Abstract
Infectious pancreatic necrosis virus (IPNV), a pathogen of salmon and trout, imposes a severe toll on the aquaculture and sea farming industries. IPNV belongs to the Aquabirnavirus genus in the Birnaviridae family of bisegmented double-stranded RNA viruses. The virions are nonenveloped with a T=13l icosahedral capsid made by the coat protein VP2, the three-dimensional (3D) organization of which is known in detail for the family prototype, the infectious bursal disease virus (IBDV) of poultry. A salient feature of the birnavirus architecture is the presence of 260 trimeric spikes formed by VP2, projecting radially from the capsid. The spikes carry the principal antigenic sites as well as virulence and cell adaptation determinants. We report here the 3.4-Å resolution crystal structure of a subviral particle (SVP) of IPNV, containing 20 VP2 trimers organized with icosahedral symmetry. We show that, as expected, the SVPs have a very similar organization to the IBDV counterparts, with VP2 exhibiting the same overall 3D fold. However, the spikes are significantly different, displaying a more compact organization with tighter packing about the molecular 3-fold axis. Amino acids controlling virulence and cell culture adaptation cluster differently at the top of the spike, i.e., in a central bowl in IBDV and at the periphery in IPNV. In contrast, the spike base features an exposed groove, conserved across birnavirus genera, which contains an integrin-binding motif. Thus, in addition to revealing the viral antigenic determinants, the structure suggests that birnaviruses interact with different receptors for attachment and for cell internalization during entry.
Birnaviruses form a distinct family of double-stranded RNA (dsRNA) viruses infecting vertebrates and invertebrates (18). Aquatic birnaviruses are the most abundant and diverse and are grouped in two separate genera: the Aquabirnavirus genus and the Blosnavirus genus, with infectious pancreatic necrosis virus (IPNV) and blotched snakehead virus (BSNV) (16) as respective type species. A third aquatic birnavirus of unassigned genus, Tellina virus 1, was recently described and found to be phylogenetically distant from the two established genera (40). However, the vast majority of aquatic birnaviruses are antigenically related to IPNV (i.e., belong to the Aquabirnavirus genus), regardless of host species or geographical origin (4, 11, 26, 39). They are implicated as etiological agents of disease in a variety of mollusks and fish species important in aquaculture, causing pathologies such as infectious pancreatic necrosis in salmonids, nephroblastoma and branchionephritis in eels, and gill necrosis in clams (21, 34, 50). Viruses in the Aquabirnavirus genus display considerable antigenic diversity and have substantial differences in biological properties such as host range and optimal replication temperature. These features contrast with the properties of other birnaviruses, in particular those infecting terrestrial species (avibirnaviruses and entomobirnaviruses). Based on reciprocal neutralization tests with polyclonal and monoclonal antibodies, nine cross-reacting serotypes (A1 to A9) have been defined for IPNV and related aquabirnaviruses (26). Serotype A2 (also known as Sp) is the most common serotype found in Europe.
Birnavirus particles are nonenveloped, displaying a single-shelled T=13l icosahedral capsid of about 70 nm in diameter, composed of 260 trimers of viral protein 2 (VP2) (5, 15, 42). Internal to the virion are VP3, which forms a ribonucleoprotein complex with the genomic RNA (9, 27, 36), and VP1, the viral RNA-dependent RNA polymerase, which is found both free and covalently attached to the genomic RNA (19). The birnavirus genome consists of two segments of dsRNA. While the smaller segment B has a single open reading frame (ORF) coding for VP1, segment A has two, a large and a small ORF, encoding the polyprotein precursor pVP2-VP4-VP3 and the nonstructural VP5, respectively. VP4 is a protease that cleaves its own N and C termini off the polyprotein, thus also releasing pVP2 (the VP2 precursor) and VP3 within the infected cell (3, 20). Subsequent serial cleavages at the C terminus of pVP2 upon particle assembly yield mature VP2 (amino acids 1 to 442 of the IPNV polyprotein) and three other peptides that remain within the virion (22). The longest of these peptides was shown to destabilize cell membranes, suggesting a role during entry (40). Maturation of the pVP2 precursor during assembly and the presence of VP3 are important for correct morphogenesis of icosahedral, T=13l virus particles.
Recombinant expression of mature VP2 alone leads to assembly of small, dodecahedral T=1 subviral particles (SVPs) containing 20 VP2 trimers (10). The crystal structures of the SVPs of infectious bursal disease virus of poultry (IBDV) were reported by several groups (15, 23, 31) as well as the three-dimensional (3D) structure of an intact T=13l IBDV virion (15).
Exposed at the virion surface, VP2 displays the humoral antigenic determinants of the virus and is the only viral protein shown to induce protective immunity. Furthermore, VP2 plays a key role during virus entry, being responsible for receptor recognition (8). Key residues controlling virulence and cell culture adaptation of IPNV were thus found to map to VP2 (46).
We report here the crystal structure of the IPNV SVP and show that, like its IBDV counterpart, it is composed of 20 VP2 trimers organized with T=1 icosahedral symmetry. IPNV VP2 displays the same fold, and the SVP is organized in the same way, as anticipated from the 43% overall amino acid sequence identity between the VP2s of the two viruses. The molecules differ significantly, however, in the loops of domain P, which forms the projections or spikes. In particular, the 3D structure shows a clustering of variable residues in the outermost loops at the top of domain P. Residues associated with virulence and cell culture adaptation map to the most peripheral of these loops—away from the 3-fold axis at the convex top of VP2. This is in contrast to IBDV, in which virulence determinants map to a central bowl at the top of the VP2 spike. These results provide new insights for understanding the determinism of antigenicity and virulence of Aquabirnavirus strains.
MATERIALS AND METHODS
Production of recombinant IPNV SVPs.
IPNV SVPs were produced using a previously described protocol (12). Briefly, the first 442 codons in the cDNA of segment A (corresponding to mature VP2) (22) of IPNV serotype Sp, French isolate 31-75 (41), were inserted into the EcoRI site of vector pFastBac1 (Gibco BRL). The resulting plasmid was used to transform a DH10Bac bacmid containing competent cells. Colonies containing the recombinant bacmid were identified by disruption of the LacZα gene. High-molecular-weight miniprep DNA was prepared from selected colonies, and this DNA was used to transfect Sf9 cells with Lipofectin. The recombinant virus was prepared and amplified by standard procedures. High-titer virus stocks of the recombinant baculovirus (108 PFU/ml) were prepared. Sf9 cells were infected at a multiplicity of infection higher than 5 in the presence of protease inhibitors, collected 100 h postinfection, and then treated with Freon 113. Purification of the particles was carried out by density gradient centrifugation in a CsCl solution.
Crystallization.
SVPs were dialyzed into a solution of 50 mM ammonium acetate, pH 7.0 and concentrated on ultrafiltration units to 10 mg/ml. Crystals were grown by vapor diffusion at acidic pH with several different alcohols as precipitant, such as ethanol, methyl-pentanediol (MPD), isopropanol, and hexanediol, in the presence of a divalent cation. The best crystals grew in a solution containing 1 M 1,6-hexanediol, 10 mM CoCl2, and 100 mM sodium acetate at pH 4.6. These crystals belong to the cubic space group I23, with a cell edge of 303.4 Å. The asymmetric unit consists of 1/12 of the SVP, i.e., 5 of the 60 VP2 polypeptides present in the particle. The icosahedral SVP is generated from this subset by the symmetry operations of the crystal, i.e., the 3-fold and 2-fold crystallographic axes. For data collection at 100 K, the crystals were transferred progressively to a cryo-protecting solution containing the same buffer in which 30% MPD was substituted for the precipitant before flash cooling in liquid ethane.
Structure determination.
The structure of the IPNV SVP was determined by molecular replacement using the coordinates of IBDV VP2 (Protein Data Bank [PDB] entry 1wcd) (38). Of note, an initial electron density map of the IPNV SVP, calculated from initial single isomorphous replacement (SIR) phases and improved by noncrystallographic symmetry averaging to 3.8-Å resolution, had been instrumental in determining the 3D structure of the IBDV counterpart, as described by Coulibaly et al. (15). The molecular replacement phases were refined by 5-fold averaging (47), and the resulting electron density was used to build the IPNV VP2 model with program O (28). Refinement was done with CNS software under strict noncrystallographic symmetry (NCS) constraints, attributing separate temperature factors for the side chain and the main chain of each residue (7). Final refinement rounds were carried out with Phenix (1) with strong NCS restraints, only one temperature factor per residue (i.e., because of the relatively limited resolution of 3.4 Å, the number of parameters refined was kept to a minimum), and a translation-libration-screw (TLS) group per VP2 polypeptide (five TLS groups in total). The weights used during the final rounds of refinement in Phenix imposed tight restraints on the geometry of the model, resulting in final root mean square deviations (RMSDs) for bond lengths of 0.005 Å and for bond angles of 0.918o compared to the dictionary values. Reflections in 12 thin shells of resolution accounting for 3% (1,992 reflections) of all reflections were excluded from the refinement for cross-validation to minimize contamination from NCS-related reflections. In the final model (R = 21.7%; Rfree = 23.3%) 92.5% of residues were in the favored region of the Ramachandran plot, with no residue in the disallowed regions (35). Crystallographic statistics are detailed in Table 1.
TABLE 1.
Data collection and refinement statistics
| Parameter | Value for Nativea |
|---|---|
| Data collection | |
| Space group | I23 |
| Cell dimensions | |
| a = b = c (Å) | 303.431 |
| Resolution (Å) | 30-3.35 (3.47-3.35) |
| Rsym | 0.080 (0.379) |
| <I>/<σI> | 14.6 (3.3) |
| Completeness (%) | 99.7 (98.3) |
| Redundancy | 4.5 (3.2) |
| Refinement | |
| Resolution (Å) | 30-3.35 |
| No. of reflections | 65,456 |
| Rwork/Rfree | 0.2171/0.2334 |
| No. of atoms | |
| Protein | 16,056 |
| Ions | 6 |
| Water | 10 |
| Avg B factor (Å2) | |
| Protein | 100.0 |
| Ions | |
| Water | 37.8 |
| RMSD | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.918 |
| NCS pairs AB, AC, AD, AE (Å) | 0.014, 0.016, 0.013, 0.021 |
Values corresponding to the highest resolution shell are shown in parentheses.
Generation of the model for T=13l IPNV particles.
The coordinates of the 13 independent VP2 polypeptides that make up the asymmetric unit of the T=13l IBDV virion were retrieved from the PDB (entry 1wce). Program Lsqkab (13) was used to superpose a VP2 trimer extracted from the IPNV SVP onto each of the four trimers present at general positions on the capsid, plus a single subunit on the trimer lying at the 3-fold icosahedral axis. The complete T=13l IPNV particle was then generated by applying the 60 icosahedral symmetry operations to this ensemble.
Illustrations.
The alignment displayed in Fig. 1C was created by ClustalW (30), and the figure was made with ESprit (24). Other figures were produced with PyMol (http://www.pymol.org/) and ConSurf (29).
FIG. 1.
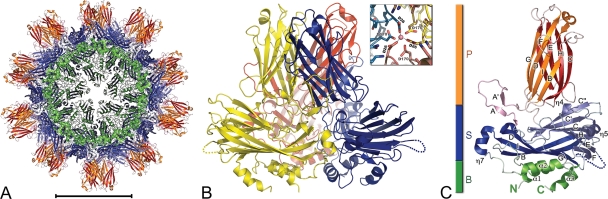
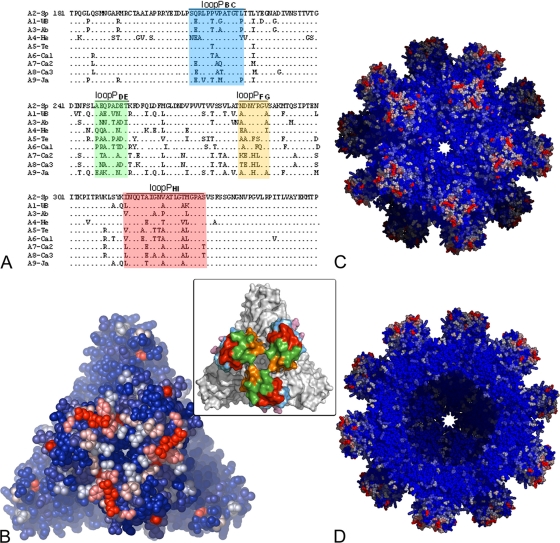
Structure of IPNV VP2. (A) The SVP is represented in a ribbon representation with base (B; green), shell (S; blue), and projection (P; rust) domains highlighted. Sixty identical copies of VP2 are organized with T=1 icosahedral symmetry. Scale bar, 100 Å. (B) The VP2 trimer, with the three protomers colored differently (blue, yellow, and red). The inset shows a Co2+ ion located at the center of the VP2 trimer and coordinated by Asp26 residues (distance, 2.2Å) from the three protomers of the trimer. Residues Asp170 bridge to consecutive Asp26 about the 3-fold axis. (C) Ribbon representation of a VP2 molecule color-coded as described in panel A. The AA′ flap is shown in pink. β-Strands are labeled according to their order in the primary sequence. N and C termini are labeled in green (B domain color). The loop connecting β-strands E and F, for which there was a break in the electron density map, is highlighted with a dotted line (also in panel B).
Protein structure accession number.
The atomic coordinates and structure factors of the T=1 IPNV particle have been deposited in the Protein Data Bank under the accession code 3IDE.
RESULTS AND DISCUSSION
The SVPs.
We have determined the structure of subviral particles of IPNV to a resolution of 3.4 Å by X-ray crystallography. The 5-fold redundancy of the icosahedral particle present in a cubic crystal form contributed to maintaining a favorable ratio of crystallographic observations to parameters of the atomic model, allowing for an adequate refinement. This would not have been the case for a less overdetermined structure at this resolution. The model shows 20 IPNV VP2 trimers interacting to make a T=1 icosahedral SVP with a marked dodecahedral shape and a maximum diameter of 260 Å (Fig. 1A). The internal and external diameters of the particle shell, without the projections, are 116 Å and 180 Å, respectively, with spikes protruding 42 Å from the surface. This arrangement is common to IPNV and IBDV SVPs and highlights the high conservation of the capsid shell.
The VP2 trimer.
One IPNV VP2 trimer, extracted from the SVP, is displayed in Fig. 1B. It shows the same organization as its IBDV counterpart, including a metal ion at the center, on the molecular 3-fold axis (Fig. 1B, inset). The presence of a central metal stabilizing each trimer is thus also a conserved feature of the birnavirus particles. The cation is located on the 3-fold axis between domains B and S, occupying the Ca2+ site observed in IBDV VP2 (23, 31), coordinated by Asp26 (IPNV numbering) from the three subunits (Fig. 1B, inset). We modeled this metal as Co2+ because this ion was present at a 10 mM concentration in the crystallization solution. The number of electrons in cobalt is indeed compatible with the observed electron density at this site. The presence of the metal was important for crystallization, perhaps reflecting a stabilization effect of the ion on the particles, which is in line with observations that, in the case of IBDV, calcium chelators destabilize the SVPs (23). In addition, the IPNV VP2 trimer has a chlorine ion also located on the 3-old molecular axis between domains S and P, coordinated to the main chain amide of Ala199 of the three VP2 subunits. A chlorine ion at this location was also identified in IBDV VP2 (31). The presence of these ions may further stabilize the VP2 trimer, but their role could also be to seal the solvent channel present along the 3-fold axis (which is clearly seen in Fig. 2B). However, the presence of such ions may also only be a consequence of the special geometry existing about the 3-fold axis and a fortuitous arrangement of chemical groups that match the coordination sphere of a circulating ion, which becomes trapped at this location.
FIG. 2.
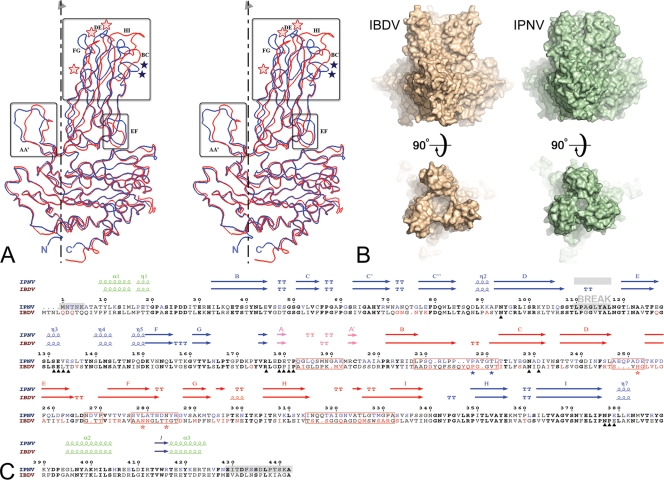
Structural comparison between IPNV and IBDV VP2. (A) Structural alignment of IPNV VP2 and IBDV VP2 subunits. The VP2 protomers were superimposed on their respective S domains and are represented as blue (IPNV) and red (IBDV) Cα traces. The molecular 3-fold axis, vertical in the figure, is represented by a broken line. Regions of structural divergence are boxed. Blue and red stars denote residues implicated in virulence of IPNV (positions 217 and 221) and IBDV (positions 253, 279, and 284), respectively. (B) Morphological differences of IPNV VP2 and IBDV VP2 spikes. The molecular surfaces of the respective VP2 trimers are shown in orthogonal views through the surface (side views at top) and from the outside (top views at bottom) of the particle. Note the more compact arrangement observed for IPNV VP2, with a narrower opening at the center of the trimer and shorter projections at the top of the spike. (C) Amino acid sequence alignment of IPNV and IBDV VP2s with their respective secondary structure elements colored according to domains as described in the legend of Fig. 1C (green, base; blue, shell; orange, projection; pink, AA′ flap). Residues in bold font are not only conserved across IPNV and IBDV sequences but are also strictly conserved within the respective genera. Residues shown in blue and red are variable between serotypes of IPNV and IBDV, respectively. Red boxes represent regions of structural dissimilarities between IPNV and IBDV also highlighted in panel A. A gray background indicates regions that were disordered in the crystal structure (including the break in density in domain S). Black triangles under the sequences mark the amino acids lining the conserved groove at the base of the spike described in the text and in Fig. 3. Blue and red stars under the sequences denote residues implicated in virulence of IPNV and IBDV, respectively, as in panel A. Note that position 231, marked with a triangle, is not in bold font because it is not aspartic acid in one of the available IPNV sequences, whereas it is aspartic in every other birnavirus that has been sequenced to date. Since this is the conserved integrin binding motif, it is possible that the particular sequence has an error at this position.
The VP2 subunit.
The tertiary structure of VP2 comprises three distinct domains termed base (B), shell (S), and projection (P), which are shown in Fig. 1C. The most conserved and most variable regions map to domains S and P, respectively. The similarity between the SVPs could be anticipated from the high sequence conservation of domain S among birnaviruses. Both domains S (IPNV residues 32 to 176 and 343 to 386) and P (residues 177 to 342, inserted within S) are folded as jelly roll β-barrels, oriented tangentially and radially, respectively, with respect to the icosahedral particle. Strands PA and PA′ form a β-arch (25) contributing a fifth strand to both β-sheets of domain P of the adjacent protomer (Fig. 1). While domain S is responsible for the trimer-trimer contacts making the particle, domain P makes the spikes, which are tower-like projections easily discernible in electron micrographs of birnavirus particles. The electron density map of the IPNV SVP displays unambiguous density for most of the polypeptide chain, allowing the visualization of residues 6 to 428 of VP2, with a break in density between amino acids 110 to 117 at the distal tip of domain S, which appears disordered (Fig. 1 and 2C). This break is most likely due to static disorder, in which the loops do not follow the symmetry of the particle, rather than to a dynamic disorder right at the 5-fold axes of symmetry of the SVP. The base domain is made by N- and C-terminal helical extensions of the polypeptide, including residues 1 to 31 and 387 to 442 (Fig. 1C and 2C), bringing together the amino and carboxy termini of the protomer. Helices α1 and α2 are amphipathic, with their hydrophobic patches hidden in a common interface. In contrast, helix α3 is rich in charged residues, with a relatively loose interaction with the S domain and with the rest of the B domain.
Structural alignment.
The individual domains B, S, and P of IPNV and IBDV superimpose well, with RMSDs of 0.8 Å, 0.6 Å, and 1.8 Å for 62, 177, and 124 equivalent Cα atoms, respectively (Fig. 2A). When the entire protomers are superposed, the RMSD is 1.2 Å for 388 equivalent residues. Furthermore, superposition of the IBDV and IPNV VP2 trimers results in a global RMS of 1.2 Å for 1,152 equivalent Cα atoms. The three domains of VP2 thus maintain their relative orientations within the trimer in the two viruses, correlating well with the extensive and mainly hydrophobic S-S and P-P interfaces.
A conserved, exposed groove at the VP2 surface.
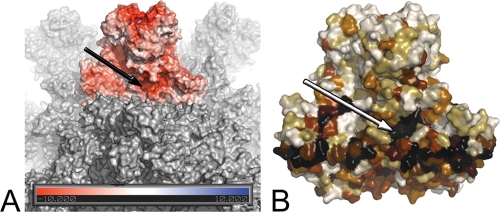
The SVP structure also reveals the presence of a conserved and exposed groove located at the S-P domain interface at the base of the spike, with an opening to solvent having dimensions of about 7 Å by 16 Å. This feature, although also present in the IBDV SVP, had not been described in previous reports. The residues lining the groove are at the base of the AA′ β-arch, involving amino acids 94, 176, 179 to 182, and 377 to 379, as well as residues 131 to 134 and 229 to 231 of the adjacent subunit in the trimer (Fig. 2C and Fig. 3). Importantly, the N229X230D231 motif at the base of the groove corresponds to a functional α4β1 integrin ligand motif recently identified as essential for IBDV binding to cells (17). The surface of the groove displays a strong negative electrostatic potential both in IPNV and IBDV (Fig. 3A). The conserved nature of the residues lining this exposed groove is indeed compatible with a functional role (Fig. 3B).
FIG. 3.
A conserved groove in birnavirus VP2 proteins. (A) Surface representation of the IPNV VP2 SVP. One trimer is colored according to the electrostatic potential on the solvent-accessible surface as calculated by the Adaptive Poisson-Boltzmann Solver (2) and displayed in PyMo (http://www.pymol.org/). Two negatively charged patches are obvious, one at the center of the trimer tip and the other at the conserved groove at the base of the trimer (arrow). (B) Representation of the sequence conservation across birnaviruses mapped onto the molecular surface of a VP2 trimer (black-brown-white gradient from most to least conserved residues). As in panel A, the arrow points to the conserved groove.
A model for the T=13l shell of the IPNV virion.
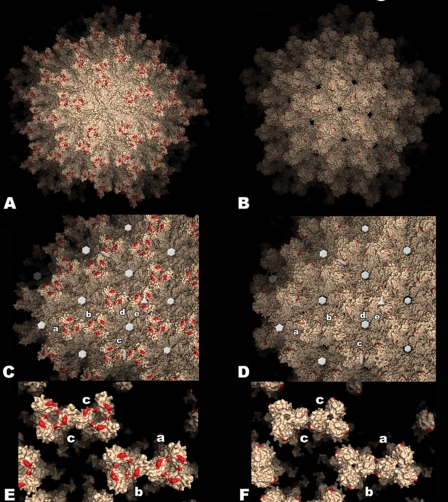
With the VP2 structure we further modeled the T=13l capsid of the IPNV virion by superimposing the VP2 trimer onto the previously described structure of the intact IBDV virion (PDB entry 1wce) (15). The T=13l particle contains five independent trimers in the icosahedral asymmetric unit, labeled a through e according to an earlier study (5). This results in a total of 260 trimers in the capsid (240 trimers a to d, lying in general positions, plus 20 e trimers at the 3-fold icosahedral axes). This model assumes that the intertrimer interactions forming the IPNV virion are similar to those observed in the IBDV virus particle. This assumption is supported by the observed high sequence and structure conservation of the shell domain of VP2 and the similarities between IBDV and IPNV SVPs. The model of the IPNV virion is shown in surface representation in Fig. 4 (right panels), displayed alongside the structure of the IBDV counterpart (Fig. 4, left panels). The extra holes present at the five- and quasi-6-fold axes of the IPNV particle are due to the break in ordered electron density in this region, and as a result loop 110 to 117 is not included in the atomic model of IPNV VP2. These residues are actually present in the particle and fill these holes, as in IBDV. The most notable differences are at the spike-spike contacts between adjacent trimers in the T=13l shell, which in the IBDV virion connect trimers a-b, d-e, and c-c (about the icosahedral 2-fold axes). The a-b and c-c contacts are maintained but are weaker in the IPNV virion. These interactions are restricted to the AA′ β-arch of adjacent trimers and do not involve the top of the spike, in contrast to the contacts in the IBDV virion. The d-e contacts, already weak in IBDV, are absent altogether in the IPNV virion (Fig. 4E and F). These differences may have implications for virus stability and also for the assembly process and correlate well with reports of a decreased mechanical resistance of IPNV compared to IBDV virions (22).
FIG. 4.
Model of the IPNV virus particle. Surface representation of the T=13l lattice of IBDV (left panels) and IPNV (right panels) virions. Red patches indicate residues implicated in cell culture adaptation and virulence in IBDV (His253, Asn279, and Thr284) and IPNV (Pro217 and Thr221). (A and B) View along a 5-fold axis of the particle. (C and D) View along a 3-fold axis. Five icosahedrally independent VP2 trimers are labeled a to e according to the nomenclature established for the IBDV virion (5). Twofold, 3-fold, 5-fold, and pseudo-6-fold axes are indicated with ellipses, triangles, pentagons, and hexagons, respectively (the holes on these axes in the IPNV virion result from the disordered loop in this region, as explained in the text). (E and F) Close-up view of contacts between spikes involving domain P. Note the weaker contacts between spikes a-b and c-c, as well as the more closed organization of the spike head in the IPNV virion.
Molecular basis for the morphological differences between birnavirus virions.
The differences in the spike contacts between VP2 trimers in virions of IPNV and IBDV result from significant alterations in domain P. The amino acids in the most dissimilar regions are highlighted with red boxes in the sequence alignment of Fig. 2C and with a colored background in Fig. 6A. These segments have different relative lengths, conferring a different overall morphology to the spikes: while in IBDV they lead to a concave shape at the top, with a central bowl as the main feature, in IPNV they result in a convex spike, with a narrow central opening. Of note, the IPNV spikes have less-pronounced petal-like outward projections, which are the hallmark of the VP2 trimer in IBDV, giving it a propeller shape when seen from the top (Fig. 2B). These “petals” are responsible for spike-spike contacts in the T=13l particle.
FIG. 6.
Sequence variability between IPNV serotypes. (A) Sequence alignment of domain P from the nine IPNV serotypes. The variable loops are colored according to the inset of Fig. 5 (shown also in panel B). (B) The IPNV VP2 trimer in an all-atom representation (as spheres). (C and D) The SVP viewed along a 5-fold axis. A blue-white-red color gradient was used for strictly conserved residues to highly variable ones as estimated by the ConSurf server. In panel D, five trimers from the front part of the particle were removed to reveal the internal icosahedral shell.
Implications for virulence determinism.
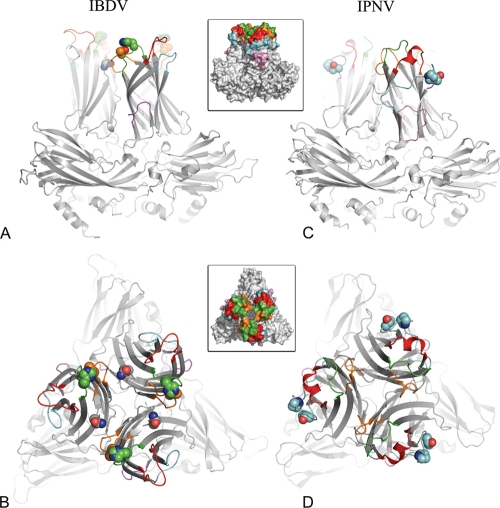
Amino acid sequence comparisons of IPNV VP2 from various field isolates of the A2 serotype, which exhibit different mortality rates in salmon fry, identified putative residues responsible for modulating virulence between strains (43, 45). This was later confirmed using a reverse genetics system, demonstrating that IPNV virulence and adaptation to cell culture are controlled by two VP2 amino acids at positions 217 and 221 in loop PBC. Highly virulent strains have Thr217 and Ala221; moderate- to low-virulence isolates have Pro217 and Ala221; variants having Thr221 are almost avirulent, irrespective of the residue at position 217 (46). To date, no other position in VP2—or in any other viral protein—has been identified as playing a role in virulence or in cell culture adaptation. Figures 2 and 5 show that the PBC loop is the most peripheral loop at the top of the VP2 spike. Because these two residues are exposed and do not participate in contacts important for the VP2 fold or in interactions between subunits that stabilize the virion, their marked effect on virulence suggests that they may be involved in attachment to the target cell.
FIG. 5.
Virulence and tropism determinants in IBDV and IPNV VP2s. Ribbon representations of IBDV (A and B) and IPNV (C and D) VP2 trimers, viewed from the side (perpendicularly to the surface of the virus particle in A and C) and from the top (down the molecular 3-fold axis of VP2 in B and D). The variable loops at the top of domain P are colored according to the insets (PAA′, pink; PBC, cyan; PDE, green; PFG, orange; PHI, red). The side chains of residues implicated in virulence and cell culture adaptation are shown as spheres. These are residues His253, Asn279, and Thr284 for IBDV and residues Pro217 and Thr221 for IPNV.
In IBDV, VP2 residues controlling virulence, cell tropism, and cell culture adaptation were also mapped to loops at the top of the P domain (6, 33, 48). These are amino acids 253 (loop PDE), 279 (strand PF, close to PFG), and 284 (loop PFG). These loops are within the central bowl of the spike instead of the periphery, as loop PBC discussed above for IPNV. In summary, as illustrated in Fig. 4 and 5, the IBDV and IPNV virions display contrasting patterns of residues controlling virulence and tropism, mapping to alternative regions at the top of the spike.
Antigenic variation.
There is considerable diversity within the Aquabirnavirus genus, with nine identified serotypes in contrast to just two in IBDV. The fact that antigenic variability is more pronounced in IPNV is probably due to their wider host range (26). Indeed, whereas IBDV infects only chicken and turkey, IPNV-related viruses have been isolated from fish belonging to at least 30 different species, as well as mollusks and crustaceans (26). Why aquabirnaviruses are able to exhibit a wider host range than their terrestrial homologues (IBDV and Drosophila X virus) remains an unsolved question. As expected, the variable residues between Aquabirnavirus serotypes map to domain P (Fig. 6A). Moreover, they are mainly clustered in the four outer loops (PFG, PDE, PHI, and PBC, listed in the order displayed in Fig. 2A, from central to peripheral with respect to the 3-fold axis). These loops are also highlighted in Fig. 6. The changes map, in particular, to the hypervariable PFG and PDE loops at the center (Fig. 6, red and green,). In the case of IBDV, in spite of its more limited antigenic diversity, antibody neutralization escape mutations map to positions 222 and 223 (loop PBC), 251 (PDE), and 313 and 322 (PHI) (44). In addition, using a reverse genetics system, it was recently confirmed that a few residues located in the peripheral PBC and PHI loops modulate the expression of neutralizing epitopes in IBDV VP2 (32). Thus, while the IPNV virulence determinants map to the periphery and residues controlling antigenicity are at the center of the spike, the inverse pattern is observed in IBDV.
Biological implications.
The reported crystal structure of a subviral IPNV particle allowed an insightful comparative analysis with the IBDV capsid. These data show that the spikes have a different shape—convex versus concave—and display a different interaction network at the surface of the virus particle. Furthermore, the residues important for virulence and tissue tropism cluster differently on the spikes. This distribution may reflect alternative interaction sites with cellular receptors, the nature of which remain to be identified, in the two viruses. The segregation of these sites into separate regions of VP2 in IPNV and IBDV contrasts with the conservation of the groove containing the integrin binding motif present at the base of the spike. The symmetry of the particle expands this groove at the virion surface because it is adjacent to the 2-fold axes relating VP2 trimers on the capsid, reminiscent of the “canyon” observed around 5-fold axes in picornavirus particles (14). Taken together, these observations suggest that birnaviruses may need to interact with two different receptors, one for attachment located at the top of the spike-which may confer also tissue tropism and modulate virulence—and a second receptor for internalization, interacting with the conserved groove. This pattern has parallels in other nonenveloped viruses, such as adenoviruses and reoviruses, in which initial attachment to a specific receptor is followed by interaction with an integrin for internalization (37, 49). The bottom line of this study is that, in spite of the detailed knowledge of the architecture of the virus particles described here and elsewhere, understanding the underlying birnavirus biology—its entry process and the mechanism of attenuation and virulence—absolutely requires now the identification of the various host partners used for cell invasion.
Acknowledgments
We thank C. Schulze-Briese and T. Tomikazi for help during data collection and G. Aumont (INRA) and C. Branlant (CNRS) for support. Diffraction data were collected at the Swiss Light Source (beamline X06SA), Paul Scherrer Institute, Villigen, Switzerland, and ESRF, Grenoble, France.
F.C. was funded by a CNRS BDI fellowship and is presently recipient of an NHMRC Career Development Award.
Footnotes
Published ahead of print on 9 December 2009.
REFERENCES
- 1.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948-1954. [DOI] [PubMed] [Google Scholar]
- 2.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birghan, C., E. Mundt, and A. E. Gorbalenya. 2000. A non-canonical lon proteinase lacking the ATPase domain employs the ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J. 19:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blake, S., J. Y. Ma, D. A. Caporale, S. Jairath, and B. L. Nicholson. 2001. Phylogenetic relationships of aquatic birnaviruses based on deduced amino acid sequences of genome segment A cDNA. Dis. Aquat. Organ. 45:89-102. [DOI] [PubMed] [Google Scholar]
- 5.Bottcher, B., N. A. Kiselev, V. Y. Stel'Mashchuk, N. A. Perevozchikova, A. V. Borisov, and R. A. Crowther. 1997. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandt, M., K. Yao, M. Liu, R. A. Heckert, and V. N. Vakharia. 2001. Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. J. Virol. 75:11974-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 8.Bruslind, L. D., and P. W. Reno. 2000. Virulence comparison of three Buhl-subtype isolates of infectious pancreatic necrosis virus in brook trout fry. J. Aquat. Anim. Health 12:301-315. [Google Scholar]
- 9.Casanas, A., A. Navarro, C. Ferrer-Orta, D. Gonzalez, J. F. Rodriguez, and N. Verdaguer. 2008. Structural insights into the multifunctional protein VP3 of birnaviruses. Structure 16:29-37. [DOI] [PubMed] [Google Scholar]
- 10.Caston, J. R., J. L. Martinez-Torrecuadrada, A. Maraver, E. Lombardo, J. F. Rodriguez, J. I. Casal, and J. L. Carrascosa. 2001. C terminus of infectious bursal disease virus major capsid protein VP2 is involved in definition of the T number for capsid assembly. J. Virol. 75:10815-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caswell-Reno, P., V. Lipipun, P. W. Reno, and B. L. Nicholson. 1989. Use of a group-reactive and other monoclonal antibodies in an enzyme immunodot assay for identification and presumptive serotyping of aquatic birnaviruses. J. Clin. Microbiol. 27:1924-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevalier, C., J. Lepault, I. Erk, B. Da Costa, and B. Delmas. 2002. The maturation process of pVP2 requires assembly of infectious bursal disease virus capsids. J. Virol. 76:2384-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 14.Colonno, R. J., J. H. Condra, S. Mizutani, P. L. Callahan, M. E. Davies, and M. A. Murcko. 1988. Evidence for the direct involvement of the rhinovirus canyon in receptor binding. Proc. Natl. Acad. Sci. U. S. A. 85:5449-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulibaly, F., C. Chevalier, I. Gutsche, J. Pous, J. Navaza, S. Bressanelli, B. Delmas, and F. A. Rey. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761-772. [DOI] [PubMed] [Google Scholar]
- 16.Da Costa, B., S. Soignier, C. Chevalier, C. Henry, C. Thory, J. C. Huet, and B. Delmas. 2003. Blotched snakehead virus is a new aquatic birnavirus that is slightly more related to avibirnavirus than to aquabirnavirus. J. Virol. 77:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delgui, L., A. Ona, S. Gutierrez, D. Luque, A. Navarro, J. R. Caston, and J. F. Rodriguez. 2009. The capsid protein of infectious bursal disease virus contains a functional alpha 4 beta 1 integrin ligand motif. Virology 386:360-372. [DOI] [PubMed] [Google Scholar]
- 18.Delmas, B., F. Kibenge, J. Leong, E. Mundt, V. Vakharia, and J. Wu. 2005. Birnaviridae, p. 561-569. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 19.Dobos, P. 1995. The molecular biology of infectious pancreatic necrosis virus (IPNV). Annu. Rev. Fish Dis. 5:25-54. [Google Scholar]
- 20.Duncan, R., E. Nagy, P. J. Krell, and P. Dobos. 1987. Synthesis of the infectious pancreatic necrosis virus polyprotein, detection of a virus-encoded protease, and fine structure mapping of genome segment A coding regions. J. Virol. 61:3655-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egusa, S. 1970. Branchionephritis prevalence among eel populations in farm-ponds in the winter 1969-1970. Fish Pathol. 5:51-56. [Google Scholar]
- 22.Galloux, M., C. Chevalier, C. Henry, J. C. Huet, B. D. Costa, and B. Delmas. 2004. Peptides resulting from the pVP2 C-terminal processing are present in infectious pancreatic necrosis virus particles. J. Gen. Virol. 85:2231-2236. [DOI] [PubMed] [Google Scholar]
- 23.Garriga, D., J. Querol-Audi, F. Abaitua, I. Saugar, J. Pous, N. Verdaguer, J. R. Caston, and J. F. Rodriguez. 2006. The 2.6-angstrom structure of infectious bursal disease virus-derived T=1 particles reveals new stabilizing elements of the virus capsid. J. Virol. 80:6895-6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouet, P., X. Robert, and E. Courcelle. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hennetin, J., B. Jullian, A. C. Steven, and A. V. Kajava. 2006. Standard conformations of beta-arches in beta-solenoid proteins. J. Mol. Biol. 358:1094-1105. [DOI] [PubMed] [Google Scholar]
- 26.Hill, B. J., and K. Way. 1995. Serological classification of infectious pancreatic necrosis (IPN) virus and other aquatic birnaviruses. Annu. Rev. Fish Dis. 5:55-77. [Google Scholar]
- 27.Hjalmarsson, A., E. Carlemalm, and E. Everitt. 1999. Infectious pancreatic necrosis virus: identification of a VP3-containing ribonucleoprotein core structure and evidence for O-linked glycosylation of the capsid protein VP2. J. Virol. 73:3484-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 29.Landau, M., I. Mayrose, Y. Rosenberg, F. Glaser, E. Martz, T. Pupko, and N. Ben-Tal. 2005. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33:W299-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 31.Lee, C. C., T. P. Ko, C. C. Chou, M. Yoshimura, S. R. Doong, M. Y. Wang, and A. H. J. Wang. 2006. Crystal structure of infectious bursal disease virus VP2 subviral particle at 2.6 Å resolution: implications in virion assembly and immunogenicity. J. Struct. Biol. 155:74-86. [DOI] [PubMed] [Google Scholar]
- 32.Letzel, T., F. Coulibaly, F. A. Rey, B. Delmas, E. Jagt, A. A. van Loon, and E. Mundt. 2007. Molecular and structural bases for the antigenicity of VP2 of infectious bursal disease virus. J. Virol. 81:12827-12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim, B. L., Y. Cao, T. Yu, and C. W. Mo. 1999. Adaptation of very virulent infectious bursal disease virus to chicken embryonic fibroblasts by site-directed mutagenesis of residues 279 and 284 of viral coat protein VP2. J. Virol. 73:2854-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo, C., Y. Hong, S. Huang, and C. Wang. 1988. The characteristics of the virus isolated from the gill of clam, Meretrix lusoria. Fish Pathol. 23:147-154. [Google Scholar]
- 35.Lovell, S. C., I. W. Davis, W. B. Arendall III, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson, and D. C. Richardson. 2003. Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins 50:437-450. [DOI] [PubMed] [Google Scholar]
- 36.Luque, D., I. Saugar, M. T. Rejas, J. L. Carrascosa, J. F. Rodriguez, and J. R. Caston. 2009. Infectious Bursal disease virus: ribonucleoprotein complexes of a double-stranded RNA virus. J. Mol. Biol. 386:891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maginnis, M. S., J. C. Forrest, S. A. Kopecky-Bromberg, S. K. Dickeson, S. A. Santoro, M. M. Zutter, G. R. Nemerow, J. M. Bergelson, and T. S. Dermody. 2006. β1 Integrin mediates internalization of mammalian reovirus. J. Virol. 80:2760-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navaza, J. 2001. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 57:1367-1372. [DOI] [PubMed] [Google Scholar]
- 39.Nicholson, B. L. 1993. Use of monoclonal antibodies in identification and characterization of fish viruses. Annu. Rev. Fish Dis. 3:241-257. [Google Scholar]
- 40.Nobiron, I., M. Galloux, C. Henry, C. Torhy, P. Boudinot, N. Lejal, B. Da Costa, and B. Delmas. 2008. Genome and polypeptides characterization of Tellina virus 1 reveals a fifth genetic cluster in the Birnaviridae family. Virology 371:350-361. [DOI] [PubMed] [Google Scholar]
- 41.Petit, S., N. Lejal, J. C. Huet, and B. Delmas. 2000. Active residues and viral substrate cleavage sites of the protease of the birnavirus infectious pancreatic necrosis virus. J. Virol. 74:2057-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pous, J., C. Chevalier, M. Ouldali, J. Navaza, B. Delmas, and J. Lepault. 2005. Structure of birnavirus-like particles determined by combined electron cryomicroscopy and X-ray crystallography. J. Gen. Virol. 86:2339-2346. [DOI] [PubMed] [Google Scholar]
- 43.Santi, N., V. N. Vakharia, and O. Evensen. 2004. Identification of putative motifs involved in the virulence of infectious pancreatic necrosis virus. Virology 322:31-40. [DOI] [PubMed] [Google Scholar]
- 44.Schnitzler, D., F. Bernstein, H. Muller, and H. Becht. 1993. The genetic basis for the antigenicity of the VP2 protein of the infectious bursal disease virus. J. Gen. Virol. 74:1563-1571. [DOI] [PubMed] [Google Scholar]
- 45.Shivappa, R. B., H. Song, K. Yao, A. Aas-Eng. O. Evensen, and V. N. Vakharia. 2004. Molecular characterization of Sp serotype strains of infectious pancreatic necrosis virus exhibiting differences in virulence. Dis. Aquat. Organ. 61:23-32. [DOI] [PubMed] [Google Scholar]
- 46.Song, H., N. Santi, O. Evensen, and V. N. Vakharia. 2005. Molecular determinants of infectious pancreatic necrosis virus virulence and cell culture adaptation. J. Virol. 79:10289-10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terwilliger, T. C. 2003. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374:22-37. [DOI] [PubMed] [Google Scholar]
- 48.van Loon, A. A., N. de Haas, I. Zeyda, and E. Mundt. 2002. Alteration of amino acids in VP2 of very virulent infectious bursal disease virus results in tissue culture adaptation and attenuation in chickens. J. Gen. Virol. 83:121-129. [DOI] [PubMed] [Google Scholar]
- 49.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 50.Wolf, K. 1988. Fish viruses and fish viral diseases. Canstock Publishing Assoc., Cornell University Press, Ithaca, NY.