Abstract
Herpesvirus nucleocapsids assemble in the nucleus and must cross the nuclear membrane for final assembly and maturation to form infectious progeny virions in the cytoplasm. It has been proposed that nucleocapsids enter the perinuclear space by budding through the inner nuclear membrane, and these enveloped nucleocapsids then fuse with the outer nuclear membrane to enter the cytoplasm. Little is known about the mechanism(s) for nuclear egress of herpesvirus nucleocapsids and, in particular, which, if any, cellular proteins are involved in the nuclear egress pathway. UL12 is an alkaline nuclease encoded by herpes simplex virus type 1 (HSV-1) and has been suggested to be involved in viral DNA maturation and nuclear egress of nucleocapsids. Using a live-cell imaging system to study cells infected by a recombinant HSV-1 expressing UL12 fused to a fluorescent protein, we observed the previously unreported nucleolar localization of UL12 in live infected cells and, using coimmunoprecipitation analyses, showed that UL12 formed a complex with nucleolin, a nucleolus marker, in infected cells. Knockdown of nucleolin in HSV-1-infected cells reduced capsid accumulation, as well as the amount of viral DNA resistant to staphylococcal nuclease in the cytoplasm, which represented encapsidated viral DNA, but had little effect on these viral components in the nucleus. These results indicated that nucleolin is a cellular factor required for efficient nuclear egress of HSV-1 nucleocapsids in infected cells.
Herpes simplex virus type 1 (HSV-1) is an enveloped DNA virus and one of the most common human pathogens, causing a wide variety of diseases such as mucocutaneous diseases, keratitis, skin diseases, and life-threatening encephalitis (48). HSV-1 virions consist of three morphologically distinct structures: the nucleocapsid containing the linear double-stranded DNA viral genome, which encodes at least 84 viral proteins, in an icosahedral capsid; the tegument, a proteinaceous layer surrounding the nucleocapsid; and the envelope, a host cell-derived lipid bilayer with viral glycoproteins on its surface and enclosing the nucleocapsid and tegument (48). After HSV-1 entry into a host cell, the de-enveloped nucleocapsid is transported to a nuclear pore and the viral genome is released into the nucleus (48). Viral DNA replication and transcription, capsid assembly, and packaging of nascent progeny virus genomes into preformed capsids takes place in the nucleus (48). Since HSV-1 nucleocapsids are too large to move through nuclear pores (14), HSV-1 evolved a mechanism for transporting nucleocapsids across the nuclear membrane (NM). Thus, progeny HSV-1 nucleocapsids acquire primary envelopes by budding through the inner NM into the space between the inner and outer NMs, the perinuclear space (30, 48). Although primary envelopment of nucleocapsids at the inner NM has been well established, transport from the perinuclear space through the cytoplasm to the extracellular space is still not completely elucidated (3, 25, 32, 33, 48, 70). It is now generally accepted that enveloped nucleocapsids in the perinuclear space fuse with the outer NM, thereby releasing de-enveloped nucleocapsids into the cytoplasm (31, 32, 59). These nucleocapsids acquire a secondary envelope at the cytoplasmic membrane, most likely at the trans-Golgi network (13, 64, 67), and mature virions are then secreted from cells by exocytosis (30). Although numerous studies have been done to elucidate the mechanism(s) by which HSV-1 virions make their way from the perinuclear space to the extracellular space, the mechanism(s) remains largely unknown.
In the present study, we investigated the transport of nascent HSV-1 nucleocapsids across the NM. It has been reported that this process involves at least six viral proteins, including UL31, UL34, UL12, Us3, glycoprotein B (gB), and gH (6, 47, 50, 55, 73), all of which except Us3 are conserved in all herpesviruses (49). In cells infected with a recombinant virus lacking UL34, UL31, or UL12, nucleocapsids remain in the nucleus and infectious titers are reduced 100- to 1,000-fold (50, 55, 69, 73), suggesting that these viral proteins are required for the primary envelopment. UL31 and UL34 form a complex that localizes at the inner NM (46, 47), and several functions of these viral proteins have been reported. These include (i) UL31 and UL34 affect maturation of virus-induced nuclear compartments, called replication compartments, in which viral DNA replication, late-gene transcription, and encapsidation take place, so nucleocapsids assemble adjacent to the NM (57) and (ii) UL31 and UL34 are required for rearranging nuclear lamina (1, 45, 57, 58), which has been hypothesized to be a barrier HSV-1 must breach to allow nucleocapsids access to the inner NM for envelopment, probably by recruiting a subset of protein kinase C isoforms (39) and/or by mislocalizing integral membrane lamin receptors, including the lamin B receptor and emerin, both of which tether lamins to the inner NM (23, 36, 53).
The functions of UL31 and UL34 appear to be modified directly or indirectly by viral Us3 serine/threonine protein kinase. Thus, Us3 phosphorylates UL31 and UL34, as well as cellular proteins associated with NMs, including emerin and laminA/C, a major component of lamina, and controls proper localization at NMs of these viral and cellular proteins (19, 23, 36-38, 40). Among the Us3 substrates, Us3 phosphorylation of UL31 appears to be involved in regulation of nuclear egress of nucleocapsids (38).
In the absence of viral Us3 protein kinase activity or upon deletion of the HSV-1 genes encoding envelope gB and gH, both of which are membrane fusion proteins that promote virus entry into cells (63), virions accumulate aberrantly in the perinuclear space, suggesting that Us3, gB, and gH promote fusion of the nascent virion envelope with the cell's outer NM (6, 47, 51). Consistent with this suggestion, it has recently been reported that Us3 phosphorylates the gB cytoplasmic tail, which may promote the fusion process (17, 71). As described above, viral proteins involved in nuclear egress of nucleocapsids are gradually being elucidated. However, data directly showing that these viral proteins are involved in the nuclear egress of nuclocapsids have been, limited and there is a lack of information about host cell factors involved in the herpesvirus nuclear egress pathway.
UL12 is a viral alkaline nuclease that has endo- and exonuclease activities (8, 15, 16, 62) and plays a role in viral DNA maturation by processing viral DNA replication intermediates and mediating strand exchange in concert with viral DNA replication factor ICP8 (10, 29, 42-44, 66). Although it has been suggested that viral DNA maturation mediated by UL12 facilitates nucleocapsid transport to the cytoplasm (55), the mechanisms(s) by which UL12 acts on nuclear egress of nucleocapsids is not known.
To identify host cell proteins that play a role in the nuclear egress pathway of HSV-1 nucleocapsids, we used a real-time imaging system to identify host cell proteins interacting with viral protein UL12 in live infected cells. We report here that the host cell nucleolin protein interacts with UL12 and is required for efficient nuclear egress of HSV-1 nucleocapsids in infected cells.
MATERIALS AND METHODS
Cells and viruses.
Vero, rabbit skin, and Plat-GP cells were described previously (65). HSV-1 wild-type strain HSV-1(F) was described previously (17, 65). Recombinant virus YK601 encoding capsid protein VP26 fused to VenusA206K FP was described previously (64).
Plasmids.
(i) Transfer plasmid pBS-VenusA206K-UL12, for generating a recombinant HSV-1 expressing the VenusA206K FP (64) and UL12 protein, was constructed as follows. The VenusA206K open reading frame (ORF) without the stop codon amplified by PCR from pBS-VenusA206K (64), the 1-kb sequence upstream of the HSV-1 UL12 start codon amplified from pBC1012 (21) by using the primers 5′-GCCGGTACCAACATCCGCGGCTTCATCGC-3′ and 5′-GCGAATTCGAGTCCACGGTAGGCCCAGC-3′, and the 1-kb sequence downstream of the HSV-1 UL12 start codon amplified from pBC1012 by using the primers 5′-GCGAATTCGAGTCCACGGTAGGCCCAGC-3′ and 5′-GCGGATCCCCATCCCCGTGTGACCGTCC-3′ were sequentially cloned into pBluescript KS(+) (Stratagene) to produce pBS-VenusA206K-UL12. The resultant plasmid consisted of VenusA206K fused to the 1-kb sequence upstream of the UL12 start codon and part of the UL12 ORF. (ii) Plasmid pMAL-UL12-N or pMAL-UL41, for generating a fusion protein of maltose-binding protein (MBP) and the N-terminal domain of UL12 (MBP-UL12-N) or the domain of UL41 (MBP-UL41), was constructed by amplifying the domain carrying UL12 codons 2 to 126 from pBC1012 or UL41 codons 239 to 489 from pBC1007 (21) and cloning the DNA fragments into pMAL-c (New England Biolabs) in frame with MBP. (iii) Plasmid pSSSP-hNucleolin, for generating a stable cell line expressing shRNA against nucleolin, was constructed as follows. The oligonucleotides 5′-TTTGGAAGACGGTGAAATTGATGGGCTTCCTGTCACCCATCAATTTCACCGTCTTCCTTTTTTG-3′ and 5′-AATTCAAAAAAGGAAGACGGTGAAATTGATGGGTGACAGGAAGCCCATCAATTTCACCGTCTTC-3′ were annealed and cloned into the BbsI and EcoRI sites of pmU6 (12, 72) (kindly provided by Hideo Iba). The BamHI-EcoRI fragment of the resultant plasmid, containing the U6 promoter and the sequence encoding shRNA against nucleolin, was cloned into the BamHI and EcoRI sites of pSSSP (12, 72) (kindly provided by Hideo Iba), which is a derivative of retrovirus vector pMX containing a puromycin resistance gene, to produce pSSSP-hNucleolin. Plasmid pSSSP-Cre encoding shRNA against Cre recombinase (12) was kindly provided by Hideo Iba. Plasmid pSSSP-GFP6 (kindly provided by Hideo Iba) encoding shRNA against green fluorescent protein (GFP) was constructed by the same procedure as pSSSP-hNucleolin, but using the oligonucleotides 5′-TTTGCCACAACGTCTATATCATGGGCTTCCTGTCACCCATGATATAGACGTTGTGGCTTTTTTG-3′ and 5′-AATTCAAAAAAGCCACAACGTCTATATCATGGGTGACAGGAAGCCCATGATATAGACGTTGTGG-3′.
Production and purification of a MBP fusion protein.
MBP fusion protein (MBP-UL12-N or MBP-UL41) was expressed in Escherichia coli transformed with pMAL-UL12-N or pMAL-UL41 and purified as described previously (20).
Antibodies.
To generate mouse polyclonal antibody to UL12 or UL41, a BALB/c mouse was immunized three times with purified MBP-UL12-N or MBP-UL41 with Freund complete or incomplete adjuvant (Sigma) and then boosted by an MBP-UL12-N or MBP-UL41 injection. The serum of the immunized mouse was used as anti-UL12 or anti-UL41 polyclonal antibody. Mouse monoclonal antibodies to nucleolin (D6), ICP8 (10A3), VP5 (3B6), and α-tubulin (DM1A) were purchased from Santa Cruz Biotechnology, Chemicon, Virusys, and Sigma-Aldrich, respectively. Rabbit polyclonal antibody and mouse monoclonal antibody (M2) to the Flag epitope were purchased from Sigma-Aldrich. Rabbit polyclonal antibody to UL48 was described previously (35).
Mutagenesis of viral genomes in E. coli and generation of recombinant HSV-1.
Recombinant virus YK651, encoding VenusA206K fused to the UL12 protein (Fig. 1), was constructed as described previously (18), except that the transfer plasmid pBS-VenusA206K-UL12 was used.
FIG. 1.
Schematic diagram of genome structure of wild-type YK304 and relevant recombinant virus domains. Line 1, linear representation of the YK304 genome. The YK304 genome has a bacmid (BAC) in the intergenic region between UL3 and UL4. Line 2, fragment of the genome domain containing the UL11, UL12, and UL13 ORFs. Line 3, UL12 ORF. Lines 4 and 6, recombinant viruses YK651 and YK653, respectively. Line 5, plasmid pYK652.
To generate recombinant virus YK653 expressing UL12 protein tagged with the Flag epitope (Flag-UL12), the two-step Red-mediated mutagenesis procedure was carried out by using E. coli GS1783 containing pYEbac102 as described previously (18), except with the primers 5′-GTTCACCCGGCGGCGCGCTCAACCACCGCTCCCCCCACGTCGTCTCGGAAATGGATTACAAGGATGACGACGATAAGAGGATGACGACGATAAGTAGGG-3′ and 5′-CGCTTAGTCACGGTGCGTCCCGGCGGACATGCTGGGCCTACCGTGGACTCCTTATCGTCGTCATCCTTGTAATCCATTTCCGAGACGACGTGGGGGGCAACCAATTAACCAATTCTGATTAG-3′. Plasmid pYEbac102 contained a complete HSV-1(F) sequence with the bacterial artificial chromosome (BAC) sequence inserted into the intergenic region between HSV UL3 and UL4 (65). Recombinant virus YK304 reconstituted from pYEbac102 has been shown to have an identical phenotype to wild-type HSV-1(F) in cell cultures and in a mouse model (65). As a result of the two-step Red-mediated mutagenesis procedure, E. coli strain GS1783 (YEbac653), containing the mutant HSV-BAC plasmid pYEbac653 encoding the UL12 protein tagged with the Flag epitope, was obtained. Recombinant virus YK653 (Fig. 1) was generated by transfection of pYEbac653 into rabbit skin cells as described previously (65). The genotypes of YK651 and YK653 were confirmed by PCR analyses with appropriate primers (data not shown).
Immunoprecipitation.
Vero cells were infected with either YK653 or wild-type HSV-1(F) at a multiplicity of infection (MOI) of 5. Infected cells were harvested at 12 h postinfection and lysed in NP-40 buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% Nonidet P-40 [NP-40]) containing a protease inhibitor cocktail (Sigma). Supernatants obtained after centrifugation of the cell lysates were precleared by incubation with protein A-Sepharose beads (GE Healthcare) at 4°C for 30 min. After a brief centrifugation, supernatants were reacted with the anti-Flag monoclonal antibody or anti-nucleolin antibody at 4°C for 1.5 h. Protein A-Sepharose beads were then added to the supernatants. Immunoprecipitates were collected by a brief centrifugation, washed extensively with NP-40 buffer, and analyzed by immunoblotting with anti-nucleolin antibody, anti-ICP8 antibody, anti-VP5 antibody, and anti-Flag antibody.
Generation of recombinant retroviruses and establishment of cell lines stably expressing shRNA against human nucleolin and GFP.
Recombinant retroviruses were generated as described previously (17). Vero cells were transduced by infection with retrovirus-containing supernatants of transfected Plat-GP cells and selected with 5 μg of puromycin/ml. Single colonies transduced by pSSSP-hNucleolin and pSSSP-GFP6 were isolated after 2 weeks and screened by immunoblotting with anti-nucleolin antibody, which led to the isolation of Vero-shNuc-4 and Vero-shNuc-5, and Vero-shGFP, respectively.
Southern blotting.
Southern blotting was performed as described previously (65).
Immunoblotting.
Vero, Vero-shNuc-4, Vero-shNuc-5, and Vero-shGFP cells were infected with HSV-1(F), YK651 or YK653 at an MOI of 5. At 12 and/or 18 h postinfection, infected cells were harvested and analyzed by immunoblotting with antibody to UL12, GFP, Flag epitope, nucleolin, and α-tubulin as described previously (21).
Immunofluorescence.
Vero cells were mock infected or infected with YK651 or YK653 at an MOI of 5. At 12 and 18 h postinfection, infected cells were fixed, permeabilized, and then stained with anti-Flag and anti-nucleolin as described previously (21). Samples were examined with a Zeiss LSM5 laser scanning confocal microscope.
Assay for cell viability.
The viability of Vero, Vero-shNuc-5, and Vero-shGFP cells was determined by using a Cell Counting Kit-8 (Dojindo) according to the manufacturer's instructions.
Cell fractionation and staphylococcal nuclease digestion.
Staphylococcal nuclease (SN) digestion was performed as described previously (24, 55). Briefly, Vero-shNuc-5 and Vero-shGFP cells infected with wild-type HSV-1(F) at an MOI of 5 for 12 h were harvested and resuspended in cold lysis buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1.5 mM MgCl2). NP-40 was added to a final concentration of 0.2%, the samples were incubated on ice for 3 min, and nuclei were collected by centrifugation. The supernatant debris was removed by further centrifugation and used as the cytoplasmic fraction. Nuclei were resuspended in digestion buffer (15 mM Tris-HCl [pH 7.4], 15 mM NaCl, 10 mM MgCl2, 1 mM CaCl2, 1 mM EDTA, 0.2 mM EGTA, 0.34 mM sucrose, 15 mM 2-mercaptoehanol). Micrococcal nuclease (Takara) was added to a concentration of 6 U/106 nuclei for the nuclear fraction or 0.2 U/106 cells for the cytoplasmic fraction, and digestion was carried out at 37°C for 30 min. For a control, purified virions prepared as described previously (65) were digested with nuclease. Digestions were stopped by addition of EDTA to 10 mM and EGTA to 5 mM. The nuclear and cytoplasmic fractions were incubated with 100 μg of proteinase K/ml and 0.5% sodium dodecyl sulfate. DNA was extracted with phenol-chloroform, precipitated with ethanol, and analyzed by Southern blotting.
Alkaline nuclease assays.
Alkaline nuclease assays were performed as described previously (44, 69). Briefly, Vero-shNuc-5 cells and Vero-shGFP cells were infected with HSV-1(F) at an MOI of 5 for 12 h, washed in NTM buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaC1, 2 mM 2-mercaptoethanol), and scraped into NTM buffer. The infected cells were pelleted by centrifugation, and cell pellets were resuspended in 20 mM Tris-HCl (pH 7.5) containing 5 mM 2-mercaptoethanol. The resuspended cells were disrupted by sonication and the samples were adjusted to 4 mM MgCl2 and 100 mM KCl. Cell debris was removed by centrifugation. The substrate for the nuclease assays was a 1-kb fragment of an HSV-1 UL41 domain amplified by PCR from pBC1007 (21) with primers 5′-GCGAATTCGGTTTGTTCGGGATGATGAA-3′ and 5′-GCGGATCCCTTCGTATCCGCCGGCGATC-3′, uniformly labeled by inclusion of [32P]dCTP in the deoxynucleoside triphosphate mix. Nuclease assays were performed in 100-μl reaction mixtures containing 25 μl of cell extract prepared as described above, 100 ng of substrate, 100 ng of M13mp18 single-stranded DNA (Takara), 20 mM Tris-HCl (pH 7.5), 40 mM NaCl, 1 mM MgC12, and 1 mM dithiothreitol. The reaction mixtures were incubated for 10 min at 37°C and then stopped by addition of 25 μl of 3.5 mg of herring testes carrier DNA/ml and 125 μl of 20% trichloroacetic acid. After 10 min on ice, the mixtures were centrifuged, and the radioactivity in 100-μl samples of the supernatant fractions was determined by scintillation counting.
Electron microscopic analysis.
Vero-shGFP and Vero-shNuc-5 cells infected with HSV-1(F) at an MOI of 5 for 12 h were fixed and processed for electron microscopic analysis as described previously (52).
Live-cell imaging.
Vero, Vero-shNuc-4, Vero-shNuc-5, and Vero-shGFP cells were infected with YK651 or YK601 at an MOI of 5. At various times postinfection, live-cell imaging was performed as described previously (64).
RESULTS
Construction and characterization of recombinant viruses expressing UL12 protein fused to a fluorescent protein or Flag epitope.
To investigate the localization of UL12 proteins in live infected cells, we constructed recombinant virus YK651, expressing UL12 fused to fluorescent protein (FP) VenusA206K (VenusA206K-UL12), and recombinant virus YK653, expressing UL12 fused to the Flag epitope (Fig. 1). Expression of the appropriate fusion protein in Vero cells infected with YK651 (VenusA206K-UL12) and YK653 (Flag-UL12) was confirmed by immunoblotting (Fig. 2A to C). Growth curves of YK651 and YK653, at an MOI of 5 or 0.01 on Vero cells, were almost identical to that of wild-type HSV-1(F) (Fig. 2D and E), indicating that tagging UL12 with VenusA206K FP or the Flag epitope had no effect on viral growth in Vero cells.
FIG. 2.
Characterization of recombinant viruses. (A to C) Immunoblots of electrophoretically separated lysates from Vero cells infected with wild-type HSV-1(F) (lane 1), YK651 (Venus-UL12) (lane 2), and YK653 (Flag-UL12) (lane 3). Infected cells were harvested 18 h postinfection and analyzed by immunoblotting with antibody to UL12 (A), GFP (B), and Flag (C). (D and E) Vero cells were infected at an MOI of 5 (D) or 0.01 (E) with wild-type HSV-1(F), YK651 (Venus-UL12), or YK653 (Flag-UL12). Total virus from cell culture supernatants and infected cells harvested at the indicated times were assayed on Vero cells.
Time-lapse studies of UL12 protein in live infected cells.
It has been reported that UL12 protein is localized diffusely throughout the nucleus in wild-type HSV-1-infected cells (11, 41, 66). To monitor the localization of UL12 in live infected cells, Vero cells infected with YK651 (VenusA206K-UL12) were examined by confocal microscopy at various times postinfection (6, 9, 12, 15, and 18 h postinfection). In agreement with previous reports (11, 41, 66), diffuse nuclear localization of VenusA206K-UL12 was observed throughout the present study (data not shown). In addition to the diffuse nuclear localization of VenusA206K-UL12, we also noted specific localization of VenusA206K-UL12 in nuclear compartments that appeared to be nucleoli throughout the present study, based on differential interference contrast (DIC) images of infected cells (data not shown).
Colocalization of UL12 and nucleolin in infected cells.
To examine whether UL12 in fact localizes in nucleoli in infected cells, Vero cells infected with YK651 (VenusA206K-UL12) were treated with antibody to nucleolin and examined by confocal microscopy. Nucleolin is a marker of the nucleolus (9). At 12 h postinfection (Fig. 3A, upper panels), the anti-nucleolin antibody colocalized with VenusA206K-UL12 in specific nuclear compartment(s), in agreement with the results above for UL12 localization in live infected cells. At 18 h postinfection, the nuclear domain containing nucleolin was marginalized and detected at the nuclear rim (Fig. 3A, lower panels). A disperse distribution of nucleolin throughout the nuclei was also observed. These features were consistent with earlier reports that HSV-1 infection induces modification of the nucleolus (2, 26, 28).
FIG. 3.
Localization of nucleolin and UL12 in infected cells. Vero cells infected with YK651 (Venus-UL12) (A) or YK653 (Flag-UL12) (B) were fixed at 12 and 18 h postinfection and permeabilized. YK651-infected cells were then treated with antibody to nucleolin, which was detected with Alexa Fluor 546-conjugated donkey anti-mouse IgG by confocal microscopy. YK653-infected cells were then treated with antibodies to Flag and nucleolin, which were detected with Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa Fluor 680-conjugated goat anti-mouse IgG, respectively, by confocal microscopy. The triangles indicate nuclear compartments that appeared to be nucleoli.
To confirm the intranuclear localization of UL12, recombinant virus YK653 expressing UL12 fused to the Flag epitope, a relatively small tag of only 8 amino acids, was used to examine localization of UL12 in infected cells. Double staining of Vero cells infected with YK653 (Flag-UL12) with anti-Flag rabbit polyclonal antibody and anti-nucleolin mouse monoclonal antibody (Fig. 3B) showed that the intranuclear localization of Flag-UL12 was similar to that of VenusA206K-UL12 (Fig. 3A) and that Flag-UL12 colocalized with nucleolin in a nuclear domain(s).
Taken together, these results indicated that UL12 specifically localized in nucleoli, as well as diffusely throughout the nuclei in infected cells.
UL12 interaction with nucleolin in HSV-1-infected cells.
A potential role(s) for nucleolin in HSV-1 replication has been previously reported (2). That report together with the results in Fig. 3, that UL12 colocalized with nucleolin in infected cells, led us to examine whether UL12 forms a complex with nucleolin in infected cells. Vero cells infected with YK653 (Flag-UL12) or wild-type HSV-1(F) were lysed and immunoprecipitated with anti-Flag or anti-nucleolin antibody, and the immunoprecipitates were analyzed by immunoblotting with antibodies to UL12, nucleolin, ICP8, and VP5. ICP8 is an HSV-1 DNA replication factor with single-stranded DNA-binding activity, and VP5 is an HSV-1 major capsid protein. As shown in Fig. 4A, the anti-Flag antibody coprecipitated nucleolin and ICP8 in lysates of cells infected with YK653 (Flag-UL12) but did not coprecipitate VP5. This result corroborated a previous report that UL12 interacts with ICP8 in infected cells (66). In contrast, the anti-Flag antibody did not coprecipitate nucleolin or ICP8 in wild-type HSV-1(F)-infected cell lysates. In a reciprocal experiment, anti-nucleolin antibody coprecipitated UL12 from wild-type HSV-1(F)-infected cell lysates but did not coprecipitate ICP8 (Fig. 4B). These results indicated that UL12 formed a complex with nucleolin in infected cells, but this complex appeared not to contain ICP8.
FIG. 4.
Identification of the UL12-nucleolin complex. (A) Vero cells infected with YK653 (Flag-UL12) (lanes 1 and 3) or wild-type HSV-1(F) (lanes 2 and 4) were lysed and immunoprecipitated with antibody to the Flag epitope. The immunoprecipitates (lanes 3 and 4) were analyzed by electrophoresis and immunoblotted with antibody to nucleolin, ICP8, Flag, and VP5. One-ninth of the Vero whole-cell lysates (WCL) used in the reaction mixtures for lanes 3 and 4 was loaded in lanes 1 and 2, respectively. (B) Vero cells infected with wild-type HSV-1(F) were lysed and immunoprecipitated with antibody to nucleolin (lanes 1 and 3) and Flag (lanes 2 and 4). The immunoprecipitates (lanes 3 and 4) were analyzed by electrophoresis and immunoblotted with antibody to UL12, ICP8, and nucleolin. One-ninth of the Vero WCLs used in the reaction mixtures for lanes 3 and 4 was loaded in lanes 1 and 2, respectively.
Effect of nucleolin knockdown on viral growth and nuclease activity.
To investigate the interaction between UL12 and nucleolin in infected cells, we generated two Vero cell lines stably expressing short hairpin RNA (shRNA) against nucleolin (Vero-shNuc-4 and Vero-shNuc-5) to knock down nucleolin expression. We also generated a control cell line (Vero-shGFP) expressing shRNA against GFP. As shown in Fig. 5A, the level of nucleolin in Vero-shNuc-5 detected by immunoblotting was significantly lower than in Vero-shGFP and normal Vero cells. Expression of these shRNAs in Vero cells had no effect on cell viability (Fig. 5B), and nucleolin knockdown was stable in Vero-shNuc-5 (data not shown). These results indicated that shRNA against nucleolin induced a considerable reduction of nucleolin expression in Vero cells without affecting cell viability.
FIG. 5.
Knockdown of nucleolin. (A) Vero-shNuc-5 cells expressing shRNA against nucleolin (lane 1), Vero-shGFP cells expressing shRNA against GFP (lane 2), and normal Vero cells (lane 3) were analyzed by immunoblotting with antibody to nucleolin (upper panel) and α-tubulin (lower panel). (B) The viability of Vero, Vero-shNuc-5, and Vero-shGFP cells relative to a control (medium only) was measured as the cell metabolic activity. Each value is the mean ± the standard deviation of three independent experiments. (C) Vero-shNuc-5, Vero-shGFP, and Vero cells infected with wild-type HSV-1(F) for 12 h were analyzed by immunoblotting with antibody to nucleolin, UL12, and α-tubulin.
To determine whether nucleolin depletion affected the total accumulation of UL12 in infected cells, Vero-shNuc-5, Vero-shGFP, and normal Vero cells were infected with wild-type HSV-1(F) and analyzed by immunoblotting with antibody to nucleolin, UL12, and α-tubulin. As shown in Fig. 5C, although nucleolin expression was downregulated in HSV-1(F)-infected Vero-shNuc-5 cells, UL12 expression was similar in all three cell lines. In addition, nucleolin depletion had no effect on the accumulation of viral structural proteins, UL41 and UL48, in infected cells (data not shown).
To determine the efficiency of viral replication in nucleolin-depleted infected cells, Vero-shNuc-5 and Vero-shGFP cells were infected with wild-type HSV-1(F) at an MOI of 5 or 0.01. Viruses were harvested at 12 and 24 h postinfection, and titers measured by standard plaque assays on Vero cells. As shown in Fig. 6A and B, the virus titer in Vero-shNuc-5 was a little lower than in Vero-shGFP at both times and MOIs: at an MOI of 5, the virus titer was 1.8-fold lower at 12 and 24 h postinfection and, at an MOI of 0.01, it was 3.2-fold lower at 12 h postinfection and 1.5-fold lower at 24 h postinfection. Similar results were obtained in a repeat experiment (data not shown).
FIG. 6.
Effect of nucleolin knockdown on viral growth and nuclease activity. (A and B) Vero-shNuc-5 (open bar) and Vero-shGFP (shaded bar) cells were infected with HSV-1(F) at an MOI of 5 (A) or 0.01 (B). At 12 and 24 h postinfection, total virus from cell culture supernatants and infected cells was harvested and assayed on Vero cells. (C) Alkaline nuclease activity in HSV-1(F)-infected Vero-shNuc-5 and Vero-shGFP cells. Nuclease activity in each cell line was calculated by subtracting the activity in mock-infected cells from that in cells infected with HSV-1(F). Each value is the mean ± the standard deviation of three independent experiments. The statistical difference of the nuclease activity between Vero-shNuc- and Vero-shGFP-infected cells is noted (*, P < 0.05).
To determine the viral nuclease activity in nucleolin-depleted infected cells, Vero-shNuc-5 and Vero-shGFP cells infected with wild-type HSV-1(F) at an MOI of 5 were lysed at 12 h postinfection and assayed for alkaline nuclease activity. As reported previously (69), mock-infected Vero-shNuc-5 and Vero-shGFP cell extracts did not have significant amounts of nuclease activity, whereas HSV-1(F) infection of these cells resulted in considerable nuclease activity (data not shown). The nuclease activity detected in HSV-1(F)-infected Vero-shGFP cell lysates was a little (1.2-fold) higher than in HSV-1(F)-infected Vero-shNuc-5 cell lysates (Fig. 6C). Similar results were also obtained at 5 or 8 h postinfection (data not shown).
Taken together, these results indicated that knockdown of nucleolin had little or no effect on UL12 accumulation, viral growth, and viral nuclease activity in infected Vero cells.
Effect of nucleolin knockdown on virion maturation.
It has been shown that viral DNA in HSV-1-infected cells that is in viral replication forms is sensitive to SN digestion, whereas unit-length and encapsidated viral DNA is resistant to SN digestion (24, 55). Cells infected with wild-type HSV-1 also have been reported to produce SN-resistant nuclear and cytoplasmic DNA-containing capsids, whereas cells infected with a UL12-null mutant virus only produced SN-resistant nuclear DNA-containing capsids, with impaired SN-sensitive DNA-containing capsids in cytoplasmic fractions (55). These observations implied that UL12 played a role in virion maturation, particularly in the efficient egress of nucleocapsids from the nucleus. Since nucleolin has been reported to interact with UL12, to investigate whether nucleolin knockdown has an effect on virion maturation, similar to that observed with UL12-null mutant virus, we carried out SN sensitivity assays in HSV-1-infected nucleolin-depleted cells. Vero-shNuc-5 and Vero-shGFP cells infected with wild-type HSV-1(F) were fractionated into nuclear and cytoplasmic fractions. Each fraction, as well as a purified virion sample, was digested with SN, and DNA was extracted from the fractions and purified virions and analyzed by Southern blotting (Fig. 7A and B). Nuclear and cytoplasmic fractions without SN digestion were also analyzed (Fig. 7C and D). As reported previously (55), both SN-resistant viral DNA, which corresponded to encapsidated virion DNA, and degraded viral DNA, which resulted from SN digestion of replicative and nonencapsidated viral DNA, were found in the nuclear fractions, whereas only SN-resistant viral DNA was found in the cytoplasmic fractions (Fig. 7A). HSV-1(F)-infected Vero-shNuc-5 and Vero-shGFP cell nuclei contained both SN-resistant and SN-sensitive viral DNA at similar levels. In contrast, the amount of SN-resistant DNA in the cytoplasm of infected Vero-shNuc-5 cells was sevenfold lower than in infected Vero-shGFP cells (Fig. 7B). Without SN digestion, there were two major DNA bands in the nuclear fraction (Fig. 7C), as reported previously (29). The upper band has been termed “well DNA” (29) and did not enter the gel. The lower band, which comigrated with virion DNA (data not shown), corresponded to encapsidated viral DNA. In the cytoplasmic fraction without SN digestion, only a single band, which corresponded to encapsidated viral DNA, was detected (Fig. 7C) and the amount of the viral DNA in the cytoplasm of infected Vero-shNuc-5 cells was fivefold lower than in infected Vero-shGFP cells (Fig. 7D), as observed with the cytoplasmic fraction with SN digestion (Fig. 7A and B). These observations showed that only encapsidated viral DNA was present in the cytoplasm of infected cells and confirmed the cell fractionation protocol used in these studies. Taken together, these results indicated that, in HSV-1(F)-infected Vero-shGFP cells, viral DNA was encapsidated and nucleocapsids were transported to the cytoplasm efficiently. However, in HSV-1(F)-infected Vero-shNuc-5 cells, viral DNA was encapsidated but the nucleocapsids failed to egress efficiently into the cytoplasm.
FIG. 7.
SN sensitivity analysis of HSV-1 DNA during viral replication. (A) Vero-shNuc-5 (lanes 1 and 3) and Vero-shGFP (lanes 2 and 4) cells infected with wild-type HSV-1(F) at an MOI of 5 were fractionated into cytoplasmic (lanes 1 and 2) and nuclear (lanes 3 and 4) fractions at 12 h postinfection, and the fractions were treated with SN. DNA from each fraction and purified virion DNA (lane 5) were analyzed by Southern blotting with UL41 DNA as the probe. (B) The amount of SN-resistant viral DNA in the cytoplasmic fractions was quantitated relative to the amount in the nuclear fractions from the data in panel A. The shGFP value was normalized to 100. (C) Vero-shNuc-5 (lanes 1 and 3) and Vero-shGFP (lanes 2 and 4) cells infected with wild-type HSV-1(F) were fractionated into cytoplasmic (lanes 1 and 2) and nuclear fractions (lanes 3 and 4) as described in panel A and were analyzed by Southern blotting with UL41 DNA as the probe without SN digestion. (D) The amount of viral DNA in the cytoplasmic fractions was quantitated relative to the amount of total viral DNA in the nuclear fractions from the data in panel C. The shGFP value was normalized to 100.
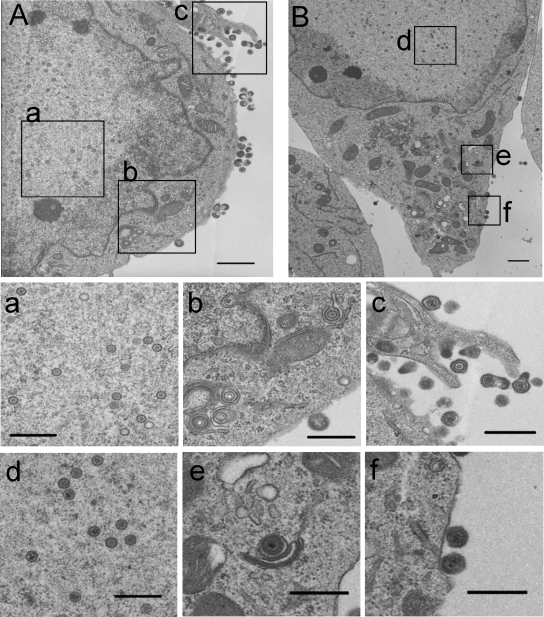
To confirm that nucleolin knockdown affects nuclear egress of nucleocapsids as indicated in the results above, we performed two sets of experiments. In the first set of experiments, Vero-shGFP and Vero-shNuc-5 cells infected with wild-type HSV-1(F) were processed for electron microscopy at 12 h postinfection, and the numbers of virus particles in the nuclear, cytoplasmic, and extracellular compartments were counted. Thin sections of HSV-1(F)-infected cells (Fig. 8) showed that nucleocapsids were readily observed within the nucleus of both infected Vero-shGFP and Vero-shNuc-5 cells. HSV-1(F)-infected Vero-shGFP cells had numerous extracellular virions attached to their surfaces, as well as a number of virions within the cytoplasm (Fig. 8A). In contrast, in infected Vero-shNuc-5 cells, the number of virions at the cell surface and in the cytoplasm was significantly decreased (Fig. 8B and Table 1 ), and the difference was statistically significant (P = 0.00000032). As shown in Table 1, 35% of the virus particles in micrographs of infected Vero-shGFP cells were found in extracellular and cytoplasmic areas, whereas significantly less (12.8%) of the virus particles in infected Vero-shNuc-5 cells were in extracellular and cytoplasmic areas.
FIG. 8.
Election microscopy of infected nucleolin knockdown cells. Electron microscopy of wild-type HSV-1(F)-infected Vero-shGFP (A) and Vero-shNuc-5 cells (B). Panels a to f show magnifications of the corresponding areas indicated in panels A and B. Scale bar: A, 700 nm; B, 1,000 nm; a to f, 500 nm.
TABLE 1.
Electron microscopic localization of HSV-1 virions in infected cells
| Type of infected cell | Avg no. of virus particles in compartment/cell ±SD (% of total particles in compartment) |
Total no. of particles/total no. of cells | ||
|---|---|---|---|---|
| Extracellular space | Cytoplasm | Nucleus | ||
| shNuc-5 | 4.6 ± 3.7 (7.5) | 3.3 ± 1.5 (5.3) | 53.8 ± 16.6 (87.2) | 617/10 |
| shGFP | 25.8 ± 8.8 (23.1) | 13.3 ± 6.4 (11.9) | 72.5 ± 33.1 (65.0) | 1116/10 |
In the second set of experiments, the effect of nucleolin knockdown on viral localization was studied by single particle analysis. It has been shown that recombinant alphaherpesviruses expressing an FP fused to the VP26 capsid protein can be studied by single-particle analysis (7, 60). Consistent with that report, we showed that cells infected by recombinant herpesviruses expressing VenusA206K-VP26 produced fluorescent puncta of mature capsids containing VenusA206K-VP26 (64). Therefore, Vero, Vero-shGFP, Vero-shNuc-4, and Vero-shNuc-5 cells were infected with YK601 (VenusA206K-VP26), examined at 12 h postinfection by confocal microscopy and reconstructed as three-dimensional Z-stacked images (Fig. 9 ). Vero-shNuc-4 is a nucleolin knockdown cell clone in which the level of nucleolin was somewhat greater than in Vero-shNuc-5 but significantly less than in Vero-shGFP cells (Fig. 9A and B). As shown in Fig. 9C, there were many discrete fluorescent puncta with VenusA206K-VP26 emission in the cytoplasm of infected normal Vero and Vero-shGFP cells. In contrast, there were fewer (three- and fourfold) fluorescent puncta with VenusA206K-VP26 emission in the cytoplasm of infected Vero-shNuc-4 and Vero-shNuc-5 cells, respectively (Fig. 9C and D). These observations were consistent with the results of the electron microscopic analysis described above (Fig. 8).
FIG. 9.
Confocal microscopy of infected nucleolin knock-down cells. (A) Vero-shNuc-4, Vero-shNuc-5, and Vero-shGFP cells infected with wild-type HSV-1(F) for 12 h were analyzed by immunoblotting with antibodies to nucleolin, UL12, and α-tubulin. (B) Quantitation of the amount of nucleolin protein relative to the amount of α-tubulin shown in panel A. The shGFP value was normalized to 100. (C) Vero (a and b), Vero-shGFP (c and d), Vero-shNuc-5 (e and f), and Vero-shNuc-4 (g and h) cells were infected with YK601 (VenusA206K-VP26) for 12 h and then examined by confocal micros- copy. Confocal Z-sections were acquired through the entire thickness of the cells. A confocal Z-stacked fluorescence image derived from the Z-sections (a, c, e, and g) and DIC (b, d, f, and h) images is shown. Insets show magnified images of the boxed areas. Scale bar, 500 nm. (D) Quantitation of virus particles in the cytoplasm of infected cells from the data in panel C.
Since VenusA206K FP is a derivative of GFP, shRNA against GFP might have downregulated expression of VenusA206K-VP26 in infected cells in these studies. However, shRNA against GFP has two nucleotide mismatches relative to VenusA206K and the level of VenusA206K-VP26 expression in infected Vero and Vero-shGFP cells was similar (data not shown). Furthermore, when another control cell line, Vero-shCre, that expresses shRNA against Cre recombinase, was used in this experiment, a similar level of fluorescent puncta was observed (data not shown).
Taken together, these two sets of experiments indicated that nucleolin knockdown impaired nuclear egress of HSV-1 nucleocapsids in infected cells.
DISCUSSION
Transport of large herpesvirus nucleocapsids across the NM appears to be a complex process, and its regulation is poorly understood. In the present study, we have shown that the cellular protein nucleolin plays a role in efficient nuclear egress of HSV-1 nucleocapsids. To our knowledge, this is the first report of a specific host cell protein acting directly in the nuclear egress pathway of herpesvirus nucleocapsids.
A possible model for nuclear egress of herpesvirus nucleocapsids would be for this process to be regulated by the interaction of cellular and viral proteins. In this regard, identification and characterization of a cellular protein(s) that interacts with a viral protein involved in the nuclear egress of nucleocapsids could provide a new experimental approach to this model. To investigate the interaction of cellular and viral proteins, a variety of methods have been used, such as the yeast two-hybrid system and a coimmunoprecipitation or pulldown system, followed by proteomics analysis. In the present study, we used real-time imaging to determine the nuclear localization of HSV-1 protein UL12, which has been suggested to be involved in the pathway for nuclear egress of nucleocapsids, since information on the subcellular localization of a viral protein in infected cells can provide insight into its function and indicate the cellular protein interacting with it. Real-time imaging of live cells was needed for these studies because subcellular localization of HSV-1 viral proteins can change as the infection progresses, so analysis using fixed cells, as has been widely used to localize viral proteins, can miss dynamic interactions. Dynamic localization of viral proteins can be studied by live imaging using a recombinant virus expressing an FP-tagged viral protein(s), which can allow visualization of the tagged protein(s) in live cells and changes in its localization as infection progresses (5, 27, 60, 64). We examined localization of HSV-1 UL12 alkaline nuclease in a live-cell imaging system using recombinant virus YK651 expressing UK12 fused to VenusA206K FP and observed the previously unreported nucleolar localization of UL12 in infected cells. This result led us to investigate whether UL12 interacts with a component(s) of the nucleolus, and we found that UL12 and nucleolin were coprecipitated by specific antibody to either protein. These results indicated that real-time imaging data can suggest cellular protein(s) that might interact with a specific viral protein and that UL12 formed a complex with nucleolin in HSV-1-infected cells. This approach can be readily applied to other HSV-1 proteins, as well as to other viruses.
UL12 has been suggested to be one of the factors involved in nuclear egress of HSV-1 nucleocapsids, based on the observation that wild-type quantities of viral DNA were produced in the nucleus of cells infected with a UL12-null mutant virus, but few DNA-containing capsids were detectable in the cytoplasm (55). Therefore, our observation that UL12 interacted with nucleolin prompted us to take a closer look at a role of nucleolin in the nuclear egress of nucleocapsids. The defect in nuclear egress of nucleocapsids was similar in Vero cells expressing shRNA against nucleolin and infected with wild-type HSV-1 in the present study to that previously reported for normal Vero cells infected with a UL12-null mutant virus (55). As previously reported for SN assays of Vero cells infected with the UL12-null mutant virus (55), the amount of SN-resistant viral DNA (encapsidated viral DNA) in the cytoplasm of wild-type HSV-1(F)-infected Vero cells in which nucleolin expression was knocked down by shRNA expression (Vero-shNuc-5 cells) was significantly lower than that in the cytoplasm of wild-type HSV-1(F)-infected Vero cells expressing control shRNA (Vero-shGFP cells), although the amount of SN-resistant viral DNA in the nucleus of both cell lines was almost equivalent. These results suggested that DNA-containing capsids in nuclei of infected Vero-shNuc-5 cells were blocked in nuclear egress and maturation in the cytoplasm. In agreement with this conclusion, electron microscopic analyses of wild-type HSV-1-infected cells and real-time imaging of cells infected with an HSV-1 recombinant virus expressing capsid protein VP26 fused to an FP showed that the number of mature capsids in the cytoplasm and at the surface of Vero-shNuc cells was significantly lower than in infected Vero-shGFP cells. From these results, we have concluded that nucleolin plays a role in the nuclear egress of HSV-1 nucleocapsids. In agreement with this conclusion, Calle et al. recently reported that nucleolin was required for efficient HSV-1 replication in HeLa cells, although the mechanism by which nucleolin affected HSV-1 growth was not reported (2). They demonstrated that nucleolin knockdown resulted in significant (10- to 23-fold) impairment of growth of wild-type HSV-1 strain 17 in HeLa cells at an MOI of 0.4 and a modest (3-fold) decrease of viral growth at an MOI of 10. In the present study, although we did not find a significant decrease in viral growth as reported by a previous study with HSV-1(F)-infected Vero-shNuc-5 cells (2), we did find that nucleolin knockdown resulted in a small (1.5- to 3.2-fold) but consistent reduction of viral growth. The effect of nucleolin knockdown on HSV-1 replication may depend on the viral strain and cell type studied.
At present we can only speculate about the mechanism(s) by which nucleolin acts in the nuclear egress of nucleocapsids. Since nucleolin interacts with UL12 and the phenotype of the HSV-1 UL12-null mutant virus with respect to the nuclear egress of nucleocapsids is, to some extent, similar to that of wild-type HSV-1(F) infection of nucleolin-depleted cells, nucleolin might interact with UL12 to regulate the role of UL12 in nuclear egress of mature nucleocapsids into the cytoplasm. It has been hypothesized that the state of the viral genome packaged into capsids affects nuclear egress of DNA-containing capsids (29, 55). Therefore, in cells infected with the UL12-null mutant virus, a defect in viral genome maturation might lead to a malfunction in the nuclear egress of nucleocapsids. In support of this hypothesis, it has been reported that smaller than unit length viral DNA molecules can be packaged into capsids in the nucleus but do not appear in the cytoplasm (68). Furthermore, viral DNA that accumulates in cells infected with the UL12-null mutant virus has been shown to have an aberrant structure compared to that in cells infected with wild-type HSV-1 (29). Since nucleolin has been reported to interact with and regulate a variety of cellular and viral enzymes (e.g., simian virus 40 large tumor-antigen DNA helicase [54], RNA-dependent RNA polymerase activity of the hepatitis C virus NS5B protein [56], telomerase [22], and recombinase Rad51 [4]) and to play multiple roles in DNA metabolism (9, 34), nucleolin might interact with UL12 and modify its ability to process viral DNA in infected cells. However, we should note that nucleolin knockdown cannot completely account for the role of UL12 in nuclear egress of nucleocapsids, based on the observation that the HSV-1 UL12-null mutant virus produced a 100- to 1,000-fold decrease in virus titer in Vero cells (55), whereas in the present study nucleolin knockdown produced only a modest (1.5- to 3.2-fold) decrease in Vero cells. Therefore, there must be UL12 function(s) in the nuclear egress of nucleocapsids that are independent of nucleolin.
Alternatively, it is more likely that nucleolin may function in the nuclear egress of nucleocapsids independently of UL12. Nucleolin is a multifunctional protein that exhibits broad localization in nucleoli, nucleoplasm, cytoplasm, and the plasma membrane; interacts with a wide variety of cellular and viral proteins; and affects various aspects of cellular functions, including transcription mediated by RNA polymerase II, rRNA gene transcription, rRNA folding, pre-rRNA maturation, assembly of ribosomal proteins, shuttling of some proteins between the nucleus and cytoplasm, mRNA stability, regulation of translation, cell adhesion, migration, and virus infection (9, 34, 61). It is possible that one or more of these functions and/or currently unreported function(s) of nucleolin may be involved in regulation of HSV-1 nuclear egress. Interestingly, we noted in the present study that nuclear domain(s) in which nucleolin concentrated was marginalized and detected at the nuclear rim in infected cells (Fig. 3). This observation is in agreement with previous reports showing that HSV-1 infection induces a disperse distribution of nucleolin throughout the nuclei and nuclear domain(s) containing nucleolin is marginalized at nuclear rim (26, 28). Therefore, a possible model of how nucleolin promotes nuclear egress of HSV-1 nucleocapsids might be that redistributed nucleolin to the inner NM somehow modifies nuclear lamina to allow nucleocapsids access to the inner NM for primary envelopment efficiently like UL34 and UL31 (1, 45, 58). To our surprise, nucleolin knockdown had no effect on cell viability (Fig. 5), although nucleolin is associated with various cellular functions as described above. These results suggest that the defect in nuclear egress of nucleocapsids by nucleolin knockdown is specific and biologically relevant. It is also possible that nucleolin plays a role in nuclear egress of nucleocapsids with UL12 functioning as a cofactor of nucleolin by an unknown mechanism. Although we could not detect the decrease in the expression of some virion proteins such as UL41 and UL48 with nucleolin knockdown (data not shown), we cannot completely eliminate the possibility that nucleolin knockdown somewhat downregulated the expression of one or some of other virion protein(s), leading to lower virion production since the number of total virions in Vero-shNuc-5 cells detected by electron microscopy was a little (1.8-fold) lower than that in Vero-shGFP cells (Table 1), and the difference was statistically significant (P = 0.0073). The lower virion production observed with nucleolin knockdown might in part explain why nucleolin knockdown reduced the accumulation of virions in the cytoplasm and at the cell surface.
In conclusion, this study has shown that the cellular protein nucleolin is required for efficient nuclear egress of HSV-1 nucleocapsids. Although the mechanism by which nucleolin acts in the nuclear egress of nucleocapsids remains to be determined, these results provide the first cellular clue that might enable the complex mechanisms of nuclear egress of herpesvirus nucleocapsids to be elucidated.
Acknowledgments
We are grateful to Hideo Iba for providing pmU6, pSSSP, pSSSP-GFP6, and pSSSP-Cre. We are grateful to Shihoko Koyama for excellent technical assistance.
This study was supported in part by Grants for Scientific Research and Grants for Scientific Research in Priority Areas from the Ministry of Education, Culture, Science, Sports and Technology (MEXT) of Japan and a grant from the Takeda Science Foundation.
Footnotes
Published ahead of print on 2 December 2009.
REFERENCES
- 1.Bjerke, S. L., and R. J. Roller. 2006. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347:261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calle, A., I. Ugrinova, A. L. Epstein, P. Bouvet, J. J. Diaz, and A. Greco. 2008. Nucleolin is required for an efficient herpes simplex virus type 1 infection. J. Virol. 82:4762-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campadelli-Fiume, G., and B. Roizman. 2006. The egress of herpesviruses from cells: the unanswered questions. J. Virol. 80:6716-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De, A., S. L. Donahue, A. Tabah, N. E. Castro, N. Mraz, J. L. Cruise, and C. Campbell. 2006. A novel interaction of nucleolin with Rad51. Biochem. Biophys. Res. Commun. 344:206-213. [DOI] [PubMed] [Google Scholar]
- 5.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farnsworth, A., T. W. Wisner, M. Webb, R. Roller, G. Cohen, R. Eisenberg, and D. C. Johnson. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 104:10187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forest, T., S. Barnard, and J. D. Baines. 2005. Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7:429-431. [DOI] [PubMed] [Google Scholar]
- 8.Francke, B., H. Moss, M. C. Timbury, and J. Hay. 1978. Alkaline DNase activity in cells infected with a temperature-sensitive mutant of herpes simplex virus type 2. J. Virol. 26:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginisty, H., H. Sicard, B. Roger, and P. Bouvet. 1999. Structure and functions of nucleolin. J. Cell Sci. 112(Pt. 6):761-772. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein, J. N., and S. K. Weller. 1998. In vitro processing of herpes simplex virus type 1 DNA replication intermediates by the viral alkaline nuclease, UL12. J. Virol. 72:8772-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstein, J. N., and S. K. Weller. 1998. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology 244:442-457. [DOI] [PubMed] [Google Scholar]
- 12.Haraguchi, T., T. Mizutani, N. Yamamichi, T. Ito, S. Minoguchi, and H. Iba. 2007. siRNAs do not induce RNA-dependent transcriptional silencing of retrovirus in human cells. FEBS Lett. 581:4949-4954. [DOI] [PubMed] [Google Scholar]
- 13.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofemeister, H., and P. O'Hare. 2008. Nuclear pore composition and gating in herpes simplex virus-infected cells. J. Virol. 82:8392-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann, P. J. 1981. Mechanism of degradation of duplex DNA by the DNase induced by herpes simplex virus. J. Virol. 38:1005-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann, P. J., and Y. C. Cheng. 1978. The deoxyribonuclease induced after infection of KB cells by herpes simplex virus type 1 or type 2. I. Purification and characterization of the enzyme. J. Biol. Chem. 253:3557-3562. [PubMed] [Google Scholar]
- 17.Kato, A., J. Arii, I. Shiratori, H. Akashi, H. Arase, and Y. Kawaguchi. 2009. Herpes simplex virus 1 protein kinase Us3 phosphorylates viral envelope glycoprotein B and regulates its expression on the cell surface. J. Virol. 83:250-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato, A., M. Tanaka, M. Yamamoto, R. Asai, T. Sata, Y. Nishiyama, and Y. Kawaguchi. 2008. Identification of a physiological phosphorylation site of the herpes simplex virus 1-encoded protein kinase Us3 which regulates its optimal catalytic activity in vitro and influences its function in infected cells. J. Virol. 82:6172-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, A., M. Yamamoto, T. Ohno, H. Kodaira, Y. Nishiyama, and Y. Kawaguchi. 2005. Identification of proteins phosphorylated directly by the Us3 protein kinase encoded by herpes simplex virus 1. J. Virol. 79:9325-9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawaguchi, Y., K. Kato, M. Tanaka, M. Kanamori, Y. Nishiyama, and Y. Yamanashi. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J. Virol. 77:2359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khurts, S., K. Masutomi, L. Delgermaa, K. Arai, N. Oishi, H. Mizuno, N. Hayashi, W. C. Hahn, and S. Murakami. 2004. Nucleolin interacts with telomerase. J. Biol. Chem. 279:51508-51515. [DOI] [PubMed] [Google Scholar]
- 23.Leach, N., S. L. Bjerke, D. K. Christensen, J. M. Bouchard, F. Mou, R. Park, J. Baines, T. Haraguchi, and R. J. Roller. 2007. Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J. Virol. 81:10792-10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lentine, A. F., and S. L. Bachenheimer. 1990. Intracellular organization of herpes simplex virus type 1 DNA assayed by staphylococcal nuclease sensitivity. Virus Res. 16:275-292. [DOI] [PubMed] [Google Scholar]
- 25.Leuzinger, H., U. Ziegler, E. M. Schraner, C. Fraefel, D. L. Glauser, I. Heid, M. Ackermann, M. Mueller, and P. Wild. 2005. Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 79:13047-13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez, M. R., E. F. Schlegel, S. Wintersteller, and J. A. Blaho. 2008. The major tegument structural protein VP22 targets areas of dispersed nucleolin and marginalized chromatin during productive herpes simplex virus 1 infection. Virus Res. 136:175-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lymberopoulos, M. H., and A. Pearson. 2007. Involvement of UL24 in herpes-simplex-virus-1-induced dispersal of nucleolin. Virology 363:397-409. [DOI] [PubMed] [Google Scholar]
- 29.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113:163-169. [DOI] [PubMed] [Google Scholar]
- 32.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9:423-429. [DOI] [PubMed] [Google Scholar]
- 33.Mettenleiter, T. C., and T. Minson. 2006. Egress of alphaherpesviruses. J. Virol. 80:1610-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mongelard, F., and P. Bouvet. 2007. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 17:80-86. [DOI] [PubMed] [Google Scholar]
- 35.Morimoto, T., J. Arii, M. Tanaka, T. Sata, H. Akashi, M. Yamada, Y. Nishiyama, M. Uema, and Y. Kawaguchi. 2009. Differences in the regulatory and functional effects of the Us3 protein kinase activities of herpes simplex virus 1 and 2. J. Virol. 83:11624-11634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris, J. B., H. Hofemeister, and P. O'Hare. 2007. Herpes simplex virus infection induces phosphorylation and delocalization of emerin, a key inner nuclear membrane protein. J. Virol. 81:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mou, F., T. Forest, and J. D. Baines. 2007. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 81:6459-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mou, F., E. Wills, and J. D. Baines. 2009. Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J. Virol. 83:5181-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purves, F. C., D. Spector, and B. Roizman. 1991. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 65:5757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Randall, R. E., and N. Dinwoodie. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 67(Pt. 10):2163-2177. [DOI] [PubMed] [Google Scholar]
- 42.Reuven, N. B., A. E. Staire, R. S. Myers, and S. K. Weller. 2003. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 77:7425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reuven, N. B., and S. K. Weller. 2005. Herpes simplex virus type 1 single-strand DNA binding protein ICP8 enhances the nuclease activity of the UL12 alkaline nuclease by increasing its processivity. J. Virol. 79:9356-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuven, N. B., S. Willcox, J. D. Griffith, and S. K. Weller. 2004. Catalysis of strand exchange by the HSV-1 UL12 and ICP8 proteins: potent ICP8 recombinase activity is revealed upon resection of dsDNA substrate by nuclease. J. Mol. Biol. 342:57-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J. Virol. 78:5564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. Herpes simplex viruses. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 49.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. The family Herpesviridae: a brief introduction, 5th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, PA.
- 50.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryckman, B. J., and R. J. Roller. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78:399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sagou, K., T. Imai, H. Sagara, M. Uema, and Y. Kawaguchi. 2009. Regulation of the catalytic activity of herpes simplex virus 1 protein kinase Us3 by autophosphorylation and its role in pathogenesis. J. Virol. 83:5773-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott, E. S., and P. O'Hare. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 75:8818-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seinsoth, S., H. Uhlmann-Schiffler, and H. Stahl. 2003. Bidirectional DNA unwinding by a ternary complex of T antigen, nucleolin and topoisomerase I. EMBO Rep. 4:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shao, L., L. M. Rapp, and S. K. Weller. 1993. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology 196:146-162. [DOI] [PubMed] [Google Scholar]
- 56.Shimakami, T., M. Honda, T. Kusakawa, T. Murata, K. Shimotohno, S. Kaneko, and S. Murakami. 2006. Effect of hepatitis C virus (HCV) NS5B-nucleolin interaction on HCV replication with HCV subgenomic replicon. J. Virol. 80:3332-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simpson-Holley, M., J. Baines, R. Roller, and D. M. Knipe. 2004. Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 78:5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simpson-Holley, M., R. C. Colgrove, G. Nalepa, J. W. Harper, and D. M. Knipe. 2005. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 79:12840-12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith, G. A., S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. U. S. A. 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srivastava, M., and H. B. Pollard. 1999. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 13:1911-1922. [PubMed] [Google Scholar]
- 62.Strobel-Fidler, M., and B. Francke. 1980. Alkaline deoxyribonuclease induced by herpes simplex virus type 1: composition and properties of the purified enzyme. Virology 103:493-501. [DOI] [PubMed] [Google Scholar]
- 63.Subramanian, R. P., and R. J. Geraghty. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. U. S. A. 104:2903-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sugimoto, K., M. Uema, H. Sagara, M. Tanaka, T. Sata, Y. Hashimoto, and Y. Kawaguchi. 2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 82:5198-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomas, M. S., M. Gao, D. M. Knipe, and K. L. Powell. 1992. Association between the herpes simplex virus major DNA-binding protein and alkaline nuclease. J. Virol. 66:1152-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turcotte, S., J. Letellier, and R. Lippe. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 79:8847-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vlazny, D. A., A. Kwong, and N. Frenkel. 1982. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc. Natl. Acad. Sci. U. S. A. 79:1423-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weller, S. K., M. R. Seghatoleslami, L. Shao, D. Rowse, and E. P. Carmichael. 1990. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J. Gen. Virol. 71(Pt. 12):2941-2952. [DOI] [PubMed] [Google Scholar]
- 70.Wild, P., M. Engels, C. Senn, K. Tobler, U. Ziegler, E. M. Schraner, E. Loepfe, M. Ackermann, M. Mueller, and P. Walther. 2005. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 79:1071-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wisner, T. W., C. C. Wright, A. Kato, Y. Kawaguchi, F. Mou, J. D. Baines, R. J. Roller, and D. C. Johnson. 2009. Herpesvirus gB-induced fusion between the virion envelope and outer nuclear membrane during virus egress is regulated by the viral US3 kinase. J. Virol. 83:3115-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamamichi, N., M. Yamamichi-Nishina, T. Mizutani, H. Watanabe, S. Minoguchi, N. Kobayashi, S. Kimura, T. Ito, N. Yahagi, M. Ichinose, M. Omata, and H. Iba. 2005. The Brm gene suppressed at the posttranscriptional level in various human cell lines is inducible by transient HDAC inhibitor treatment, which exhibits antioncogenic potential. Oncogene 24:5471-5481. [DOI] [PubMed] [Google Scholar]
- 73.Ye, G. J., and B. Roizman. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. U. S. A. 97:11002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]