Abstract
The high-affinity in vivo interaction between soluble HIV-1 envelope glycoprotein (Env) immunogens and primate CD4 results in conformational changes that alter the immunogenicity of the gp120 subunit. Because the conserved binding site on gp120 that directly interacts with CD4 is a major vaccine target, we sought to better understand the impact of in vivo Env-CD4 interactions during vaccination. Rhesus macaques were immunized with soluble wild-type (WT) Env trimers, and two trimer immunogens rendered CD4 binding defective through distinct mechanisms. In one variant, we introduced a mutation that directly disrupts CD4 binding (368D/R). In the second variant, we introduced three mutations (423I/M, 425N/K, and 431G/E) that disrupt CD4 binding indirectly by altering a gp120 subdomain known as the bridging sheet, which is required for locking Env into a stable interaction with CD4. Following immunization, Env-specific binding antibody titers and frequencies of Env-specific memory B cells were comparable between the groups. However, the quality of neutralizing antibody responses induced by the variants was distinctly different. Antibodies against the coreceptor binding site were elicited by WT trimers but not the CD4 binding-defective trimers, while antibodies against the CD4 binding site were elicited by the WT and the 423I/M, 425N/K, and 431G/E trimers but not the 368D/R trimers. Furthermore, the CD4 binding-defective trimer variants stimulated less potent neutralizing antibody activity against neutralization-sensitive viruses than WT trimers. Overall, our studies do not reveal any potential negative effects imparted by the in vivo interaction between WT Env and primate CD4 on the generation of functional T cells and antibodies in response to soluble Env vaccination.
The HIV-1 Envs mediate the entry of the virus into target cells and are the only virally encoded proteins exposed on the surface of the virus. HIV-1 Env is the sole target for neutralizing antibodies (Abs) and therefore is an important component of a vaccine designed to elicit protective antibody responses (4, 20). The viral spike is a trimer comprised of three heterodimers of the exterior envelope glycoprotein, gp120, noncovalently attached to the transmembrane protein, gp41. The gp120 subunit binds the primary receptor, CD4 (7), to form or to expose the gp120 coreceptor binding elements, which interact with the viral coreceptor, primarily CCR5 (1, 9, 12, 45). The highly conserved coreceptor binding site (CoRbs) overlaps the gp120 bridging sheet and also contains both proximal and distal elements of V3 (18, 32, 43, 45).
In attempts to mimic the native trimeric structure of Env present on the virus, various forms of soluble Env trimers were designed (reviewed in reference 14). One design consists of cleavage-defective trimers derived from the primary R5 isolate YU2 that possess a heterologous trimerization motif derived from T4 bacteriophage fibritin (F; YU2 gp140-F) (3, 21, 34, 40, 50, 51). We recently demonstrated that the immunization of monkeys, but not rabbits, with gp140-F trimers resulted in the generation of Abs directed against the CoRbs of gp120 capable of cross-neutralizing HIV-2 (15). CoRbs-directed Abs (also referred to as CD4-induced, or CD4i, Abs) also were elicited in rabbits transgenic for human CD4 (15). Taken together, these data strongly suggest that Env interacts with high-affinity primate CD4 in vivo, resulting in the formation, or exposure, of a highly immunogenic gp120 determinant that overlaps the CoRbs. Early in infection, the frequency of HIV-1-infected individuals with significant antibody responses against the CoRbs is high (8, 33), and CoRbs-directed antibody responses are elicited abundantly in humans inoculated with Env-based immunogens (15). Collectively, these data suggest that the recognition of the HIV-1 CoRbs by naïve B cells is greatly increased when Env is presented in complex with high-affinity primate CD4, leading to a productive Ab response against this epitope (41). With rare exceptions, the majority of CoRbs-directed monoclonal antibodies (MAbs) do not neutralize HIV-1 primary viruses in vitro, bringing into question the utility of this region as a relevant neutralization target (23, 31, 47, 49). Strategies aimed to diminish vaccine-elicited B-cell responses to the CoRbs, and shift responses toward more accessible neutralization targets, represent one approach to improve the design of Env-based vaccine candidates. The selective manipulation of Env immunogens to decrease their CD4 binding capacity may reduce the elicitation of CoRbs-directed Abs and circumvent potential occlusion effects of the conserved CD4 binding site caused by CD4 itself.
In addition to the potential effects of in vivo Env-CD4 interactions on the Ab repertoire elicited by Env-based immunogens, interactions between Env and CD4 also may have consequences on CD4+ T-cell responses. CD4 is an important costimulatory molecule expressed on several subsets of T cells and antigen-presenting cells, and interactions with Env were shown to alter the function of CD4-expressing T cells in a number of in vitro systems (13, 37, 44). The elimination of the Env-CD4 interaction in the context of vaccination may be beneficial to improve the elicitation of helper T-cell responses and effective neutralizing Ab responses. In vivo evaluation in subjects possessing high-affinity CD4 (i.e., rhesus macaques or humans) of CD4 binding-competent and CD4 binding-deficient Env immunogens so far have not been described.
To address these questions, we designed Env trimer variants rendered CD4 binding defective through two distinct mechanisms. In the first variant, the interaction between CD4 and HIV-1 Env was directly disrupted by the introduction of a mutation (368D/R) in the CD4 binding loop of the gp120 outer domain (29). This alteration abolishes the initial binding of CD4 and most CD4 binding site (CD4bs)-directed MAbs (42) to variant forms of gp120 and would be expected to do the same in the soluble stable timer context. The aim of the second variant was to decrease the CD4 binding affinity while preserving the antigenicity of the CD4bs (48). This variant was generated in the soluble gp140-F trimers by the introduction of three point mutations, 423I/M, 425N/K, and 431G/E, in the β20 strand region of gp120. These mutations were suggested to favor a helix rather than the β20/21 antiparallel strands visible in the gp120 structure (23, 31, 47, 49). In the monomeric context, mutations in the β20 strand region of gp120 abolish binding by CoRbs-directed Abs, presumably because the bridging sheet cannot form (48). The introduction of the 423I/M, 425N/K, and 431G/E mutations in the trimer context therefore should disrupt the normally high-affinity gp120-CD4 interaction, while recognition by CD4bs Abs would not be affected. Indeed, a recent study provides a mechanistic basis for the impact of these mutations on CD4 binding (52). This study revealed that CD4 interacts with gp120 by a two-step binding mechanism in which the first step involves a direct, but low-affinity, CD4 interaction with the gp120 outer domain, while the second step requires a conformational change in gp120 to fully stabilize the high-affinity gp120-CD4 interaction.
Here, we exploit this two-step model to generate novel CD4 binding-defective soluble trimers that, unlike the 368D/R trimers, possess a CD4bs surface that retains recognition by well-described CD4bs Abs. By immunizing rhesus macaques with the wild-type (WT) and CD4 binding-defective trimer variants, we demonstrate that similar levels of Env-specific Ab and T-cell responses were elicited in the three groups, suggesting that in vivo interactions between CD4 binding-competent (WT) Env and CD4 do not measurably affect T-cell responses against Env in this immunization regimen. However, the quality of the Ab response was markedly different between the groups. As hypothesized, CoRbs-directed Abs were elicited only in animals inoculated with WT trimers and not in those inoculated with 368D/R or 423I/M, 425N/K, and 431G/E trimers (hereafter referred to as 368 and 423/425/431 trimers, respectively). Importantly, we show that the 423/425/431 trimers retain the capacity to elicit binding and neutralizing CD4bs-directed Abs. In conclusion, the results generated in this study suggest that CD4 engagement by the WT soluble Env trimers did not impair the overall magnitude of the elicited Env-specific antibody or T-cell responses. Furthermore, our data provide new insights into the characteristics of Env that impact immunogenicity. The data also provide a potential path forward for the design of Env immunogens that have the capacity to elicit neutralizing Abs against the conserved gp120 CD4 binding surface while eliminating both the elicitation of nonneutralizing CoRbs-directed Abs and the potential occlusion of the CD4 binding surface of gp120 by the engagement of the primary virus receptor, CD4.
MATERIALS AND METHODS
Construction of Env immunogens.
The construction of the plasmid expressing cleavage-defective YU2-derived gp140 trimers possessing a heterologous foldon (F) trimerization motif [YU2gp140(-/FT)], herein referred to as WT gp140-F or WT trimers, was described previously (51). The two variant trimer glycoproteins were generated by the site-directed mutagenesis (Stratagene) of the WT gp140-F sequence. In brief, the 368 trimer was generated by modifying the codon for amino acid (aa) 368 from aspartic acid to arginine (368D/R), while the 323/325/431 variant trimer was generated by altering codons 423, 425, and 431 from isoleucine to methionine (423I/M), asparagine to lysine (425N/K), and glycine to glutamic acid (431G/E), respectively.
Expression and purification of Env immunogens.
All proteins were produced by the transient transfection of Freestyle 293F suspension cells (Invitrogen) as previously described (15). In brief, cells were transfected at a density of 1.1 × 106/ml in Gibco Freestyle293 expression medium using 293Fectin according to the manufacturer's instructions (Invitrogen). Supernatants were collected 4 days after transfection. Following collection, all supernatants were centrifuged at 3,500 × g to remove cells or cell debris, filtered through a 0.22-μm filter, and supplemented with Complete EDTA-free protease inhibitor cocktail (Roche) and penicillin-streptomycin (Invitrogen) and stored at 4°C until further purification. WT, 368, and 423/425/431 trimers used for immunizations were purified by a three-step process. First, the proteins were captured via glycans by lentil-lectin affinity chromatography (GE Healthcare). After extensive washes with PBS supplemented with 0.5 M NaCl, the proteins were eluted with 1 M methyl-α-d-mannopyranoside and captured in the second step via the His tag by nickel chelation chromatography (GE Healthcare). After being washed with 40 mM imidazole (IM) and 0.5 M NaCl in PBS, proteins were eluted with 300 mM IM in PBS. Trimers then were separated from lower-molecular-weight forms by gel filtration chromatography using a Superdex200 26/60 prep-grade column by the ÄKTA fast protein liquid chromatography system (GE Healthcare). Env probes used in the B-cell enzyme-linked immunospot (ELISpot) assay were purified by lentil-lectin and nickel chelation chromatography but not subjected to subsequent gel filtration chromatography.
Biochemical analysis of Env immunogens.
The capacity of respective trimer variants to bind CD4 was assessed as previously described (15). In brief, 0.25 μg/ml sCD4 (Progenics) was coincubated with various concentrations of the respective trimer variant. Free sCD4, not bound to trimers, subsequently was captured on a plate coated with the Env-competitive, anti-CD4 Ab RPA-T4 (ebiosciences). After extensive washes with PBS-0.05% Tween 20, bound CD4 was detected with the noncompetetive, biotinylated, anti-CD4 Ab OKT-4 (eBiosciences), followed by streptavidin-conjugated horseradish peroxidase (HRP) and the addition of a colorimetric peroxide enzyme immunoassay substrate (3,3′,5,5′-tetramethylbenzidine [TMB]; Bio-Rad). The induced colorimetric change was measured at 450 nm after stopping the reaction with 1 M H2SO4. The antigenic surface of respective trimers was assessed by coating wells with 200 ng of either trimer variant in 100 μl PBS. After blocking, MAbs were added at concentrations ranging from 20 to 0.26 ng/ml. MAb binding to each trimer was probed by an HRP-conjugated anti-human immunoglobulin G (IgG) (Jackson ImmunoResearch) and developed by the addition of TMB+ substrate chromogen (Dako). The reaction was stopped with 1 M H2SO4, and the colorimetric change was measured at 450 nm. The binding of the MAb 447-52D was used to control for the amount of protein coated on the plate. 447-52D binds with high affinity to the V3 region of gp120, and neither the 368 nor the 423/425/431 mutations are expected to influence the binding of this MAb. MAbs b12, F105, b6, and F91 target the CD4bs of gp120 and have overlapping binding footprints, but only b12 is broadly neutralizing. MAbs 17b, 48D, E51, and 2.1C target the CoRbs region.
SPR kinetic analysis.
All kinetic reactions were performed at room temperature (RT) on a Biacore3000 surface plasmon resonance (SPR) spectrometer. An indirect capture method was used to determine the kinetic constants of the interaction between the different trimers and CD4. To prepare the reference and test cell surfaces, anti-his MAb (25 μg/ml in 10 mM acetate acid, pH 5.0; R&D Systems) was immobilized on a CM5 chip by the amine-coupling method by following the manufacturer's protocol. His-tagged trimers at a concentration of 10 μg/ml in HBS-EP buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20) were captured on the chip and analyte. Four-domain sCD4 serially diluted in HBS-EP buffer at concentrations ranging from 10 to 2.56 μM was flowed over the chip. Association was allowed for 6 min at 30 μl/min. Dissociation was determined by washing off bound analyte during the next 6 min. In each reaction cycle the chip surface was regenerated with one injection (30 s) of 10 mM glycine, pH 2.5, at 50 μl/min. When possible, the kinetic rate constants were obtained by fitting the curves to a 1:1 Langmuir binding model. The affinity constant for 423/425/431 was obtained through a steady-state analysis of the binding data.
Animals.
Fifteen female rhesus macaques of Chinese origin, 5 to 6 years old, were housed in the Astrid Fagraeus laboratory at the Swedish Institute for Infectious Disease Control. Housing and care procedures were in compliance with the provisions and general guidelines of the Swedish Animal Welfare Agency. All procedures were approved by the Local Ethical Committee on Animal Experiments. The animals were housed in pairs in 4-m3 cages and enriched to give them the chance to express their physiological and behavioral needs. They were habituated to the housing conditions for more than 6 weeks before the start of the experiment and were subjected to positive reinforcement training to reduce the stress associated with experimental procedures. All inoculations and blood samplings were performed under sedation with ketamine at 10 mg/kg (100 mg/ml Ketaminol; Intervet, Sweden). The macaques were weighed and examined for the swelling of lymph nodes and spleen at each immunization or sampling occasion. Before entering the study, all animals were confirmed negative for simian immunodeficiency virus (SIV), simian T-cell lymphotropic virus, and simian retrovirus type D. The 15 rhesus macaques (three groups of five animals each) were inoculated five times with the immunogens described above. Immunizations were performed at weeks 0, 4, 8, 12, and 18 by the intramuscular (i.m.) route of injection. All protein immunizations were administered in combination with 500 μg CpG ODN 2395 (Coley Pharmaceutical Group) and 75 μg Abisco-100 (Isconova AB) as the adjuvant. Protein doses were 200 μg per animal for the first inoculation and 100 μg for the subsequent injections. The immunogens were given in a total volume of 1 ml, divided equally between the left and right hind leg. Blood samples were taken before and 2 weeks after each immunization.
Analysis of Env-binding Abs in plasma.
HIV-1 Env-specific IgG was measured by ELISA as previously described (28). Briefly, insect cell-produced YU2 gp120 protein was coated onto Nunc Maxisorp microtiter plates at 1 μg/ml in 50 mM carbonate buffer, pH 9.6, overnight (ON) at 4°C. After being blocked in PBS containing 2% milk, serum samples were added and incubated for 2 h at RT. The gp120-specific IgG was detected by adding secondary biotinylated anti-human IgG Ab (Jackson ImmunoResearch) and was streptavidin conjugated to horseradish peroxidase (Mabtech) followed by O-phenylenediamine (OPD; Sigma, Schnelldorf, Germany). Between each incubation step, the plates were washed six times with PBS-0.05% Tween 20. Reactions were terminated by adding 2.5 M H2SO4, and the optical density (OD) was read at 492 nm. The half-maximum binding titer (OD50) for each sample was calculated by interpolation from mean OD50 values calculated from controls using the formula [(ODmax − ODmin)/2) + ODmin].
B-cell ELISpot assay.
To determine the frequency of antigen-specific memory B cells, total peripheral blood mononuclear cells (PBMCs) were cultured at 2 × 106 cells/ml and stimulated with CpG ODN 10103 (5 μg/ml; Coley), Pokeweed mitogen (PWM) (10 μg/ml; Sigma), and Staphylococcus aureus Cowan (SAC) (1:10,000; Sigma) for 3 days in 48-well plates. To detect Ab-secreting cells, MAIPSWU10 96-well plates (Millipore) were coated with 10 μg/ml anti-human IgG (Fcγ) (Jackson ImmunoResearch), and PBMCs were transferred to the plates in dilution series and incubated overnight at 37°C in 5% CO2. The plates then were washed with PBS containing 0.05% Tween and incubated with biotinylated gp140-F, gp120-F, gp120ΔV123-F, or β-galactosidase (11), followed by washing and incubation with streptavidin-alkaline phosphatase (ALP; Mabtech). The reactions were developed using 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium substrate (Sigma-Aldrich) and stopped by a wash in water. Spots corresponding to Ab-secreting cells were counted using an ImmunoSpot analyzer (Cellular Technology Ltd.). The results were converted to Ab-secreting cells per million cultured PBMCs (ASC/106 PBMC). Percentages of gp41 and variable region 1-, 2-, and 3-specific memory B cells were calculated using the formulas [(gp140−gp120)/gp140] × 100 and [(gp120-gp120ΔV123-F)/gp140] × 100, respectively.
PBMC isolation and intracellular cytokine staining.
PBMCs were separated from EDTA blood by Ficoll-Paque Plus (GE Healthcare), frozen in fetal calf serum (FCS) in 10% dimethylsulfoxide (DMSO), and stored at −150°C. Frozen PBMCs were thawed and rested ON in medium at 37°C with 5% CO2. Cell viability was consistently >90%. Cells were added at 1 × 106 cells/well in duplicate in medium alone (RPMI 1640 medium containing 10% FCS, 2 mM l-glutamine, 100 U/ml penicillin, 100 mM streptomycin, 2% HEPES [all from Invitrogen Life Technologies]) or in medium containing 1 μg/ml staphylococcal enterotoxin B (SEB) or 2.5 μg/ml Env peptides. The Env peptide pool consisted of 15mer peptides overlapping by 10 aa and spanning the full-length gp140-F sequence (New England Peptide LLC). All cultures contained anti-CD28 and anti-CD49d at 1 μg/ml with brefeldin A (10 μg/ml) and GolgiStop (0.7 μg/ml) (Becton Dickinson). Cells were incubated for 6 h in 5% CO2 at 37°C, washed, and surface stained with CD8-allophycocyanin (CD8-APC) and CD4-peridinin chlorophyll protein (PerCP)-Cy5.5. Cells then were permeabilized using the cytofix/cytoperm kit (Becton Dickinson) and stained with CD3-Cy7-APC, interleukin-2-phycoerythrin (IL-2-PE), gamma interferon-fluorescein isothiocyanate (IFN-γ-FITC), and tumor necrosis factor alpha (TNF-α)-Cy7-PE. All Abs were from BD-Pharmingen and were previously titrated for the optimal staining of rhesus macaque PBMCs. Cell viability was assessed following staining with the Live/Dead Fixable Violet Dead Cell Stain kit (ViViD) (Molecular Probes). Samples were collected on a FACS CANTO II (BD Immunocytometry Systems) and analyzed using FlowJo software (Tree Star).
Virus neutralization assays.
Plasma from immunized animals was tested for virus neutralization against a panel of diverse HIV-1 isolates. Neutralization assays were performed using a single round of infection with the HIV-1 Env pseudovirus assay and TZM-bl target cells as previously described (24, 39). Env pseudoviruses were prepared by cotransfecting 293T cells with an Env expression plasmid containing a full-length gp160 env gene and an env-deficient HIV-1 backbone vector (pSG3ΔEnv). To determine the dilution that resulted in a 50% reduction in relative luminescence units (RLU), plasma was serially diluted and neutralization dose-response curves were fit by nonlinear regression using a four-parameter hill slope equation programmed into JMP statistical software (JMP 5.1; SAS Institute, Inc.). The results are reported as the serum neutralization ID50, which is the reciprocal of the serum dilution producing 50% virus neutralization. In some cases, serum IgG was fractionated, and neutralization results are reported at the 50% inhibitory concentration of IgG (IC50). Diverse HIV-1 isolates, including viruses from clades A, B, and C, were used in the neutralization assays. Clade B viruses included a panel of Env pseudoviruses that were characterized recently and recommended for use in assessing neutralization by HIV-1 immune sera (24). Several investigators also provided replication-competent viruses or functional Env plasmids for pseudoviruses. Dana Gabuzda (Dana Farber Cancer Institute) provided the Env plasmids for YU2 and MuLV. Env plasmids for SF162 and JRFL were provided by Leonidas Stamatatos (Seattle Biomedical Research Institute) and James Binley (Torrey Pines Institute), respectively. The clade A DJ263.8 sequence was cloned from the original PBMC-derived virus provided by Francine McCutchan and Vicky Polonis (U.S. Military HIV Research Program), and the clade C MW965 Env plasmid was obtained from the AIDS Research and Reagent Repository. The isolation of the Env BaL.01 plasmids was described recently by us (39), and the SS1196.1 Env was previously described (24).
Ab adsorption studies.
The method for Ab adsorption and elution has been described previously (25, 26). Each protein was covalently coupled to 1.1 μm tosyl-activated magnetic beads (MyOne Tosylactivated Dynalbeads; Invitrogen) according to the manufacturer's instructions. Beads were used at a concentration of 40 mg/ml. Generally, 40 mg beads was incubated with 2 mg of protein. The antigenic integrity of each protein after bead coupling was verified by flow cytometry using the known MAbs b12, 2G12, 447, and 17b and polyclonal HIVIG. For adsorptions, the respective IgG fraction from plasma was isolated using a Pierce Affinity Pak Protein G column (Pierce). The IgG fractions, in Dulbecco's modified essential medium (DMEM)-10% fetal bovine serum, were incubated with 400 to 500 μl of beads at RT for 30 min. After IgG adsorption, beads were removed with a magnet followed by centrifugation and were stored in PBS-0.2% bovine serum albumin (BSA)-0.02% sodium azide buffer at 4°C. The concentrations of the remaining fractions of IgG were measured using a commercial radial immunodiffusion (RID) kit that quantifies human IgG (Binding Site Corp.), and the IgG was used for ELISA and neutralization assays. Influenza hemagglutinin (HA)-coated beads and blank beads were used as controls.
Statistical analyses.
In all group comparisons, statistical significances were determined by one- or two-way analysis of variance of log-transformed data using GraphPad Prism software version 5 and were considered significant at P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***).
RESULTS
Characterization of WT and CD4 binding-defective soluble Env trimers.
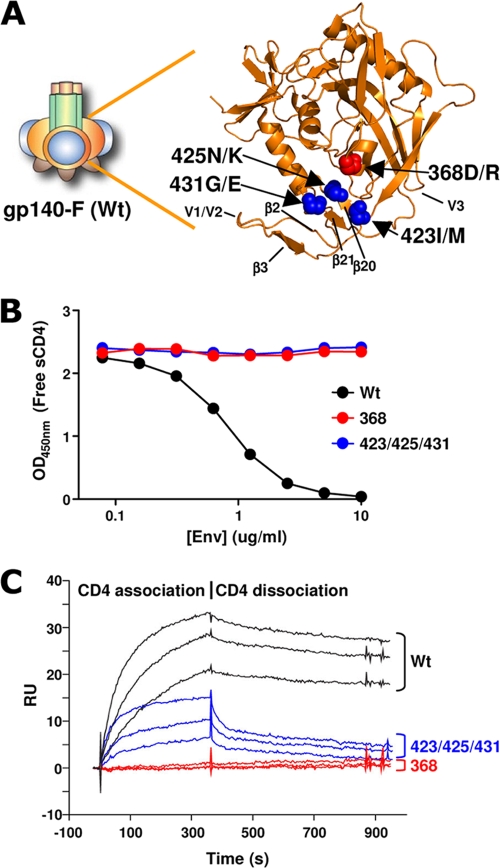
Prior to the inoculation of WT and variant trimers into rhesus macaques, we sought to characterize the CD4 binding profiles and antigenic properties of these trimeric immunogens by in vitro binding assays. The first variant, the 368 trimers, harbored a D/R substitution of aa 368 in gp120. This mutation was expected to impair its ability to bind CD4 but not its capacity to bind CoRbs-directed Abs (29). The second variant, the 423/425/431 trimers, harbored three substitutions in the β20 strand and intervening sequences just upstream of β21 to impart a helical propensity to this region. These two strands radiate from the gp120 outer domain and comprise half of the bridging sheet. These mutations were predicted to disrupt binding by CoRbs Abs but not affect binding by CD4bs-directed Abs. The locations of the 368D/R (red) and the 423I/M, 425N/K, and 431G/E (blue) substitutions are shown on the background of the WT YU2 gp120 core carbon trace in Fig. 1A. The locations of the mutations in relation to the CD4 binding footprint of the YU2 gp120 core protein are shown in Fig. S1A in the supplemental material.
FIG. 1.
Design and CD4 binding properties of WT and CD4 binding-defective HIV-1 Env trimers. (A) Schematic representation of gp140-F. gp140-F consists of a trimer of gp120 (gp120 core, orange; variable regions 1 and 2, blue; variable region 3, dark brown) covalently linked to gp41 (green) and a C-terminal, heterologous, trimerization motif, foldon (light brown). CD4 binding-defective trimers were designed and generated from WT gp140-F on the YU2 background, as depicted in the atomic resolution image of the YU2 gp120 core protein (Protein Data Bank no. 1G9N [22]). The carbon trace of the core protein is colored orange, and the locations of the 368D/R mutation (red) and the 423I/M, 425N/K, 431G/E mutations (blue) are shown as spheres. (B) WT, 368, and 423/425/431 trimer variants (colored black, red, and blue, respectively) were evaluated for their capacity to bind sCD4 in a competition ELISA format. sCD4 (0.25 μg/ml) was coincubated with the indicated concentrations of the respective trimer variant. Remaining noncomplexed sCD4 was detected by capture with an anti-CD4 antibody (competing with Env for sCD4 binding). (C) Binding curves of the CD4 interaction with the respective trimer variant by surface plasmon resonance spectrometry. The time frame for association and dissociation is shown.
To confirm the CD4 binding properties of the purified WT, 368, and 423/425/431 trimers, we measured their binding to soluble CD4 (sCD4) in a competition ELISA. The assay is based on RPA-T4, an anti-CD4 capture Ab that competes with Env for sCD4 binding. After incubation with increasing concentrations of soluble Env trimers, CD4 binding is detected by measuring the amount of captured sCD4 using a noncompeting anti-CD4 Ab (15). WT trimers bound with high affinity to sCD4 and competed with the RPA-T4 capturing of sCD4 in a dose-dependent manner. In contrast, there was no detectable sCD4 competition by either the 368 or the 423/425/431 variants (Fig. 1B). To assess if the two sets of mutations (368 and 423/425/431) disrupted binding by Env trimers to sCD4 by distinct mechanisms, we studied binding kinetics by surface plasmon resonance (Biacore). The kinetic analysis confirmed that sCD4 binds WT Env with a relatively fast on-rate (1.2 × 105 M−1 s−1) and slow off-rate (3.1 × 10−4 s−1), resulting in high-nanomolar affinity (2.7 nM) (Fig. 1C). The two-step mechanism of CD4 binding to gp120 (52) begins with the interaction of CD4 with the gp120 conformationally stable outer domain, followed by a subsequent rearrangement of the inner domain and the formation of the bridging sheet. This is consistent with the slow off-rate observed for the WT trimers in the Biacore analysis (Fig. 1C). In contrast, the two trimer variants, 368 and 423/425/431, demonstrated altered binding kinetics to CD4. As expected, the nonconservative D/R mutation in the center of the gp120-CD4 binding region abolished CD4 binding by reducing the association (on-rate) of sCD4 to a level undetectable by SPR (Fig. 1C). In contrast, the 423/425/431 variant displayed an on-rate that was roughly equivalent to that of the WT trimer (1.0 × 105 M−1 s−1), but this initial interaction was followed by an extremely rapid off-rate, suggesting that the 423/425/431 variant is metastable and rapidly dissociates. Due to the rapid off-rate, an affinity constant for sCD4 binding to the 423/425/431 variant could not be accurately determined using the 1:1 Langmuir binding model. Accordingly, we estimated the overall affinity by steady-state analysis and found it to be 138 nM, approximately 50-fold lower than that for the WT (complete SPR data and steady-state analysis are shown in Fig. S1B in the supplemental material). These data are consistent with a first binding step by CD4 that does not require a conformational change in gp120. This is followed by a second step that requires a conformational change in gp120, which the 423/425/431 variant is unable to undergo.
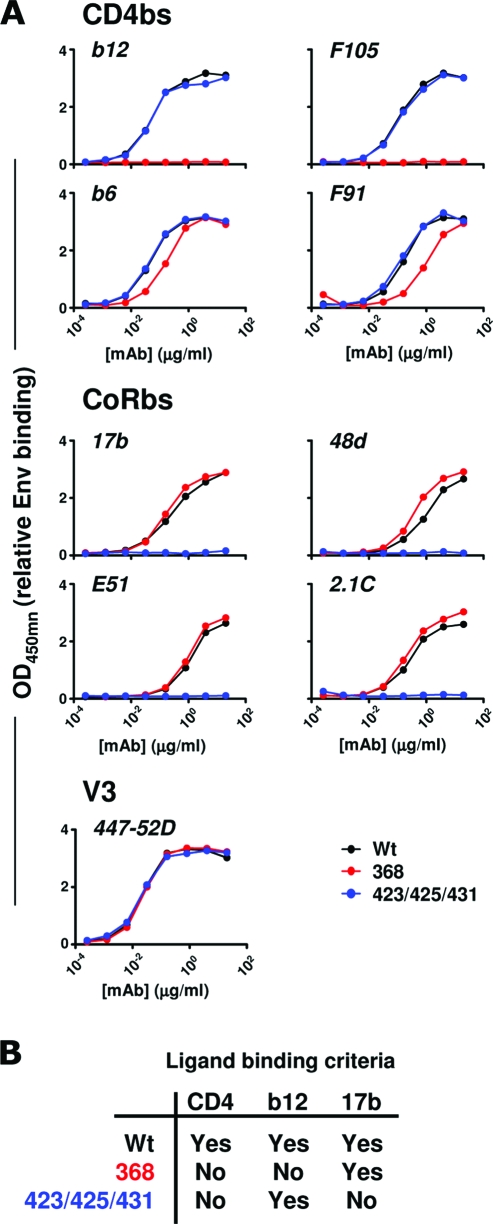
The three trimer variants also displayed distinct Ab binding profiles that were consistent with analyses of these mutations in the context of gp120 monomers (42, 48). The 368D/R mutation disrupted recognition by several CD4bs-directed MAbs, including b12 and F105, while recognition by F91 and b6 by the 368 trimers was slightly reduced but not abolished (Fig. 2A, upper). In contrast, the 423/425/431 trimers retained binding by all CD4bs MAbs with binding titers similar to those of the WT trimers. However, the recognition of the 423/425/431 trimers by CoRbs-directed Abs 17b, 48d, E51, and 2.1c was completely abolished, which is consistent with the inability of this variant to undergo the necessary conformational changes to form the bridging sheet (48). WT and 368 trimers exhibited comparable binding to the CoRbs-directed Abs (Fig. 2A, middle). Thus, both the 368 and 423/425/431 trimers lacked the capacity to stably bind human CD4, while they differed markedly in their capacity to bind Abs directed against the primary receptor and coreceptor binding regions. As a control for the concentration of protein coated in the wells, we used a V3-directed MAb, 447-52D. Similar binding titers were measured for all three trimer variants using this MAb (Fig. 2A, lower). In summary, the two CD4 binding-defective trimer variants were antigenically distinct and fulfilled the ligand binding criteria for this study; neither variant bound CD4 stably, the 368 variant bound 17b but not b12, and the 423/425/431 variant bound the broadly neutralizing MAb b12 (5) but not 17b (Fig. 2B).
FIG. 2.
Antigenic analysis of WT and CD4 binding-defective trimer variants. (A) Antigenic analysis of trimer variants. A panel of CD4bs-directed (b12, b6, F105, and F91) and CoRbs-directed (17b, E51, 48d, and 2.1C) MAbs and the V3-directed MAb 447-52D were evaluated for their ability to bind WT (black), 368 (red), and 423/425/431 (blue) trimers. The antibody concentrations used to generate the binding curves are shown. (B) Trimer variant selection criteria for the study.
Binding Ab and memory B-cell responses in macaques inoculated with WT and CD4 binding-defective Env trimers.
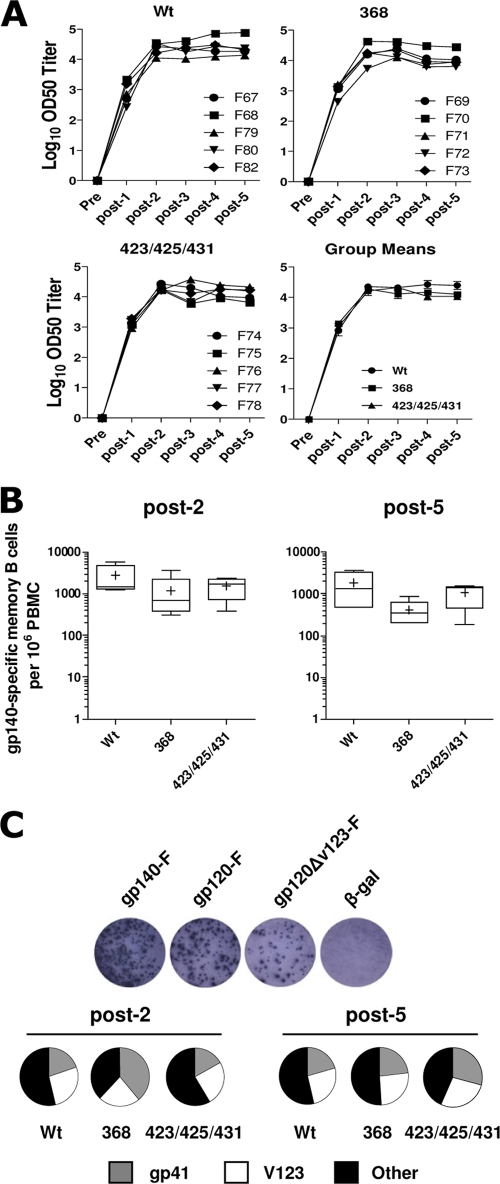
The three variant trimers, WT, 368, and 423/425/431, were inoculated into rhesus macaques divided into three groups of five animals each. The immunogens were administered in an adjuvant consisting of Abisco-100 and CpG ODN 2395, and inoculations were performed five times at monthly intervals. Since CD4 is an important accessory molecule expressed on several subsets of immune cells, we first asked if the overall humoral immune response to the CD4 binding-competent (WT) and binding-defective Envs (368 and 423/425/431) was different. Plasma samples from individual animals were collected before and 2 weeks following each immunization and tested for binding to YU2 gp120 protein by ELISA (Fig. 3A). Data are presented as half-maximum binding titers as described in Materials and Methods. All animals in the three groups mounted primary Ab responses 2 weeks following a single Env immunization. The titers were boosted by the second immunization, resulting in peak responses in all three groups, and these responses were maintained following the subsequent immunizations. Mean ELISA titers are shown in the lower right panel of Fig. 3A, and titration curves of all individual animals are presented in the supplementary material (see Fig. S2 in the supplemental material). These data demonstrate that immunization with WT or CD4 binding-defective Env trimers elicited comparable levels of Env-specific Abs.
FIG. 3.
Analysis of the Env-specific antibody and memory B-cell responses from macaques inoculated with WT or CD4 binding-defective trimers. (A) Plasma from individual animals was collected before (pre) and 2 weeks following each immunization (post-1 to post-5), and the level of HIV-1 gp120-specific serum IgG was measured by ELISA. OD50 titers from individual animals are shown in the upper panel, and group means are shown in the lower panel. (B) Frequency of gp140-specific memory B cells within PBMCs following two and five immunization with HIV-1 envelope glycoproteins. (C) The frequencies of memory B cells specific for defined structural determinants of Env were determined using gp140-F-, gp120-F-, and gp120-F-ΔV123-biotinylated trimeric Envs as differential probes in the ELISpot assay. Results from representative wells where individual spots correspond to Ab-secreting cells (ASC) are shown in the upper panel. Values were calculated by subtractive analysis as described in Materials and Methods and are presented as pie charts. The results are shown as mean percentages of memory B cells specific for gp41 (gray), V1 to V3 (white), and other determinants (black) within total gp140 specific memory B cells. Data are shown as means from five individual animals per group.
Env-specific memory B-cell responses in animals inoculated with WT, 368, and 423/425/431 trimers also were examined. For this analysis, we employed a recently described highly sensitive, Env-specific B-cell ELISpot assay (11) to evaluate frequencies of Env-specific memory B cells in peripheral blood mononuclear cells following the second and fifth immunization. Total PBMCs collected 2 weeks after each inoculation were stimulated in vitro to differentiate memory B cells into ASC, as shown in Fig. S3A in the supplemental material. The frequency of Env-specific B cells, presented as Env-specific memory B cells per 106 PBMCs, was comparable between the three groups (P > 0.05) (Fig. 3B). Furthermore, to determine the frequencies of memory B cells specific for defined structural determinants of Env, we employed three different biotinylated trimeric Env probes: full-length gp140-F, gp120-F (a probe lacking the gp41 determinants of gp140-F), and gp120-F-ΔV1/2/3 (a probe lacking both gp41 and variable regions 1 to 3). By enumerating ASC reactive with the different probes, the percent gp41- and V1- to 3-specific cells was determined by subtractive analysis as described previously (11). The results, shown as the percent region-specific responses of total gp140-F-specific ASC, suggest that there were no significant differences (P > 0.05) in the proportion of gp41- or V1- to V3-reactive cells between animals inoculated with WT, 368, or 423/425/431 trimers after the second or fifth immunization (Fig. 3C), while significant differences in the number of ASC detected with the three probes were detected within each group (see Fig. S3B in the supplemental material).
CD4 binding capacity does not affect trimer-elicited Env-specific T-cell responses.
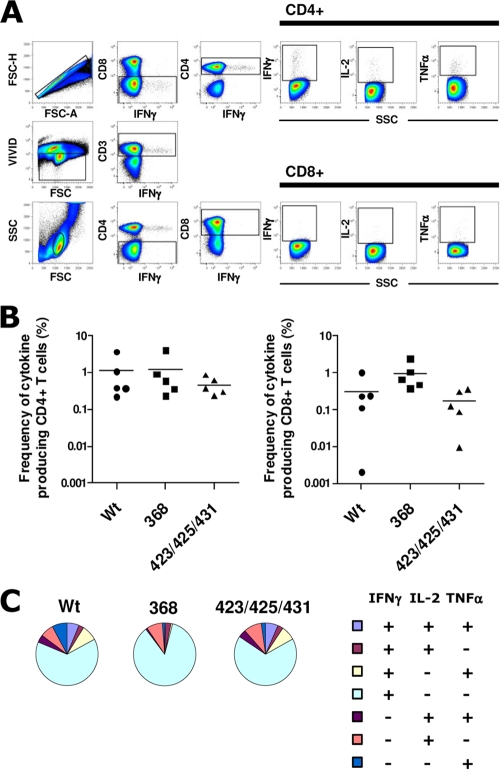
It was previously suggested that interactions between Env and T cells expressing high-affinity primate CD4 interfere with vaccine-induced immune responses (13, 38), but the impact of Env-CD4 interactions during immunization so far were not investigated in humans or monkeys. To investigate the impact of Env trimer-CD4 in vivo interactions on the elicitation of Env-specific T cells, we analyzed T-cell responses in the animals inoculated with WT, 368, or 423/425/431 trimers. PBMCs collected 2 weeks after the fifth immunization were analyzed by multiparameter flow cytometry to determine the magnitude and function of Env-specific CD4+ and CD8+ T cells. The gating strategy from one representative animal from the WT trimer-immunized group is shown in Fig. 4A. The frequencies of cytokine-producing (IFN-γ, IL-2, or TNF-α) CD4+ and CD8+ T cells were measured after the restimulation of total PBMCs with overlapping 15-mer peptides spanning the full-length YU2 gp140-F aa sequence. Total Env-specific CD4+ and CD8+ T-cell cytokine responses (combined frequencies of IL-2-, IFN-γ-, or TNF-α-producing cells) are presented for each individual animal (Fig. 4B). High frequencies of Env-specific CD4+ T-cell responses were observed in all animals in the three groups. Env-specific CD8+ T-cell responses also were detected, although at lower frequencies; these responses are consistent with the reported ability of Iscom-based adjuvants to induce cross-presentation (36). No significant differences (P > 0.05) in the magnitude of Env-specific CD4+ and CD8+ T cells were observed among the animals immunized with WT, 368, or 423/425/431 trimers.
FIG. 4.
Env-specific T-cell responses in macaques immunized with HIV-1 Env trimer variants. (A) Frequency of cytokine production (IFN-γ, IL-2, or TNF-α) among CD4+ and CD8+ T cells after the restimulation of total PBMCs with overlapping 15-mer peptides covering YU2 gp140. The gating strategy is shown for one animal (F79, from the WT trimers group) 2 weeks after five immunizations. (B) Frequencies of Env-specific T cells from individual samples taken 2 weeks after the fifth immunization were determined. Data from five individual animals per group are shown as the percent cytokine-producing cells after medium subtraction. (C) Total Env-specific CD4 T cells were subdivided into seven distinct populations producing any combination of IFN-γ, IL-2, or TNF-α. Data are presented as mean frequencies (n = 5 animals per group) of each subpopulation.
The quality of Env-specific T-cell responses was analyzed further for potential heterogeneity in the composition of cytokine responses based on the average data derived from five individual animals. In all three groups, Env-specific CD4+ T cells predominantly consisted of single cytokine-producing cells, mostly IFN-γ. CD4+ T cells producing one or two of the cytokines (IFN-γ, IL-2, or TNF-α) were represented equally in the animals in all three groups, with a slightly lower fraction of cells producing all three cytokines in the 368 trimer-inoculated animals (Fig. 4C). We conclude that comparable Env-specific T-helper 1 (Th1) and CD8+ T-cell responses were obtained in the three groups, suggesting that the in vivo interactions between Env and endogenous primate CD4 using the current vaccine regimen does not markedly influence vaccine-induced Env-specific T-cell responses.
Neutralizing Ab responses elicited by WT and CD4 binding-defective trimers is qualitatively different.
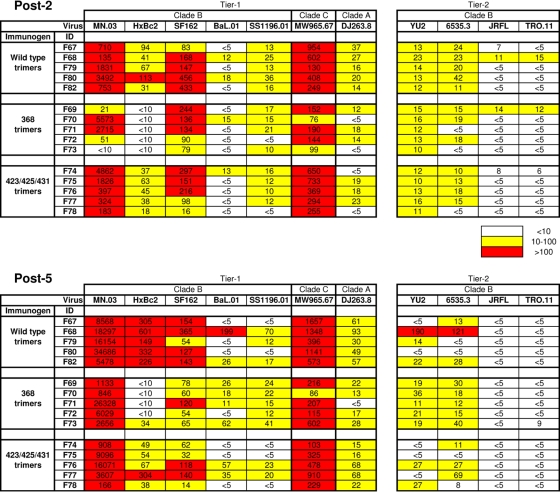
A central aim of this study was to directly compare the quality of NAb responses elicited by the WT and the two CD4bs-defective trimer variants in nonhuman primates. To examine NAb responses, we used a panel of recombinant viruses pseudotyped with Envs derived from the tier 1 isolates MN, HxBc2, SF162, BaL, and SS1196 (clade B), as well as MW965 (clade C) and DJ263 (clade A), and Envs derived from the tier 2 isolates YU2, 6535.3, JRFL, and TRO.11. The data are presented as 50% plasma inhibitory dilution (ID50) values. Potent ID50 neutralizing titers against MN were observed in animals in all three groups, with group mean ID50 titers of approximately 1,000 after two immunizations and roughly 10,000 after five immunizations (Fig. 5). Potent neutralizing activity also was observed in all animals against SF162 and the clade C virus MW965. In contrast, neutralization against HxBc2 was observed only in the WT and 423/425/431 trimer-immunized animals and not in animals immunized two times with the 368 trimers. After five inoculations, only one animal (F73) from this group displayed weak HxBc2 neutralization, while plasma from all WT and 423/425/431 trimer-immunized animals neutralized this virus. Sporadic neutralizing activity was observed against SS1196 and BaL and the clade A virus DJ263.8, as well as against the tier 2 viruses YU2 and 6535.3, and weak or no neutralization was observed against JRFL and TRO.11. Increases in ID50 neutralization titers after five compared to two immunizations were observed against MN, HxBc2, and Mw965, and, interestingly, the increase against the latter two viruses was observed only in WT trimer-inoculated animals (Fig. 5). These data suggested that there were qualitative differences in the neutralizing Ab responses elicited by the three trimer variants.
FIG. 5.
Neutralization against a panel of Env pseudoviruses. Neutralizing activity in plasma collected 2 weeks after the second and fifth immunizations was measured using a panel of HIV-1 isolates. Data from individual animals are shown as the reciprocal dilution giving 50% neutralization (ID50 titer).
Elicitation of CoRbs-directed Abs by WT trimers but not by CD4 binding-defective trimers.
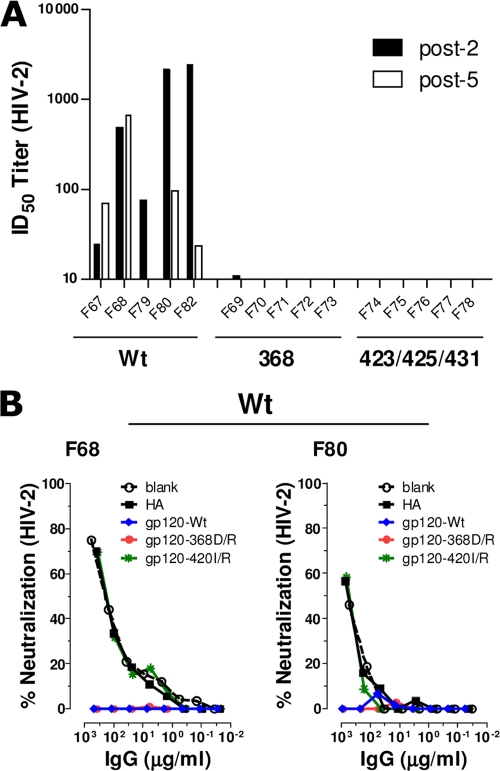
The differences in neutralizing Ab responses elicited by WT, 368, and 423/425/431 trimers prompted us to perform epitope-mapping studies of an attempt to define the Ab subspecificities that mediated neutralization against selected viruses. We first asked if the three trimer immunogens differed in their capacity to elicit Abs against the CoRbs. For this, we employed a neutralization assay that is based on the inhibition of an HIV-2 Env-pseudotyped virus that was engineered to be highly sensitive to CoRbs-directed Abs in the presence of subinhibitory concentrations of soluble CD4 (8). Data are presented as ID50 neutralizing titers after two and five inoculations. Neutralizing activity against HIV-2 was observed only in plasma from monkeys inoculated with WT trimers and not in animals inoculated with the 368 or 423/425/431 trimer (Fig. 6A). Thus, the two CD4 binding-defective immunogens did not elicit anti-CoRbs neutralizing Abs. Mean ID50 titers after two inoculations with WT trimers was approximately 100, and this response was not increased by subsequent immunizations.
FIG. 6.
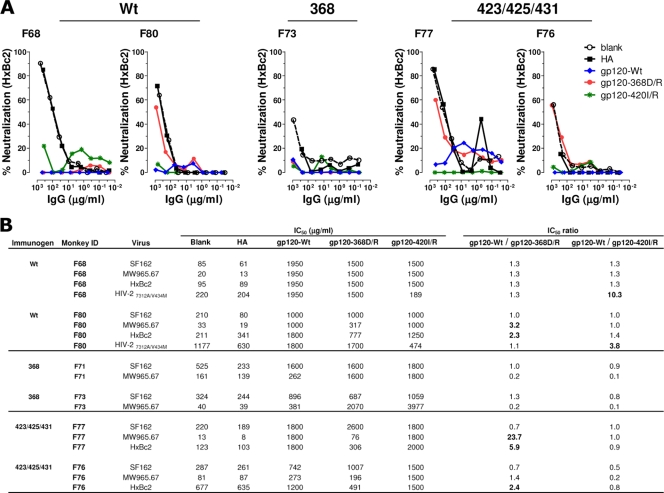
Neutralization mediated by CoRbs-directed Abs. (A) Plasma samples were collected 14 days after the second (post-2) and fifth (post-5) immunizations and tested for neutralization against the HIV-2 Env virus. Data from individual animals are presented as the reciprocal plasma dilution showing 50% neutralization (ID50 titer). (B) The neutralizing antibody specificity in plasma samples with detectable HIV-2 neutralization activity was mapped using differential solid-phase adsorption on beads coated with blank or influenza HA (control), YU2gp120-WT, YU2gp120-368D/R, and YU2gp120-420I/R proteins. Plasma IgG samples were adsorbed onto magnetic beads coated with the indicated proteins, and the residual antibodies were tested for neutralization against HIV-2. Shown are the neutralization curves of adsorbed plasmas from two animals from the WT group (F68 and F80). No HIV-2 neutralizing activity was observed in animals from the 368 or the 423/425/431 group (Fig. 6A).
As an independent confirmation that the neutralizing activity measured in the HIV-2 assay was mediated by Abs directed against the CoRbs, we selected two WT trimer-immunized animals with the highest HIV-2 neutralizing titers (F68 and F80) for analysis by selective adsorption as previously described (2, 10, 17, 25, 26, 35). In brief, the IgG fractions from the plasma samples were incubated with paramagnetic beads; either control beads (blank) or beads coated with influenza HA protein, gp120-WT, gp120-368D/R, or gp120-420I/R. The reactivities not adsorbed by the beads remain in the flowthrough; the gp120-368D/R-coated beads do not adsorb CD4bs-directed Abs, while the gp120-420I/R-coated beads do not adsorb Abs directed against the CoRbs. The flowthrough material from the F68 and F80 IgG fractions was analyzed for the neutralization of HIV-2 using the assay described above (Fig. 6B). In both analyzed samples, the neutralization activity was completely depleted with the gp120-WT beads and the gp120-368D/R beads but not with control beads or beads coated with gp120-420I/R. These results confirm that Abs directed against the HIV-1 CoRbs are elicited by WT trimers, and that this activity is responsible for the neutralizing activity against HIV-2.
Elicitation of binding and neutralizing CD4bs-directed Abs by WT and 423/425/431 trimers.
A striking result from the neutralization studies was the lack of neutralizing activity against HxBc2 in animals inoculated with the 368 trimers (Fig. 5). Since the 368D/R mutation disrupts binding by several CD4bs-directed MAbs (42), a distinct possibility was that WT and 423/425/431 trimers, but not the 368 trimers, elicited CD4bs-directed Abs that were responsible for a major fraction of this activity. We considered it less likely that this neutralization was mediated by CoRbs Abs, as these were not elicited by the 423/425/431 trimers (Fig. 6). To assess the elicitation of CD4bs-directed antibodies at the binding level, we used a b12 cross-competition assay. We used core gp120 as the protein on the binding surface to diminish off-target effects. This assay clearly showed that all animals inoculated with WT or 423/425/431 trimers elicited b12 cross-competitive Abs, which were detectable at several dilutions of the plasmas. In contrast, animals inoculated with 368D/R exhibited only b12 cross-competitive Abs at the lowest dilution of the plasmas, suggesting that the concentration of Abs capable of binding the WT CD4bs in these animals was considerably lower than that in the other two groups (see Fig. S5 in the supplemental material).
We next determined if CD4bs-directed Abs were responsible for HxBc2 neutralization in animals that exhibited the highest neutralization titers against this virus. We performed selective adsorptions on plasma collected after five inoculations using the same approach as that used for Fig. 6B. In addition to F68 and F80 from the WT trimer-inoculated group, we selected F76 and F77, two animals from the group inoculated with 423/425/431 trimers. We also selected F73, the only animal with detectable neutralizing activity against HxBc2 in the group of animals inoculated with the 368 trimers. The flowthrough fractions from these adsorptions were tested for the neutralization of HxBc2 (Fig. 7A) and selected additional viruses (Fig. 7B). The ratios of neutralization IC50s for the adsorbed fractions were used as an indicator for the content of site-specific neutralizing Abs. For example, successful adsorption by gp120-WT beads, but not the gp120-368D/R beads, indicates the presence of CD4bs-directed neutralizing Abs (a high gp120-WT/gp120-368D/R ratio). Likewise, a high gp120-WT/gp120-420I/R ratio indicates the presence of CoRbs Abs (Fig. 7B). For the two animals inoculated with WT trimers, these ratios demonstrate that CoRbs-directed Abs contributed to the neutralization of HIV-2 but not to the neutralization of HxBc2 (Fig. 7). In contrast, the gp120-368D/R-coated beads did not adsorb the activity in F80, F76, and F77, yielding gp120-Wt/gp120-368D/R IC50 ratios of 2.3, 5.9, and 2.4, respectively, suggesting that CD4bs-directed Abs are responsible for a major fraction of the neutralizing activity against HxBc2 in these animals. The activity responsible for HxBc2 neutralization in animals F68 and F73 was not identified by the current adsorptions.
FIG. 7.
Mapping of neutralizing Ab responses elicited by WT or CD4 binding-defective Env. The neutralizing antibody specificity in plasma IgG samples from WT (F68, F80)-, 368D/R (F71, F73)-, and 423/425/431 (F76, F77)-immunized animals were mapped using differential adsorption on beads coated with blank, influenza HA, gp120-WT, gp120-368D/R, or gp120-420I/R protein. (A) Neutralization curves of plasma IgG from two animals inoculated with WT trimers (F68 and F80), one animal inoculated with 368 trimers (F73), and two animals inoculated with 423/425/431 trimers (F76 and F77) previously adsorbed with the indicated protein-coated beads. The effect of adsorption was analyzed in the flowthrough fraction for neutralization against HxBc2. (B) Plasma IgG IC50s and IC50 ratios for neutralization against an extended panel of viruses, including SF162, MW965, HxBc2, and HIV-2, are summarized. Ratios equal to or greater than 2 are shown in boldface.
The selectively adsorbed IgG fractions from F68 and F80 (WT trimers), F71 and F73 (368 trimers), and F76 and F77 (423/425/431 trimers) also were tested for their potency to neutralize SF162 and MW965, against which high neutralization titers were observed for the unfractionated plasma samples. As summarized in Fig. 7B, CD4bs Abs accounted for a major fraction of MW965 neutralization in F80 and F77 (IC50 ratios of 3.2 and 23.7, respectively) but to a lesser extent in F76 (IC50 ratio of 1.4), one of the animals in which CD4bs Abs mediated HxBc2 neutralization. In contrast, neither CD4bs nor CoRbs Abs appeared to contribute to the neutralization of SF162. While not examined here, the activity responsible for neutralizing SF162 likely is V3 mediated, as shown in our previous studies of macaques inoculated with soluble trimers (28). These data demonstrate that the WT and 423/425/431 trimers, but not the 368 trimers, have the capacity to elicit CD4bs-directed Abs, and that some of the CD4bs-directed antibodies are capable of neutralizing relatively sensitive viruses, such as HxBc2 and MW965, as shown in the animals selected for the adsorption analysis. Thus, the 423/425/431 trimers display a different immunogenicity profile than the WT trimers, in that they do not elicit CoRbs-directed Abs but retain the capacity to elicit neutralizing CD4bs-directed Abs, as shown in selected animals.
DISCUSSION
In the current study, we directly compared WT trimers to two CD4 binding-defective trimer variants to determine the impact of the high-affinity in vivo interaction between Env and CD4 on vaccine-elicited immune responses. To our knowledge, this is the first comparative analysis of CD4 binding-competent and CD4 binding-defective Env immunogens analyzed in nonhuman primates, a highly relevant model for human vaccine testing. Furthermore, the novel design of the 423/425/431 trimer variant, which eliminated CD4 binding in an indirect manner by exploiting the two-step binding mechanism of the gp120-CD4 interaction, is the first example of such an Env immunogen and its analysis in nonhuman primates.
This study follows directly on our previous report in which we investigated the neutralizing Ab response elicited by soluble HIV-1 Env trimers in macaques, WT rabbits, and rabbits transgenic for human CD4 (15). In that analysis, we demonstrated that the elicitation of CoRbs-directed Abs requires the presence of high-affinity primate CD4, suggesting that some fraction of CD4-binding-competent Env immunogens interacts with primate CD4 in vivo, affecting the quality of the elicited B-cell response. The elicitation of CoRbs-directed Abs also was observed in humans immunized with purified monomeric gp120 in the Vaxgen phase III clinical trial (15), likely a result of gp120 interactions with human CD4. Here, we show that CoRbs-directed Abs were elicited in all animals immunized with WT trimers but not in the animals immunized with CD4-binding-defective trimer variants. These results definitively confirm that the CoRbs on gp120 is not efficiently recognized by the naïve B-cell repertoire unless it is presented in complex with host CD4. We also demonstrated that both the direct disruption of the initial interaction between the gp120 outer domain and CD4 and the disruption of the conformational change required for stabilizing the Env-CD4 interaction completely abolished the elicitation of CoRbs-directed Abs.
The role of different types of CoRbs-directed Abs in virus neutralization and protection against infection remains something of an enigma. The detection of CoRbs Abs prior to autologous virus neutralization in HIV-1-infected patients suggests that CoRbs-directed Abs generally are nonneutralizing (16). This is supported by data from the phase III Vaxgen clinical trial where no protection was observed (30) despite high titers of CoRbs-directed Abs (15). Furthermore, the access of two intact Ab molecules to the conserved CoRbs on the functional virus spike was shown to be sterically restricted (23), while Ab subdomains derived from these same two Abs can access this region and inhibit viral entry (6). These data argue against a role of CoRbs-directed Abs for virus neutralization. However, CoRbs-directed Abs recently were shown to contribute to the neutralization of some primary viruses in vitro, at least when present at high titers (10). It is possible that some yet-to-be-defined class of CoRbs-directed Abs exists that can contribute to virus neutralization during natural infection. Such specificities were suggested in two recent mapping studies (17, 26). Well-characterized Env immunogens, such as the 423/425/431 trimers described here, which selectively lack the capacity to elicit CoRbs-directed Abs but retain the ability to elicit other classes of Abs, will be important for future investigations aimed at determining the role of CoRbs-directed Abs in virus neutralization and in vivo protection.
In addition to the formation of the CoRbs epitope, conformational changes induced by CD4 ligation may affect the exposure of other Env epitopes, altering the Env-specific B-cell repertoire and neutralizing Ab responses. Recent electron tomographic analyses of HIV-1 suggested dramatic conformational changes in the quarternary structure of the liganded forms of the trimer compared to the native, unliganded form (27). Other reports show the stabilization and/or exposure of the V3 region on gp120 when it is in complex with CD4 (19, 41). Increased breadth of V3-mediated neutralization also was reported against a panel of HIV-1 reference strains in the presence of sCD4 using both vaccine-elicited sera and human V3-specific MAbs (46). An increase in the exposure of the V3 region on the soluble gp140-F trimers also may occur upon CD4 binding, as suggested by an increased proportion of V3-specific compared to total Env-specific Abs in rabbits transgenic for human CD4 compared to that of WT rabbits, as shown in our previous studies (15). However, whether CD4 binding alters the exposure of other potential neutralizing epitopes on Env besides V3 remains unknown.
To investigate the impact of in vivo CD4 binding on the Ab response elicited by the three trimer variants, we used several complementary approaches. Striking differences were observed at the level of antibody subspecificities, in particular the capacity of the WT trimers, but not the 368 and 423/425/431 trimers, to elicit CoRbs-directed Abs, and the capacity of WT and 423/425/431, but not the 368 trimers, to elicit either binding or weakly neutralizing CD4bs Abs. For HxBc2, it was possible to examine only one animal immunized with the 368 trimers in adsorption studies (F73, the only animal in this group that displayed weak HxBc2 neutralization). No evidence of CD4bs neutralizing Abs was detected in this animal. Although not formally shown, the absence of CD4bs-directed neutralizing Abs in all animals immunized with 368 trimers may explain the overall poor HxBc2 neutralization seen in this group. In contrast, both animals inoculated with 423/425/431 trimers generated CD4bs-directed Abs that contributed to the neutralization of both HxBc2 and MW965. Furthermore, all animals in the WT and 423/425/431 trimer-inoculated groups elicited CD4bs-directed binding Abs as detected by a b12 cross-competition assay. These results suggest that optimally designed CD4-binding-defective immunogens provide a means to minimize the potential occlusion of broadly neutralizing epitopes caused by in vivo ligation to high-affinity primate CD4. This may be an important consideration as Env immunogens progress from preclinical small-animal studies into human trials. No apparent advantage of the 423/425/431 trimers over the WT trimers in their capacity to stimulate neutralizing Abs against the CD4bs was detected in our studies, but further modifications to the 423/425/431 trimers may improve the performance of this immunogen.
Another observation was that the animals inoculated with WT trimers exhibited higher ID50 neutralizing titers against HxBc2 and MW965 after the fifth immunization than after the second immunization, while no increase in neutralizing titers was observed between the second and the fifth inoculation in the 423/425/431 trimer group. These data suggest that some undefined Ab specificity, or specificities, were selectively boosted in the animals inoculated with WT trimers. The boost effect suggests that some element(s) of the soluble trimers is selectively exposed upon CD4 binding, perhaps the V3 region. In that regard, we did not detect differences in the frequencies of memory B cells specific for the V123 regions elicited by WT and the CD4-binding-defective soluble trimers by the differential B-cell ELISpot assay. However, this analysis would not reveal qualitative differences, for example, potential differences in antibody affinity elicited by the immunogens. Since the boost effect was present only in the WT trimer-inoculated animals, it also is possible that this represents some class of CoRbs-directed Abs that is not detected by the HIV-2 cross-neutralization assay, since no boost effect was observed between the second and fifth inoculations in this assay (Fig. 6).
One interpretation of the neutralization data is that the WT trimers are superior to the 423/425/431 trimers, as they elicit higher neutralizing titers against a subset of the viruses tested. However, because these viruses are sensitive to neutralizing specificities that are not effective against many primary circulating isolates (i.e., V3-directed responses), these responses may not reflect those required to accomplish broad neutralization. Therefore, our interpretation of the data generated in this study is that the WT trimers are not markedly superior to the 423/425/431 trimers in terms of vaccine-relevant broadly protective responses.
A potential consequence of in vivo CD4 binding during immunization is that the ligation of Env to cell surface-expressed CD4 stimulates signaling events that negatively impact T-cell function. CD4 plays an important role as an accessory molecule to activate T-cell proliferation in response to cognate interactions between the T-cell receptor and major histocompatibility complex class II-restricted antigen presentation. Studies using full-length membrane-bound, but not soluble, HIV-1 Env trimers showed that CD4-dependent Env interactions suppressed CD4+ T-cell activation and proliferation in vitro (13), suggesting that the elimination of Env interaction with CD4 in the context of vaccination is beneficial to better elicit functional T-cell help and more potent neutralizing Ab responses. In the present study, we investigated the potential effects of CD4 ligation by a comprehensive analysis of Env-specific immune responses elicited by WT and CD4 binding-defective trimers. We show that the titers of circulating Env-specific binding Abs were similar between the three groups, as were the frequencies of Env-specific memory B cells and the magnitude and functionality of Env-specific CD4+ and CD8+ T cells. These results suggest that the disruption of the Env-CD4 interaction is not critical for the induction of potent immune responses against Env. Although not addressed here, our data also argue against a role for Env-CD4 interactions in promoting immune dysfunction during HIV-1 infection. However, due to the multitude of differences between natural infection and active Env protein vaccination, cautious extrapolation between the two systems is advised.
It remains to be determined if in vivo CD4 interactions limit the elicitation of antibodies directed against the CD4 binding site as a result of potential occlusion effects in an animal system both possessing and lacking primate CD4 (i.e., huCD4 transgenic small animals). In this regard, our data demonstrate that modifications to the CD4 binding site should be avoided or approached with care to not disrupt the antigenic surface of the CD4bs, as observed for the 368D/R mutation. To circumvent potential CD4 occlusion effects and to limit the elicitation of nonneutralizing CoRbs-directed responses, rationally designed CD4 binding-defective Env immunogens that retain the capacity to stimulate Ab responses against the CD4bs may remain worthy of further pursuit.
Supplementary Material
Acknowledgments
We thank Mats Spångberg, Helene Fredlund, and the personnel at the Astrid Fagraeus laboratory at the Swedish Institute for Infectious Disease Control for expert assistance. We also thank Joseph Sodroski for helpful discussions, Linda Stertman and Karin Lövgren Bengtsson at Isconova AB for the generous gift of Abisco-100, James Robinson for providing MAbs 17b, 48d, F91, 1.5E, E51, and 2.1C, Susan Zolla-Pazner for MAb 447-52D, Marshal Posner for MAb F105, and Dennis Burton for MAbs b6 and b12.
This study was supported by grants from the International AIDS Vaccine Initiative (G.K.H. and K.L.), the Swedish International Development Agency/Department of Research Cooperation (G.K.H. and K.L.), the intramural research program of the Vaccine Research Center, NIAID, NIH (R.T.W. and J.R.M.), and by the Karolinska Institutet (G.K.H.).
Footnotes
Published ahead of print on 2 December 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Binley, J. M., E. A. Lybarger, E. T. Crooks, M. S. Seaman, E. Gray, K. L. Davis, J. M. Decker, D. Wycuff, L. Harris, N. Hawkins, B. Wood, C. Nathe, D. Richman, G. D. Tomaras, F. Bibollet-Ruche, J. E. Robinson, L. Morris, G. M. Shaw, D. C. Montefiori, and J. R. Mascola. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. Nabel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233-236. [DOI] [PubMed] [Google Scholar]
- 5.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 6.Chen, W., Z. Zhu, Y. Feng, and D. S. Dimitrov. 2008. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc. Natl. Acad. Sci. USA 105:17121-17126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 8.Decker, J. M., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major coreceptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 10.Dey, B., K. Svehla, L. Xu, D. Wycuff, T. Zhou, G. Voss, A. Phogat, B. K. Chakrabarti, Y. Li, G. Shaw, P. D. Kwong, G. J. Nabel, J. R. Mascola, and R. T. Wyatt. 2009. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 5:e1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dosenovic, P., B. Chakrabarti, M. Soldemo, I. Douagi, M. N. Forsell, Y. Li, A. Phogat, S. Paulie, J. Hoxie, R. T. Wyatt, and G. B. Karlsson Hedestam. 2009. Selective expansion of HIV-1 envelope glycoprotein-specific B cell subsets recognizing distinct structural elements following immunization. J. Immunol. 183:3373-3382. [DOI] [PubMed] [Google Scholar]
- 12.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 13.Fernando, K., H. Hu, H. Ni, J. A. Hoxie, and D. Weissman. 2007. Vaccine-delivered HIV envelope inhibits CD4(+) T-cell activation, a mechanism for poor HIV vaccine responses. Blood 109:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsell, M., W. R. Schief, and R. T. Wyatt. 2009. Immunogenicity of HIV-1 envelope glycoprotein oligomers. Curr. Opin. HIV AIDS 4:380-387. [DOI] [PubMed] [Google Scholar]
- 15.Forsell, M. N., B. Dey, A. Morner, K. Svehla, S. O'Dell, C. M. Hogerkorp, G. Voss, R. Thorstensson, G. M. Shaw, J. R. Mascola, G. B. Karlsson Hedestam, and R. T. Wyatt. 2008. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 4:e1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, E. S., P. L. Moore, I. A. Choge, J. M. Decker, F. Bibollet-Ruche, H. Li, N. Leseka, F. Treurnicht, K. Mlisana, G. M. Shaw, S. S. Karim, C. Williamson, and L. Morris. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray, E. S., N. Taylor, D. Wycuff, P. L. Moore, G. D. Tomaras, C. K. Wibmer, A. Puren, A. DeCamp, P. B. Gilbert, B. Wood, D. C. Montefiori, J. M. Binley, G. M. Shaw, B. F. Haynes, J. R. Mascola, and L. Morris. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83:8925-8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, C. C., S. N. Lam, P. Acharya, M. Tang, S. H. Xiang, S. S. Hussan, R. L. Stanfield, J. Robinson, J. Sodroski, I. A. Wilson, R. Wyatt, C. A. Bewley, and P. D. Kwong. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlsson Hedestam, G. B., R. A. Fouchier, S. Phogat, D. R. Burton, J. Sodroski, and R. T. Wyatt. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6:143-155. [DOI] [PubMed] [Google Scholar]
- 21.Kim, M., Z. S. Qiao, D. C. Montefiori, B. F. Haynes, E. L. Reinherz, and H. X. Liao. 2005. Comparison of HIV type 1 ADA gp120 monomers versus gp140 trimers as immunogens for the induction of neutralizing antibodies. AIDS Res. Hum. Retrovir. 21:58-67. [DOI] [PubMed] [Google Scholar]
- 22.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 23.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Y., S. A. Migueles, B. Welcher, K. Svehla, A. Phogat, M. K. Louder, X. Wu, G. M. Shaw, M. Connors, R. T. Wyatt, and J. R. Mascola. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y., K. Svehla, M. K. Louder, D. Wycuff, S. Phogat, M. Tang, S. A. Migueles, X. Wu, A. Phogat, G. M. Shaw, M. Connors, J. Hoxie, J. R. Mascola, and R. Wyatt. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83:1045-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, J., A. Bartesaghi, M. J. Borgnia, G. Sapiro, and S. Subramaniam. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mörner, A., I. Douagi, M. N. Forsell, C. Sundling, P. Dosenovic, S. O'Dell, B. Dey, P. D. Kwong, G. Voss, R. Thorstensson, J. R. Mascola, R. T. Wyatt, and G. B. Karlsson Hedestam. 2009. Human immunodeficiency virus type 1 env trimer immunization of macaques and impact of priming with viral vector or stabilized core protein. J. Virol. 83:540-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olshevsky, U., E. Helseth, C. Furman, J. Li, W. Haseltine, and J. Sodroski. 1990. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 64:5701-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitisuttithum, P., P. Gilbert, M. Gurwith, W. Heyward, M. Martin, F. van Griensven, D. Hu, J. W. Tappero, and K. Choopanya. 2006. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 194:1661-1671. [DOI] [PubMed] [Google Scholar]
- 31.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 32.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 33.Robinson, J. E., D. H. Elliott, E. A. Martin, K. Micken, and E. S. Rosenberg. 2005. High frequencies of antibody responses to CD4 induced epitopes in HIV infected patients started on HAART during acute infection. Hum. Antibodies 14:115-121. [PubMed] [Google Scholar]
- 34.Sanders, R. W., M. Vesanen, N. Schuelke, A. Master, L. Schiffner, R. Kalyanaraman, M. Paluch, B. Berkhout, P. J. Maddon, W. C. Olson, M. Lu, and J. P. Moore. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76:8875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sather, D. N., J. Armann, L. K. Ching, A. Mavrantoni, G. Sellhorn, Z. Caldwell, X. Yu, B. Wood, S. Self, S. Kalams, and L. Stamatatos. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnurr, M., M. Orban, N. C. Robson, A. Shin, H. Braley, D. Airey, J. Cebon, E. Maraskovsky, and S. Endres. 2009. ISCOMATRIX adjuvant induces efficient cross-presentation of tumor antigen by dendritic cells via rapid cytosolic antigen delivery and processing via tripeptidyl peptidase II. J. Immunol. 182:1253-1259. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz, O., M. Alizon, J. M. Heard, and O. Danos. 1994. Impairment of T-cell receptor-dependent stimulation in CD4+ lymphocytes after contact with membrane-bound HIV-1 envelope glycoprotein. Virology 198:360-365. [DOI] [PubMed] [Google Scholar]
- 38.Seagal, J., E. Spectorman, J. M. Gershoni, and G. F. Denisova. 2001. Use of human CD4 transgenic mice for studying immunogenicity of HIV-1 envelope protein gp120. Transgenic Res. 10:113-120. [DOI] [PubMed] [Google Scholar]
- 39.Shu, Y., S. Winfrey, Z. Y. Yang, L. Xu, S. S. Rao, I. Srivastava, S. W. Barnett, G. J. Nabel, and J. R. Mascola. 2007. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine 25:1398-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srivastava, I. K., L. Stamatatos, E. Kan, M. Vajdy, Y. Lian, S. Hilt, L. Martin, C. Vita, P. Zhu, K. H. Roux, L. Vojtech, D. C. Montefiore, J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2003. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J. Virol. 77:11244-11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thali, M., C. Furman, D. D. Ho, J. Robinson, S. Tilley, A. Pinter, and J. Sodroski. 1992. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 66:5635-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 44.Weinhold, K. J., H. K. Lyerly, S. D. Stanley, A. A. Austin, T. J. Matthews, and D. P. Bolognesi. 1989. HIV-1 GP120-mediated immune suppression and lymphocyte destruction in the absence of viral infection. J. Immunol. 142:3091-3097. [PubMed] [Google Scholar]
- 45.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 46.Wu, X., A. Sambor, M. C. Nason, Z. Y. Yang, L. Wu, S. Zolla-Pazner, G. J. Nabel, and J. R. Mascola. 2008. Soluble CD4 broadens neutralization of V3-directed monoclonal antibodies and guinea pig vaccine sera against HIV-1 subtype B and C reference viruses. Virology 380:285-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang, S. H., N. Doka, R. K. Choudhary, J. Sodroski, and J. E. Robinson. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retrovir. 18:1207-1217. [DOI] [PubMed] [Google Scholar]
- 48.Xiang, S. H., P. D. Kwong, R. Gupta, C. D. Rizzuto, D. J. Casper, R. Wyatt, L. Wang, W. A. Hendrickson, M. L. Doyle, and J. Sodroski. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 76:9888-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiang, S. H., L. Wang, M. Abreu, C. C. Huang, P. D. Kwong, E. Rosenberg, J. E. Robinson, and J. Sodroski. 2003. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology 315:124-134. [DOI] [PubMed] [Google Scholar]
- 50.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74:5716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, X., J. Lee, E. M. Mahony, P. D. Kwong, R. Wyatt, and J. Sodroski. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.