Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) contains the major glycoprotein, GP5, as well as three other minor glycoproteins, namely, GP2a, GP3, and GP4, on the virion envelope, all of which are required for generation of infectious virions. To study their interactions with each other and with the cellular receptor for PRRSV, we have cloned each of the viral glycoproteins and CD163 receptor in expression vectors and examined their expression and interaction with each other in transfected cells by coimmunoprecipitation (co-IP) assay using monospecific antibodies. Our results show that a strong interaction exists between the GP4 and GP5 proteins, although weak interactions among the other minor envelope glycoproteins and GP5 have been detected. Both GP2a and GP4 proteins were found to interact with all the other GPs, resulting in the formation of multiprotein complex. Our results further show that the GP2a and GP4 proteins also specifically interact with the CD163 molecule. The carboxy-terminal 223 residues of the CD163 molecule are not required for interactions with either the GP2a or the GP4 protein, although these residues are required for conferring susceptibility to PRRSV infection in BHK-21 cells. Overall, we conclude that the GP4 protein is critical for mediating interglycoprotein interactions and, along with GP2a, serves as the viral attachment protein that is responsible for mediating interactions with CD163 for virus entry into susceptible host cell.
Porcine reproductive and respiratory syndrome virus (PRRSV) is endemic in pork-producing countries worldwide. Infection of pigs with the virus can result in PRRS disease, leading to significant economic losses to the swine industry. PRRSV causes respiratory disorders leading to pneumonia and is responsible for mortality observed in young piglets. The virus also infects pregnant sows, causing several reproductive disorders resulting in abortion, infertility, mummified fetuses, stillborn piglets, etc. PRRSV, along with equine arteritis virus (EAV), simian hemorrhagic fever virus (SHFV), and lactate dehydrogenase-elevating virus (LDV), is grouped in the family Arteriviridae and the order Nidovirales (43) based on their similar genome organization and replication strategy. PRRSV is classified into two genotypes, genotype I (European genotype) and genotype II (North American genotype). These two genotypes share approximately 60% genome sequence homology (20, 25).
The genome of PRRSV is approximately 15.4 kb in length. It has nine open reading frames (ORFs). ORF 1a and ORF 1ab, which is accessed by ribosomal frameshifting during protein synthesis (43), together span approximately 75% of the genome from the 5′ end. These ORFs produce polyproteins that are processed by different viral proteases encoded in the ORF1a region to generate a total 13 or 14 nonstructural proteins (Nsp) named Nsp1α, Nsp1β, Nsp2 to -6, Nsp7α, Nsp7β, and Nsp8 to -12 (29, 36, 47). The Nsps are involved in processing of the viral polyproteins, genome replication, and transcription (36). ORFs 2a, 2b, and 3 to 7 encompass approximately 25% of the genome at the 3′ end, and they produce the viral structural proteins, namely, glycoprotein 2a (GP2a), nonglycosylated protein 2b (or E), GP3, GP4, GP5, the matrix protein (M), and the nucleocapsid protein (N), respectively (54). Of the structural proteins, GP2a, GP3, GP4, and GP5 are N glycosylated and are present on the viral envelope (9), as are the nonglycosylated M protein and 2b proteins. The M and GP5 proteins are known to form heterodimers (35). The 2b (or E) protein possesses ion-channel-like properties and may function as a viroporin on the envelope (30). GP5 is the most abundant glycoprotein found on the surface of the virion and hence is named the major envelope glycoprotein, whereas the GP2a, GP3, and GP4 proteins, which are also present on the surface of the virion in less abundant quantities, are termed the minor envelope glycoproteins. Although early studies reported that the GP3 protein is not a structural component of the North American (genotype II) PRRS virions (24, 34), recent studies from our laboratory have shown that it is present on the virion envelope (10), an observation consistent with the European (genotype I) PRRS virions (49). All of the major and minor envelope proteins are required for generation of infectious PRRSV (53). Previous studies with both PRRSV and EAV have shown that the minor envelope glycoproteins GP2a, GP3, and GP4 and the unglycosylated 2b protein form a heterotetrameric complex in infected cells and that formation of such a complex is required for the transport of these proteins from the endoplasmic reticulum (ER) to the Golgi apparatus in infected cells prior to virion assembly (52, 53). Furthermore, the GP5 and M proteins of EAV and the homologous proteins of LDV have been shown to form a heterodimer through disulfide bonds, which is required for viral infectivity (2, 19, 42, 43). This heterodimeric GP5-M protein interaction is required for proper posttranslational processing of the proteins, and the N-linked glycosylation of GP5 is not required for GP5-M heterodimer formation (35). Although the envelope glycoproteins have been shown to form multimeric complexes, the specific interactions among these glycoproteins and how such interactions result in formation of the large multimeric complex are currently unknown.
PRRSV has tropism for porcine alveolar macrophage (PAM) cells, and sialoadhesin expressed on the surface of these cells has been shown to be a receptor for PRRSV (12-14). Under in vitro infection conditions, PRRSV has been found to infect and replicate to high levels only in MARC-145 cells (27), a derivative of African green monkey kidney cell line MA-104. However, the MARC-145 cells do not express sialoadhesin, indicating that molecules other than sialoadhesin are involved in entry of PRRSV in MARC-145 cells. Although several other molecules, such as vimentin, CD151, and heparin sulfate, have been identified as potential entry molecules for PRRSV (28, 41), recently porcine CD163 isolated from PAM cells was found to confer susceptibility to PRRSV infection to nonpermissive cells (5). Additionally, CD163 is also expressed in MARC-145 cells, suggesting that it may be responsible for entry of PRRSV in these cells. The GPs on the virion envelope must then interact with CD163 to mediate virus entry into these cells. At this time, it is unknown which of the viral envelope glycoproteins directly interact with CD163 to mediate virus entry into susceptible cells. Since GP5 protein is present on the PRRSV envelope in abundant amounts, it was presumed to play an important role in interacting with the cell surface receptor for PRRSV entry. However, studies in which the ectodomains of the GP5 and M proteins of EAV were replaced with the ectodomains of the corresponding proteins from PRRSV or LDV in an EAV infectious clone, the resulting chimeric EAV did not exhibit the altered tissue tropism, indicating that the GP5 protein may not be the PRRSV envelope protein that interacts with the receptor (17, 50). This finding is suggestive of the involvement of other PRRSV envelope glycoproteins, especially the minor envelope glycoproteins of PRRSV, in receptor interaction and virus entry. However, to date, the PRRSV glycoprotein(s) that interacts with the receptor CD163 has not been identified.
In this report, we have described studies to delineate the interactions among the four envelope glycoproteins of PRRSV as well as their interactions with the porcine CD163 receptor. Our results have revealed that a strong interaction exists between the GP4 and GP5 proteins, although weak interactions among the other glycoproteins have also been demonstrated. In addition, we have observed that only GP2a and GP4 proteins interact with CD163. A truncated CD163 molecule lacking 223 amino acid (aa) residues from its carboxy terminus was also found to interact with these two glycoproteins, indicating that the carboxy-terminal residues of CD163 are not involved in interactions with the GP2a or GP4 proteins.
MATERIALS AND METHODS
Cell culture, viruses, and reagents.
Baby hamster kidney-21 (BHK-21) cells were maintained in minimal essential medium (MEM) containing 5% fetal bovine serum (FBS) and 100 units of penicillin, 20 units of kanamycin, and 20 units of streptomycin (PKS) antibiotics per milliliter of medium. MARC-145 cells were maintained as described before (1, 46). vTF7-3 virus (21), a recombinant vaccinia virus that expresses the bacteriophage T7 RNA polymerase in the cytoplasm of infected cells, was grown and titrated in BHK-21 cells as described before (1) and stored at −80°C in small aliquots. The infectious clone-derived FL-12 virus was grown in MARC-145 cells as described before (1, 46). Endoglycosidase H (endo H) was purchased from New England Biolabs (NEB) and used at concentrations described earlier (1, 46). Protein molecular mass markers were purchased from Bio-Rad.
Antibodies.
The porcine anti-CD163 monoclonal antibody was purchased from AbD Serotec USA (Raleigh, NC). The monoclonal antibody (SDOW17) against the PRRSV nucleocapsid (38) was purchased from National Veterinary Services Laboratories (Ames, IA). The anti-GP5 rabbit polyclonal antibody was kindly provided by Carl Gagnon (University of Quebec, Montreal, Canada). Alexa Fluor-488 goat anti-rabbit and anti-mouse immunoglobulin G (IgG) antibodies were purchased from Molecular Probes and used as secondary antibodies for fluorescence microscopy. Monospecific antibodies against GP2a, GP3, and GP4 were generated by coimmunization of rabbits with two highly immunogenic peptides for each protein as described previously (10). The details of production and characterization of the GP3 monospecific antibody were reported earlier (10). The selection of peptides, their synthesis, and production of anti-GP2a and anti-GP4 monospecific polyclonal antibodies were done by methods similar to those reported previously (10, 11). The amino acid residues of the selected peptides for GP2a are CEMVSRRMYRTMEKA and CKAGQAAWKQVVSEAT. The amino acid residues of the selected peptides for GP4 are CLFYASEMSEKGFK and CFTSYVQHVKEFTQR. The peptides were synthesized and conjugated to a carrier protein (keyhole limpet hemocyanin [KLH]) either by adding a cysteine residue at the amino terminus (underlined) or via activated 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) chemistry to its N-terminal region. The antibodies were produced by Sigma and were characterized in our laboratory. Sera collected at day 77 postimmunization were used in all experiments.
Plasmid construction.
The coding regions of the minor GPs were cloned from the infectious clone FL-12 of PRRSV (46). The genes for the individual minor GPs were amplified by PCR using gene-specific primers (Table 1) and cloned in pGEM3 vector (Promega Biotech, Madison, WI) under the control of the T7 RNA polymerase promoter or in pcDNA3.1(+) vector (Invitrogen) under the control of the cytomegalovirus (CMV) promoter and T7 RNA polymerase promoter. The FLAG tag (DYKDDDDK) was fused at the carboxy terminus of GP2a by PCR using primers GP2a-FLAG-EcoRI-Rev and GP2a-NheI-For and cloned in pGEM3 vector that was modified to contain the CMV promoter in addition to the T7 RNA polymerase promoter. The ORF for GP3 and GP4 was PCR amplified and cloned in pGEM3 vector by use of EcoRI and BamHI restriction sites. The GP5 ORF was cloned in pGEM3 by use of EcoRI and HindIII restriction sites. The nucleotide sequences of the plasmids encoding various GPs were determined to ensure that the correct clones were used in the studies.
TABLE 1.
Primers used in this study
| Primer | Nucleotide sequencea |
|---|---|
| GP2a-EcoRI-For | ATATATGAATTCGCCACCATGAAATGGGGTCCATGC |
| GP2a-SphI-Rev | ATATATGCATGCTCACCGTGAGTTCGAAGG |
| GP2a-NheI-For | ATATGCTAGCGCCGCCACCATGAAATGGGGTCCATGC |
| GP2a-FLAG-EcoRI-Rev | ATATGAATTCTCACTCGAGCTTGTCATCGTCGTCCTTGTAGTCCATCCGTGAGTTCGAAGGAAAAATTGC |
| GP3-EcoRI-For | ATATAGAATTCGCCACCATGGCTAATAGCTGTGC |
| GP3-BamHI-Rev | TATATGGATCCCTATCGCCGCGCGGC |
| GP4-EcoRI-For | ATATAGAATTCGCCACCATGGCTGCGCCCCTTC |
| GP4-BamHI-Rev | TATATGGATCCTCAAATTGCCAGTAAGATG |
| GP5-EcoRI-For | GCCGGAATTCGGAGCCGCCGCCACCATGTTGGGGAGATGCTTGAC |
| GP5-HindIII-XhoI-Rev | ATCACTCGAGAAGCTTCTAAAGACGACCCCATTGTTC |
| CD163-HindIII-For | ATATAAGCTTATGGACAAACTCAGAATGGTGCTAC |
| CD163-XhoI-Rev | ATATCTCGAGTCATTGTACTTCAGAGTGGTCTCCTG |
Primer sequences are in the 5′ to 3′ direction. Restriction enzyme sites in the primers are underlined.
The ORF of the porcine CD163 receptor was amplified from the total RNA isolated from PAM cells by reverse transcription-PCR (RT-PCR) using specific primers (Table 1) and cloned in pGEM-T vector (Promega Corporation). Four clones of the CD163 receptor with inserts of the correct size were further transferred from the pGEM-T vector by restriction enzyme digestion with HindIII and XhoI and cloned by blunt-end ligation in pcDNA3.1(+) vector.
Transfection.
Plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen) as described earlier (1). Briefly, BHK-21 cells were plated in six-well culture dishes at 24 h before transfection. The cells were first infected with vTF7-3 (21) at a multiplicity of infection (MOI) of 5. The DNA-Lipofectamine 2000 complexes prepared according to the manufacturer's recommendations were added to the infected and washed BHK-21 cells and incubated at 37°C for 4 h. Following incubation, the cells were washed and incubated in Dulbecco's modified Eagle's medium (DMEM) containing 2% FBS and antibiotics as described before (1). The transfected cells were processed for radiolabeling or immunofluorescent staining as described below.
Metabolic labeling, IP, co-IP, and analysis of proteins.
The plasmid-transfected cells at 16 to 18 h posttransfection were washed twice in phosphate-buffered saline (PBS) and incubated with cysteine-methionine-free DMEM for 1 h prior to radiolabeling with 33 μCi of Expre35S35S protein labeling mix (Perkin-Elmer) per ml of methionine-cysteine-free DMEM without serum for 4 h. Following radiolabeling, the cells were washed with cold PBS and the cell lysate was prepared in radioimmunoprecipitation assay (RIPA) buffer as described before (1) or in coimmunoprecipitation (co-IP) buffer (1% NP-40, 0.5% Triton X-100, 50 mM Tris-HCl [pH 7.6], 500 mM NaCl, 2 mM EDTA, and 1× protease inhibitor cocktail) as described before (8). The cell lysate was clarified by centrifugation at 16,000 × g for 5 min. The supernatant was used for immunoprecipitation with the appropriate antibody at 4°C for 8 to 10 h. A slurry of approximately 3.0 mg of protein A-Sepharose (GE Healthcare Bioscience AB) washed and resuspended in 100 μl RIPA buffer was added and incubated further for 2 h at 4°C. The protein A-Sepharose beads with bound immune complexes were washed in RIPA buffer or co-IP buffer three times, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, and 10% glycerol), and boiled for 5 min. After centrifugation at 16,000 × g for 1 min, the proteins in the clarified supernatant were resolved by SDS-10% or -12% PAGE. Following electrophoresis, the gels were processed for fluorography as described previously (1). Endo H treatment of immunoprecipitated proteins was as described before (1).
Fluorescence microscopy.
The transfected cells were fixed either with 4% paraformaldehyde for 30 min at room temperature or with methanol-acetone solution (1:1) for 30 min at −20°C. The cells were blocked with 5% bovine serum albumin (BSA) in PBS containing 0.05% Tween20 for 30 min at room temperature, and immunofluorescence staining was performed with appropriate primary and secondary antibodies as described previously (6, 7) and visualized under an Olympus FV500/IX81 inverted laser scanning confocal microscope. The images were captured with a charge-coupled-device camera.
RESULTS
Expression of PRRSV minor glycoproteins in transfected cells.
The minor glycoproteins GP2a, GP3, and GP4 of PRRSV are 256 amino acids (aa), 254 aa, and 178 aa long, respectively. The unglycosylated forms of these proteins have predicted sizes of approximately 29 kDa, 29 kDa, and 20 kDa, respectively, whereas the predicted size of the unglycosylated FLAG-tagged GP2a used in later studies is approximately 30 kDa. Each of these proteins also has several predicted N-linked glycosylation sites as well as amino-terminal signal sequences (Fig. 1A). We had previously shown that the major glycoprotein GP5, when expressed ectopically in transfected cells, remained in the endoplasmic reticulum or in the cis-Golgi region (1). To examine expression of the minor glycoproteins in transfected cells, plasmids encoding these glycoproteins under the control of the T7 RNA polymerase promoter were transfected into BHK-21 cells that had been infected with vTF7-3, which expresses the T7 RNA polymerase. Expression of these proteins could be readily detected (Fig. 1B) by radiolabeling and immunoprecipitation with monospecific polyclonal antibodies prepared against peptides containing immunodominant epitopes (11) from each of these glycoproteins. The molecular masses of the fully mature glycoproteins (identified by white dots on the left sides of the lanes) were estimated to be approximately 32 kDa, 42 kDa, and 29 kDa, respectively, for the GP2a, GP3, and GP4 proteins. Although the major species of GP3 and GP4 corresponded to the fully glycosylated and mature forms of the proteins (white dots in lanes 4 and 6), the fully glycosylated GP2a (white dot in lane 2) represented only a small fraction, while a smaller GP2a was detected as the major protein species (top black dot in lane 2). The electrophoretic mobility of this protein appeared to be consistent with GP2a lacking glycans at one of the sites, and it may represent a partially glycosylated form of the protein. In addition, it should be noted that these proteins contain signal sequences at the amino terminus, which appear to be cleaved off to generate the mature proteins and therefore may be smaller than the expected nonglycosylated proteins. Furthermore, based on the predicted size, it appears that these proteins possess anomalous mobilities in the gels. Since the predominant protein species in cells transfected with the GP2a expression plasmid was smaller than the fully glycosylated mature GP2a protein, we examined whether incompletely glycosylated forms of GP2a and possibly other minor glycoproteins were also synthesized in MARC-145 cells, which are normally used for propagation of PRRSV. Additionally, since the major species of GP2a possessed an electrophoretic mobility similar to that of GP4 (Fig. 1B, lanes 2 and 6), we also examined the expression of a carboxy-terminal FLAG-tagged GP2a protein (GP2a-Fl) for use in later studies. Our results revealed that the predominant protein in MARC-145 cells transfected with GP2a-Fl-encoding plasmid also corresponded to the partially glycosylated form of the protein (Fig. 1C), as observed earlier in BHK-21 cells. Furthermore, the GP2a-Fl-tagged protein could be readily distinguished from the GP4 protein based on the mobility in the gel, a property that we have used in our protein interaction studies reported below. Interestingly, in MARC-145 cells, these three proteins produced a ladder-like pattern (Fig. 1C, lanes 2, 4, and 6), indicating that the proteins were also incompletely glycosylated in these cells. However, the fully mature glycoproteins (white dots in Fig. 1C, lanes 2, 4, and 6), with mobilities similar to those observed in BHK-21 cells, were readily detected in each case. Ladder-like patterns of the glycoproteins were also seen in MARC-145 cells infected with PRRSV FL-12 (data not shown). It is possible that the ladder-like pattern of the viral glycoproteins may represent various degraded forms of the proteins. However, our observation that single and multiple N-linked glycosylation mutant proteins comigrate with the protein bands seen in the ladder-like pattern (data to be reported elsewhere) and the fact that endo H digestion (Fig. 1D) has resulted in detection of single species of these proteins (except for GP2a, for which the signal-cleaved form of the protein is also detected) argue against the possibility that the ladder-like pattern represents degraded proteins.
FIG. 1.
Expression of individual minor envelope glycoproteins of PRRSV in transfected cells. (A) Schematic representation of GP2a, GP3, and GP4 proteins, with their sizes in amino acids shown. Potential N-linked glycosylation sites (with residue number and letter Y above of the rectangular boxes), predicted signal sequences (SS), and transmembrane regions (TM) are shown. The amino (N) and carboxy (C) termini are identified. (B) BHK-21 cells were infected with vTF7-3 and subsequently mock transfected or transfected with plasmids encoding individual envelope glycoproteins as shown above the lanes. The cells were radiolabeled with 35S protein labeling mix as described in Materials and Methods, cell extracts were prepared and immunoprecipitated with monospecific polyclonal antibodies as shown below the lanes, and the proteins were detected by SDS-10% PAGE and fluorography. The fully glycosylated proteins are identified by white dots, whereas the partially glycosylated forms of the proteins are identified by black dots, on the left sides of the lanes. Relative mobilities of molecular mass markers in kilodaltons (kDa) are shown on the right. (C) Expression of the GPs in MARC-145 cells. The experiment was done as described for panel B, but the plasmid encoding GP2a with a FLAG tag was used in place of GP2a and the fusion protein was detected with anti-FLAG antibody. The full-length mature GPs are identified with white dots in lanes 2, 4, and 6, whereas the partially glycosylated forms of the proteins are identified by black dots in the lanes. Relative mobilities of molecular mass markers in kDa are shown on the right. (D) Endo H sensitivity of the GPs. The proteins expressed in BHK-21 cells as described above were recovered by immunoprecipitation, treated without (−) or with (+) endo H, and detected by electrophoresis as described above. Mock-transfected cells immunoprecipitated with the antibodies and treated without endo H are shown in lanes 1, 4, and 7. White dots show the fully glycosylated proteins, and black dots show the protein species generated by the enzyme treatment. The protein band identified by an asterisk is most likely the signal-cleaved deglycosylated form of GP2a. Relative mobilities of molecular mass markers in kDa are shown on the right.
The intracellular localization of the minor glycoproteins in BHK-21 cells was further studied by examining their sensitivity to endo H digestion and also by confocal microscopy. All of the fully and partially glycosylated proteins in BHK-21 cells were found to contain only high-mannose-type glycans, as they were sensitive to digestion by endo H, yielding species that corresponded to the protein backbones only (Fig. 1D). It should be noted that a smaller protein species (identified by asterisk on the right of lane 3) was also detected. This protein is most likely the signal-cleaved GP2a without glycans. Immunofluorescent staining of cells expressing the individual glycoproteins with monospecific antibodies also showed that most of the minor glycoproteins were localized in the cytoplasm in association with the ER (data not shown). Similar results were also obtained in MARC-145 cells. Overall, our results show that the minor glycoproteins (GP2a, GP3, and GP4) synthesized in transfected cells are localized in the ER, similar to what was previously described for the major envelope protein GP5 (1, 53).
GP5 protein interacts strongly with GP4 protein of PRRSV.
To examine whether the envelope glycoproteins interact with each other, we cotransfected BHK-21 cells with two plasmids at a time encoding GP2a-Fl, GP3, GP4, or GP5. Approximately 16 h after transfection, the cells were radiolabeled and the cell extracts were examined for protein interactions by co-IP assays using a monospecific antibody against one of the proteins. The results (Fig. 2A) show that the GP2a polyclonal antibody was able to coimmunoprecipitate GP3 (lane 6) or GP4 (lane 7) protein when they were expressed together. Since the GP2a antibody did not immunoprecipitate GP3 (lane 3) or GP4 (lane 4) protein when these proteins were expressed alone in transfected cells, coimmunoprecipitation of GP3 or GP4 by GP2a antibody in cotransfected cells indicates that GP2a interacts with both of these proteins. No detectable level of interaction between GP2a and GP5 was observed (lane 8) under similar conditions in repeat experiments. Similarly, by the use of anti-GP3 polyclonal antibody, GP2a (Fig. 2B, lane 9) and GP4 (lane 10) proteins could be coimmunoprecipitated with GP3 in cotransfected cells, suggesting that GP3 interacts with GP2a and GP4. Interaction between GP3 and GP5 was not observed (lane 11) under these conditions. By the use of monospecific anti-GP5 antibody, GP4 protein could be efficiently immunoprecipitated from cells cotransfected with plasmids encoding the GP4 and GP5 proteins (Fig. 2C, lane 11), indicating strong interaction between these two proteins. GP2a protein could be consistently detected at low levels when coexpressed with GP5 and immunoprecipitated with anti-GP5 antibody (lane 9). However, undetectable to very low levels of GP3 protein could be seen in some but not all experiments when GP3 was coexpressed with GP5 and immunoprecipitated with anti-GP5 antibody (lane 10). Similar results were also obtained using the anti-GP4 polyclonal antibody (data not shown). Overall, the results from these co-IP studies suggest that GP2a protein interacts with the GP3, GP4, and GP5 proteins; GP3 interacts with GP2a and GP4 but not with GP5; GP4 also interacts with all the three glycoproteins; and GP5 interacts with the GP4 and GP2a proteins. Although the extent of interaction between the GPs is difficult to estimate from the co-IP studies, it is clear that the interaction of GP4 with GP5 appears to be much stronger than the interactions between the other GPs.
FIG. 2.
Interaction of PRRSV GPs. (A) Examination of GP interactions using GP2a antibody. BHK-21 cells were infected with vTF7-3 virus and mock transfected (lane 1) or transfected with plasmids encoding the GPs as shown below the panel. − and + indicate transfection without or with the plasmid shown on the left. At 16 h posttransfection, the cells were radiolabeled with 35S protein labeling mix as described in Materials and Methods, cell extracts were prepared and immunoprecipitated with monospecific polyclonal antibodies as shown below the lanes, and the proteins were detected by SDS-12% PAGE and fluorography. The full-length mature GPs are identified with white dots, whereas the partially glycosylated forms of the proteins are identified by black dots, on the left sides of the lanes. Relative mobilities of molecular mass markers in kDa are shown on the right. (B) Examination of GP interactions using anti-GP3 antibody. The experiment was performed as described for panel A but using anti-GP3 antibody. (C) Examination of GP interactions using anti-GP5 antibody. The experiment was performed as described for panel A but using anti-GP5 antibody. Relative mobilities of molecular mass markers in kDa are shown on the right.
GP4 mediates interactions resulting in detection of a multiprotein complex.
To further examine whether multiprotein glycoprotein complexes can be detected by co-IP assay, we transiently expressed multiple glycoproteins of PRRSV in BHK-21 cells and attempted to pull down all the interacting glycoproteins using one monospecific antibody by co-IP. We chose to use GP3 antibody in this study, as the use of this antibody in BHK-21 cell extracts resulted in fewer background bands (Fig. 1B). We observed that the GP3 antibody was not able to pull down GP5 protein when both of these proteins were coexpressed (Fig. 3, lane 6), confirming the results obtained previously. Furthermore, this antibody was able to pull down small amounts of GP2a protein but not GP5 protein when these three proteins were coexpressed (lane 7), indicating that GP3 interaction with GP2a is not sufficient to pull down GP5 protein in co-IP assay. However, when the GP3, GP4, and GP5 proteins were coexpressed, all the three proteins could be specifically immunoprecipitated with anti-GP3 antibody (lane 8), indicating that GP4 protein was important for generation of this tripartite glycoprotein complex. Additionally, when all the four glycoproteins were coexpressed and immunoprecipitation was carried out with GP3 antibody, all four proteins could be detected (lane 9). The results suggest that GP4 most likely mediates the interactions among PRRSV glycoproteins to generate the multiprotein glycoprotein complex.
FIG. 3.
Interaction of GP5 with GP4 is necessary for multiprotein complex formation. BHK-21 cells were infected with vTF7-3 and subsequently mock transfected (lane 1) or transfected with individual or combinations of plasmids encoding various GPs as shown below each lane. Cells were radiolabeled as described in Materials and Methods and immunoprecipitated with antibodies as shown below the lanes. The proteins were detected by SDS-12% PAGE and fluorography. Various GPs are identified on the right. The fully glycosylated GPs are identified with white dots on the left side of the lanes. GP2a (1N), the monoglycosylated form of GP2a, is identified with black dots on the left sides of the lanes. Relative mobilities of molecular mass markers in kDa are shown on the left.
Cloning, expression, and functionality of porcine CD163, the cellular receptor for PRRSV.
It was previously reported that the expression of porcine CD163 in a nonpermissive cell confers susceptibility to PRRSV infection (5). A recent study (48) suggested that CD163 acts internally to uncoat the virus genome and that expression of both CD163 and sialoadhesin is required for efficient infection by PRRSV. However, in the same study (48), it was also shown that expression of CD163 alone in BHK-21 cells, which are otherwise nonpermissive to PRRSV infection, led to productive infection, indicating that CD163 alone is sufficient to establish a productive replicative cycle of the virus. Therefore, PRRSV must bind to cells expressing CD163 to initiate the viral replication process. To determine which of the PRRSV envelope glycoproteins interact with CD163, we first cloned in pcDNA3.1(+) vector the full-length cDNA of CD163 from porcine alveolar macrophage (PAM) cells by RT-PCR amplification of total RNA from the cells using CD163-specific primers (Table 1). Four clones were used to examine the expression of the encoded protein by transfection, radiolabeling, and immunoprecipitation with porcine anti-CD163 antibody. We observed that three of the clones expressed proteins (Fig. 4A, lanes 3 to 5) that could be specifically immunoprecipitated with anti-CD163 antibody and possessed an electrophoretic mobility (∼130 kDa) corresponding to the full-length CD163 (5). A smaller protein product of ∼100 kDa encoded in another clone was also detected by the CD163 antibody (Fig. 4A, lane 2). Sequence analysis of the clones showed that CD163 in the clone that produced the smaller protein contained a premature termination codon at aa position 893, resulting in a truncated protein of 892 aa corresponding to the observed molecular mass. This truncated protein lacks the carboxy-terminal cytoplasmic domain, the transmembrane (TM) domain, and the ninth repeat unit of the scavenger receptor cysteine-rich (SRCR) protein domain (33). We have termed this truncated protein CD163ΔTM. The CD163 cDNAs in the other clones contained full-length sequences of 1,115 amino acids with greater than 99% sequence identity with the reported sequence of the porcine CD163 (5, 40).
FIG. 4.
Expression and functional analysis of CD163. (A) Expression of CD163 in transfected cells. BHK-21 cells were infected with vTF7-3 and transfected with pcDNA3.1 (E. Vector, lane 1) or CD163 cDNA containing clones (no. 1, 2, 3, and 4 [lanes 2 to 5, respectively]). Cells were radiolabeled for 4 h at 16 h posttransfection, and the radiolabeled proteins were analyzed by immunoprecipitation with porcine anti-CD163 monoclonal antibody, resolved by SDS-10% PAGE, and detected by fluorography. Mobilities of full-length CD163 and its truncated form (CD163ΔTM) are shown on the right. Relative mobilities of molecular mass markers in kDa are shown on the left. (B) Susceptibility of BHK-21 cells expressing CD163 encoded in the clones to PRRSV infection. Cells were transfected with the plasmid clones as shown (EV, empty vector control), and expression of CD163 was driven by the CMV promoter in pcDNA3.1 vector. At 48 h after transfection, cells were infected with PRRSV and expression of N protein was examined using the monoclonal antibody SDOW17 and secondary antibody conjugated to Alexa-488. (C) Immunofluorescent staining of BHK-21 cells transfected with empty vector (panels a and b), full-length CD163 receptor encoding clone 2 (panels c and d), or the CD163ΔTM-encoding clone (panels e and f). Cell surface or cytoplasmic staining was performed with CD163 monoclonal antibody and Alexa-488 conjugated secondary antibody.
To determine if the cloned CD163 confers susceptibility to PRRSV infection, we transfected the plasmids into BHK-21 cells, which are nonpermissive to PRRSV infection. Subsequently, at about 48 h posttransfection the cells were infected with PRRSV. Synthesis of N protein, which is indicative of PRRSV entry, transcription, and replication was examined in these cells by immunofluorescent staining of the cells at 48 h postinfection with anti-N monoclonal antibody SDOW17 (38). Synthesis of the N protein was readily detected in the cells transfected with clones encoding the full-length CD163 (Fig. 4B), indicating that the CD163 encoded in these clones conferred susceptibility to PRRSV infection. The N protein was not detected in cells transfected with the clone encoding CD163ΔTM, suggesting that this truncated protein is nonfunctional in conferring PRRSV susceptibility to the cells. The inability of CD163ΔTM to confer PRRSV susceptibility to the cells is not due to low levels of expression of the protein, since under similar transfection conditions, CD163ΔTM is detected at least at levels similar to or greater than that of the full-length protein (Fig. 4A). The results indicate that the carboxy-terminal 223 amino acids of CD163 are required for the function of the protein in conferring PRRSV susceptibility. It is possible that CD163ΔTM may have defective cellular localization or improper folding, leading to loss of its function.
Immunofluorescence microscopic examination of cells transfected with one of the full-length CD163 clones showed that CD163 protein was localized on the plasma membrane (Fig. 4C, panel c) as well as in the cytoplasm (Fig. 4C, panel d). Cells transfected with the clone encoding CD163ΔTM showed no surface expression of the protein (Fig. 4C, panel e), although the protein was expressed in the cytoplasm (Fig. 4C, panel f), indicating that CD163ΔTM is defective in plasma membrane localization. Empty vector-transfected cells did not exhibit any immunofluorescent staining on the plasma membrane or in the cytoplasm (Fig. 4C, panels a and b).
The GP2a and GP4 proteins of PRRSV interact with the porcine CD163 receptor.
We next wanted to examine which of the PRRSV envelope glycoproteins interact with the receptor CD163. To perform these studies, the CD163 along with each of the four envelope glycoproteins was coexpressed in transfected cells, and interactions were examined by co-IP with porcine CD163 monoclonal antibody. This antibody did not immunoprecipitate the individual glycoproteins when these glycoproteins were expressed alone (Fig. 5A, lanes 7 to 10), demonstrating that the antibody does not immunoprecipitate the proteins nonspecifically. However, when the individual glycoproteins were coexpressed with CD163, GP2a and GP4 proteins could be specifically immunoprecipitated with anti-CD163 antibody (lanes 11 and 13) in multiple repeat experiments. Interestingly, both the mature GP2a and partially glycosylated GP2a proteins (identified by white and black dots, respectively, in lane 11) could be immunoprecipitated with the CD163 antibody (lane 11), indicating that both forms of GP2a interact with CD163. The use of GP-specific antibodies also showed that CD163 protein could be immunoprecipitated with anti-GP2a or anti-GP4 antibodies (data not shown). The GP3 and GP5 proteins could not be detected by immunoprecipitation with anti-CD163 antibody (lanes 12 and 14) from cotransfected cells. These results suggest that the GP3 and GP5 proteins do not interact with the CD163 or that their interactions cannot be detected under conditions in which interactions with GP2a and GP4 could be readily detected.
FIG. 5.
Interaction of CD163 with PRRSV GPs. (A) BHK-21 cells were infected with vTF7-3 and transfected with various GPs alone or in combination with CD163 (clone 2). The cells were radiolabeled, and the proteins were immunoprecipitated with antibodies as shown below the lanes. Immunoprecipitated proteins were detected by SDS-10% PAGE and fluorography. Mobilities of molecular mass marker proteins in kDa are shown on the right. CD163 and the fully glycosylated viral GPs are identified on the left. Fully glycosylated GPs are identified with white dots in the fluorograms on the left side of each lane. The monoglycosylated GP2a is identified with black dots on the left side in lanes 3 and 11. (B). Interaction of GP2a and GP4 with CD163ΔTM. The experiment was performed as described for panel A using the clone encoding the CD163ΔTM protein.
Further, we performed a co-IP assay in which CD163ΔTM was coexpressed with the viral GPs (Fig. 5B). By using the CD163 monoclonal antibody, we were able to pull down GP2a or GP4 protein, indicating that the 223 amino acids from the carboxy terminus of CD163 that are missing in CD163ΔTM are not required for its interaction with the GP2a or GP4 protein.
DISCUSSION
PRRSV contains one major glycoprotein (GP5) and three minor glycoproteins (GP2a, GP3, and GP4) on the virion envelope. All of these glycoproteins are required for generation of infectious virions, indicating that they play critical roles in virion assembly and/or interactions with the cell surface receptor for virus entry and/or uncoating. Previous studies have reported that GP2a, GP3, GP4, and the unglycosylated envelope protein 2b form a multiprotein complex in cells expressing these proteins (53). In this study, we have examined the interactions of the envelope glycoproteins among themselves as well as the interactions of these proteins with the cell surface receptor, CD163. Overall, our studies lead us to conclude that (i) the GP4 and GP2a proteins interact with the other GPs; (ii) GP4 mediates the formation of the multiprotein complex between the minor GPs and the major envelope GP, GP5; and (iii) GP4 along with GP2a interact with CD163. These studies for the first time show that the GP4 protein is the key glycoprotein of PRRSV that is responsible for formation of the multiprotein complex and, along with GP2a, may serve as the viral attachment protein, allowing virus binding to the cell surface receptor during infection.
Expression of the individual minor glycoproteins in transfected cells resulted in detection of these proteins by the corresponding monospecific antibodies. Several partially glycosylated forms of the proteins could be readily detected by the antibodies. Particularly striking was the detection of a ladder-like pattern of GP3 in MARC-145 cells (Fig. 1C), which most likely corresponds to the partially glycosylated forms of the protein. Partially glycosylated forms were also observed for the GP2a and GP4 proteins in both BHK-21 and MARC-145 cells. For GP2a, the size of the major species of the protein corresponds to that lacking one glycan moiety. GP2a contains two N-glycosylation sites, at residues 178 and 184. The reason(s) for the synthesis of predominantly a GP2a molecule with one glycan moiety is not clear at this time. However, it is possible that one of the N-glycosylation sites is glycosylated inefficiently or with reduced frequency, resulting in a predominantly monoglycosylated GP2a. It has been suggested that N-glycosylation sites are less likely to be used if they have not passed through the oligosaccharyltransferase (OST) active site prior to translation termination (22). The estimated distance between the peptidyl transferase site of a ribosome attached to the translocation pore on the ER membrane and the active site of OST complex roughly corresponds to 75 amino acid residues (3, 22, 23, 51). Since cotranslational and posttranslational glycosylation on a single protein have been demonstrated (4) and such modifications occur by distinct mammalian OST isoforms (39), we speculate that the glycosylation site at residue 184 in GP2a is most likely modified inefficiently and/or modified by posttranslational glycosylation. It is possible that glycan addition at the sites in the GP3 and GP4 proteins is also inefficient and may occur through co- and posttranslational mechanisms, resulting in ladder-like patterns of these proteins as seen in Fig. 1. We are currently examining whether the PRRSV GPs are glycosylated through cotranslational as well as posttranslational mechanisms.
It has been previously reported for the genotype II Lelystad virus that the envelope minor glycoproteins and 2b (or E) protein together form a heterotetrameric complex and that this complex formation is crucial for intracellular targeting of these proteins from the ER through the Golgi complex to the plasma membrane (53). Our studies not only confirm these observations but also show that the major envelope glycoprotein GP5 interacts with GP4 strongly. Although direct interaction between GP3 and GP5 could not be demonstrated, a multiprotein complex comprising all four envelope glycoproteins could be readily detected by immunoprecipitation with anti-GP3 antibody (Fig. 3). Our studies suggest that GP4 mediates the formation of the large multiprotein complex between the minor and the major envelope proteins of PRRSV. In cells expressing GP5 alone or in the presence of M protein, GP5 does not acquire endo-H resistance even after 4 h of synthesis (1), but in virus-infected cells or in the extracellular virions, the GP5 becomes endo H resistant (1). These results indicate that the multiprotein complex generated by the interactions of the envelope proteins may be critical for the transport of these proteins from the ER to the Golgi complex and finally to the plasma membrane for incorporation into virus particles, consistent with previous findings that the minor envelope glycoproteins of Lelystad virus form a multimeric complex for their transport to the plasma membrane (53).
Although multiprotein complexes can be detected in transfected cells, the molar ratio of the proteins in such complexes is unknown. Since the GP5 and M proteins are the major envelope proteins, they are likely distributed uniformly throughout the virion envelope. Recent cryo-electron tomographic studies (44) reveal that PRRSV particles display mostly smooth envelope outlines, for which the authors suggest that the small ectodomains of GP5 and M proteins are possibly not large enough to be detected as visible spikes on the virion envelope. On the other hand, the authors also detected a few features protruding from the virion envelope, which most likely correspond to the large multimeric complexes made from the bulkier but less abundant minor envelope glycoproteins such as GP2a (44). We reason that the protruding features seen on the PRRSV virion envelope represent the multimeric complexes of the minor envelope glycoproteins with or without the GP5 and M proteins, and these are likely to be directly involved in interactions with the cell surface receptor during PRRSV infection.
The data presented here support the contention that the multimeric complexes of the minor glycoproteins with or without the M and GP5 proteins are involved in direct interaction with the cell surface receptor. Since the GP5 and M proteins are present in abundant amounts on the virion envelope, these two proteins were considered to play a major role in receptor interaction. However, this contention was challenged by the results from the studies using chimeric viruses (17, 50), which suggested that the GP5 and M proteins do not play a role in receptor interaction. Our studies show that only the GP2a and GP4 proteins interact with CD163. It appears that GP4 is a critical viral envelope protein that not only mediates interactions with other GPs on the virion envelope but also, along with GP2a, mediates interactions with CD163 for virus entry. Since the minor envelope proteins are not required for particle formation (53) and there are only a few large multimeric glycoprotein complexes present on the virion envelope (44), we suggest that the major function of the minor glycoprotein complex on the virion envelope is to interact specifically with the cell surface receptor for virus entry. On the other hand, the GP5 and M heterodimeric complexes, which are present in large amounts and are uniformly distributed throughout the virion envelope (44), play major roles in virion assembly. The GP5-M complexes on the virion envelope may also play additional roles in nonspecific interactions with heparin-like receptors on PAM cells, since M protein has been shown to bind to such molecules (15). The nonspecific interaction would allow initial virus binding to the cell surface followed by specific interaction with the receptor through the GP2a and GP4 proteins for receptor-mediated entry of the virion.
It is possible, then, that GP2a and GP4, by containing viral receptor-interacting domains, would potentially be involved in the establishment of protective immunity against PRRSV infection. Viral receptor-interacting proteins and domains are known to induce highly neutralizing antibodies and contribute important targets for vaccine and therapeutic development (16). While a single report so far suggests that GP4 contains at least one neutralizing epitope (37), the full potential of GP4 and GP2a to induce PRRSV-neutralizing antibodies and T-cell immunity should be further investigated. This becomes particularly important in light of recent studies indicating that viral receptor-interacting domains may be directly involved in the induction of broadly reacting neutralizing antibodies, as shown for hepatitis C virus (45), severe acute respiratory syndrome (SARS) coronavirus (18), influenza virus (31), paramyxovirus (26), and others. These broadly reactive, cross-neutralizing, and possibly cross-protective antibodies may be of central importance for protection against highly variable viruses such as PRRSV (32).
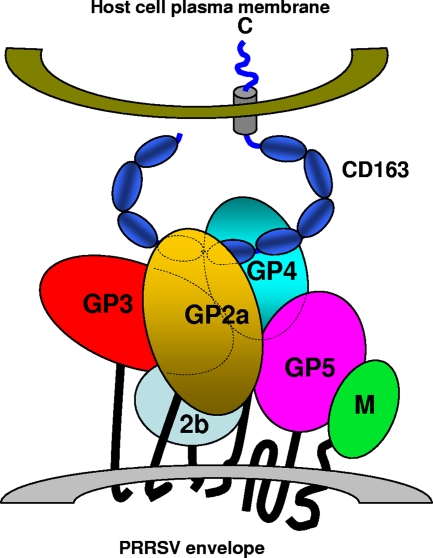
Based on the results presented in this communication, we propose a tentative model for the multiprotein complex on the PRRSV envelope and its interaction with the cell surface receptor CD163 (Fig. 6). Although the involvement of proteins 2b and M in the formation of this complex or interaction with CD163 has not been examined in the current study, several of these interactions have already been confirmed. Further studies are being conducted to test this preliminary model of GP-CD163 interaction.
FIG. 6.
A preliminary model of the PRRSV envelope protein complex and its interaction with CD163 on the host cell plasma membrane. For the sake of simplicity and convenience, the GPs are depicted as globular structures with lines spanning the viral envelope. CD163 is shown as a structure with an extracellular region having repeating units (corresponding to the nine SRCR domains) projecting from the plasma membrane of a host cell. The carboxy-terminal cytoplasmic domain and the transmembrane domains are also shown. The dotted lines in the schematic represent the regions of the proteins that are masked by GP2a.
Acknowledgments
We thank You Zhou and Terri Fangman of the microscopy core facility of Center for Biotechnology, University of Nebraska-Lincoln, for their help with fluorescence microscopy studies. We thank Jonathan Cooney for helping with various laboratory procedures, Debasis Panda for comments on the manuscript, and Carl Gagnon (University of Quebec, Montreal, Canada) for providing anti-GP5 polyclonal antibody.
The research was supported by grants from the USDA-NRICGP (project no. 2008-00903), USDA-AFRI (project no. 2009-01654), and National Pork Board (NPB no. 08-248 and no. 08-253).
Footnotes
Published ahead of print on 25 November 2009.
REFERENCES
- 1.Ansari, I. H., B. Kwon, F. A. Osorio, and A. K. Pattnaik. 2006. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J. Virol. 80:3994-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasuriya, U. B., and N. J. MacLachlan. 2004. The immune response to equine arteritis virus: potential lessons for other arteriviruses. Vet. Immunol. Immunopathol. 102:107-129. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Dor, S., N. Esterman, E. Rubin, and N. Sharon. 2004. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology 14:95-101. [DOI] [PubMed] [Google Scholar]
- 4.Bolt, G., C. Kristensen, and T. D. Steenstrup. 2005. Posttranslational N-glycosylation takes place during the normal processing of human coagulation factor VII. Glycobiology 15:541-547. [DOI] [PubMed] [Google Scholar]
- 5.Calvert, J. G., D. E. Slade, S. L. Shields, R. Jolie, R. M. Mannan, R. G. Ankenbauer, and S. K. Welch. 2007. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 81:7371-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das, S. C., D. Nayak, Y. Zhou, and A. K. Pattnaik. 2006. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J. Virol. 80:6368-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das, S. C., D. Panda, D. Nayak, and A. K. Pattnaik. 2009. Biarsenical labeling of vesicular stomatitis virus encoding tetracysteine-tagged M protein allows dynamic imaging of M protein and virus uncoating in infected cells. J. Virol. 83:2611-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das, S. C., and A. K. Pattnaik. 2005. Role of the hypervariable hinge region of phosphoprotein P of vesicular stomatitis virus in viral RNA synthesis and assembly of infectious virus particles. J. Virol. 79:8101-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dea, S., C. A. Gagnon, H. Mardassi, B. Pirzadeh, and D. Rogan. 2000. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch. Virol. 145:659-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lima, M., I. H. Ansari, P. B. Das, B. J. Ku, F. J. Martinez-Lobo, A. K. Pattnaik, and F. A. Osorio. 2009. GP3 is a structural component of the PRRSV type II (US) virion. Virology 390:31-36. [DOI] [PubMed] [Google Scholar]
- 11.de Lima, M., A. K. Pattnaik, E. F. Flores, and F. A. Osorio. 2006. Serologic marker candidates identified among B-cell linear epitopes of Nsp2 and structural proteins of a North American strain of porcine reproductive and respiratory syndrome virus. Virology 353:410-421. [DOI] [PubMed] [Google Scholar]
- 12.Delputte, P. L., S. Costers, and H. J. Nauwynck. 2005. Analysis of porcine reproductive and respiratory syndrome virus attachment and internalization: distinctive roles for heparan sulphate and sialoadhesin. J. Gen. Virol. 86:1441-1445. [DOI] [PubMed] [Google Scholar]
- 13.Delputte, P. L., and H. J. Nauwynck. 2004. Porcine arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J. Virol. 78:8094-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delputte, P. L., W. Van Breedam, I. Delrue, C. Oetke, P. R. Crocker, and H. J. Nauwynck. 2007. Porcine arterivirus attachment to the macrophage-specific receptor sialoadhesin is dependent on the sialic acid-binding activity of the N-terminal immunoglobulin domain of sialoadhesin. J. Virol. 81:9546-9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delputte, P. L., N. Vanderheijden, H. J. Nauwynck, and M. B. Pensaert. 2002. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J. Virol. 76:4312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delrue, I., P. L. Delputte, and H. J. Nauwynck. 2009. Assessing the functionality of viral entry-associated domains of porcine reproductive and respiratory syndrome virus during inactivation procedures, a potential tool to optimize inactivated vaccines. Vet. Res. 40:62. [DOI] [PubMed] [Google Scholar]
- 17.Dobbe, J. C., Y. van der Meer, W. J. Spaan, and E. J. Snijder. 2001. Construction of chimeric arteriviruses reveals that the ectodomain of the major glycoprotein is not the main determinant of equine arteritis virus tropism in cell culture. Virology 288:283-294. [DOI] [PubMed] [Google Scholar]
- 18.Du, L., Y. He, Y. Zhou, S. Liu, B. J. Zheng, and S. Jiang. 2009. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 7:226-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faaberg, K. S., C. Even, G. A. Palmer, and P. G. Plagemann. 1995. Disulfide bonds between two envelope proteins of lactate dehydrogenase-elevating virus are essential for viral infectivity. J. Virol. 69:613-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsberg, R. 2005. Divergence time of porcine reproductive and respiratory syndrome virus subtypes. Mol. Biol. Evol. 22:2131-2134. [DOI] [PubMed] [Google Scholar]
- 21.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gavel, Y., and G. von Heijne. 1990. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 3:433-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glabe, C. G., J. A. Hanover, and W. J. Lennarz. 1980. Glycosylation of ovalbumin nascent chains. The spatial relationship between translation and glycosylation. J. Biol. Chem. 255:9236-9242. [PubMed] [Google Scholar]
- 24.Gonin, P., H. Mardassi, C. A. Gagnon, B. Massie, and S. Dea. 1998. A nonstructural and antigenic glycoprotein is encoded by ORF3 of the IAF-Klop strain of porcine reproductive and respiratory syndrome virus. Arch. Virol. 143:1927-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanada, K., Y. Suzuki, T. Nakane, O. Hirose, and T. Gojobori. 2005. The origin and evolution of porcine reproductive and respiratory syndrome viruses. Mol. Biol. Evol. 22:1024-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashiguchi, T., M. Kajikawa, N. Maita, M. Takeda, K. Kuroki, K. Sasaki, D. Kohda, Y. Yanagi, and K. Maenaka. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. U. S. A. 104:19535-19540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 28.Kim, J. K., A. M. Fahad, K. Shanmukhappa, and S. Kapil. 2006. Defining the cellular target(s) of porcine reproductive and respiratory syndrome virus blocking monoclonal antibody 7G10. J. Virol. 80:689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroese, M. V., J. C. Zevenhoven-Dobbe, J. N. Bos-de Ruijter, B. P. Peeters, J. J. Meulenberg, L. A. Cornelissen, and E. J. Snijder. 2008. The nsp1alpha and nsp1 papain-like autoproteinases are essential for porcine reproductive and respiratory syndrome virus RNA synthesis. J. Gen. Virol. 89:494-499. [DOI] [PubMed] [Google Scholar]
- 30.Lee, C., and D. Yoo. 2006. The small envelope protein of porcine reproductive and respiratory syndrome virus possesses ion channel protein-like properties. Virology 355:30-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim, A. P., C. E. Chan, S. K. Wong, A. H. Chan, E. E. Ooi, and B. J. Hanson. 2008. Neutralizing human monoclonal antibody against H5N1 influenza HA selected from a Fab-phage display library. Virol. J. 5:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez, O. J., and F. A. Osorio. 2004. Role of neutralizing antibodies in PRRSV protective immunity. Vet. Immunol. Immunopathol. 102:155-163. [DOI] [PubMed] [Google Scholar]
- 33.Madsen, M., H. J. Moller, M. J. Nielsen, C. Jacobsen, J. H. Graversen, T. van den Berg, and S. K. Moestrup. 2004. Molecular characterization of the haptoglobin.hemoglobin receptor CD163. Ligand binding properties of the scavenger receptor cysteine-rich domain region. J. Biol. Chem. 279:51561-51567. [DOI] [PubMed] [Google Scholar]
- 34.Mardassi, H., P. Gonin, C. A. Gagnon, B. Massie, and S. Dea. 1998. A subset of porcine reproductive and respiratory syndrome virus GP3 glycoprotein is released into the culture medium of cells as a non-virion-associated and membrane-free (soluble) form. J. Virol. 72:6298-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mardassi, H., B. Massie, and S. Dea. 1996. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 221:98-112. [DOI] [PubMed] [Google Scholar]
- 36.Meulenberg, J. J. 2000. PRRSV, the virus. Vet. Res. 31:11-21. [DOI] [PubMed] [Google Scholar]
- 37.Meulenberg, J. J., A. P. van Nieuwstadt, A. van Essen-Zandbergen, and J. P. Langeveld. 1997. Posttranslational processing and identification of a neutralization domain of the GP4 protein encoded by ORF4 of Lelystad virus. J. Virol. 71:6061-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson, E. A., J. Christopher-Hennings, T. Drew, G. Wensvoort, J. E. Collins, and D. A. Benfield. 1993. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 31:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruiz-Canada, C., D. J. Kelleher, and R. Gilmore. 2009. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136:272-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez-Torres, C., P. Gomez-Puertas, M. Gomez-del-Moral, F. Alonso, J. M. Escribano, A. Ezquerra, and J. Dominguez. 2003. Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Arch. Virol. 148:2307-2323. [DOI] [PubMed] [Google Scholar]
- 41.Shanmukhappa, K., J. K. Kim, and S. Kapil. 2007. Role of CD151, a tetraspanin, in porcine reproductive and respiratory syndrome virus infection. Virol. J. 4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snijder, E. J., J. C. Dobbe, and W. J. Spaan. 2003. Heterodimerization of the two major envelope proteins is essential for arterivirus infectivity. J. Virol. 77:97-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 44.Spilman, M. S., C. Welbon, E. Nelson, and T. Dokland. 2009. Cryo-electron tomography of porcine reproductive and respiratory syndrome virus: organization of the nucleocapsid. J. Gen. Virol. 90:527-535. [DOI] [PubMed] [Google Scholar]
- 45.Tarr, A. W., A. M. Owsianka, J. M. Timms, C. P. McClure, R. J. Brown, T. P. Hickling, T. Pietschmann, R. Bartenschlager, A. H. Patel, and J. K. Ball. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43:592-601. [DOI] [PubMed] [Google Scholar]
- 46.Truong, H. M., Z. Lu, G. F. Kutish, J. Galeota, F. A. Osorio, and A. K. Pattnaik. 2004. A highly pathogenic porcine reproductive and respiratory syndrome virus generated from an infectious cDNA clone retains the in vivo virulence and transmissibility properties of the parental virus. Virology 325:308-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Aken, D., J. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2006. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 87:3473-3482. [DOI] [PubMed] [Google Scholar]
- 48.Van Gorp, H., W. Van Breedam, P. L. Delputte, and H. J. Nauwynck. 2008. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 89:2943-2953. [DOI] [PubMed] [Google Scholar]
- 49.van Nieuwstadt, A. P., J. J. Meulenberg, A. van Essen-Zanbergen, A. Petersen-den Besten, R. J. Bende, R. J. Moormann, and G. Wensvoort. 1996. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. J. Virol. 70:4767-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verheije, M. H., T. J. Welting, H. T. Jansen, P. J. Rottier, and J. J. Meulenberg. 2002. Chimeric arteriviruses generated by swapping of the M protein ectodomain rule out a role of this domain in viral targeting. Virology 303:364-373. [DOI] [PubMed] [Google Scholar]
- 51.Whitley, P., I. M. Nilsson, and G. von Heijne. 1996. A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 271:6241-6244. [DOI] [PubMed] [Google Scholar]
- 52.Wieringa, R., A. A. de Vries, J. van der Meulen, G. J. Godeke, J. J. Onderwater, H. van Tol, H. K. Koerten, A. M. Mommaas, E. J. Snijder, and P. J. Rottier. 2004. Structural protein requirements in equine arteritis virus assembly. J. Virol. 78:13019-13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wissink, E. H., M. V. Kroese, H. A. van Wijk, F. A. Rijsewijk, J. J. Meulenberg, and P. J. Rottier. 2005. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J. Virol. 79:12495-12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, W. H., Y. Fang, R. R. Rowland, S. R. Lawson, J. Christopher-Hennings, K. J. Yoon, and E. A. Nelson. 2005. The 2b protein as a minor structural component of PRRSV. Virus Res. 114:177-181. [DOI] [PMC free article] [PubMed] [Google Scholar]