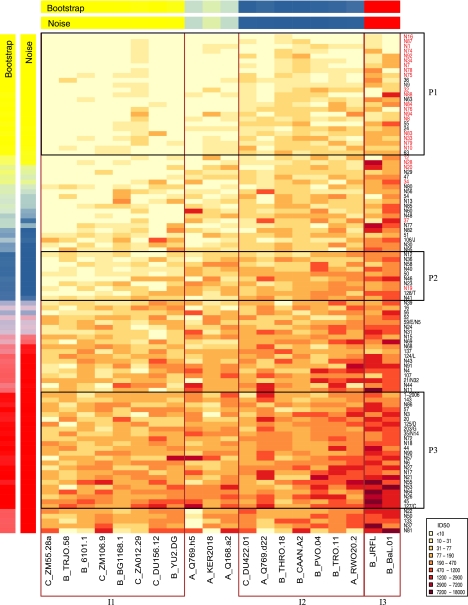
To find patterns of neutralization reactivity in this data set, and determine potential common specificities of neutralization, we performed a clustering analysis based on the ID50 values for the 103 sera and 20 isolates. This analysis is shown as a heat map in Fig. 2, in which darker red colors indicate higher ID50 values, and the data are arranged to highlight patterns of similar neutralization profiles. The data was clustered using k means, a procedure for clustering into a fixed number, k, of groups. In this procedure, the Euclidean distance between the vector of log10 ID50 values (i.e., the set of neutralization values in one row or column) is calculated relative to candidate group location vectors, and each serum is assigned to the cluster with the closest group location. New group means are formed on the basis of these group assignments, and this procedure is iterated until group identities do not change. This procedure was iterated 20,000 times with random initial mean vectors to find the most compact clusters. The same strategy was used to organize the isolates into serological susceptibility patterns. We added two statistical measures to assess how robust the clusters were and to determine how many clusters (k) were statistically supported in the data. To assess the impact of limited sampling, bootstrap analysis was performed by sampling the rows (or columns) with replacement 10,000 times and obtaining the fraction of times the serum (or isolate) belonged to the same cluster. The resulting degree of consensus is shown in the row or column labeled “Bootstrap” in Fig. 2. To assess the impact of assay-to-assay variability, experimental “noise” was modeled from experimental data. Replicate ID50 values were log10 transformed, then normalized by subtracting the per-isolate or per-serum geometric means, yielding a normal distribution with a standard deviation of 0.166. Values were sampled from the distribution and added to the real data, then the k means process was repeated 1,000 times. Stability of categories for these data is shown in the row and column labeled “Noise” in Fig. 2. Stability of categories for these data is shown in the row and column labeled “Noise” in Fig. 2. Three clusters (k = 3) was the maximum number such that each cluster was comprised only of serum (or isolates) that were assigned to that cluster at a consistency of more than 90% by both measures of stability. Thus, each cluster shown in the boxes in Fig. 2 represents a relatively robust grouping that would be expected to be preserved upon repeated experiments, or if different but comparable sets of sera or isolates were studied, and provides groupings of similar neutralization profiles for both sera and isolates.
FIG. 2.
Heat map and clustering analysis of serum. ID50 values of 103 sera against 20 isolates are shown. Each row of the heat map shows ID50 values for a single serum, and columns show virus isolates. Darker colors represent stronger neutralization (see key). The vertical order of sera is based on geometric mean titer; placement of clusters within this ranking uses the mean titer for all cluster members. The bars labeled “Bootstrap” or “Noise” show the results of statistical analysis of clustering. Both are visualized by mixing red, yellow, and blue corresponding to the relative frequencies of matched group assignments. A bright red, yellow, or blue color is a categorization that is unambiguous. Sera or isolates are grouped if they have a categorization of 90% or greater consistency by both the bootstrap and noise tests. Boxes highlight the clusters. Sera in red type are from LTNP. Clusters of patient sera are labeled P1, P2, and P3, while clusters of isolates are labeled I1, I2, and I3.
Serum cluster P1 included the majority of LTNP sera and a few additional low potency/breadth sera (Fig. 2). Serum cluster P3 included 24 sera with the greatest potency and breadth of neutralization. In addition to showing sera and isolate k means clusters, we ordered the columns and rows in the heat map according to their geometric mean values to better visualize like patterns in the columns and rows. Five sera have higher geometric means than serum cluster 3 so are placed at the bottom of Fig. 2. Among the sera with intermediate activity, one robust cluster was defined as follows: sera in this cluster (P2) do not neutralize the isolates most difficult to neutralize (geometric mean ID50, 18 for P2 sera versus I3 isolates) but do neutralize isolates in the other two isolate clusters (geometric mean titers 297 and 110 for P2 sera versus I3 and I2, respectively). Thus, the patient clusters are defined not only by overall breadth or potency, but also by which isolates are neutralized. Of the clinical variables measured, only viral load was significantly associated with membership in a cluster: median VL is lower in patients with sera in cluster P1, which contains most of the LTNP, than in those with non-P1 sera (Bonferroni corrected P < 0.001).
Figure 2 also shows that the panel of 20 diverse viral isolates we used to study breadth could be categorized into three clusters. Isolate cluster 3 (I3) consists of two B clade Envs, JRFL and BaL.01, which are the most neutralization-sensitive in the panel. Each of the other two clusters contains isolates of different clades. Four of five clade C isolates are in the most resistant cluster I1; these isolates are known to be sensitive to clade C sera but more resistant to clade B sera (11, 17). Of note, genetically closely related viruses were not always found within the same neutralization-susceptibility cluster. For example, isolates Q769.d22 and Q769.h5, both clade A, contain two different envelope proteins from the same patient but do not appear in the same cluster. These isolates are known to have differing sensitivities to autologous plasma and to MAb (6).
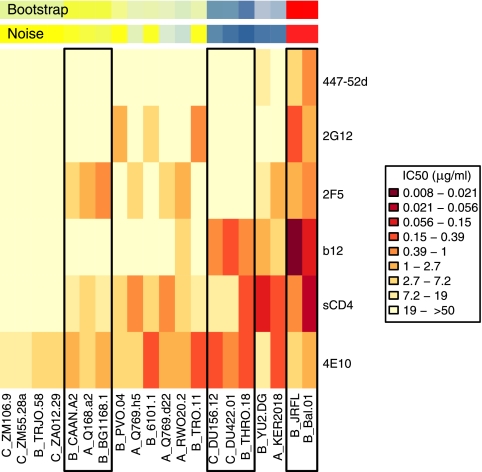
We also analyzed neutralization titers of soluble CD4 (sCD4) and the commonly used monoclonal antibodies (MAb) 2G12, 4E10, 2F5, b12, and 447-52d against the same 20 isolates. The epitopes targeted by these MAb are well defined (7). It was possible that the isolate clustering of sera shown in Fig. 2 was driven by responses that are directed mainly to epitopes thought to confer neutralization breadth. If this were the case, we hypothesized that commonalities between neutralization mediated by MAb and by sera might be observed. Clustering analysis of MAbs (Fig. 3) was performed as described above. The clustering based on patterns of neutralization by MAbs was less statistically robust than that calculated with serum titers, with no clusters found at 90% bootstrap confidence. Figure 3 shows the three clusters of isolates that appeared in only 75% of bootstrap and noise replicates. These isolate clusters (with the exception of the sensitive JRFL/Bal.01 cluster I3) were completely interspersed with those defined in the serum analysis. To further examine the relationship of serum and MAb reactivity, we compared membership of an Env in an isolate cluster as defined by serum titers (Fig. 2) with its sensitivity to each individual MAb. While membership in isolate cluster I3 correlated with neutralization titers of two of the MAb, b12 and 447-52d (P < 0.01 for each for both JRFL and BaL.01), membership in clusters I1 and I2 as defined by serum titers did not correlate with sensitivity to any one MAb. These data suggest that the clustering based on serum titers was not defined by any single epitope matching the MAb specificities.
FIG. 3.
Heat map and clustering analysis of monoclonal antibodies and sCD4. Each row of the heat map shows IC50 values for a single reagent, and columns show virus isolates. See legend to Fig. 2 for an explanation. Boxes show isolates assigned to clusters at the 75% level. Neutralization data are from references 16 and 17 and this study.
This clustering analysis allowed us to discern patterns of neutralization reactivity that are distinct from clade and from sensitivity to known cross-neutralizing MAbs. The appearance of isolates from multiple clades in clusters I1 and I2 is consistent with prior analyses of MAbs (5) and sera (11, 14) in which sequences from the same clade were distributed among multiple clusters. In general, the clade of a virus does not predict its sensitivity to patient sera and is not directly equivalent to a neutralization serotype (13, 21, 26, 34). Furthermore, the discordance between the results for MAbs and for sera may suggest that the clustering based on serum titers was not dominated by reactivities that are similar to those of the MAbs. It is unclear at present whether these differences are mediated by targeting of conserved epitopes that are not yet identified, epitopes similar to those of MAbs but with differences in neutralization patterns, or multiple specificities. These findings are potentially consistent with antibody cloning (30) and serum mapping (4, 8, 11, 18, 19, 29) studies which found that in some sera, multiple specificities are responsible for the breadth of neutralization. Future studies of this cohort using additional isolates may allow determination of possible neutralization serotypes, and viral sequence motifs that are signatures of neutralization cluster, as for the clade C sera analyzed in reference 14, or neutralization sensitivity.
The clinical data suggest that extended exposure to antigen may be beneficial for the development of broad NAbs. We observed that viral load has a modest positive correlation with neutralization breadth (Fig. 1C), as was also seen in cohorts in the United States (29) and Kenya (28). Conversely, LTNP with a VL of <50 rarely had breadth (Fig. 1B), again consistent with other reports (1, 27). Length of exposure also seems to play a role in the development of broad NAbs: Sather et al. (29) noted an association of breadth with the duration of infection in a seroconversion cohort. These associations are modest, and several slow progressors had low NAbs; thus, antigen may be necessary but not sufficient for the development of broad NAbs. Long-term exposure to HIV antigen has been shown to directly impact immunoglobulin G (IgG) development: Scheid et al. (30) found that Env-specific IgG genes were highly mutated compared to other IgG genes in patients with broad NAbs, implying multiple rounds of selection and hypermutation in response to persistence or turnover of viral antigen. Collectively, these data suggest that a vaccine may need to supply viral antigen for long periods of time, via multiple dosing or a replication-competent vector, to allow antibody maturation and development of a broad neutralizing response.
The prevalence and titers of NAbs in chronically HIV-infected patients provide encouragement for the development of vaccines that elicit protective humoral immunity. We found that that 20% of progressor patients make broad NAbs; although our cohort is enriched for slow progressors, similar frequencies were noted in other, less-selected cohorts (11, 29, 32). The fact that so many patients make broad NAbs, even in the setting of B-cell dysfunction caused by HIV (25), demonstrates the ability of the human immune system to generate such NAbs. An appropriate vaccine given to immunocompetent individuals could potentially elicit broad NAbs at higher frequencies. Furthermore, most patients neutralized at least some isolates with titers in the hundreds in the TZM-bl assay. Data from a recent passive transfer experiment in a low-dose-challenge simian-HIV (SHIV) macaque model demonstrated a protective effect of neutralizing titers of 1:200 in the TZM-bl assay (12). Thus, these examples show that it is possible to elicit NAbs at sufficient levels and breadth to potentially contribute to the protective efficacy of an HIV vaccine.
Acknowledgments
We thank Krisha McKee, Mark Louder, Stephen Schmidt, and Adjoa Smalls-Mantey for technical assistance and Nancy Cogliano, Catherine Rehm, Gregg Roby, and Sara Stallings for coordinating samples and patient visits.
This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, a grant from the NIH Intramural AIDS Targeted Antiviral Program (M.C.), and by grant no. 38619 from the Bill and Melinda Gates Foundation (B.T.K.).
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.Bailey, J. R., K. G. Lassen, H. C. Yang, T. C. Quinn, S. C. Ray, J. N. Blankson, and R. F. Siliciano. 2006. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J. Virol. 80:4758-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, J. R., T. M. Williams, R. F. Siliciano, and J. N. Blankson. 2006. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J. Exp. Med. 203:1357-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beirnaert, E., P. Nyambi, B. Willems, L. Heyndrickx, R. Colebunders, W. Janssens, and G. van Der Groen. 2000. Identification and characterization of sera from HIV-infected individuals with broad cross-neutralizing activity against group M (env clade A-H) and group O primary HIV-1 isolates. J. Med. Virol. 62:14-24. [PubMed] [Google Scholar]
- 4.Binley, J. M., E. A. Lybarger, E. T. Crooks, M. S. Seaman, E. Gray, K. L. Davis, J. M. Decker, D. Wycuff, L. Harris, N. Hawkins, B. Wood, C. Nathe, D. Richman, G. D. Tomaras, F. Bibollet-Ruche, J. E. Robinson, L. Morris, G. M. Shaw, D. C. Montefiori, and J. R. Mascola. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blish, C. A., R. Nedellec, K. Mandaliya, D. E. Mosier, and J. Overbaugh. 2007. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. AIDS 21:693-702. [DOI] [PubMed] [Google Scholar]
- 7.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. Nabel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233-236. [DOI] [PubMed] [Google Scholar]
- 8.Dhillon, A. K., H. Donners, R. Pantophlet, W. E. Johnson, J. M. Decker, G. M. Shaw, F. H. Lee, D. D. Richman, R. W. Doms, G. Vanham, and D. R. Burton. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 81:6548-6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doria-Rose, N. A., R. M. Klein, M. M. Manion, S. O'Dell, A. Phogat, B. Chakrabarti, C. W. Hallahan, S. A. Migueles, J. Wrammert, R. Ahmed, M. Nason, R. T. Wyatt, J. R. Mascola, and M. Connors. 2009. Frequency and phenotype of HIV envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J. Virol. 83:188-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham, B. S. 2002. Clinical trials of HIV vaccines. Annu. Rev. Med. 53:207-221. [DOI] [PubMed] [Google Scholar]
- 11.Gray, E. S., N. Taylor, D. Wycuff, P. L. Moore, G. D. Tomaras, C. K. Wibmer, A. Puren, A. DeCamp, P. B. Gilbert, B. Wood, D. C. Montefiori, J. M. Binley, G. M. Shaw, B. F. Haynes, J. R. Mascola, and L. Morris. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83:8925-8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hessell, A. J., P. Poignard, M. Hunter, L. Hangartner, D. M. Tehrani, W. K. Bleeker, P. W. Parren, P. A. Marx, and D. R. Burton. 2009. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 15:951-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kostrikis, L. G., Y. Cao, H. Ngai, J. P. Moore, and D. D. Ho. 1996. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J. Virol. 70:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulkarni, S. S., A. Lapedes, H. Tang, S. Gnanakaran, M. G. Daniels, M. Zhang, T. Bhattacharya, M. Li, V. R. Polonis, F. E. McCutchan, L. Morris, D. Ellenberger, S. T. Butera, R. C. Bollinger, B. T. Korber, R. S. Paranjape, and D. C. Montefiori. 2009. Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C HIV-1 Env clones from India. Virology 385:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambotte, O., G. Ferrari, C. Moog, N. L. Yates, H. X. Liao, R. J. Parks, C. B. Hicks, K. Owzar, G. D. Tomaras, D. C. Montefiori, B. F. Haynes, and J. F. Delfraissy. 2009. Heterogeneous neutralizing antibody and antibody-dependent cell cytotoxicity responses in HIV-1 elite controllers. AIDS 23:897-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, M., J. F. Salazar-Gonzalez, C. A. Derdeyn, L. Morris, C. Williamson, J. E. Robinson, J. M. Decker, Y. Li, M. G. Salazar, V. R. Polonis, K. Mlisana, S. A. Karim, K. Hong, K. M. Greene, M. Bilska, J. Zhou, S. Allen, E. Chomba, J. Mulenga, C. Vwalika, F. Gao, M. Zhang, B. T. Korber, E. Hunter, B. H. Hahn, and D. C. Montefiori. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 80:11776-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Y., S. A. Migueles, B. Welcher, K. Svehla, A. Phogat, M. K. Louder, X. Wu, G. M. Shaw, M. Connors, R. T. Wyatt, and J. R. Mascola. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Y., K. Svehla, M. K. Louder, D. Wycuff, S. Phogat, M. Tang, S. A. Migueles, X. Wu, A. Phogat, G. M. Shaw, M. Connors, J. Hoxie, J. R. Mascola, and R. Wyatt. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83:1045-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahalanabis, M., P. Jayaraman, T. Miura, F. Pereyra, E. M. Chester, B. Richardson, B. Walker, and N. L. Haigwood. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naive human immunodeficiency virus controllers. J. Virol. 83:662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mascola, J. R., M. K. Louder, S. R. Surman, T. C. Vancott, X. F. Yu, J. Bradac, K. R. Porter, K. E. Nelson, M. Girard, J. G. McNeil, F. E. McCutchan, D. L. Birx, and D. S. Burke. 1996. Human immunodeficiency virus type 1 neutralizing antibody serotyping using serum pools and an infectivity reduction assay. AIDS Res. Hum. Retrovir. 12:1319-1328. [DOI] [PubMed] [Google Scholar]
- 22.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, D. S. Burke, and the National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 23.Migueles, S. A., A. C. Laborico, W. L. Shupert, M. S. Sabbaghian, R. Rabin, C. W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, and M. Connors. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061-1068. [DOI] [PubMed] [Google Scholar]
- 24.Migueles, S. A., C. M. Osborne, C. Royce, A. A. Compton, R. P. Joshi, K. A. Weeks, J. E. Rood, A. M. Berkley, J. B. Sacha, N. A. Cogliano-Shutta, M. Lloyd, G. Roby, R. Kwan, M. McLaughlin, S. Stallings, C. Rehm, M. A. O'Shea, J. Mican, B. Z. Packard, A. Komoriya, S. Palmer, A. P. Wiegand, F. Maldarelli, J. M. Coffin, J. W. Mellors, C. W. Hallahan, D. A. Follman, and M. Connors. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moir, S., and A. S. Fauci. 2009. B cells in HIV infection and disease. Nat. Rev. Immunol. 9:235-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, J. P., Y. Cao, J. Leu, L. Qin, B. Korber, and D. D. Ho. 1996. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J. Virol. 70:427-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereyra, F., M. M. Addo, D. E. Kaufmann, Y. Liu, T. Miura, A. Rathod, B. Baker, A. Trocha, R. Rosenberg, E. Mackey, P. Ueda, Z. Lu, D. Cohen, T. Wrin, C. J. Petropoulos, E. S. Rosenberg, and B. D. Walker. 2008. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 197:563-571. [DOI] [PubMed] [Google Scholar]
- 28.Piantadosi, A., D. Panteleeff, C. A. Blish, J. M. Baeten, W. Jaoko, R. S. McClelland, and J. Overbaugh. 2009. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J. Virol. 83:10269-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sather, D. N., J. Armann, L. K. Ching, A. Mavrantoni, G. Sellhorn, Z. Caldwell, X. Yu, B. Wood, S. Self, S. Kalams, and L. Stamatatos. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheid, J. F., H. Mouquet, N. Feldhahn, M. S. Seaman, K. Velinzon, J. Pietzsch, R. G. Ott, R. M. Anthony, H. Zebroski, A. Hurley, A. Phogat, B. Chakrabarti, Y. Li, M. Connors, F. Pereyra, B. D. Walker, H. Wardemann, D. Ho, R. T. Wyatt, J. R. Mascola, J. V. Ravetch, and M. C. Nussenzweig. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636-640. [DOI] [PubMed] [Google Scholar]
- 31.Shu, Y., S. Winfrey, Z. Y. Yang, L. Xu, S. S. Rao, I. Srivastava, S. W. Barnett, G. J. Nabel, and J. R. Mascola. 2007. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine 25:1398-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simek, M. D., W. Rida, F. H. Priddy, P. Pung, E. Carrow, D. S. Laufer, J. K. Lehrman, M. Boaz, T. Tarragona-Fiol, G. Miiro, J. Birungi, A. Pozniak, D. McPhee, O. Manigart, E. Karita, A. Inwoley, W. Jaoko, J. Dehovitz, L. G. Bekker, P. Pitisuttithum, R. Paris, L. M. Walker, P. Poignard, T. Wrin, P. E. Fast, D. R. Burton, and W. C. Koff. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, S., J. S. Kennedy, K. West, D. C. Montefiori, S. Coley, J. Lawrence, S. Shen, S. Green, A. L. Rothman, F. A. Ennis, J. Arthos, R. Pal, P. Markham, and S. Lu. 2008. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 26:1098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber, J., E. M. Fenyo, S. Beddows, P. Kaleebu, A. Bjorndal, and the WHO Network for HIV Isolation and Characterization. 1996. Neutralization serotypes of human immunodeficiency virus type 1 field isolates are not predicted by genetic subtype. J. Virol. 70:7827-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]