Abstract
The central domain of the 200-kDa Lassa virus L protein is a putative RNA-dependent RNA polymerase. N- and C-terminal domains may harbor enzymatic functions important for viral mRNA synthesis, including capping enzymes or cap-snatching endoribonucleases. In the present study, we have employed a large-scale mutagenesis approach to map functionally relevant residues in these regions. The main targets were acidic (Asp and Glu) and basic residues (Lys and Arg) known to form catalytic and binding sites of capping enzymes and endoribonucleases. A total of 149 different mutants were generated and tested in the Lassa virus replicon system. Nearly 25% of evolutionarily highly conserved acidic and basic side chains were dispensable for function of L protein in the replicon context. The vast majority of the remaining mutants had defects in both transcription and replication. Seven residues (Asp-89, Glu-102, Asp-119, Lys-122, Asp-129, Glu-180, and Arg-185) were selectively important for mRNA synthesis. The phenotype was particularly pronounced for Asp-89, Glu-102, and Asp-129, which were indispensable for transcription but could be replaced by a variety of amino acid residues without affecting genome replication. Bioinformatics disclosed the remote similarity of this region to type IIs endonucleases. The mutagenesis was complemented by experiments with the RNA polymerase II inhibitor α-amanitin, demonstrating dependence of viral transcription from the cellular mRNA pool. In conclusion, this paper describes an N-terminal region in L protein being important for mRNA, but not genome synthesis. Bioinformatics and cell biological experiments lend support to the hypothesis that this region could be part of a cap-snatching enzyme.
Lassa virus is a segmented negative-strand RNA virus of the family Arenaviridae. It belongs to the Old World complex of the arenaviruses, which also includes the prototype virus of the family, lymphocytic choriomeningitis virus (LCMV). Lassa virus is endemic in large areas of West Africa, where its natural reservoir host, the rodent Mastomys natalensis (25, 38), is prevalent. Transmission of the virus to humans causes Lassa fever, a hemorrhagic fever that is estimated to affect some 100,000 people annually (32). Human-to-human transmission may give rise to nosocomial epidemics with high case fatality rates (7). Currently, no vaccine exists for use in humans. Specific treatment is limited to ribavirin, a broad-spectrum nucleoside analogue, which is mainly effective during the first days of illness (31).
The arenavirus genome consists of two single-stranded RNA segments, each containing two genes in opposite directions, a coding strategy called “ambisense” (1). The S RNA segment encodes the nucleoprotein (NP) and the glycoprotein precursor. The L RNA encodes the small matrix protein Z (41, 45) and the 200-kDa protein L (44). The minimal viral trans-acting factors required for replication and transcription of the genome are NP and L protein (17, 26, 28). L protein mediates the synthesis of two RNA species: mRNA terminating in the intergenic region and noncapped genomic or antigenomic RNA representing a full-length copy of the genome (10, 35). The central domain of the protein from amino acid residues 1000 to 1500 harbors the RNA-dependent RNA polymerase (RdRp) (8, 11, 19, 29, 47). Enzymatic functions residing in the N terminus and C terminus of the L protein are unknown. Neither biochemical assays nor three-dimensional structures are currently available for the L protein or any of its domains.
Arenaviral mRNA contains 4 to 5 nontemplated nucleotides, including the cap at 5′ end (35, 42, 43). It is believed that this oligonucleotide is cleaved off from 5′ end of cellular mRNAs by a virus-encoded endonuclease and serves as primer for viral mRNA synthesis, a mechanism known as “cap snatching.” This mechanism is well established for influenza virus (24), which also belongs to the group of segmented negative-strand RNA viruses and shares principal features of the replication cycle with arenaviruses. Recently, the influenza virus endonuclease has been identified in the N terminus of PA by structural analysis and mutagenesis (4, 16, 50). It is well conceivable that Lassa virus L protein also harbors an endonuclease for cap snatching.
We have previously used large-scale mutagenesis to characterize the Lassa virus RdRp domain (19). In the present paper, we have extended this approach to N terminus and C terminus of the L protein. To identify residues important for mRNA synthesis, including those forming the catalytic site of a putative endonuclease, all conserved acidic and basic amino acid residues in the N terminus and C terminus were subjected to mutagenesis. The functionality of the L gene mutants in RNA synthesis and gene expression was studied in the context of the Lassa virus replicon system (17). The mutagenesis experiments were complemented by a set of experiments with the RNA polymerase II (Pol II) inhibitor α-amanitin to explore the dependence of viral transcription from the cellular mRNA level.
MATERIALS AND METHODS
Lassa virus replicon system.
Plasmids pCITE-NP and pCITE-L expressing Lassa virus NP and L protein under the control of a T7 RNA polymerase promoter have been described previously (17). Minigenome (MG) plasmid pLAS-MG contains the T7 promoter followed by a single additional G residue, the 5′ noncoding region, a chloramphenicol acetyltransferase (CAT) gene, an intergenic region, a Renilla luciferase (Ren-Luc) gene in reverse orientation, and the 3′ noncoding region (18). For transfection, the MG, including the T7 promoter, was amplified for 25 cycles with Phusion DNA polymerase (Finnzymes), 3.3 ng of linearized pLAS-MG as a template, and vector-specific primer pUC-fwd and primer LVS-3400− (CGCACAGTGGATCCTAGGCTATTGGA) to generate a functional 3′ end. Amplified MG was purified by using a PCR purification kit (Macherey & Nagel) and quantified spectrophotometrically.
Mutagenesis of the L gene.
The functional cassette of pCITE-L (T7 RNA polymerase promoter, internal ribosome entry site, and L gene) was amplified by mutagenic PCR, and the resulting PCR products were used for transfection without prior cloning as described previously (19). PCR was performed with Phusion DNA polymerase (Finnzymes). First, two fragments were amplified for 30 cycles by using 10 ng of linearized pCITE-L as a template and primer combination pUC-fwd/L-mut− and L-mut+/pUC-rev, respectively. (L-mut− was reverse complementary to the corresponding L-mut+ primer.) The sequences of L-mut− and L-mut+ primers (n = 302) can be obtained on request. PCR products were gel purified and fused together in a second PCR containing aliquots of both fragments as a template and primers pUC-fwd and pUC-rev. For generation of hemagglutinin (HA)-tagged L gene PCR product, the pUC-rev primer was replaced by primer L-HA-rev adding a C-terminal HA tag to the L gene. Mutant L gene constructs were purified by using a PCR purification kit (Macherey & Nagel) and quantified spectrophotometrically. The presence of the artificial mutation was ascertained by sequencing the final PCR product. In each mutagenesis PCR, the wild-type L gene and an inactive mutant with a mutation in the catalytic site of the RdRp were amplified in parallel using primers binding to the L gene around amino acid position 1334. The wild-type and mutant PCR products served as positive and negative controls, respectively, for the transfection experiment.
Transfections.
BSR T7/5 cells (2) stably expressing T7 RNA polymerase were grown in Glasgow's minimal essential medium (GMEM; GIBCO) supplemented with 5% fetal calf serum (FCS). Every second passage, 1 mg of Geneticin (GIBCO) per ml of medium was added to the cells. One day before transfection, BSR T7/5 cells were seeded at a density of 1 × 105 cells per well of a 24-well plate. Transfections were performed with Lipofectamine 2000 (Invitrogen) at 3 μl/μg of DNA in GMEM without FCS. BSR T7/5 cells in a well of a 24-well plate were transfected with 250 ng of MG, 250 ng of L gene PCR product, 300 ng of pCITE-NP, and 10 ng of pCITE-FF-luc (expression construct for firefly luciferase) as a transfection control. Medium was replaced 4 h after transfection by fresh medium complemented with FCS. One day after transfection, total RNA was prepared for Northern blotting or cells were lysed in 100 μl of passive lysis buffer (Promega) per well, and 20 μl of the lysate was assayed for firefly luciferase and Ren-Luc activity using the Promega dual-luciferase reporter assay system as described by the manufacturer. Ren-Luc levels were first corrected with the firefly luciferase levels (resulting in standardized relative light units [sRLU]) to compensate for differences in transfection efficiency or cell density. Northern blot data were not corrected with firefly luciferase values as the transcription/replication phenotype of mutants did not depend on the expression level of the internal control (see Fig. S1A in the supplemental material).
To consider the full activity range of the replicon system, which covers 2 to 3 log units (17) and to render the data from different experiments comparable, the sRLU values were log transformed and then normalized with respect to the wild type (100%) and negative controls (0%) as follows: relative log activity = (log[sRLUmutant] − log[sRLUnc])/(log[sRLUwt] − log[sRLUnc]), where “nc” stands for negative control and “wt” stands for wild type. On average, four (range, 2 to 10) independent transfection experiments were performed for each mutant. The interexperimental variability was 9.2% relative log units (mean standard deviation calculated with log activity values of 583 independent transfections for 149 mutants). Depending on their log activity level, the mutants were grouped into three classes: inactive mutants (<30% log activity; roughly corresponding to 0.5 to 3% of the wild-type sRLU value), mutants with clearly detectable but reduced activity (30% to 80% log activity; roughly corresponding to 3 to 40% of the wild-type sRLU value), and mutants with wild-type activity (>80% log activity; roughly corresponding to 40 to 300% of the wild-type sRLU value). For a list of all mutants and raw data, see Table S1 in the supplemental material.
Immunoblot analysis of L protein.
BSR T7/5 were inoculated with modified vaccinia virus Ankara expressing T7 RNA polymerase (MVA-T7) (46) at a multiplicity of infection (MOI) of 5 for 1 h before transfection. Cells in two wells of a 24-well plate were transfected with 1 μg of mutant or wild-type L gene tagged with a HA sequence. They were harvested 24 h after transfection and lysed in cytoplasmic lysis buffer containing Complete protease inhibitor mix (Roche). Nuclei were pelleted by centrifugation, and the cytoplasmic lysate was mixed with 4× NuPAGE LDS sample buffer (Invitrogen) and dithiothreitol (DTT). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membrane (Schleicher & Schuell), and visualized by staining with Fast Green FCF (Roth) for 5 min. Membranes were blocked with 1× Roti-Block (Roth) overnight at room temperature and then incubated with anti-HA (1:10,000 H-6908; Sigma-Aldrich) in phosphate-buffered saline (PBS)-0.2× Roti-Block for 1 h at room temperature. After a washing step, blots were incubated with horseradish peroxidase-coupled secondary antibodies (Dianova) for 1 h at room temperature. Protein bands were visualized by chemiluminescence using SuperSignal West Pico or Femto substrate (Pierce) and X-ray film (Kodak).
Northern blot analysis.
Total RNA of transfected or infected cells was purified by using TRIzol LS reagent (Invitrogen). RNA (5 to 10 μg) was separated in a 1.5% agarose-formaldehyde gel and transferred onto a Hybond N+ membrane (Amersham Pharmacia Biotech). Blots were prehybridized in 50% deionized formamide-0.5% SDS-5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-5× Denhardt's solution for 1 h at 68°C. Hybridization was done in the same buffer with an antisense 32P-labeled riboprobe targeting the Ren-Luc gene for detection of MG or the NP gene of Lassa or Mopeia virus for detection of authentic virus RNA at 68°C for 16 h. Filters were washed several times, and RNA bands were visualized by autoradiography using a PhosphorImager Typhoon 9210 (Amersham Biosciences). Intensity profiles were generated for each lane, and signals were quantified by using TINA software (Raytest, Straubenhardt, Germany).
Virus infection.
Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum. One day before infection, cells were seeded at a density of 3 × 105 cells per well of a 6-well plate. Cells were inoculated with Lassa virus strain AV (14), Mopeia virus strain 21366 (48), or vesicular stomatitis virus (VSV) strain Indiana at an MOI of 0.01 to 3 for 1 h. Cells were washed with phosphate buffered saline (PBS), and fresh medium with or without 10 μg/ml α-amanitin was added. Control cells were mock infected. Infections with Lassa virus were carried out under biosafety level 4 conditions.
Infectious particles assay.
Vero cells in 24-well plates were inoculated with 200 μl of serial 10-fold dilutions of supernatant from cells infected with VSV, Lassa virus, or Mopeia virus. The inoculum was removed after 1 h and replaced by a 1% methylcellulose medium overlay. VSV particles were quantified by plaque assay. After 1 day of incubation, cells were fixed with 4% formaldehyde, stained with 1% crystal violet in methanol, and washed with water until plaques became visible. Lassa and Mopeia virus particles were quantified by immunofocus assay. After 5 days of incubation, cells were fixed with 4% formaldehyde, permeabilized with 0.5% Triton X-100, blocked with 10% FCS, and washed. Infected cell foci were detected by using Lassa and Mopeia virus NP-specific monoclonal antibodies (21), peroxidase-labeled anti-mouse immunoglobulin G antibody (Jackson ImmunoResearch), and 3,3′,5,5′-tetramethylbenzidine (TMB). Titers are expressed as plaque (focus)-forming units (PFU) per milliliter.
Cell growth assay.
Cell growth and viability under α-amanitin treatment were determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazoliumbromide (MTT) method. MTT solution (5 mg/ml in PBS) was added 1:3 to the supernatant of cells. The cells were incubated for 90 min at 37°C. The supernatant was completely removed, and the cells were fixed with 4% formaldehyde for 20 min. Tetrazolium crystals in the cells were dissolved in 500 μl isopropanol, and the extinction was measured at 560 nm.
IP of mRNA.
Total RNA was purified from Lassa virus-infected Vero cells 24 h after infection by using TRIzol LS reagent, and mRNA was precipitated with a mouse monoclonal antibody to cap structures containing 2,2,7-trimethylguanosine (m3G-cap) or 7-monomethylguanosine (m7G-cap) (Synaptic Systems). RNA (15 μg) was dissolved in 250 μl IPPL buffer (150 mM NaCl, 10 mM Tris [pH 7.5], 1 mM EDTA, 0.05% NP-40) supplemented with 1 mM DTT and 30 U RNasin. The m3G-cap/m7G-cap antibody (10 μl) was bound to protein G-Sepharose beads (50 μl of a 50% suspension in 20% ethanol; Sigma) at 4°C overnight with agitation. Antibody-coupled beads were washed three times with ice-cold PBS and three times with ice-cold IPPL buffer, mixed with 250 μl RNA-IPPL buffer solution, and incubated at 4°C for 8 h with agitation. After centrifugation, supernatant was removed and the beads were washed three times with ice-cold IPPL buffer. Fifty percent of the supernatant from the first immunoprecipitation (IP) was subjected to a second IP round after adding fresh DTT, 30 U RNasin, and antibody-coupled beads. Supernatant and bead pellet from the first and second IP were supplemented with 20 μg carrier RNA (Qiagen), and RNA was purified by using TRIzol LS reagent and analyzed by Northern blotting. The fraction of precipitated RNA was calculated from the signal intensities for NP mRNA and antigenomic RNA in precipitate and supernatant.
RESULTS
Alanine mutagenesis of conserved basic and acidic amino acid residues.
Lassa virus may essentially employ two strategies for mRNA synthesis: expression of capping enzymes for cap synthesis like VSV (39) or cap snatching from cellular mRNA like influenza virus (24). Either of these functions may reside in the N- and C-terminal parts of L protein. Cap snatching could involve endo- or exoribonucleases, while mRNA capping would involve RNA triphosphatase, guanylyltransferase, and (guanine-7)-methyltransferase (9). Catalytic motifs of virtually all superfamilies of ribonucleases are composed of acidic and/or basic residues, namely, Asp, Glu, Lys, or Arg (36, 51). Likewise, these residues form the catalytic and binding sites of mRNA capping enzymes (15, 27). Mutational and structural studies indicate that Glu, Lys, and Phe are key residues for cap binding by influenza virus PB2 (6, 13), while Asp, Glu, and Lys are key residues for endonucleolytic cleavage by influenza virus PA (4, 16, 50).
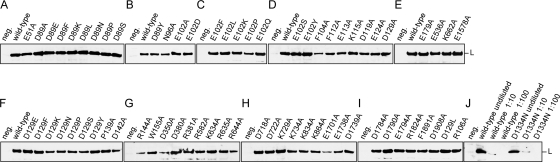
To identify regions in Lassa virus L protein that might play a role in cap snatching or synthesis, all highly conserved Asp (n = 18), Glu (n = 18), Lys (n = 22), Arg (n = 16), and Phe (n = 19) residues outside of the RdRp domain were replaced by alanine and the mutants were tested in the replicon system (Fig. 1). Mutant L genes were generated by using a previously described PCR-based mutagenesis technique that circumvents cloning before transfection, facilitating analysis of large numbers of mutants (19). In the initial screening experiment, expression of the MG-carried reporter gene coding for Renilla luciferase (Ren-Luc) was chosen as the readout. Activity of Ren-Luc reflects the level of mRNA synthesis by L protein. Depending on the Ren-Luc level, the mutants were grouped into three classes: inactive mutants, mutants with clearly detectable but reduced activity, and mutants with wild-type-like activity. (For a list of all mutants and corresponding data generated in this paper, see Table S1 in the supplemental material.) Alanine mutation of 31 (42%) of the charged residues led to complete loss of Ren-Luc expression, mutation of 25 (34%) sites led to an intermediate phenotype, and mutation of 18 (24%) sites had no major influence (Fig. 1). No considerable differences were seen between acidic or basic residues. At three positions, Lys-334, Lys-555, and Lys-581, alanine mutants were more active than the wild type (190 to 300% of wild-type sRLU level; see the table in the supplemental material). Only 3 of the 19 Phe mutants were inactive, while the others showed reduced or full activity (Fig. 1). Immunoblot analysis of all inactive L protein mutants demonstrates that they are expressed in full length and lack of activity is not due to reduced protein stability (Fig. 2). (Table S1 in the supplemental material provides links to easily localize a certain mutant in the figure.) Residues important for Ren-Luc expression were not clustered in one domain but are scattered throughout the protein. In conclusion, the Ren-Luc data suggest that both N- and C-terminal parts of the L protein are involved in mRNA synthesis.
FIG. 1.
Alanine mutations introduced in Lassa virus L protein. The activity of the respective mutants in the replicon system as measured by expression of Ren-Luc is indicated as follows: (i) white typeface on black background, inactive mutant; (ii) black typeface on gray background, mutant with reduced activity; (iii) black typeface on white background, mutant with wild-type activity. (Upper panel) Asp, Glu, Lys, and Arg mutants; (lower panel) Phe mutants. The localization of conserved domains L1 to L4 is shown at the bottom (47).
FIG. 2.
Immunoblot analysis of HA-tagged L protein mutants. BSR T7/5 cells were infected with MVA-T7 and transfected with PCR products expressing L protein mutants containing a C-terminal HA tag. L protein in cytoplasmic lysate was separated by SDS-PAGE, blotted, and detected with anti-HA antibody. A semiquantitative standard based on titration of lysate is shown in panel J. Negative (neg.) control cells were infected with MVA-T7 but not transfected.
Selective impact of mutations on transcription and replication.
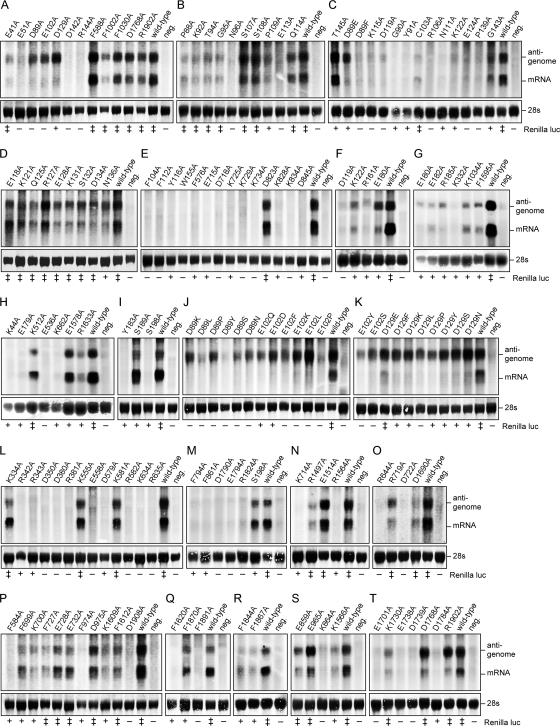
The Lassa virus L protein mediates the synthesis of two RNA species: first, capped mRNA terminating within the intergenic region; second, uncapped antigenomic RNA being a full-length copy of the genomic RNA template (10, 35). This dual role in RNA synthesis is reproduced in the minireplicon system. It is conceivable that L protein mutants with a defect in mRNA synthesis (Ren-Luc-negative mutants) are still capable of antigenome synthesis, or vice versa, Ren-Luc-positive mutants may show a selective defect in antigenome synthesis. To distinguish these phenotypes, L protein mutants were expressed in the context of the replicon system, and total cellular RNA was prepared and subjected to Northern blotting (Fig. 3). (Table S1 in the supplemental material provides links to easily localize a certain mutant in the figure.) The first set included all charged-to-alanine (n = 74) and Phe-to-alanine mutants (n = 19) tested so far in the Ren-Luc assay (mutants 1 to 93 in Table S1 in the supplemental material). The signal intensities on the Northern blot were measured and used to calculate the steady-state level of Ren-Luc mRNA in relation to that of antigenomic RNA (Fig. 4). To render data from different experiments comparable, the mRNA/antigenome ratio was normalized with the wild-type data (see Fig. S2 in the supplemental material for details and examples). All mutants showing wild-type activity in the luciferase assay expressed Ren-Luc mRNA and antigenome in a ratio comparable to that of wild-type L protein. RNA signals were present for some, but not all, mutants with reduced Ren-Luc activity. Note that the luciferase assay is more sensitive than Northern blotting, and, thus, mRNA of mutants with strongly reduced Ren-Luc activity may not be detectable by Northern blotting (see Fig. S1B in the supplemental material for a scatter plot of Ren-Luc activity versus mRNA/antigenome ratio). Nearly all mutants with reduced Ren-Luc activity either expressed Ren-Luc mRNA and antigenome in a ratio comparable to wild type or did not express detectable amounts of both RNA species. However, six mutants were found showing a clearly reduced mRNA level in relation to the antigenomic RNA level (D89A, E102A, and D129A mutants in Fig. 3A; D119A and K122A mutants in Fig. 3C and F; R185A mutant in Fig. 3G and Fig. 4, upper panel; and see Fig. S2 in the supplemental material). This phenotype was particularly pronounced for the two D89A and E102A mutants, which were negative in Ren-Luc assay, while the antigenomic RNA level was comparable to wild type. The phenotype was similar for the D129A mutant, which expressed antigenomic RNA at wild-type level in conjunction with a strongly reduced, though still detectable, level of luciferase (4% of the wild-type sRLU level). The phenotype was less pronounced for the D119A (Ren-Luc-negative), K122A (8% of wild-type sRLU level), and R185A (7% of wild-type sRLU level) mutants. These mutants showed selective reduction in the mRNA level coupled with an overall reduction in RNA synthesis. This phenotype was reproducible in independent transfection experiments.
FIG. 3.
Influence of L protein mutations on transcription and replication. RNA synthesized by L protein mutants in the context of the replicon system was analyzed by Northern blotting. Antigenomic RNA and Ren-Luc mRNA were detected using a radiolabeled riboprobe hybridizing to the Ren-Luc gene. The activity of the mutants in luciferase assay is indicated below the blot (−, inactive mutant; +, mutant with reduced activity; ‡, mutant with wild-type activity). Mutations in L protein are indicated above the blot. Negative (neg.) control cells expressed minigenome, NP, and an L protein mutant with a mutation in the catalytic site of the RdRp. The methylene blue-stained 28S rRNA is shown below the blots as a marker for gel loading and RNA transfer. Each panel represents an independent transfection experiment.
FIG. 4.
Level of Ren-Luc mRNA in relation to antigenome level. Signals on Northern blots in Fig. 3 were quantified via intensity profiles (for details and examples, see Fig. S2 in the supplemental material). All mutants with detectable signals are shown. Those with significant reduction of the mRNA/antigenome ratio compared to the wild type (<40%) are indicated by black bars. The wild-type mRNA/antigenome ratio was set at 1 for each experiment to render independent blots comparable. Careful examination of the intensity profiles revealed residual signals at the mRNA position (about 10% of the wild-type mRNA signal) for mutants completely negative in the Ren-Luc assay. These signals were not seen in the negative control lanes. The precise nature of this RNA is not clear, as the luciferase data indicate that functional mRNA is not expressed.
To further substantiate the effect seen with the D89A, E102A, and D129A mutants and to explore structural requirements with respect to the side chain at these positions, Asp-89, Glu-102, and Asp-129 were replaced by a range of amino acid residues of different chemical types (mutants 94 to 117 in Table S1 in the supplemental material). Protein stability of all these mutants was verified by immunoblot analysis (Fig. 2). Asp-89 was exchanged with Glu, Asn, Lys, Ser, Phe, Tyr, Leu, and Pro (Fig. 3C and J; Fig. 4, lower panel). The D89F, D89Y, and D89L mutants hardly expressed RNA, indicating that hydrophobic or large residues at position 89 are not tolerable for overall activity of L protein. The remaining mutants clearly expressed antigenome but showed a defect in mRNA synthesis. The only mutant with residual Ren-Luc expression was D89E (6% of wild-type sRLU level); all others were completely defective. Glu-102 was exchanged with Asp, Gln, Lys, Ser, Phe, Tyr, Leu, and Pro (Fig. 3J and K; Fig. 4, lower panel). Minimal Ren-Luc expression was only seen with the E102D and E102Q mutants (5% of wild-type sRLU level). Antigenome synthesis was compatible with all exchanges, except Pro. Asp-129 was exchanged with Glu, Asn, Lys, Ser, Phe, Tyr, Leu, and Pro (Fig. 3K; Fig. 4, lower panel). The D129E mutant expressed Ren-Luc close to the wild-type level (45% of wild-type sRLU level); the D129L mutant was completely defective, while the other mutants expressed low levels of Ren-Luc similar to D129A mutant. None of the exchanges interfered with antigenome synthesis. In conclusion, Asp-89 and Glu-102 are indispensable for mRNA expression but not relevant for antigenome synthesis. Even changes to chemically closely related residues (D89E and E102D) are associated with nearly complete loss of the former function. Asp-129 may be replaced at least by Glu. Other side chains strongly reduce mRNA expression, while antigenome synthesis is not affected at all.
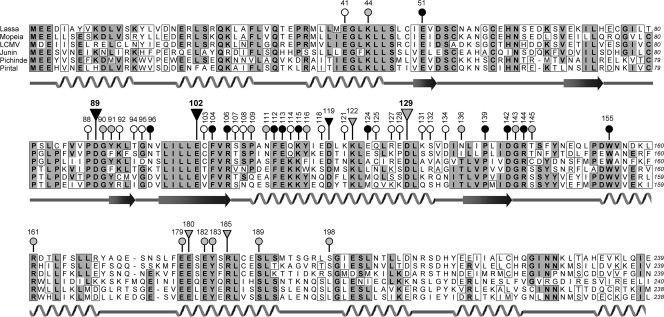
Since the initial screening indicated that residues between positions 89 and 185 play a specific role in mRNA synthesis, mutagenesis was extended to a further 32 residues in this region (mutants no. 118 to 149 in Table S1 in the supplemental material). Most of these residues are not conserved. Exchange with alanine did not interfere with Ren-Luc expression in 16 (50%) mutants, 12 (38%) mutants showed reduced expression, and only 4 (12%) mutants were completely defective. Protein stability of the defective mutants was verified by immunoblot analysis. Conserved positions tended to be more susceptible to mutagenesis. To search for specific defects in transcription, Northern blot analysis was performed for all mutants. Only one mutant (E180A; 13% of wild-type sRLU level) showed a phenotype similar to that of the D119A, K122A, and R185A mutants, namely, selective reduction of the mRNA level associated with overall suppression of RNA synthesis (Fig. 3F and G; Fig. 4, lower panel; see Fig. S2 in the supplemental material). Taken together, analysis of virus RNA species generated by L protein mutants led to the identification of a region in the N terminus of L protein (L1 domain) being important for mRNA rather than antigenome synthesis. Residues involved in this function are Asp-89, Glu-102, Asp-119, Lys-122, Asp-129, Glu-180, and Arg-185. Acidic side chains at positions 89, 102, and 129 are crucial for activity. Figure 5 summarizes the mutagenesis data for the L1 domain.
FIG. 5.
Amino acid sequence alignment of the N-terminal part of L protein (L1 domain) of Old and New World arenaviruses and summary of the mutagenesis experiments. Positions subjected to mutagenesis are marked above the sequence. The data from Northern blotting are coded as follows: (i) large inverted triangle, mutant with reduced mRNA expression but wild-type levels of antigenome; (ii) small inverted triangle, mutant with selective reduction of the mRNA level associated with overall reduction of RNA synthesis; (iii) circle, mutant with wild-type mRNA/antigenome ratio or no RNA signals at all. The Ren-Luc data are coded as follows: (i) black head, inactive mutant; (ii) gray head, mutant with reduced activity; (iii) white head, mutant with wild-type activity. The predicted secondary structure of the domain is shown below the alignment.
Influence of host cell mRNA synthesis on arenavirus replication.
The mutagenesis experiments facilitated mapping of residues relevant for mRNA synthesis, but do not provide a clue as to their precise role. It is conceivable that they are part of capping enzymes or involved in cap snatching. If a virus uses cap snatching, its multiplication is dependent on cellular mRNA as a source for cap structures and, thus, should be prone to inhibition by α-amanitin, a specific inhibitor of DNA-dependent RNA polymerase II (Pol II) (3). This has been shown for influenza virus (30) and the New World arenaviruses Junin and Tacaribe (33, 34). To extend these data, Vero cells were infected with Lassa virus or the closely related Mopeia virus and treated with 10 μg/ml α-amanitin. Treatment reduced the virus titer in supernatant by about 2 log units, irrespective of inoculation dose or duration of infection (Table 1). Microscopic evaluation of the cells revealed no evidence for cytotoxicity of treatment, although the MTT test value was reduced by 40%, indicating some cell growth inhibition. As a control, cells were infected with VSV, which encodes its own capping enzymes (39). As expected, no effect of α-amanitin on VSV titer was observed (Table 1), although microscopy revealed a strong cytopathic effect due to VSV replication.
TABLE 1.
Effect of α-amanitin treatment on virus replication in Vero cells
| Virus | MOI | Duration of α-amanitin treatment (h postinfection) | Mean % virus titer in supernatant of treated vs. untreated cells (range) |
|---|---|---|---|
| Lassa | 1 | 24 | 3.60 (2.20-5.00) |
| 0.01 | 24 | 0.86 (0.71-1.00) | |
| 0.01 | 48 | 1.03 (0.85-1.22) | |
| Mopeia | 0.01 | 48 | 0.43 (0.37-0.49) |
| VSV | 0.1 | 24 | 74.68 (74.36-75.00) |
| 0.1 | 48 | 141.67 (133.33-150.00) |
If the antiarenaviral effect of α-amanitin is indeed via reduction of the pool of cellular mRNA available for cap snatching, it should manifest in a selective reduction of virus transcripts. To test this hypothesis, Vero cells were infected with Mopeia virus at an MOI of 3 and treated with 10 μg/ml α-amanitin. Cells were harvested at 6-h intervals, total cellular RNA was subjected to Northern blotting, and NP mRNA and antigenome were detected by using an NP gene-specific probe (Fig. 6A). Quantitative evaluation of the RNA signals revealed an increasing reduction in the level of mRNA relative to antigenome level up to a maximum of 60% after 24 h in treated versus nontreated cells (Fig. 6B and C). These data were confirmed by analysis of Lassa virus RNA levels 24 h postinfection. At all MOIs tested (0.01, 1, and 3), the level of mRNA was specifically reduced relative to the antigenome level by about 60% in α-amanitin-treated cells (Fig. 6A and C). In conclusion, depletion of host cell mRNAs by α-amanitin inhibits multiplication of Lassa virus and Mopeia virus but not that of VSV, known to express capping enzymes. The effect is associated with selective reduction of viral mRNA synthesis.
FIG. 6.
Effect of α-amanitin treatment on Lassa virus RNA synthesis. (A) Northern blot analysis. Vero cells were infected with Mopeia virus at an MOI of 3 (left) or with Lassa virus at different MOIs (right) and treated with 10 μg/ml α-amanitin or left untreated. Total RNA was prepared from Mopeia virus-infected cells at the indicated intervals and from Lassa virus-infected cells 24 h after infection and subjected to Northern blotting. Hybridization was performed with a 32P-labeled antisense probe of NP gene. The methylene blue-stained 28S rRNA is shown below the blot as a semiquantitative marker for gel loading and RNA transfer. (B) Representative intensity profiles from Northern blot lanes in panel A used for quantification of signals. (C) Quantitative evaluation of S RNA and NP mRNA steady-state levels. Signals on the Northern blot were quantified via intensity profiles as shown in panel B, and the NP mRNA/S RNA ratio was calculated for α-amanitin-treated and untreated cells. The NP mRNA/S RNA ratio in untreated cells was set at 100%. p.i., postinfection.
Determination of the fraction of capped viral RNA in cells treated with α-amanitin.
Based on the assumption that cellular cap structures are required for viral transcripts, we wondered if Lassa virus RNA produced in the presence of α-amanitin still contains cap structures, or if L protein starts producing a larger fraction of uncapped “mRNA.” Vero cells were infected with Lassa virus at a MOI of 1 and treated with 10 μg/ml α-amanitin. After 24 h, total cellular RNA was prepared and mRNA was immunoprecipitated by using an antibody against m3G-cap/m7G-cap structures. Precipitate and supernatant were subjected to Northern blotting, and virus RNA was detected by using an NP gene-specific probe (Fig. 7A). Analysis of an aliquot of the total RNA used as starting material revealed that the level of mRNA was specifically reduced relative to the antigenome level by about 65% in α-amanitin-treated cells. After two rounds of IP, about 85% of NP mRNA was precipitated from both treated and nontreated cells (Fig. 7B). Surprisingly, a fraction of antigenomic RNA and 28S rRNA was precipitated as well, which may be due to cross-reactivity of the antibody. Neither virus RNA nor 28S rRNA was precipitated if the antibody was omitted or an unrelated antibody was used for the procedure. These data indicate that depletion of cellular mRNA pool does not lead to preferential synthesis of uncapped “mRNA” by Lassa virus.
FIG. 7.
Effect of α-amanitin treatment on capping of Lassa virus mRNA. (A) IP of capped RNA. Vero cells were infected with Lassa virus at a MOI of 1 and treated with 10 μg/ml α-amanitin. After 24 h, total cellular RNA was prepared and capped RNA was immunoprecipitated in two successive rounds by using an antibody against m3G-cap/m7G-cap structures. An unrelated antibody (anti-NP) was used as a control. Precipitate and supernatant were subjected to Northern blotting, and virus RNA was detected by using an NP gene-specific probe. The methylene blue-stained 28S rRNA is shown below the blot as a semiquantitative marker for gel loading and RNA transfer. T, total RNA; P1, pellet after first round of IP; S1, supernatant after first round of IP; P2, pellet after second round of IP; S2, supernatant after second round of IP. (B) Cumulative fractions of NP mRNA and S RNA precipitated after the first and second rounds of IP. The fraction was calculated based on the signal intensities on the blot shown in panel A and the amount of material subjected to each round of IP.
DISCUSSION
This paper describes the identification of an N-terminal region in Lassa virus L protein being important for mRNA, but not antigenome, synthesis. Residues involved in this function are Asp-89, Glu-102, Asp-119, Lys-122, Asp-129, Glu-180, and Arg-185. Acidic side chains at positions 89, 102, and 129 are crucial for activity. Depletion of host cell mRNAs by α-amanitin inhibits multiplication of Lassa and Mopeia virus, and this effect is associated with selective reduction of viral mRNA synthesis.
The Lassa virus L protein plays a central role in replication and transcription of the viral genome. It is presumably a multidomain protein harboring several enzymatic functions. Structural and biochemical evidence for any of these functions is still missing. We have recently provided circumstantial evidence for the presence of an RdRp domain in the central part of the molecule by combining bioinformatics with large-scale mutagenesis (19, 47). In the present paper, we have extended the large-scale mutagenesis approach to the N terminus and C terminus of the L protein. The main targets for mutagenesis have been positively or negatively charged residues known to be part of catalytic sites of capping enzymes or nucleases (15, 27, 36, 51). About 75% of all alanine mutants reduced or abolished Ren-Luc expression. However, the vast majority of these mutations affected both transcription and replication. Expression analysis of completely defective mutants revealed no evidence for destabilization of the protein, an effect that has been reported for the N terminus of influenza PA (16). The replicon system is quite robust to variability in intracellular L protein levels. A 10-fold reduction in the amount of transfected L protein expression construct was still associated with high activity; only a 25-fold reduction was deleterious (see Fig. S3 in the supplemental material). Thus, the drop in steady-state level needed to render the replicon system inactive should be clearly visible in the Western blot, even if one considers that changes in protein stability may be masked to a certain extent by the high levels of expression obtained with the MVA-T7-based system.
Therefore, the mutations presumably affect enzymatic activities involved in both replication and transcription or interfere with inter- or intramolecular interactions or structural integrity important for the overall function of L protein. On the other hand, it is surprising that nearly 25% of evolutionarily highly conserved acidic and basic side chains are dispensable for function of L protein in the replicon context. The three K334A, K555A, and K581A mutants showed even higher activity than the wild type. Many residues with dispensable side chain such as Glu-41, Lys-334, Lys-512, Lys-555, Lys-581, Arg-719, Glu-728, and Glu-732 are located within highly conserved sequence stretches. The function and reason for conservation of these residues are not clear. They may play a role in processes not covered by the replicon system: e.g., primary transcription, packaging of RNPs into virions during assembly, or interaction with cellular proteins not relevant in an in vitro overexpression system.
The most interesting observation of this paper is the identification of seven residues in the N terminus of L protein, Asp-89, Glu-102, Asp-119, Lys-122, Asp-129, Glu-180, and Arg-185, which are specifically important for viral mRNA synthesis. The phenotype was most clear for Asp-89, Glu-102, and Asp-129, which are indispensable for transcription but can be replaced by a variety of amino acid residues without affecting viral genome replication. The finding that Asp-89 and Glu-102 cannot even be replaced by chemically closely related amino acid residues without a more than 1-log decrease in activity (as measured in luciferase units) suggests these residues are critically involved in catalysis or substrate binding. There are a number of possibilities for the precise role of the N-terminal part of L protein, the so-called conserved L1 domain (from position 1 to around position 300), in viral mRNA synthesis. The L1 domain may be a capping enzyme similar to that in VSV L protein (39), a cap binding site similar to that found in influenza virus PB2 (13), or an endonuclease similar to that in influenza virus PA (4, 50). Finally, it is also conceivable that it represents a 3′→5′ exonuclease, as found in severe acute respiratory syndrome coronavirus (SARS CoV) nsp14 (37), which is able to process RNA to final products of some nucleotides that could serve as primers for mRNA synthesis.
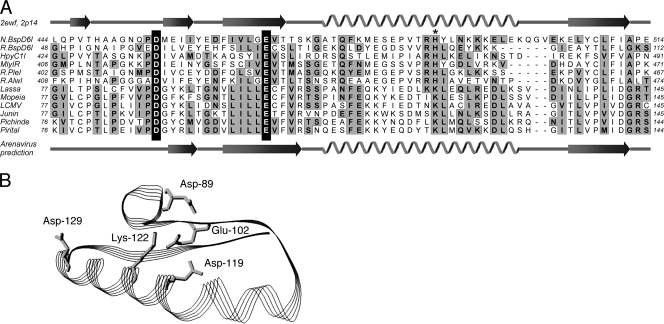
To substantiate any of these hypotheses, we employed a bioinformatics approach. The region encompassing residues 75 to 150 of the L1 domain was subjected to threading analysis using a range of public servers. (“Threading” means calculating the probability that the L1 domain may adopt the fold of existing protein structures.) One analysis, with mGenTHREADER (22), disclosed a remote “structural” similarity between the L1 fragment and the catalytic site of type IIs or nicking endonucleases, featuring a PD-E catalytic site motif (23) (Fig. 8A). Most interestingly, the key catalytic residues (Asp and Glu) of the endonucleases (20, 49) exactly correspond to Asp-89 and Glu-102 of the L1 domain, while a histidine assumed to be an additional catalytic residue (23) corresponds to Lys-122. The obvious similarity at the primary and secondary structure levels even facilitated three-dimensional modeling (Pdb Viewer 3.7) (12) of the L1 fragment using the structure of nicking endonuclease N.BspD6I (23) as a template (Fig. 8B). In that model, Asp-89, Glu-102, Asp-119, and Lys-122 are spatially close together, while Asp-129 is further apart. The former residues may form a catalytic site, which is well consistent with the mutagenesis data demonstrating their relevance for mRNA synthesis. Most recently, the structure of influenza virus endonuclease in the N terminus of PA has been solved (4, 50). This structure also resembles a type II endonuclease and shows a PD-E-K catalytic site motif. The folding model of the L1 fragment corresponds to the folding of the catalytic site of influenza virus endonuclease. In conclusion, bioinformatics supports the hypothesis that the L1 domain contains an endonuclease in analogy to the N terminus of influenza virus PA.
FIG. 8.
Similarity between L1 fragment of Lassa virus L protein (positions 77 to 145) and type IIs or nicking endonucleases. (A) Threading analysis using mGenTHREADER (22) identified structural similarity of the L1 fragment with structure 2ewf of the nicking endonuclease N.BspD6I (23). The alignment was extended to related type IIs and nicking endonucleases as well as further Old and New World arenaviruses. Secondary structure elements in 2ewf (N.BspD6I) and 2p14 (R.BspD6I) and the predicted secondary structure of arenavirus L protein are shown, respectively, above and below the alignment. Key catalytic residues of the endonucleases (Asp and Glu) (20, 23, 49) align with Asp-89 and Glu-102 of the L1 domain (highlighted in white on black background). An additional putative catalytic residue (His; marked with an asterisk) (23) aligns with Lys-122 of L protein. (B) Three-dimensional model of the L1 fragment shown in panel A using the structure of N.BspD6I (23) as a template. Modeling and ribbon drawing were performed with Pdb Viewer 3.7 (12). Side chains of residues relevant for mRNA but not antigenome synthesis are shown.
Further data are in line with this hypothesis. Arena-, orthomyxo-, and bunyaviruses are phylogenetically related (47) and share many features of the replication cycle. Like orthomyxo- and bunyaviruses, for which cap snatching has been shown (5, 24, 40), the 5′ end of arenavirus mRNA contains a stretch of nontemplated nucleotides (35, 42, 43) which may be derived from cellular mRNA. Replication of the Junin and Tacaribe arenaviruses is strongly inhibited by depleting the cellular mRNA pool with α-amanitin, while replication of VSV, which expresses capping enzymes, is not affected at all (33, 34, 39). We have shown that α-amanitin also inhibits replication of Lassa virus and Mopeia virus and that virus mRNA synthesis is particularly sensitive to treatment. The virus mRNA level was specifically reduced by 60% compared to the antigenome level, suggesting that downregulation of viral mRNA and gene expression is at least partially responsible for the 2-log reduction in virus titer. This is consistent with the hypothesis that cap structures of host cell mRNAs are a substrate for virus mRNA synthesis. However, it cannot be excluded that inhibition of Pol II-mediated transcription results in inhibition of host cell genes required for one or several activities associated with the function of the Lassa virus polymerase. We also tested if the fraction of viral mRNA (i.e., RNA species terminating in the intergenic region) containing cap structures decreases when the cap pool is depleted by α-amanitin. The IP experiments clearly show that the fractions of capped virus mRNA in treated and nontreated cells are identical (about 85%). Assuming a cap-snatching mechanism, this result suggests that transcription initiation via a capped primer is a prerequisite for mRNA synthesis.
In conclusion, this paper demonstrates that the L1 domain of Lassa virus contains residues that are critically involved in mRNA, but not antigenome, synthesis. Bioinformatics and cell biological experiments lend support to the hypothesis that these residues could be part of the catalytic site of an endonuclease. Structural and biochemical data are required to prove this hypothesis.
Supplementary Material
Acknowledgments
This work was supported by a grant from the “Vereinigung der Freunde des Tropeninstituts Hamburg e.V.” to M.L. and VIZIER integrated project grant LSHG-CT-2004-511960 of the European Union 6th Framework. The Bernhard-Nocht-Institute is supported by the Bundesministerium für Gesundheit and the Freie und Hansestadt Hamburg.
Footnotes
Published ahead of print on 9 December 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Auperin, D. D., V. Romanowski, M. Galinski, and D. H. Bishop. 1984. Sequencing studies of Pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. J. Virol. 52:897-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushnell, D. A., P. Cramer, and R. D. Kornberg. 2002. Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc. Natl. Acad. Sci. U. S. A. 99:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dias, A., D. Bouvier, T. Crepin, A. A. McCarthy, D. J. Hart, F. Baudin, S. Cusack, and R. W. Ruigrok. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914-918. [DOI] [PubMed] [Google Scholar]
- 5.Duijsings, D., R. Kormelink, and R. Goldbach. 2001. In vivo analysis of the TSWV cap-snatching mechanism: single base complementarity and primer length requirements. EMBO J. 20:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fechter, P., L. Mingay, J. Sharps, A. Chambers, E. Fodor, and G. G. Brownlee. 2003. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J. Biol. Chem. 278:20381-20388. [DOI] [PubMed] [Google Scholar]
- 7.Fisher-Hoch, S. P., O. Tomori, A. Nasidi, G. I. Perez-Oronoz, Y. Fakile, L. Hutwagner, and J. B. McCormick. 1995. Review of cases of nosocomial Lassa fever in Nigeria: the high price of poor medical practice. BMJ 311:857-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller-Pace, F. V., and P. J. Southern. 1989. Detection of virus-specific RNA-dependent RNA polymerase activity in extracts from cells infected with lymphocytic choriomeningitis virus: in vitro synthesis of full-length viral RNA species. J. Virol. 63:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuichi, Y., and A. J. Shatkin. 2000. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55:135-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcin, D., and D. Kolakofsky. 1990. A novel mechanism for the initiation of Tacaribe arenavirus genome replication. J. Virol. 64:6196-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcin, D., and D. Kolakofsky. 1992. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J. Virol. 66:1370-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 13.Guilligay, D., F. Tarendeau, P. Resa-Infante, R. Coloma, T. Crepin, P. Sehr, J. Lewis, R. W. Ruigrok, J. Ortin, D. J. Hart, and S. Cusack. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15:500-506. [DOI] [PubMed] [Google Scholar]
- 14.Günther, S., P. Emmerich, T. Laue, O. Kühle, M. Asper, A. Jung, T. Grewing, J. ter Meulen, and H. Schmitz. 2000. Imported lassa fever in Germany: molecular characterization of a new lassa virus strain. Emerg. Infect. Dis. 6:466-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hakansson, K., A. J. Doherty, S. Shuman, and D. B. Wigley. 1997. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell 89:545-553. [DOI] [PubMed] [Google Scholar]
- 16.Hara, K., F. I. Schmidt, M. Crow, and G. G. Brownlee. 2006. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Virol. 80:7789-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hass, M., U. Gölnitz, S. Müller, B. Becker-Ziaja, and S. Günther. 2004. Replicon system for Lassa virus. J. Virol. 78:13793-13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hass, M., M. Westerkofsky, S. Müller, B. Becker-Ziaja, C. Busch, and S. Günther. 2006. Mutational analysis of the Lassa virus promoter. J. Virol. 80:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hass, M., M. Lelke, C. Busch, B. Becker-Ziaja, and S. Günther. 2008. Mutational evidence for a structural model of the Lassa virus RNA polymerase domain and identification of two residues, Gly1394 and Asp1395, that are critical for transcription but not replication of the genome. J. Virol. 82:10207-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins, L. S., C. Besnier, and H. Kong. 2001. The nicking endonuclease N.BstNBI is closely related to type IIs restriction endonucleases MlyI and PleI. Nucleic Acids Res. 29:2492-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hufert, F. T., W. Ludke, and H. Schmitz. 1989. Epitope mapping of the Lassa virus nucleoprotein using monoclonal anti-nucleocapsid antibodies. Arch. Virol. 106:201-212. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. T. 1999. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 287:797-815. [DOI] [PubMed] [Google Scholar]
- 23.Kachalova, G. S., E. A. Rogulin, A. K. Yunusova, R. I. Artyukh, T. A. Perevyazova, N. I. Matvienko, L. A. Zheleznaya, and H. D. Bartunik. 2008. Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit. J. Mol. Biol. 384:489-502. [DOI] [PubMed] [Google Scholar]
- 24.Krug, R. M. 1981. Priming of influenza viral RNA transcription by capped heterologous RNAs. Curr. Top. Microbiol. Immunol. 93:125-149. [DOI] [PubMed] [Google Scholar]
- 25.Lecompte, E., E. Fichet-Calvet, S. Daffis, K. Koulemou, O. Sylla, F. Kourouma, A. Dore, B. Soropogui, V. Aniskin, B. Allali, S. Kouassi Kan, A. Lalis, L. Koivogui, S. Günther, C. Denys, and J. ter Meulen. 2006. Mastomys natalensis and Lassa fever, West Africa. Emerg. Infect. Dis. 12:1971-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, K. J., I. S. Novella, M. N. Teng, M. B. Oldstone, and J. C. de La Torre. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74:3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lima, C. D., L. K. Wang, and S. Shuman. 1999. Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell 99:533-543. [DOI] [PubMed] [Google Scholar]
- 28.Lopez, N., R. Jacamo, and M. T. Franze-Fernandez. 2001. Transcription and RNA replication of Tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 75:12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukashevich, I. S., M. Djavani, K. Shapiro, A. Sanchez, E. Ravkov, S. T. Nichol, and M. S. Salvato. 1997. The Lassa fever virus L gene: nucleotide sequence, comparison, and precipitation of a predicted 250 kDa protein with monospecific antiserum. J. Gen. Virol. 78:547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mark, G. E., J. M. Taylor, B. Broni, and R. M. Krug. 1979. Nuclear accumulation of influenza viral RNA transcripts and the effects of cycloheximide, actinomycin D, and alpha-amanitin. J. Virol. 29:744-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCormick, J. B., I. J. King, P. A. Webb, C. L. Scribner, R. B. Craven, K. M. Johnson, L. H. Elliott, and R. Belmont-Williams. 1986. Lassa fever. Effective therapy with ribavirin. N. Engl. J. Med. 314:20-26. [DOI] [PubMed] [Google Scholar]
- 32.McCormick, J. B., I. J. King, P. A. Webb, K. M. Johnson, R. O'Sullivan, E. S. Smith, S. Trippel, and T. C. Tong. 1987. A case-control study of the clinical diagnosis and course of Lassa fever. J. Infect. Dis. 155:445-455. [DOI] [PubMed] [Google Scholar]
- 33.Mersich, S. E., M. E. Leon, and C. E. Coto. 1979. Cell nucleus participation in the multiplication of the arenavirus Tacaribe. FEMS Microbiol. Lett. 6:205-207. [Google Scholar]
- 34.Mersich, S. E., E. B. Damonte, and C. E. Coto. 1981. Induction of RNA polymerase II activity in Junin virus-infected cells. Intervirology 16:123-127. [DOI] [PubMed] [Google Scholar]
- 35.Meyer, B. J., and P. J. Southern. 1993. Concurrent sequence analysis of 5′ and 3′ RNA termini by intramolecular circularization reveals 5′ nontemplated bases and 3′ terminal heterogeneity for lymphocytic choriomeningitis virus mRNAs. J. Virol. 67:2621-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mian, I. S. 1997. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 25:3187-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minskaia, E., T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard, and J. Ziebuhr. 2006. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 103:5108-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monath, T. P., V. F. Newhouse, G. E. Kemp, H. W. Setzer, and A. Cacciapuoti. 1974. Lassa virus isolation from Mastomys natalensis rodents during an epidemic in Sierra Leone. Science 185:263-265. [DOI] [PubMed] [Google Scholar]
- 39.Ogino, T., and A. K. Banerjee. 2007. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 25:85-97. [DOI] [PubMed] [Google Scholar]
- 40.Patterson, J. L., B. Holloway, and D. Kolakofsky. 1984. La Crosse virions contain a primer-stimulated RNA polymerase and a methylated cap-dependent endonuclease. J. Virol. 52:215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. U. S. A. 100:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polyak, S. J., S. Zheng, and D. G. Harnish. 1995. 5′ termini of Pichinde arenavirus S RNAs and mRNAs contain nontemplated nucleotides. J. Virol. 69:3211-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raju, R., L. Raju, D. Hacker, D. Garcin, R. Compans, and D. Kolakofsky. 1990. Nontemplated bases at the 5′ ends of Tacaribe virus mRNAs. Virology 174:53-59. [DOI] [PubMed] [Google Scholar]
- 44.Singh, M. K., F. V. Fuller-Pace, M. J. Buchmeier, and P. J. Southern. 1987. Analysis of the genomic L RNA segment from lymphocytic choriomeningitis virus. Virology 161:448-456. [DOI] [PubMed] [Google Scholar]
- 45.Strecker, T., R. Eichler, J. Meulen, W. Weissenhorn, H. D. Klenk, W. Garten, and O. Lenz. 2003. Lassa virus Z protein is a matrix protein sufficient for the release of virus-like particles. J. Virol. 77:10700-10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutter, G., M. Ohlmann, and V. Erfle. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9-12. [DOI] [PubMed] [Google Scholar]
- 47.Vieth, S., A. E. Torda, M. Asper, H. Schmitz, and S. Günther. 2004. Sequence analysis of L RNA of Lassa virus. Virology 318:153-168. [DOI] [PubMed] [Google Scholar]
- 48.Wulff, H., B. M. McIntosh, D. B. Hamner, and K. M. Johnson. 1977. Isolation of an arenavirus closely related to Lassa virus from Mastomys natalensis in south-east Africa. Bull. W. H. O. 55:441-444. [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, Y., K. D. Lunnen, and H. Kong. 2001. Engineering a nicking endonuclease N.AlwI by domain swapping. Proc. Natl. Acad. Sci. U. S. A. 98:12990-12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan, P., M. Bartlam, Z. Lou, S. Chen, J. Zhou, X. He, Z. Lv, R. Ge, X. Li, T. Deng, E. Fodor, Z. Rao, and Y. Liu. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909-913. [DOI] [PubMed] [Google Scholar]
- 51.Zuo, Y., and M. P. Deutscher. 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.