Abstract
The human retrovirus XMRV (xenotropic murine leukemia virus-related virus) is associated with prostate cancer, but a causal relationship has not been established. Here, we have used cultured fibroblast and epithelial cell lines to test the hypothesis that XMRV might have direct transforming activity but found only rare transformation events, suggestive of indirect transformation, even when the target cells expressed the human Xpr1 cell entry receptor for XMRV. Characterization of cells from three transformed foci showed that all were infected with and produced XMRV, and one produced a highly active transforming virus, presumably generated by recombination between XMRV and host cell nucleic acids. Given the sequence similarity of XMRV to mink cell focus-forming (MCF) viruses and the enhanced leukemogenic activity of the latter, we tested XMRV for related MCF-like cytopathic activities in cultured mink cells but found none. These results indicate that XMRV has no direct transforming activity but can activate endogenous oncogenes, resulting in cell transformation. As part of these experiments, we show that XMRV can infect and be produced at a high titer from human HT-1080 fibrosarcoma cells that express TRIM5α (Ref1), showing that XMRV is resistant to TRIM5α restriction. In addition, XMRV poorly infects NIH 3T3 cells expressing human Xpr1 but relatively efficiently infects BALB 3T3 cells expressing human Xpr1, showing that XMRV is a B-tropic virus and that its infectivity is regulated by the Fv1 mouse locus.
The association of human prostate cancer with mutations that impair the function of the antiviral defense protein RNase L suggested a role for virus in prostate cancer. Indeed, analysis of cDNA from prostate tumors by use of a DNA microarray (Virochip) containing conserved DNA sequences from all known virus families indicated the presence of a novel gammaretrovirus in 40% of prostate cancer patients having homozygous R462Q mutations in RNase L (35). Cloning and sequencing of the virus revealed a close similarity to mouse xenotropic retroviruses; thus, the new virus was named XMRV (xenotropic murine leukemia virus-related virus) (35). Importantly, XMRV has been found integrated into human genomic DNA from tumor-bearing prostatic tissue samples of 11 patients, showing that XMRV can indeed infect humans and is not a laboratory contaminant (7, 13). Although an initial study found XMRV only in tumor stromal cells (35), recent studies have found XMRV in the prostate carcinoma cell line 22Rv1 (14) and in malignant epithelial cells in prostate tumors (34).
XMRV lacks a host cell-derived oncogene, but examples of oncogenic activity in Env proteins from other retroviruses (1, 6, 16, 24) raise the possibility that the Env protein of XMRV might also be oncogenic. Such activity could be a result of interaction of the XMRV Env protein with the virus entry receptor Xpr1 (7, 14), which shows similarity to a yeast protein involved in G protein-coupled signal transduction (2), or interaction with other cellular proteins that do not function as virus entry receptors, as is the case for jaagsiekte sheep retrovirus (JSRV) Env (interacting protein unknown) (16) and the Env protein of spleen focus-forming virus, which interacts with and activates the erythropoietin receptor and the receptor tyrosine kinase Stk (24). Detection of XMRV oncogenic activity would strengthen the argument for a role for XMRV in prostate cancer.
In addition, while XMRV shows the highest sequence similarity to the mouse xenotropic retroviruses, it is also similar to the mink cell focus-forming (MCF) retroviruses of mice, which are highly leukemogenic due to their ability to multiply reinfect cells, leading to more-frequent activation of cellular oncogenes (36). MCF viruses were first defined by their ability to induce foci of altered cells in mink cell layers (11). Initially, it was unclear whether these foci were the result of cell transformation or cytopathic effects of the virus (11), but it is clear now that these foci result from cytopathic effects related to the ability of MCF viruses to multiply reinfect cells in what can be a receptor-independent manner, leading to cell apoptosis (23, 36, 37). It was thus important to determine if XMRV has similar properties and might be able to more frequently activate cellular oncogenes.
Here, we have found that while XMRV lacks direct transforming activity in the fibroblast and epithelial cell lines tested and does not induce cytopathic effects typical of multiple reinfection by MCF viruses, it is able to induce rare transformed foci in a rat fibroblast cell line. Interestingly, in one case, transformation led to the production of a highly active oncogenic retrovirus.
MATERIALS AND METHODS
Cell culture and viruses.
Cells were propagated in Dulbecco's modified Eagle's medium (DMEM) with 7% fetal bovine serum (FBS), with the exception that PG-4 cat cells (9) were grown in McCoy's medium with 15% FBS. HTX cells are a pseudodiploid subclone of HT-1080 cells (ATCC CCL 121). The LAPSN retroviral vector (22) contains the human placental alkaline phosphatase (AP) cDNA driven by the viral long terminal repeat (LTR) promoter and the neomycin phosphotransferase cDNA driven by the simian virus 40 early promoter. LAPSN vector infection was measured by staining cells for heat-stable placental AP 2 days after infection, as described previously (8). 10A1 amphotropic murine leukemia virus (AM-MLV) was obtained from NIH 3T3 cells transfected with the permuted 10A1 virus DNA clone pB6 (26) after the plasmid was cut with SalI and the DNA was religated to generate intact 10A1 provirus circles. Prior to use, virus-containing medium from cells was filtered through 0.45-μm-pore-size surfactant-free cellulose acetate filters. All retrovirus infections were performed by exposing subconfluent cells to virus in the presence of 4 μg Polybrene per ml.
XMRV preparations.
Several sources of XMRV were used: XMRV from 22Rv1 cells (ATCC CRL-2505) (14), XMRV+LAPSN virus from HTX/LAPSN cells infected with virus from 22Rv1 cells, XMRV from LNCaP or HTX cells infected with virus produced from HEK 293 cells following transfection with the XMRV-VP62 molecular clone (a kind gift from Robert Silverman) (7), and XMRV+LAPSN virus from HTX/LAPSN cells infected with virus from XMRV-VP62-transfected 293 cells. The XMRV titers in these preparations were found to range from 3 × 106 to 107 focus-forming units (FFU) per ml when measured by S+L− assay. In virus preparations containing the LAPSN vector, the LAPSN titers were found to range from 2 × 107 to 6 × 107 AP+ FFU per ml when measured by using PG-4 S+L− cells as targets for infection.
S+L− assay for replication-competent retroviruses.
PG-4 cells were seeded at 2 × 105 cells per 6-cm dish (diameter = 52 mm) or 105 cells per well (diameter = 35 mm) of 6-well plates. The next day, the cells were fed with fresh medium containing 4 μg Polybrene per ml and test samples were added. Foci resulting from the rescue and spreading of the transforming virus present in the PG-4 cells by replication-competent virus in the test sample were counted when well developed (3 to 6 days after infection). LAPSN virus in the test sample was quantitated by staining the PG-4 cells for AP and counting AP+ foci immediately after the transformed foci were counted.
RESULTS
XMRV does not directly transform 208F rat fibroblasts.
Cultured 208F rat fibroblasts (27) grow to form a flat monolayer that will persist without cell overgrowth for more than a month when fed every 3 to 4 days with DMEM plus 5% FBS. These cells exhibit foci of piled-up, rounded, dividing cells in response to transfection or infection with a variety of oncogenes. To test for oncogenic activity of XMRV, we exposed 208F cells to 0.25 ml of XMRV+LAPSN virus made from HTX/LAPSN cells infected with XMRV produced by 22Rv1 prostate carcinoma cells (14). The presence of the LAPSN vector in the XMRV+LAPSN virus preparation provided a marker to easily measure virus infection rates by staining cells for AP. Indeed, staining for AP 2 days after virus exposure revealed that nearly 100% of the 208F cells exposed to the virus but none of the unexposed cells expressed AP, indicating efficient virus infection. The LAPSN vector was also found to be produced by the XMRV+LAPSN-infected 208F cells at a titer of 2 × 103 AP+ FFU per ml of medium exposed to the cells overnight when measured 6 days after virus exposure. Because the LAPSN vector encodes no viral proteins, production of the vector implies that all of the XMRV viral proteins were expressed in the 208F cells.
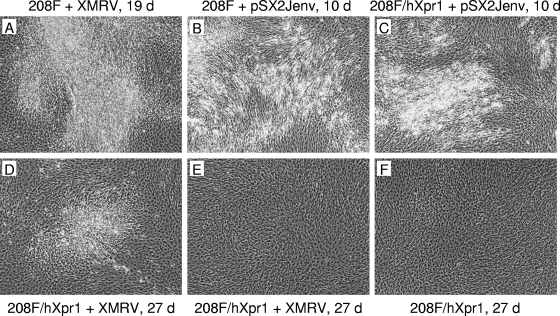
Microscopic observation of the 208F cells for 20 days after infection revealed one focus of transformed cells (Fig. 1A) in three 6-cm-diameter dishes of cells infected with XMRV+LAPSN virus and none in three 6-cm-diameter dishes of uninfected cells. The focus of transformed cells appeared late after infection (∼2 weeks) and consisted primarily of piled up cells that were not very refractile, unlike foci induced by the JSRV Env gene, for example, which appear in as little as 3 days and are highly refractile (Fig. 1B and C). The appearance of only one transformed focus in cells that were almost all infected indicates that XMRV lacks direct transforming activity but may mediate low-frequency indirect transformation, perhaps by activation of cellular oncogenes.
FIG. 1.
Morphology of transformed foci in 208F fibroblasts. 208F and 208F/LhXpr1SN cells seeded the day before at 1 × 105 to 2 × 105 cells per 6-cm dish were infected with XMRV+LAPSN virus (produced by HTX/LAPSN cells infected with virus from 22Rv1 cells) (A, D, E), were transfected with the pSX2Jenv plasmid by using the calcium phosphate method (B, C), or were treated with culture medium only (F). The cells were trypsinized and replated in multiple 6-cm dishes 1 day (A, D to F) or 10 days (B, C) after treatment. Areas with (A to D) and without (E and F) foci are shown. Cell layers were photographed at the indicated times after replating. d, days.
We repeated this experiment by using the same stock of XMRV+LAPSN virus and several related viruses, including NZB clone 9-1 xenotropic murine leukemia virus (25) plus the LAPSN vector, Friend MCF strain 98D virus (4), and AM-MLV (20). The NZB+LAPSN virus was made, like the XMRV+LAPSN virus, by infecting cells transduced with the LAPSN vector with NZB virus and by harvesting the mixed virus from these cells. The 208F cells were exposed to 0.5 ml of each virus stock (>106 infectious units) or control medium. Microscopic observation of the infected and uninfected 208F cells for 18 days revealed no foci of transformed cells (three 10-cm-diameter dishes for each condition). AP staining of the cells at this time revealed that 95% of the XMRV+LAPSN-infected cells, 80% of the NZB+LAPSN-infected cells, and none of the uninfected cells expressed AP, indicating efficient infection by these viruses. The MCF and amphotropic virus stocks did not contain the LAPSN vector, and we did not monitor infection, but previous results indicate that infection by these viruses should have occurred (data not shown).
Generation of 208F rat fibroblasts expressing human Xpr1.
The finding that 208F cells are efficiently infected by the LAPSN vector pseudotyped by XMRV shows that 208F cells express a functional Xpr1 receptor for XMRV, but we wanted to test whether the human Xpr1 (hXpr1) protein might be required for transformation by XMRV. Therefore, we generated 208F cells expressing hXpr1 by exposing 208F cells to the retroviral vector LhXpr1SN (2), produced in the absence of replication-competent retrovirus by using PT67 retrovirus packaging cells (18), and by growing the 208F cells in G418 to select for the presence of the neomycin phosphotransferase gene carried by the vector. To test for functional expression of the hXpr1 protein, we measured XMRV+LAPSN infection of 208F cells and 208F cells expressing the LhXpr1SN vector (Table 1). The titer of the LAPSN vector was, on average, 42% ± 8% (standard error [SE]) higher on 208F/LhXpr1SN cells than on 208F cells. To control for factors that affect infection independent of receptor function, we also compared the XMRV+LAPSN titers normalized to 10A1+LAPSN vector titers and found an average increase of 34% ± 4% (SE) on 208F/LhXpr1SN cells in comparison to the level for 208F cells. This increase is statistically significant (P < 0.02) and indicates that the LhXpr1SN vector encodes a functional Xpr1 receptor in the transduced 208F cells.
TABLE 1.
Human Xpr1 expression in 208F cells increases their susceptibility to XMRV infectiona
| Virus | Expt | Titer (AP+ FFU/ml) of LAPSN vector on cells |
Ratio | XMRV ratio normalized to 10A1 ratio | |
|---|---|---|---|---|---|
| 208F/hXpr1 | 208F | ||||
| XMRV+LAPSN | 1 | 4.55 × 106 | 2.90 × 106 | 1.57 | 1.27 |
| 2 | 2.54 × 106 | 1.90 × 106 | 1.34 | 1.34 | |
| 3 | 4.16 × 106 | 3.09 × 106 | 1.35 | 1.42 | |
| 10A1+LAPSN | 1 | 1.68 × 107 | 1.35 × 107 | 1.24 | |
| 2 | 7.40 × 106 | 7.40 × 106 | 1.00 | ||
| 3 | 1.23 × 107 | 1.30 × 107 | 0.95 | ||
Cells were seeded at 5×104 cells per well in 6-well plates, were infected the next day with appropriate dilutions of XMRV+LAPSN virus harvested from HTX/LAPSN+XMRV cells (XMRV was from 22Rv1 cells) or 10A1+LAPSN virus from Mus dunni/LAPSN+10A1 cells (21), and were stained for AP 2 days after infection. Results are from three independent experiments.
Because 208F cells are relatively susceptible to XMRV infection, and the infection rate was only modestly increased by transduction of the 208F cells with the LhXpr1SN vector, we also tested the activity of the LhXpr1SN vector in mouse cells that are resistant to xenotropic virus entry. The XMRV+LAPSN virus did not infect NIH 3T3 or BALB 3T3 mouse cells (<1 AP+ FFU per ml), but transduction of these cells with the identical frozen LhXpr1SN vector preparation used to make the 208F/LhXpr1SN cells, followed by selection of the cells in G418, rendered the NIH 3T3 and BALB 3T3 cells susceptible to infection by the XMRV+LAPSN virus (titers of 90 and 2 × 104 AP+ FFU per ml, respectively [means of results from two experiments]). The BALB 3T3/LhXpr1SN cells were >200-fold more infectible than were the NIH 3T3/LhXpr1SN cells, indicating that XMRV is a B-tropic retrovirus and that XMRV infection is partially restricted in NIH 3T3 cells. These results show that the LhXpr1SN vector encodes a functional Xpr1 protein.
XMRV does not directly transform 208F rat fibroblasts expressing human Xpr1.
To test the effect of hXpr1 expression on 208F cell transformation by XMRV, the 208F/LhXpr1SN cells were exposed to 0.5 ml of XMRV+LAPSN, NZB+ LAPSN, or MCF viruses, all of which can use hXpr1 for cell entry (2, 14), or no virus as a negative control. Microscopic observation for over 1 month revealed four foci of transformed cells in three 6-cm-diameter dishes of XMRV+LAPSN-infected cells and no foci in any of the other cultures. Figure 1 shows an example of a focus in an XMRV-infected culture (Fig. 1D), the flat appearance of the cells in other areas of the dish (Fig. 1E), and the flat appearance of uninfected cells (Fig. 1F). The foci produced in the 208F/LhXpr1SN cells, like the transformed focus described above for XMRV-infected 208F cells (Fig. 1A), were visible only long after infection (≥2 weeks) and were composed primarily of piled-up cells that were not very refractile. In total, our results show that XMRV and NZB xenotropic viruses, and likely the MCF and amphotropic viruses, are unable to directly transform 208F cells even if the cells express hXpr1, although XMRV could induce rare foci of transformed cells. In addition, we saw no evidence of transformation by hXpr1.
XMRV does not transform Madin Darby canine kidney (MDCK) epithelial cells.
MDCK cells exhibit prototypical epithelial cell morphology, grow in tight clusters, and remain as a monolayer after they reach confluence. These cells have been used to assay transformation by multiple oncogenes, including the JSRV env gene (17), and may be more appropriate than fibroblasts as a model for transformation of prostate epithelial cells. We exposed subconfluent MDCK cells to XMRV+LAPSN virus or control medium and fed the cells with DMEM plus 5% FBS every 3 to 4 days for 20 days. There were no morphological differences between the control and virus-exposed cultures (three 6-cm-diameter dishes each), and no transformed foci were observed. AP staining 2 days after virus exposure revealed that 10 to 20% of the cells exposed to XMRV+LAPSN virus and none of the uninfected cells expressed AP, showing that the cells exposed to XMRV+ LAPSN virus had been infected. The LAPSN vector was also found to be produced by the infected MDCK cells at a titer of 2 × 104 AP+ FFU per ml of medium exposed to the cells overnight when measured 6 days after virus exposure, showing that the XMRV viral proteins (including Env) were expressed in the XMRV+LAPSN-exposed MDCK cells. Thus, XMRV can infect and replicate in but does not transform MDCK cells.
Plasmids expressing XMRV or XMRV Env do not transform 208F cells with or without coexpression of hXpr1.
We considered the possibility that the promoter in XMRV might not express XMRV Env at high enough levels to elicit cell transformation. To address this issue, we cloned the XMRV env coding region from the XMRV-VP62 infectious molecular clone (7) (GenBank accession no. EF185282) into the pSX2 expression vector in place of the existing 10A1 amphotropic env coding region to make pSX2Xenv. In this plasmid, XMRV Env expression is driven by the strong Moloney murine leukemia virus LTR promoter, and we have shown that insertion of the JSRV env coding region into the same site in the pSX2 vector yields a plasmid (pSX2Jenv) that can transform 208F cells in just a few days (28). We performed two separate transfections by using either calcium phosphate (3) or Transit-LT1 transfection reagent (Mirus Bio), and by using either 208F or 208F/LhXpr1SN cells as targets for transfection, to test the transforming activity of pSX2Xenv; a mixture of equal amounts of pSX2Xenv and the pLhXpr1SN plasmid; and pXMRV-VP62. Control transfections included the pSX2 plasmid, encoding the nontransforming 10A1 amphotropic Env protein, and the pSX2Jenv plasmid, encoding the highly transforming JSRV Env protein, as a positive control. We also included a small amount of the pLAPSN vector plasmid (5% of the test plasmids by mass) to allow estimation of transfection efficiency by AP staining. No transformed foci were observed for pSX2 or for the plasmids expressing XMRV Env or the complete XMRV cDNA clone, while many foci were observed for the pSX2Jenv plasmid, encoding JSRV Env (Table 2). AP staining of the cell layers revealed similar transfection rates in each experiment for the pSX2, pSX2Xenv, and pSX2Xenv-plus-pLhXpr1SN plasmids. There was a significant increase in transfection rate for the pSX2Jenv plasmid, suggesting that transformation by the pSX2Jenv plasmid enhances stable transfection. There was a more dramatic increase in AP+ foci in cells transfected with pXMRV-VP62, most likely reflecting a spread of the LAPSN vector due to XMRV replication and a concomitant mobilization of the LAPSN vector.
TABLE 2.
XMRV expression plasmids do not transform 208F or 208F/LhXpr1SN cellsa
| Transfection method | Test plasmid(s) | No. of foci in cell line |
|||
|---|---|---|---|---|---|
| Transformed |
AP+ |
||||
| 208F | 208F/ LhXpr1SN | 208F | 208F/ LhXpr1SN | ||
| CaPO4 | pSX2 | 0 | 0 | 18 | 22 |
| pSX2Xenv | 0 | 0 | 20 | 20 | |
| pSX2Xenv, pLhXpr1SN | 0 | 0 | 18 | 21 | |
| pXMRV-VP62 | 0 | 0 | 2,000 | 1,600 | |
| pSX2Jenv | 720 | 980 | 164 | 172 | |
| Transit-LT1 | pSX2 | 0 | 0 | 190 | 340 |
| pSX2Xenv | 0 | 0 | 420 | 640 | |
| pSX2Xenv, pLhXpr1SN | 0 | 0 | 310 | 350 | |
| pXMRV-VP62 | 0 | ND | 1,600 | ND | |
| pSX2Jenv | 3,800 | 3,600 | 810 | 960 | |
Transformed foci were counted 17 (CaPO4) or 18 (Transit-LT1) days after transfection, and AP+ foci were counted 17 (CaPO4) or 19 (Transit-LT1) days after transfection. Numbers represent the total numbers of transformed and AP+ foci induced by 10 μg test plasmids plus 0.5 μg pLAPSN plasmid DNA, used for each CaPO4 transfection (3) or by 5 μg test plasmids plus 0.25 μg pLAPSN plasmid DNA, used for each Transit-LT1 transfection (Mirus Bio). Plates that exhibited no transformed foci remained negative for at least 24 (CaPO4) and 30 (Transit-LT1) days after transfection. ND, no data.
To prove that the pSX2 and pXMRV-VP62 plasmids expressed functional Env protein, we transfected these plasmids into NIH 3T3 cells that express Moloney MLV Gag-Pol proteins (19) and were transduced with the LAPSN vector (LGPS/LAPSN cells). Table 3 shows that the pSX2Xenv, pXMRV-VP62, pSX2, and pSX2Jenv plasmids have similar abilities to rescue the LAPSN vector from LGPS/LAPSN cells but that the negative-control plasmid pUC19 had no activity. In summary, despite encoding functional Env protein, none of the XMRV expression plasmids were able to transform 208F cells or 208F cells expressing hXpr1.
TABLE 3.
Production of functional Env proteins by XMRV plasmidsa
| Transfected plasmid | LAPSN vector titer (no. of AP+ FFU/ml) |
|---|---|
| pUC19 | <2 |
| pSX2Xenv | 5.5 × 103 |
| pXMRV-VP62 | 6.6 × 103 |
| pSX2 | 1.3 × 104 |
| pSX2Jenv | 4.2 × 103 |
LGPS/LAPSN cells (NIH 3T3 thymidine kinase-negative cells expressing Moloney MLV Gag-Pol proteins [19] and containing the LAPSN vector) were transfected with the indicated plasmids by calcium phosphate coprecipitation (5). One day after transfection, the cells were fed, and 2 days after transfection, the culture medium was harvested and filtered through 0.45-μm-pore-size filters to remove cells and debris, and the medium samples were assayed for LAPSN vector titer by infection of HTX cells. Results are means for two independent experiments.
XMRV lacks the cytopathic activity of MCF viruses.
The cytopathic effects of MCF viruses in cultured mink Mv1Lu cells (ATCC CCL-64) correlate with their ability to multiply reinfect (superinfect) the cells, thereby inducing apoptosis (23, 37). Multiple reinfections also result in more-frequent oncogene activation in hematolymphoid cells of mice infected with MCF viruses, resulting in frequent induction of leukemias. Given the similarity of XMRV to xenotropic and MCF retroviruses, we tested whether XMRV could also induce cytopathic effects in mink cells as an indicator of an ability to multiply reinfect the cells. Mink cells were seeded at 1 × 105 to 2 × 105 per 6-cm dish and, on the next day, were infected with over 106 infectious units of XMRV+LAPSN, Friend MCF strain 98D (4), or NZB xenotropic (25) virus. The cells were then grown, and every time they became confluent, they were trypsinized and split 1:10. In two independent experiments, cytotoxicity (a reduction in the number of attached cells and the appearance of floating cells) was first observed in the MCF-infected cells at days 3 and 5 and in the NZB-infected cells at days 11 and 12, and no toxicity was observed in the XMRV-infected cells or in the uninfected cells passaged in parallel for 40 days after infection. AP staining of the XMRV+LAPSN-infected cells at 12 days (experiment 1) or 36 days (experiment 2) after infection revealed 50 to 70% AP+ cells, showing that the cells had been efficiently infected. After the first toxicity was observed in the MCF and NZB virus-infected cells, these populations nearly completely died over a period of a week, although ultimately, clones of cells that were resistant to the cytotoxicity of each virus grew out. We found that the cells that became resistant to MCF virus cytotoxicity were producing the MCF virus and thus were still infected (data not shown), but we have not determined the nature of the acquired resistance to cytotoxicity. These results document the novel finding that NZB xenotropic virus is cytotoxic in mink cells, although it is less potent than the MCF virus, while XMRV shows no cytotoxicity in mink cells. On the basis of previously described correlations between cytotoxicity and reinfection (23, 36, 37), we conclude that XMRV is less able to multiply reinfect the mink cells than are the MCF and NZB retroviruses.
XMRV does not transform mink cells.
Mv1Lu lung epithelial cells grow to form a flat contact-inhibited monolayer and have been used to assay for transformation by mammalian sarcoma viruses (12). To test whether XMRV could induce transformed foci in the mink cells, we allowed XMRV+LAPSN-infected mink cells to reach confluence and continued to feed the cells with DMEM plus 5% FBS every 3 to 4 days for over a month. In two experiments, we did not detect any foci indicative of transformation in XMRV-infected cultures.
Further characterization of rare 208F cell transformation by XMRV.
We considered the possibility that rare transformation of 208F cells by XMRV originally obtained from 22Rv1 cells was due to contamination with a rare transforming virus, so we generated a clonal stock of XMRV from the VP62 plasmid clone of XMRV by transfection of 293 cells, infected HTX/LAPSN cells with this virus, and assayed 1 ml of virus from the HTX/LAPSN+XMRV VP62 cells for transformation of 208F cells. One focus of transformation was detected in a 10-cm dish, similar to the results described above, by using XMRV originally from 22Rv1 cells, indicating that a clonal stock of XMRV can also induce rare transformed foci in 208F cells. We also tested XMRV directly from 22Rv1 cells for transforming activity. The use of 2 ml of this virus in an assay involving 8 10-cm dishes of 208F cells revealed five transformed foci, showing that neither the growth of XMRV in HTX cells nor the presence of the LAPSN vector was required for rare transformation events to occur.
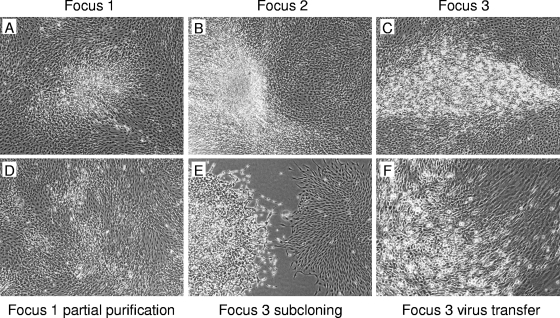
To characterize the properties of transformed foci that arose after XMRV+LAPSN virus infection, we isolated cells from three foci (Fig. 2A, B, and C). Cloning rings were first used to isolate cells from the foci along with nearby nontransformed 208F cells, and then the cells were plated at a limiting dilution to enable isolation of clonal transformed cell lines. Cells from all three foci retained a stable transformed phenotype after cloning (Fig. 2D and E and data not shown), including cells from the very weakly transformed focus 1 (Fig. 2A and D). Cells from foci 1 and 2 showed a lack of contact inhibition, resulting in piled-up cell growth, while focus 3 showed a highly transformed phenotype (Fig. 2D and E and data not shown). Cells from all foci produced replication-competent virus by an S+L− assay (Table 4), indicating infection by and production of XMRV. Cells from all foci stained positive for AP (not shown) and produced virus encoding AP (Table 4), indicating infection by and production of the LAPSN vector as well. Assay of medium from these cells for the presence of virus that could transform 208F cells (Table 4) showed that cells from foci 1 and 2 did not produce transforming virus but that focus 3 cells produced a relatively high titer of a virus that rapidly transformed 208F cells (Fig. 2F), with morphological changes in the 208F cells being visible within about 4 days.
FIG. 2.
Morphology of transformed foci of 208F cells and purification of the transformed cells. Panels A to C show transformed foci at 27 days (focus 1) or 15 days (foci 2 and 3) after plating of XMRV-infected 208F cells (see Table 4 for infection details). Panel D shows the mixed population of transformed and nontransformed cells 1 week after plating of cells from focus 1 that were obtained by using a cloning ring. Panel E shows two colonies of cells produced by plating low numbers of mixed cells from focus 3. The left colony is clearly transformed and resembles the cells in the original focus (C), while the right colony appears to be a colony of untransformed parental 208F cells. Panel F shows a focus of transformed cells (lower-left part of picture) induced by infection of 208F cells with virus from focus 3 cells.
TABLE 4.
Characteristics of and virus production by cells derived from transformed 208F focia
| 208F focus | Presence of hXpr1 | Infecting XMRV virus | Virus production level (FFU/ml) |
||
|---|---|---|---|---|---|
| S+L− foci | AP+ foci | Transforming virus | |||
| 1 | Yes | 22Rv1 | 4 × 103 | 2 × 104 | <1 |
| 2 | No | VP62 | 600 | 104 | <1 |
| 3 | No | 22Rv1 | 2.5 × 104 | 9 × 105 | 3 × 104 |
Medium was harvested 12 to 24 h after feeding of confluent layers of cells obtained from transformed foci of 208F cells (foci 2 and 3) or 208F/LhXpr1SN cells (focus 1) infected with the indicated viruses. The medium was assayed for replication-competent virus by an S+L− assay. After the S+L− foci were counted microscopically, the cells were stained for AP, and AP+ foci were counted in the same dishes. The presence of transforming virus in medium harvested from cells obtained from the transformed foci was measured by exposing 208F cells to 1 ml of virus and by checking for transformed foci for 1 month after infection. For focus 3, transformed foci were visible within 5 days of infection and were counted at 10 days after infection with diluted virus.
DISCUSSION
Given the potential role of XMRV in prostate cancer and the ability of some retroviruses to directly transform cells, it was important to test whether XMRV is capable of direct transformation. However, we found no transformation by XMRV in canine kidney (MDCK) and mink lung epithelial cells and found only rare transformation events in 208F rat fibroblasts. Although 208F cells express a functional Xpr1 cell entry receptor for XMRV, we also tested whether expression of human Xpr1 in these cells might lead to increased transformation but found no increase. The few foci that we did observe in 208F fibroblasts with or without hXpr1 arose long after XMRV infection (≥2 weeks) and typically had a piled-up appearance (Fig. 1A and D), unlike the transformed foci induced by oncogenes such as JSRV env (Fig. 1B and C) and the fos oncogene (28), which arise in a few days and are composed of rounded separated cells that can readily detach from the cell layer. These results indicate that XMRV has no direct transforming activity but may be able to activate cellular oncogenes at low rates, leading to transformation.
In contrast to rare transformation of 208F fibroblasts by XMRV, we did not see any transformation of MDCK and mink lung epithelial cells by XMRV. These epithelial cell types were less efficiently infected by XMRV than were 208F rat fibroblasts; therefore, the rates of virus integration and oncogene activation might have been too low for detection of transformation. Alternatively, the 208F cells may simply be more sensitive to transformation. It is also surprising that we did not see focus formation by NZB, MCF, or amphotropic viruses in 208F cells. While this may be due to intrinsic differences between XMRV and the other viruses, an alternative explanation is that differences in infection rates are responsible for this result. In none of these cases have we tried to detect transformation by greatly increasing the amount of virus and number of target cells used.
During testing for hXpr1 activity as a receptor for XMRV, we found that XMRV+LAPSN virus could efficiently infect BALB 3T3 cells expressing hXpr1 but was 200-fold less able to infect NIH 3T3 cells expressing hXpr1, characteristics of B-tropic murine retroviruses. N and B tropism is governed by the mouse Fv1 locus and depends on sequences in the viral capsid protein. Consistent with the conclusion that XMRV is B tropic, the XMRV capsid residues at all positions shown to affect N and B tropism (residues 92, 105, 109, 110, 114, and 117) match those of the typical B-tropic murine retrovirus WNB5 MLV (15). B-tropic murine retroviruses are also typically resistant to Ref1 (TRIM5α) restriction in human cells. HT-1080 cells express high levels of TRIM5α (15), and the XMRV+LAPSN virus can efficiently infect and replicate in HTX cells (a subclone of HT-1080 cells), showing that XMRV is also resistant to TRIM5α restriction.
XMRV shows no cytotoxicity in mink cells, unlike MCF virus, which caused dramatic death starting at 3 to 5 days after infection, and NZB xenotropic virus, which caused dramatic death starting at 11 to 12 days after infection. Cell death caused by MCF viruses has been shown to result from the ability of MCF to multiply reinfect cells, indicating that XMRV is not able to reinfect cells to the same extent. However, it is true that multiple XMRV proviruses can be found in some prostate carcinoma cells (14), so oncogene activation by XMRV is not precluded.
Transformed 208F cell foci were not observed in control uninfected cells or in cells infected with NZB, MCF, or amphotropic retroviruses, so transformed foci produced by XMRV do not appear to be artifactual. Several XMRV preparations were used in these experiments, including virus from 22Rv1 cells, which may be heterogeneous due to the multiple integrated copies of XMRV in these cells, and virus containing the LAPSN vector, which could contribute to transformation. However, transformed foci were also observed with the use of presumably homogenous virus made from the XMRV-VP62 plasmid clone and with the use of virus from 22Rv1 cells that does not contain the LAPSN vector. In addition, production of transformed foci was not dependent on the cell type used to grow XMRV; foci were observed with the use of XMRV from 22Rv1 cells and from HTX cells infected with virus from 293 cells transfected with the XMRV-VP62 plasmid clone. Together, these results indicate that XMRV itself is responsible for rare focus formation in the 208F cells.
The rare transformed foci induced by XMRV and XMRV+LAPSN virus preparations that we have observed are likely the result of virus integration near and activation of oncogenes in the infected cells. Such events are very well documented for animals infected with many types of retroviruses (33) and have been documented for humans receiving retroviral vectors to treat genetic disease (10). Such events might also contribute to prostate cancer in humans but would require multiple integration events resulting from active virus replication in prostate cells. Interestingly, cells from one of the transformed foci that we studied (focus 3) are producing a highly active transforming virus in addition to the XMRV and LAPSN viruses used to infect these cells (Table 4). Many such transforming viruses have been observed and characterized for animals (33) and have also been generated in a few cases in cultured cells (29-32). We are currently cloning and will sequence the acutely transforming virus to understand its genesis, specifically, whether it acquired an oncogene from the 208F rat cells in which the transformed focus appeared.
In summary, the lack of direct transforming activity and an inability to readily reinfect cells, leading to more-frequent oncogene activation, argue against two models for XMRV oncogenesis in the prostate. However, our results indicate that XMRV, like other retroviruses, might still induce cancer by low-frequency insertional activation of oncogenes or by generation of highly active transforming viruses.
Acknowledgments
We thank Robert Silverman for providing the plasmid containing the XMRV-VP62 infectious clone of XMRV.
This work was supported by funding from the Fred Hutchinson Cancer Research Center (A.D.M.), a Pilot grant from the Northwest Genome Engineering Consortium grant UL1 DE19582 (M.J.M.), NCI grants U54 CA132381 and U54 CA132383 (C.J.H.), and a Minority Access to Research Careers fellowship (C.J.H.).
Footnotes
Published ahead of print on 9 December 2009.
REFERENCES
- 1.Alian, A., D. Sela-Donenfeld, A. Panet, and A. Eldor. 2000. Avian hemangioma retrovirus induces cell proliferation via the envelope (env) gene. Virology 276:161-168. [DOI] [PubMed] [Google Scholar]
- 2.Battini, J. L., J. E. Rasko, and A. D. Miller. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. U. S. A. 96:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chesebro, B., and K. Wehrly. 1985. Different murine cell lines manifest unique patterns of interference to superinfection by murine leukemia viruses. Virology 141:119-129. [DOI] [PubMed] [Google Scholar]
- 5.Corsaro, C. M., and M. L. Pearson. 1981. Enhancing the efficiency of DNA-mediated gene transfer in mammalian cells. Somatic Cell Genet. 7:603-616. [DOI] [PubMed] [Google Scholar]
- 6.Dirks, C., F. M. Duh, S. K. Rai, M. I. Lerman, and A. D. Miller. 2002. Mechanism of cell entry and transformation by enzootic nasal tumor virus. J. Virol. 76:2141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong, B., S. Kim, S. Hong, J. Das Gupta, K. Malathi, E. A. Klein, D. Ganem, J. L. Derisi, S. A. Chow, and R. H. Silverman. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. U. S. A. 104:1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields-Berry, S. C., A. L. Halliday, and C. L. Cepko. 1992. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc. Natl. Acad. Sci. U. S. A. 89:693-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haapala, D. K., W. G. Robey, S. D. Oroszlan, and W. P. Tsai. 1985. Isolation from cats of an endogenous type C virus with a novel envelope glycoprotein. J. Virol. 53:827-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 11.Hartley, J. W., N. K. Wolford, L. J. Old, and W. P. Rowe. 1977. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc. Natl. Acad. Sci. U. S. A. 74:789-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson, I. C., M. M. Lieber, and G. J. Todaro. 1974. Mink cell line Mv1Lu (CCL 64). Focus formation and the generation of “nonproducer” transformed cell lines with murine and feline sarcoma viruses. Virology 60:282-287. [DOI] [PubMed] [Google Scholar]
- 13.Kim, S., N. Kim, B. Dong, D. Boren, S. A. Lee, J. Das Gupta, C. Gaughan, E. A. Klein, C. Lee, R. H. Silverman, and S. A. Chow. 2008. Integration site preference of xenotropic murine leukemia virus-related virus, a new human retrovirus associated with prostate cancer. J. Virol. 82:9964-9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knouf, E. C., M. J. Metzger, P. S. Mitchell, J. D. Arroyo, J. R. Chevillet, M. Tewari, and A. D. Miller. 2009. Multiple integrated copies and high-level production of the human retrovirus XMRV (xenotropic murine leukemia virus-related virus) from 22Rv1 prostate carcinoma cells. J. Virol. 83:7353-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lassaux, A., M. Sitbon, and J. L. Battini. 2005. Residues in the murine leukemia virus capsid that differentially govern resistance to mouse Fv1 and human Ref1 restrictions. J. Virol. 79:6560-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, S. L., and A. D. Miller. 2007. Oncogenic transformation by the jaagsiekte sheep retrovirus envelope protein. Oncogene 26:789-801. [DOI] [PubMed] [Google Scholar]
- 17.Liu, S. L., and A. D. Miller. 2005. Transformation of Madin-Darby canine kidney epithelial cells by sheep retrovirus envelope proteins. J. Virol. 79:927-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, A. D., and F. Chen. 1996. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J. Virol. 70:5564-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, A. D., J. V. Garcia, N. von Suhr, C. M. Lynch, C. Wilson, and M. V. Eiden. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 65:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, A. D., M. F. Law, and I. M. Verma. 1985. Generation of helper-free amphotropic retroviruses that transduce a dominant-acting, methotrexate-resistant dihydrofolate reductase gene. Mol. Cell. Biol. 5:431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, A. D., and G. Wolgamot. 1997. Murine retroviruses use at least six different receptors for entry into Mus dunni cells. J. Virol. 71:4531-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, D. G., R. H. Edwards, and A. D. Miller. 1994. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. U. S. A. 91:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nanua, S., and F. K. Yoshimura. 2004. Differential cell killing by lymphomagenic murine leukemia viruses occurs independently of p53 activation and mitochondrial damage. J. Virol. 78:5088-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishigaki, K., C. Hanson, T. Jelacic, D. Thompson, and S. Ruscetti. 2005. Friend spleen focus-forming virus transforms rodent fibroblasts in cooperation with a short form of the receptor tyrosine kinase Stk. Proc. Natl. Acad. Sci. U. S. A. 102:15488-15493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill, R. R., C. E. Buckler, T. S. Theodore, M. A. Martin, and R. Repaske. 1985. Envelope and long terminal repeat sequences of a cloned infectious NZB xenotropic murine leukemia virus. J. Virol. 53:100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ott, D., R. Friedrich, and A. Rein. 1990. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J. Virol. 64:757-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quade, K. 1979. Transformation of mammalian cells by avian myelocytomatosis virus and avian erythroblastosis virus. Virology 98:461-465. [DOI] [PubMed] [Google Scholar]
- 28.Rai, S. K., F. M. Duh, V. Vigdorovich, A. Danilkovitch-Miagkova, M. I. Lerman, and A. D. Miller. 2001. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. U. S. A. 98:4443-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rapp, U. R., and G. J. Todaro. 1978. Generation of new mouse sarcoma viruses in cell culture. Science 201:821-824. [DOI] [PubMed] [Google Scholar]
- 30.Rapp, U. R., and G. J. Todaro. 1980. Generation of oncogenic mouse type C viruses: in vitro selection of carcinoma-inducing variants. Proc. Natl. Acad. Sci. U. S. A. 77:624-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rapp, U. R., and G. J. Todaro. 1978. Generation of oncogenic type C viruses: rapidly leukemogenic viruses derived from C3H mouse cells in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 75:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasheed, S., M. B. Gardner, and R. J. Huebner. 1978. In vitro isolation of stable rat sarcoma viruses. Proc. Natl. Acad. Sci. U. S. A. 75:2972-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg, N., and P. Jolicoeur. 1997. Retroviral pathogenesis: oncogenesis, p. 478-523. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 34.Schlaberg, R., D. J. Choe, K. R. Brown, H. M. Thaker, and I. R. Singh. 2009. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc. Natl. Acad. Sci. U. S. A. 106:16351-16356. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Urisman, A., R. J. Molinaro, N. Fischer, S. J. Plummer, G. Casey, E. A. Klein, K. Malathi, C. Magi-Galluzzi, R. R. Tubbs, D. Ganem, R. H. Silverman, and J. L. Derisi. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Wensel, D. L., W. Li, and J. M. Cunningham. 2003. A virus-virus interaction circumvents the virus receptor requirement for infection by pathogenic retroviruses. J. Virol. 77:3460-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshimura, F. K., T. Wang, and S. Nanua. 2001. Mink cell focus-forming murine leukemia virus killing of mink cells involves apoptosis and superinfection. J. Virol. 75:6007-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]