Abstract
Pre-mRNAs of the influenza A virus M and NS genes are poorly spliced in virus-infected cells. By contrast, in influenza C virus-infected cells, the predominant transcript from the M gene is spliced mRNA. The present study was performed to investigate the mechanism by which influenza C virus M gene-specific mRNA (M mRNA) is readily spliced. The ratio of M1 encoded by a spliced M mRNA to CM2 encoded by an unspliced M mRNA in influenza C virus-infected cells was about 10 times larger than that in M gene-transfected cells, suggesting that a viral protein(s) other than M gene translational products facilitates viral mRNA splicing. RNase protection assays showed that the splicing of M mRNA in infected cells was much higher than that in M gene-transfected cells. The unspliced and spliced mRNAs of the influenza C virus NS gene encode two nonstructural (NS) proteins, NS1(C/NS1) and NS2(C/NS2), respectively. The introduction of premature translational termination into the NS gene, which blocked the synthesis of the C/NS1 and C/NS2 proteins, drastically reduced the splicing of NS mRNA, raising the possibility that C/NS1 or C/NS2 enhances viral mRNA splicing. The splicing of influenza C virus M mRNA was increased by coexpression of C/NS1, whereas it was reduced by coexpression of the influenza A virus NS1 protein (A/NS1). The splicing of influenza A virus M mRNA was also increased by coexpression of C/NS1, though it was inhibited by that of A/NS1. These results suggest that influenza C virus NS1, but not A/NS1, can upregulate viral mRNA splicing.
The influenza C virus genome consists of seven single-stranded RNA segments of negative polarity. RNA segment 6 (M gene) of C/Yamagata/1/88 is 1,181 nucleotides in length and has a single open reading frame (positions 27 to 1148) capable of encoding a polypeptide of 374 amino acids with a predicted Mr of 42,000 (9, 35). However, the predominant mRNA transcript of this RNA segment lacks nucleotides 755 to 982 and encodes a 242-amino-acid matrix (M1) protein with an Mr of 27,000. Elimination of the intron results in the introduction of a termination codon (consisting of nucleotides 753, 754, and 983) after amino acid residue 242 (9, 35). Unspliced mRNA from RNA segment 6 is synthesized in infected cells, though at very low levels (13% of spliced mRNA) (9). This mRNA species is capable of coding for a 374-amino-acid protein (P42) containing an additional 132 amino acids from the C terminus of M1 (11), which is cleaved by signal peptidase to generate CM2, composed of the C-terminal 115 amino acids, in addition to the M1′ protein, composed of the N-terminal 259 amino acids (12, 26). The CM2 protein forms a voltage-activated ion channel permeable to chloride ions (13). Recently, it has been suggested that CM2 also has pH-modulating activity (2).
In influenza A virus-infected cells, the colinear transcript from segment 7 encodes M1, and the M1 mRNA also undergoes alternative splicing to generate spliced M2 mRNA and mRNA3. The colinear transcript and spliced mRNA from influenza A virus segment 8 are translated into NS1 and NS2 (NEP), respectively. It has been reported that the steady-state level of spliced viral transcripts is only 10% of that of unspliced viral transcripts in influenza A virus-infected cells (15, 16). The inefficient splicing of viral pre-mRNAs can be understood partly by the fact that influenza A virus NS1 protein is associated with spliceosomes and inhibits pre-mRNA splicing (6, 7, 17). cis-acting sequences in the NS1 transcript also negatively regulates splicing (22). The splicing of influenza A virus M1 mRNA is controlled by the rate of nuclear export (34). The alternative splicing of influenza A virus M1 mRNA is regulated by the binding of the viral polymerase complex and cellular splicing factor SF2/ASF (28, 29). In the case of influenza C virus, however, the mechanism by which influenza C virus M mRNA is efficiently spliced has not yet been clarified.
Influenza C virus RNA segment 7 (NS gene) encodes two nonstructural proteins, the 246-amino-acid NS1 protein (C/NS1), encoded by a colinear mRNA transcript of the gene, and the 182-amino-acid NS2 protein (C/NS2), encoded by a spliced mRNA (1, 8, 18) (see Fig. 5). Using transfected cells, Paragas et al. (25) have demonstrated that C/NS2 possesses nuclear export activity. We recently demonstrated that C/NS2 plays a role in the nuclear export of vRNP in influenza C virus-infected cells, and that C/NS2 is incorporated into virions, where it associates with vRNP inside the viral envelope (14). However, the role of C/NS1 in the viral life cycle remains to be determined.
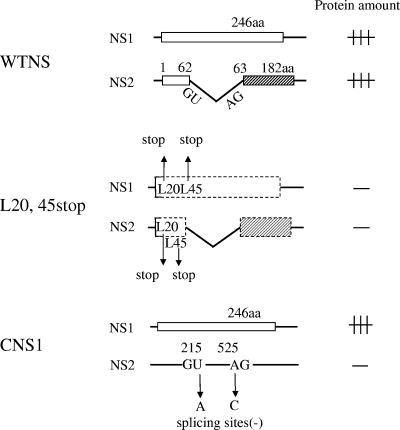
FIG. 5.
Schematic representation of the NS gene construct. Boxes show open reading frames. (WTNS construct) A 246-amino-acid (aa) NS1 protein encoded by a colinear mRNA transcript of influenza C virus RNA segment 7 (NS gene) and a 182-amino-acid NS2 protein encoded by a spliced mRNA are expressed in cells transfected with pME18S-WTNS. (L20,45stop construct) Premature termination codons were introduced into Leu residues 20 and 45 of the N-terminal 62-amino-acid region, which is common to both the NS1 and NS2 coding regions, to eliminate the expression of both the NS1 and NS2 proteins. (CNS1 construct) Plasmid pME18S-CNS1, which expressed NS1 alone, was constructed by substituting the donor (214-GU-215) and acceptor (525-AG-526) for splicing into 214-GA-215 and 525-CG-526, respectively, thereby resulting in the abolition of transcription to the NS2 mRNA.
In this study, we investigated the mechanism by which influenza C virus M gene-specific mRNA is efficiently spliced. The results indicate that influenza C virus NS1 protein can facilitate the splicing of viral mRNAs.
MATERIALS AND METHODS
Viruses and cells.
The Yamagata/1/88 strain of influenza C virus was grown in the amniotic cavities of 9-day-old embryonated hen's eggs (36). COS-1, CV-1, and 293T cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum.
Plasmid construction.
The plasmid pCNS-5′3′F, which contains a full-length cDNA of the influenza C virus NS gene, was constructed as follows. The DNA fragment corresponding to positions 1 to 935 (mRNA sense) of RNA segment 7 of Yamagata/1/88 was prepared by PCR utilizing plasmid pCN5-8 (8), containing nucleotides 5 to 923 of the Yamagata/1/88 virus NS gene, as a template and two primers, a plus-sense primer (5′-dGGCTCGAGAGCAGAAGCAGGGGTACTTTTCCAAAATGTC) containing an XhoI site (underlined) followed by sequence corresponding to positions 1 to 31 and a minus-sense primer (5′-dGGTCTAGAAGCAGGAGCAAGGGGTTTTTTAACTTTGGAATAATAACTTA) containing an XbaI site (underlined) followed by sequence corresponding to positions 935 to 895. The PCR product was digested with XhoI and XbaI and then cloned into the XhoI and XbaI sites of pBluescript II KS (+) (Stratagene) to generate pCNS-5′3′F. pCNS-5′3′F was then digested with XhoI and XbaI and subcloned into the XhoI and XbaI sites of the transient expression vector pME18S (32) to generate pME18S-WTNS, which can express the NS1 and NS2 proteins. To introduce a premature termination of translation into the NS gene, Leu residues at positions 20 and 45 were substituted into stop codons by PCR using pCNS-5′3′F as a template and mutated primers (sequences are available on request), and the resulting PCR product was digested with XhoI and XbaI and then subcloned into the XhoI and XbaI sites of pME18S to generate pME18S-L20,45stop. Plasmid pME18S-CNS1, which expressed C/NS1 alone, was constructed by substituting the donor (214-GU-215) and acceptor (525-AG-526) sites for splicing into 214-GA-215 and 525-CG-526, respectively, thereby resulting in the abolition of transcription to the C/NS2 mRNA. pT3/PR8-M containing cDNA of the M gene of the influenza A virus PR/8/34 strain, a generous gift from M. Enami, was digested with EcoRI and XbaI, and the resulting fragment was subcloned into the EcoRI and XbaI sites of pME18S to generate pME18S-AM for the analysis of influenza A virus M gene-derived mRNA. pT3/PR8-NS1 containing the NS1 coding region of the influenza A virus PR/8/34 strain (5), also provided by M. Enami, was digested with EcoRI and XbaI, and the resulting fragment was subcloned into the EcoRI and XbaI sites of pME18S to generate pME18S-ANS1 for the expression of the NS1 protein of influenza virus strain A/PR/8/34. The nucleotide sequences of all mutant cDNAs were confirmed by sequencing.
RNA purification and RNase protection assay.
Total RNA was extracted from both infected and transfected cells using the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. An RNase protection assay was performed by using RNase protection assay kit RPA III (Ambion) as described previously (9), with some modifications. Briefly, a 33P-labeled influenza C virus RNA 6-specific RNA probe (vRNA sense) was synthesized by in vitro transcription of pCM5-3′F (9) using a T3 promoter and hybridized with total RNA from infected cells or cells transfected with pME18S-CM, which contained a full-length copy of the influenza C virus M gene (11), at 42°C overnight in 40 mM PIPES buffer containing 0.4 M sodium acetate, 1 mM EDTA, and 80% formamide. Hybrids were digested with RNase A (0.08 U) and RNase T1 (3 U) at 37°C for 30 min and then analyzed on a 4% polyacrylamide gel containing 4 M urea. A 33P-labeled influenza C virus NS gene-specific RNA probe (vRNA sense) was synthesized by in vitro transcription of pCNS-5′3′F using a T7 promoter for the analysis of influenza C virus NS mRNA, and an influenza A virus M gene-specific probe (vRNA sense) was synthesized using pT3/PR8-M and a T3 promoter for influenza A virus M mRNA analysis.
Antibodies.
Mouse monoclonal antibody (MAb) L2 against matrix protein M1 (31) has been described previously. Anti-glutathione S-transferase (GST)/CM2 rabbit serum was produced as described previously (9). Anti-GST/NS2, rabbit antiserum against the C-terminal region (residues 63 to 182) of C/NS2, and anti-GST/NS1, rabbit antiserum against the C-terminal region (residues 225 to 246) of C/NS1, were also produced as described previously (1). Primer sequences for the GST fusion proteins are available on request.
Metabolic labeling and immunoprecipitation of transfected and infected cells.
Subconfluent monolayers of COS-1 cells in 3.5-cm petri dishes were transfected with either pME18S-CM (11) or pME18S-WTNS (1 μg/plate) using the Lipofectamine procedure and then incubated at 37°C. At 48 h posttransfection (p.t.), cells were labeled with 50 μCi/ml [35S]methionine (ARC) for 1 h in methionine-deficient Dulbecco's modified Eagle's medium. COS-1 cells infected with the C/Yamagata/1/88 virus at a multiplicity of infection (MOI) of approximately 10 PFU/cell were labeled with [35S]methionine (50 μCi/ml; ARC) for 30 min in methionine-deficient Dulbecco's modified Eagle's medium at 26 h postinfection (p.i.). The transfected and infected cells were then disrupted in 0.01 M Tris-HCl (pH 7.4) containing 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 0.15 M NaCl, and a cocktail of protease inhibitors (10) and immunoprecipitated as described previously (30) utilizing anti-GST/CM2 serum and anti-M1 MAb (L2) or anti-GST/NS2 and anti-GST/NS1, respectively. The immunoprecipitates obtained were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 17.5% gels containing 4 M urea under reducing conditions and then processed for analysis by fluorography (36).
Indirect immunofluorescent staining.
HMV-II cells infected with C/Yamagata/1/88 and COS-1 cells transfected with pME18S-CNS1 were fixed with carbon tetrachloride at various times after infection and transfection, respectively. The cells were then stained by an indirect method using anti-GST/NS1 serum as the primary antibody and fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Seikagaku Kogyo) as the secondary antibody. Photomicroscopy was performed on an Olympus VANOX microscope.
Statistical analysis.
Values were expressed as means ± standard deviations of n independent determinations. Comparison between groups of experimental data was performed using Student's t test or one-way analysis of variance, as appropriate, with an SPSS software package (v. 17).
RESULTS
The splicing efficiency of influenza C virus M gene-specific mRNA (M mRNA) in infected cells was higher than that in M gene-transfected cells.
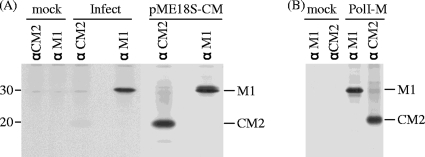
We previously showed that, in influenza C virus-infected cells, the predominant mRNA derived from RNA segment 6 (M gene) is spliced mRNA (9). To examine whether M mRNA is intrinsically spliced at a higher rate in cells or whether the splicing rate is enhanced by a viral protein(s), we first analyzed M gene-transfected cells using radioimmunoprecipitation (RIP). COS-1 cells either infected with C/Yamagata/1/88 or transfected with the transient expression vector pME18S, containing full-length cDNA of the influenza C virus M gene (pME18S-CM), were labeled with [35S]methionine for 60 min at 26 h p.i. or 48 h p.t., and cell lysates were immunoprecipitated with anti-GST/CM2 serum or anti M1 MAb (L2). The resulting immunoprecipitates were analyzed by SDS-PAGE, followed by fluorography (Fig. 1A). Densitometric analysis showed that the ratio of the immunoprecipitated M1 derived from spliced M mRNA to CM2 derived from unspliced M mRNA in influenza C virus-infected cells was about 10 times higher than that in M gene-expressing cells. Furthermore, to confirm these results, we also performed RIP experiments using another cell line, CV-1, infected with either C/Yamagata/1/88 or simian virus 40 recombinant virus containing M gene cDNA, which expressed the M gene alone. The ratio of M1 to CM2 synthesized in the influenza C virus-infected cells was also 10 times higher than that in the M gene-expressing cells (data not shown). Taken together, these results suggest that some viral protein(s) other than the M gene product possesses the ability to enhance the splicing efficiency of mRNA derived from the influenza C virus M gene. To rule out the possibility that the differences in the splicing efficiency of viral transcripts between infected and transfected cells resulted from differences between the RNA polymerase II-driven transcript and authentic viral transcript, M gene expression in transfected cells, in which viral transcripts were generated by using RNA polymerase I-driven vector containing influenza C virus M gene cDNA, was analyzed by RIP as described above (Fig. 1B). The ratio of M1 to CM2 in the cells transfected with the RNA polymerase I-driven vector set, in which authentic viral M mRNAs were transcribed, was similar to that in the cells transfected with pME18S-CM. Therefore, the above-mentioned possibility could be ruled out. As the transfection of many kinds of plasmids may reduce the efficiency of cotransfection in the same cell, pME18S-CM expressing the RNA polymerase II-driven transcript was used in the subsequent experiments.
FIG. 1.
M gene translational products synthesized in infected and M gene-transfected cells. (A) COS-1 cells mock infected (lane mock), infected with C/Yamagata/1/88 (lane Infect) at an MOI of 10 PFU/cell, or transfected with the transient expression vector pME18S, containing the full-length cDNA of the influenza C virus M gene (lane pME18S-CM), were labeled with [35S]methionine for 60 min at 26 h p.i. or at 48 h p.t., and cell lysates were immunoprecipitated with anti-GST/CM2 serum (lane αCM2) or anti M1 MAb (L2) (lane αM1). The resulting immunoprecipitates were analyzed by SDS-PAGE on 17.5% gels containing 4 M urea under reducing conditions, followed by fluorography. The positions of molecular weight markers are shown to the left. (B) 293T cells mock transfected (lane mock) or transfected with an RNA polymerase I-driven vector containing influenza C virus M gene cDNA, pPolI/M, and plasmids expressing polymerases (PB2, PB1, and P3) and NP, pcDNA/PB2-AA, pcDNA/PB1-AA, pcDNA/P3-AA, and pCAGGS.MCS/NP-AA, respectively (20) (lane PolI-M), were labeled with [35S]methionine for 60 min at 48 h p.t., and cell lysates were immunoprecipitated with anti-GST/CM2 serum (lane αCM2) or anti M1 MAb (L2) (lane αM1).
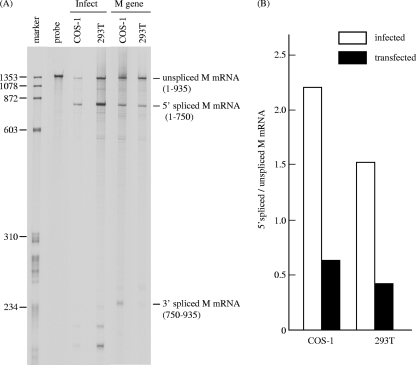
To further verify the possibility that a viral protein other than the M gene translational product facilitates the splicing of M mRNA, the splicing efficiency of M mRNA in COS-1 cells either infected with C/Yamagata/1/88 or transfected with pME18S-CM was investigated by RNase protection assay using an antisense M gene-specific RNA probe as described previously (9) (Fig. 2). We have previously demonstrated that RNase protection assays with total RNA and poly(A)+ RNA yielded results virtually identical to each other, indicating that the majority of the RNA 6-specific transcripts present in influenza C virus-infected cells are polyadenylated and suggesting that the amount of full-length template RNA is very small compared with that of unspliced mRNA (9). Therefore, total RNA extracted from the cells was utilized for RNase protection assays in this study. As shown in Fig. 2A, the spliced mRNA of the M gene (spliced M mRNA) was predominant in infected COS-1 cells, whereas the major mRNAs of the M gene in the M gene-transfected cells were unspliced. Densitometric analysis showed that the ratio of 5′ spliced M mRNA to unspliced M mRNA was about four times larger in infected cells (2.20) than in transfected cells (0.63) (Fig. 2B). This result also suggests that a viral protein other than the translational product of the M gene facilitates the splicing of M mRNA. To further examine this, another cell line, 293T, was either infected with C/Yamagata/1/88 or transfected with pME18S-CM and then analyzed by RNase protection assay. The obtained data demonstrated that the splicing efficiency of M mRNA in infected cells (1.52) was also about four times higher than that in cells expressing the M gene (0.42). These data strongly suggest that a viral protein other than the M gene translational products possesses the ability to increase the splicing efficiency of M mRNA.
FIG. 2.
RNase protection assay of influenza C virus M gene-derived mRNAs in infected and M gene-transfected cells. (A) Total RNA was extracted from COS-1 and 293T cells infected with C/Yamagata/1/88 at an MOI of 10 PFU/cell at 24 h p.i. (lane Infect) and from cells transfected with pME18S-CM at 48 h p.t. (lane M gene) and analyzed by RNase protection assay using a 33P-labeled influenza C virus RNA segment 6 (M gene)-specific RNA probe (vRNA sense). The protected fragments were separated on a 4% polyacrylamide gel containing 4 M urea. The HaeIII digest of ψX174 DNA was 5′ end labeled and used as a size marker (marker lane). The probe lane shows an undigested probe. (B) The ratios of 5′ spliced M mRNA to unspliced M mRNA in infected (open box) and transfected (closed box) COS-1 and 293T cells were calculated after densitometry of panel A.
Kinetics of M mRNA splicing in infected cells.
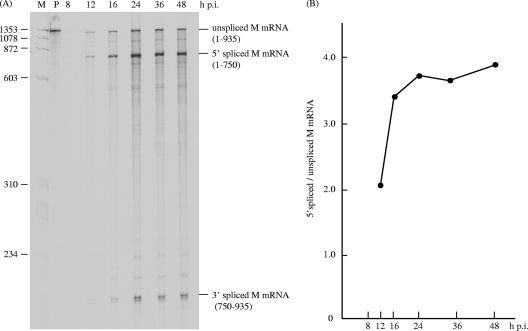
If the hypothesis described above is indeed correct, the splicing of M mRNA will increase as the expression of viral proteins increases. To determine the kinetics of M mRNA splicing, total RNA was extracted from C/Yamagata/1/88 virus-infected cells at various times of infection and processed for RNase protection assay using an antisense M RNA probe (Fig. 3A). As shown in Fig. 3B, the ratio of 5′ spliced M mRNA to unspliced M mRNA was 2.03 at 12 h p.i., increasing to 3.37 at 16 h p.i. and reaching a maximum of 3.71 at 24 h p.i. This finding suggests that the accumulation of some viral proteins in the late phase of infection increased the splicing efficiency of M mRNA.
FIG. 3.
Kinetics of M mRNA splicing in influenza C virus-infected cells. (A) RNA was extracted from C/Yamagata/1/88 virus-infected cells at various times postinfection and processed for RNase protection assay using an antisense M gene-specific RNA probe. Lanes M and P show the 5′ end-labeled HaeIII digest of ψX174 DNA as a size marker and an undigested probe, respectively. (B) The ratios of 5′ spliced M mRNA to unspliced M mRNA in each lane of panel A are shown by closed circles.
The influenza C virus NS gene translational product may upregulate the splicing of viral mRNAs.
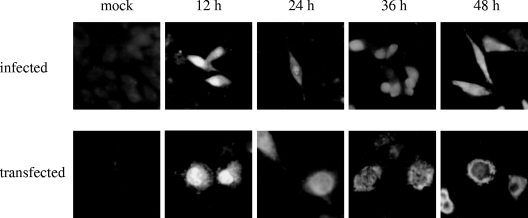
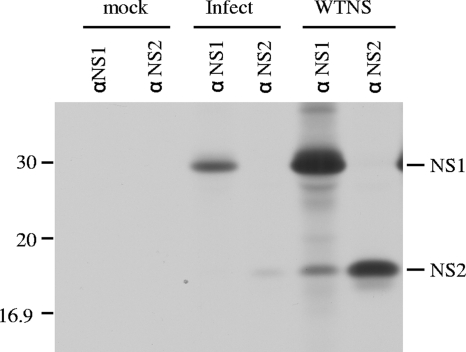
Marschall et al. (18) demonstrated that influenza C virus NS1 protein is distributed in the nucleus and cytoplasm of MDCK cells infected with persistent virus variant C/AA-pi. To further examine the intracellular localization of the NS1 protein, HMV-II cells infected with C/Yamagata/1/88 were fixed at various time points after infection and analyzed by immunofluorescent staining using anti-GST/NS1 serum (Fig. 4, upper panel). We also observed that NS1 localized in the nucleus in the early phase of infection and was distributed in both the nucleus and cytoplasm in the late phase of infection. These data raise the possibility that influenza C virus NS1 protein plays a role in viral mRNA splicing that occurs in the nucleus. To examine whether NS1 is involved in the splicing of mRNA derived from the NS gene (NS mRNA), influenza C virus NS gene cDNA was subcloned into a transient expression vector, pME18S (designated pME18S-WTNS) (Fig. 5), and the NS gene translational products, the NS1 and NS2 proteins, were overexpressed in COS-1 cells transfected with pME18S-WTNS. The cells were labeled with [35S]methionine for 30 min at 48 h p.t. and immunoprecipitated with either anti-GST/NS1 or anti-GST/NS2 serum. The resulting immunoprecipitates in two independent experiments were analyzed in comparison with those from influenza C virus-infected cells labeled at 15 h p.i., and a representative result is shown in Fig. 6. The ratio of NS2 protein encoded by spliced NS mRNA to NS1 protein derived from unspliced NS mRNA was 2.7 times larger in transfected COS-1 cells overexpressing the NS gene products than that in infected cells. These data suggest that overexpression of influenza C virus NS1 and/or NS2 protein may enhance the splicing efficiency of its own mRNA (i.e., NS mRNA).
FIG. 4.
Indirect immunofluorescent staining of infected and transfected cells. HMV-II cells mock infected (mock) or infected with C/Yamagata/1/88 (upper panel) and COS-1 cells mock transfected (mock) or transfected with pME18S-CNS1 (lower panel) were fixed with carbon tetrachloride at 12, 24, 36, and 48 h after infection or transfection, respectively. The cells were then stained by an indirect method using anti-GST/NS1 serum.
FIG. 6.
NS1 and NS2 synthesis in influenza C virus-infected and NS gene-transfected cells. COS-1 cells mock infected (lane mock), infected with C/Yamagata/1/88 (Infect lanes) at an MOI of 10 PFU/cell, or transfected with pME18S-WTNS (WTNS lanes) were labeled with [35S]methionine and immunoprecipitated with anti-GST/NS1 serum (αNS1 lanes) or anti-GST/NS2 serum (αNS2 lanes). The resulting immunoprecipitates were analyzed by SDS-PAGE, followed by fluorography.
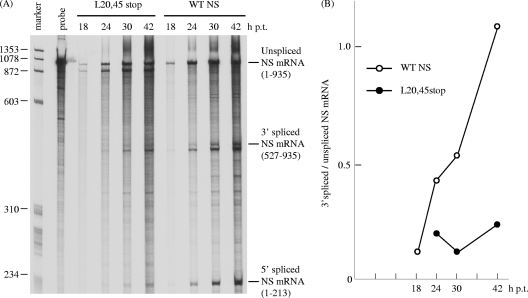
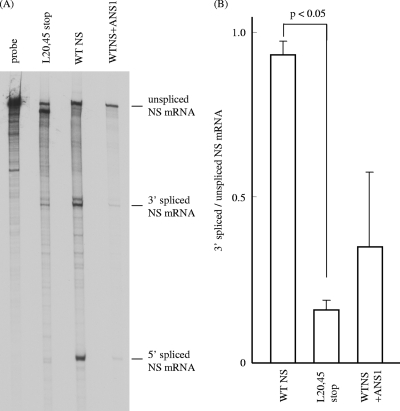
If this is the case, splicing of NS mRNA will increase as NS1 or NS2 accumulates in the later phase of infection. To examine this, COS-1 cells were transfected with pME18S-WTNS and the unspliced and spliced NS mRNAs were quantitated at various times posttransfection by RNase protection assay using an antisense NS RNA probe. The splicing rate of NS mRNA increased with time after transfection (Fig. 7A, lane WTNS). Densitometric analysis showed that the ratio of 3′ spliced NS mRNA to unspliced NS mRNA at 42 h p.t. (1.089) was about 10 times higher than that at 18 h p.t. (0.117) (Fig. 7B). This result indicates that a translational product of the NS gene, NS1 or NS2, may facilitate the splicing of NS mRNA. To confirm this, we introduced premature termination codons into Leu residues 20 and 45 of the N-terminal 62-amino-acid region, which is common to both the NS1 and NS2 coding regions (L20,45stop), to eliminate the expression of both the NS1 and NS2 proteins (Fig. 5). pME18S containing NS cDNA with the premature termination codons (pME18S-L20,45stop) was introduced into COS-1 cells, and the splicing rate of NS mRNA therein was examined as described above (Fig. 7A, lane L20,45stop). First we confirmed that neither NS1 nor NS2 was detected by RIP and immunofluorescent staining (data not shown). The amount of spliced NS mRNA was drastically reduced in cells transfected with pME18S-L20,45stop at all times after transfection compared to that in pME18S-WTNS-transfected cells, with the splicing rate of NS mRNA in pME18S-L20,45stop-transfected cells at 42 h p.t. (0.229) only 21% of that in WTNS-expressing cells (1.089) (Fig. 7B). This result strongly suggests that the NS gene translational products are required for the efficient splicing of NS mRNA. To further test this possibility, cytoplasmic RNA was isolated from WTNS-expressing cells and pME18S-L20,45stop-transfected cells according to the instructions accompanying the RNeasy Mini kit (Qiagen) and subjected to RNase protection assay. Representative data from two independent experiments are shown in Fig. 8A, and the splicing ratio of NS mRNA in the cells transfected with each NS construct in the two experiments was compared (Fig. 8B). The ratio of 3′ spliced to unspliced NS mRNA in pME18S-L20,45stop-transfected cells at 48 h p.t. (mean ± standard deviation = 0.160 ± 0.015) was significantly lower than that in WTNS-expressing cells (0.932 ± 0.045) (P < 0.05). This result also supports the idea that the NS gene translational products are required for the efficient splicing of NS mRNA. RNase protection assay with the cytoplasmic RNA from COS-1 cells cotransfected with pME18S-WTNS and pME18S-ANS1 expressing the NS1 protein of the influenza A virus PR/8/34 strain demonstrated that the splicing rate of influenza C virus NS mRNA (0.349 ± 0.240) was drastically reduced by 63% (Fig. 8, lane WTNS+ANS1) compared with that in WTNS-expressing cells, but there was no significant difference between two sets of transfected cells. This result reveals that this transfection system is useful and reliable since the inhibitory effect of influenza A virus NS1 protein on mRNA splicing, which has been reported (6, 7, 17) and is generally accepted, was clearly detected using this system.
FIG. 7.
Kinetics of NS mRNAs in transfected cells. (A) COS-1 cells were transfected with either pME18S-WTNS (WTNS lanes) or pME18S-L20,45stop (L20,45stop lanes) in which a premature termination of translation was introduced into the Leu residues at positions 20 and 45 of the influenza C virus NS coding region. The unspliced and spliced NS mRNAs at the indicated times posttransfection were quantitated by RNase protection assay using a 33P-labeled influenza C virus RNA segment 7 (NS gene)-specific RNA probe (vRNA sense). The probe lane contains the undigested probe. (B) Ratios of 3′ spliced NS mRNA to unspliced NS mRNA in cells transfected with pME18S-WTNS (open circles) or pME18S-L20,45stop (closed circles).
FIG. 8.
RNase protection assay of NS mRNAs in transfected cells. (A) Cytoplasmic RNA was isolated from COS-1 cells transfected with either pME18S-WTNS (WTNS lane) or pME18S-L20,45stop (L20,45stop lane) or cotransfected with pME18S-WTNS and pME18S-ANS1 that expressed the NS1 protein of influenza A/PR/8/34 (WTNS+ANS1 lane) at 48 h p.t. and analyzed by RNase protection assay as described in Materials and Methods. The representative data of two independent experiments are shown. (B) The splicing ratios of 3′ spliced to unspliced NS mRNAs in cells transfected with each NS construct in the two experiments whose results are shown in panel A were analyzed statistically.
Influenza C virus NS1 protein may upregulate viral mRNA splicing.
We recently demonstrated that influenza C virus NS2 protein (C/NS2) was localized in the nucleus only immediately after synthesis and thereafter was predominantly found in the cytoplasm during infection (14), suggesting that C/NS2 is not likely to be involved in mRNA splicing, which occurs in the nucleus. Actually, in influenza C virus M gene-expressing cells, coexpression of C/NS2 showed no significant effect on the splicing efficiency of M mRNA (data not shown). On the other hand, the influenza C virus NS1 protein (C/NS1) was distributed in the nucleus during the early phase of infection and thereafter retained in the nucleus, even though some was exported into the cytoplasm during the late phase of infection, as described above (Fig. 4, upper panel). From this, we hypothesized that C/NS1 potentially facilitates viral mRNA splicing that occurs in the nucleus. To study this possibility, we generated a pME18S vector expressing C/NS1 alone (pME18S-CNS1) by introducing mutations into the donor and acceptor splicing sites of NS mRNA (Fig. 5) and confirmed by RIP that the cells transfected with pME18S-CNS1 expressed C/NS1 but not C/NS2. We further analyzed COS-1 cells transfected with pME18S-CNS1 by indirect immunofluorescent staining as described in Materials and Methods. C/NS1 was localized predominantly in the nucleus at 12 h p.t. and distributed in both the nucleus and the cytoplasm from 24 to 36 h p.t. Some portion of the NS1 protein was retained in the nucleus even at 48 h p.t., although most of the NS1 protein had been exported to the cytoplasm (Fig. 4, lower panel). These data suggest that C/NS1 intrinsically localizes in the nucleus. We next investigated the role of C/NS1 in the splicing of M gene-specific mRNA by cotransfection with pME18S-CNS1. COS-1 cells were either transfected with pME18S-CM alone or cotransfected with both pME18S-CM and pME18S-CNS1, and the splicing rate of M mRNA was examined by RNase protection assay using an antisense M gene-specific RNA probe (Fig. 9A). The splicing ratio of 5′ spliced to unspliced M mRNA increased in a dose-dependent manner with increasing expression of C/NS1 by up to 80% (Fig. 9B, lane CM + 2.0 CNS1; 2.07) compared with that in the cells expressing the influenza C virus M gene alone (Fig. 9B, lane CM; 1.15). By contrast, when COS-1 cells transfected with 0.25 μg of pME18S-CM were cotransfected with 0.5 μg of pME18S-ANS1, the splicing rate of influenza C virus M mRNA was reduced by 40% compared with that observed in cells transfected with 0.25 μg of pME18S-CM alone (data not shown). These data indicate that the influenza C virus NS1 protein does not possess the splicing-inhibitory effect observed in the influenza A virus NS1 protein. Rather, it is suggested that the influenza C virus NS1 protein is able to enhance the splicing efficiency of viral mRNAs.
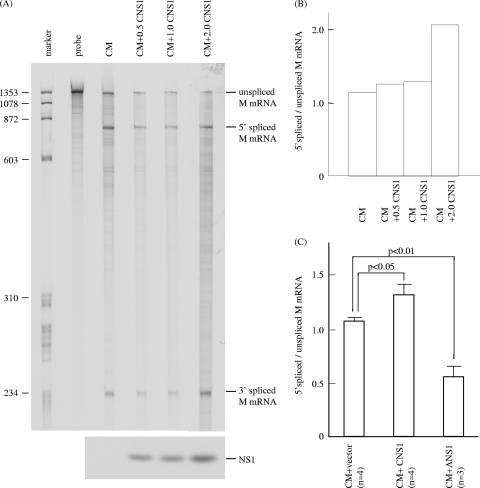
FIG. 9.
RNase protection assay of M mRNAs in influenza C virus M gene-transfected cells coexpressing influenza C virus NS1 protein. (A) pME18S-CM (0.5 μg) was introduced into COS-1 cells either together with 0.5, 1.0, or 2.0 μg of pME18S-CNS1 (right three lanes) or without pME18S-CNS1 (CM lane) and analyzed by RNase protection assay using an antisense M-specific RNA probe. The probe lane contains the undigested probe (upper panel). The expression of the C/NS1 protein in cells transfected as described above was analyzed by RIP with anti-GST/NS1 serum (lower panel). (B) Ratios of 5′ spliced to unspliced M mRNA in the transfected cells shown in panel A. (C) The ratios of 5′ spliced to unspliced M mRNA in COS-1 cells cotransfected with pME18S-CM and the pME18S vector as a control (CM+vector bar) or pME18S-CM and pME18S-CNS1 (CM+CNS1 bar) in four independent RNase protection assay experiments and cotransfected with pME18S-CM and pME18S-ANS1 (CM+ANS1 bar) in three independent experiments were compared.
To confirm this notion, COS-1 cells cotransfected with both pME18S-CM and pME18S-CNS1 at a ratio of 1:2 were analyzed in four independent RNase protection assay experiments (Fig. 9C). The ratios of 5′ spliced to unspliced M mRNA in the cells (1.333 ± 0.188) was significantly higher than that (1.079 ± 0.078) in cells cotransfected with pME18S-CM and the pME18S vector as a control (P < 0.05), indicating that the influenza C virus NS1 protein is able to enhance the splicing efficiency of influenza C virus M mRNA. By contrast, the splicing ratio observed in cells cotransfected with pME18S-CM and pME18S-ANS1 in three independent experiments (0.561 ± 0.196) was significantly lower than that in cells expressing influenza C virus M mRNA alone (1.079 ± 0.078) (P < 0.01), confirming that influenza A virus NS1 inhibits mRNA splicing.
Influenza C virus NS1 protein can facilitate the splicing efficiency of influenza A virus M gene-specific mRNA.
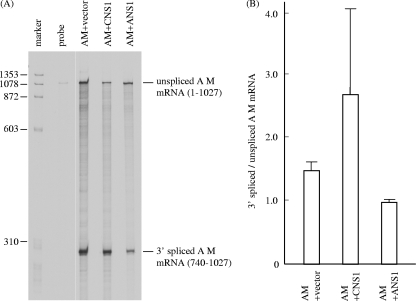
We further examined whether C/NS1 can upregulate the splicing of mRNAs of influenza A virus, in which the functions of A/NS1 have already been extensively studied. pME18S-AM (0.33 μg) containing cDNA of the M gene of influenza A/PR/8/34 was introduced into COS-1 cells with either 0.67 μg of pME18S alone, 0.67 μg of pME18S-CNS1, or 0.67 μg of pME18S-ANS1, and mRNAs derived from the influenza A virus M gene were analyzed by RNase protection assay using an antisense probe specific for the influenza A virus M gene in two independent experiments (Fig. 10). The splicing rate of 3′ spliced to unspliced mRNAs of the influenza A virus M gene was increased by 81% in cells coexpressing influenza C virus NS1 (lane AM+CNS1; 2.672 ± 1.678) compared with that in cells expressing the influenza A virus M gene alone (lane AM+vector; 1.472 ± 0.267), whereas it was reduced by 34% by the coexpression of influenza A virus NS1 (lane AM+ANS1; 0.974 ± 0.034). However, there were no statistically significant differences in splicing rates between these transfected cells. These data further demonstrate that influenza C virus NS1 facilitates mRNA splicing, whereas influenza A virus NS1 inhibits it.
FIG. 10.
RNase protection assay of influenza A virus M mRNAs in influenza A virus M gene-transfected cells coexpressing either influenza C virus NS1 or influenza A virus NS1. (A) pME18S-AM (0.33 μg), which can express the M gene-derived mRNA of influenza A virus PR/8/34 strain, was introduced into COS-1 cells with either 0.67 μg pME18S alone (AM+vector lane), 0.67 μg of pME18S-CNS1 (AM+CNS1 lane), or 0.67 μg of pME18S-ANS1, which can express the NS1 protein of influenza A/PR/8/34 (AM+ANS1 lane). The influenza A virus M mRNAs were analyzed by RNase protection assay using an antisense probe specific for the influenza A virus M gene. The probe lane shows an undigested probe. Representative data from two independent experiments are shown. (B) The splicing ratios of 3′ spliced to unspliced AM mRNAs in the cells cotransfected with each construct in the two experiments whose results are shown in panel A were compared.
DISCUSSION
In influenza A virus-infected cells, splicing is controlled so that the steady-state amount of spliced mRNAs is only 5 to 10% of that of unspliced mRNAs (15, 16). The mechanisms by which influenza A virus NS pre-mRNAs are poorly spliced have been investigated and the following confirmed. Influenza A virus NS1 protein associates with spliceosomes and inhibits pre-mRNA splicing (6, 17). Two cis-acting sequences in the NS1 transcript (positions 153 to 465 in the intron and positions 775 to 860 in the 3′ exon region) inhibit splicing (22). The splicing of influenza A virus M1 pre-mRNA is controlled by the rate of nuclear export (34). The alternative splicing of influenza A virus M1 pre-mRNA is regulated by the binding of the viral polymerase complex and cellular splicing factor SF2/ASF (28, 29). By contrast, influenza C virus M gene-specific mRNA (M mRNA) is efficiently spliced in influenza C virus-infected cells (9). In this study, we examined the mechanism by which influenza C virus M mRNA is efficiently spliced and the regulatory mechanism of the splicing of NS gene-specific mRNA (NS mRNA).
The introduction of a premature translational termination into the influenza C virus NS gene, thereby blocking the synthesis of the influenza C virus NS1 (C/NS1) and NS2 (C/NS2) proteins, drastically reduced the splicing rate of NS mRNA (Fig. 7 and 8). We further examined whether C/NS1 potentially facilitates viral mRNA splicing. The splicing rate of M mRNA of influenza C virus was increased by coexpression with C/NS1, whereas it was reduced by coexpression with influenza A virus NS1 protein (A/NS1) (Fig. 9). The splicing of influenza A virus M gene-specific mRNA was also increased by coexpression with C/NS1, though it was inhibited by coexpression with A/NS1 (Fig. 10). These results suggest that influenza C virus NS1 can facilitate viral mRNA splicing but in no way inhibit it, which is in striking contrast to the inhibitory effect of influenza A virus NS1 on pre-mRNA splicing (6, 7, 17).
As shown in Fig. 7 and 8, NS mRNA was rarely spliced when neither C/NS1 nor C/NS2 was expressed. However, in cells transfected with pME18S-CM, influenza C virus M mRNA was spliced to some extent (Fig. 2 and 9). These data raise the possibility that influenza C virus NS mRNA possesses a cis element(s) that negatively regulates splicing. Alternatively, the possibility exists that M mRNA has a cis-acting sequence(s) that upregulates splicing.
Shih and Krug (29) suggested that the cellular splicing factor SF2/ASF binds to the purine-rich splicing enhancer sequence located in the 3′ exon of the influenza A virus M1 mRNA and activates splicing to yield M2 mRNA on the basis that the amount of M2 protein synthesized in different cell lines infected with influenza A virus is proportional to the amount of cellular SF2/ASF splicing factor present in the cell line. In the case of influenza C virus-infected cells, there was no substantial difference in the splicing rate of influenza C virus M mRNA among MDCK, HMV-II (data not shown), COS-1, and 293T cells (Fig. 2), suggesting that the cellular SF2/ASF splicing factor is not likely to regulate influenza C virus M mRNA splicing.
We have not yet determined whether the potential effect of C/NS1 on splicing has specificity for viral transcripts or is general. To address this question, either an in vitro splicing system using model pre-mRNA or cotransfection of a C/NS1-expressing vector with constructs expressing model pre-mRNA should be performed in the future. The mechanism for splicing enhancement by C/NS1 also remains to be determined. We speculate that C/NS1 may interact with some host proteins involved in splicing, thereby leading to an upregulation in splicing, or that C/NS1 may bind to pre-mRNA, increasing its accessibility to the spliceosome.
It has been recently reported that the expression of influenza A virus NS1 did not alter the stability of colinear or spliced NS mRNAs encoding NS1 and NEP (NS2), respectively (7). In this study, the expression of C/NS1 increased the ratio of spliced to unspliced mRNAs, indicating that the expression of C/NS1 enhances the splicing of viral mRNA. However, if C/NS1 has the potential to increase the stability of spliced mRNAs rather than unspliced mRNAs, similar results might also be obtained. To rule out this possibility, it remains to be determined whether the expression of C/NS1 could alter the stability of unspliced or spliced mRNAs.
The spliced mRNA of the influenza C virus M gene encodes the M1 protein, which plays an important role in virus formation and determines virion morphology (20, 21, 23). Therefore, it is speculated that the mechanism for efficient splicing of M mRNA, which provides the M1 protein necessary for virus assembly in a redundant amount, has been maintained in the influenza C virus. By contrast, unspliced mRNA from the influenza C virus M gene encodes the CM2 ion channel, which is permeable to chloride ions (13), and also has pH-modulating activity (2). The influenza A virus M2 protein, encoded by the spliced mRNA derived from segment 7, plays an important role in uncoating the influenza A virus by inducing the translocation of protons from the endosomal lumen into the virion interior to disrupt acid-labile M1-vRNP interactions, thereby releasing vRNP into the cytoplasm (3, 19). M2 ion channel activity is also thought to raise the pH of the trans-Golgi network, preventing conformational changes of the acid-sensitive hemagglutinin (HA) of some H7 and H5 subtypes (4, 24, 33). Although the role of the influenza C virus CM2 ion channel in virus replication remains to be determined, it is conceivable that the overexpression of the CM2 protein has a deleterious effect on virus replication since the fact that a high level of influenza A virus M2 protein expression inhibits the rate of intracellular transport of the influenza A virus HA protein and other integral membrane glycoproteins has been demonstrated (27). If this is the case, efficient splicing of M mRNA may control the amount of CM2 synthesized to optimize virus replication. Therefore, we speculate that efficient splicing of M mRNA leads to a high level of M1 expression and the reduced expression of CM2, thereby creating conditions that are optimal for virus replication.
In this study, we provided evidence that C/NS1 facilitates the splicing of M mRNA. Furthermore, C/NS1 may regulate the splicing efficiency of its own NS mRNA during infection, controlling the amount of C/NS1 and C/NS2 proteins in infected cells. C/NS2 plays an important role in the nuclear export of vRNP (14, 25), is also associated with vRNP in the later stages of infection in virus-infected cells, and is incorporated into virions (14), suggesting that C/NS2 is involved not only in the sorting of vRNP into the assembly site but also in virus assembly. Therefore, it is likely that there is a mechanism by which an appropriate amount of C/NS2 is provided during infection to accomplish these functions. Actually, the rate of synthesis of C/NS2 reached a maximum by 14 h p.i. and the maximum rate of C/NS2 synthesis continued up to 48 h p.i., with C/NS2 continuing to be accumulated in the infected cells up to 3 days p.i. (14). By contrast, the rate of C/NS1 synthesis reached a maximum by 14 h p.i., continued at the maximum rate up to 24 h p.i. before decreasing by 30 h p.i. (data not shown). These findings can be explained by the mechanism in which the expressed C/NS1 promotes the splicing of its own mRNA, resulting in the lower expression of C/NS1 and the greater expression of C/NS2 in the later phase of infection. In conclusion, C/NS1, which enhances the splicing of viral mRNA, may regulate both the expression level of the M gene-derived M1 and CM2 proteins and that of the NS gene-derived NS1 and NS2 proteins, thereby leading to optimal virus replication.
Acknowledgments
We thank M. Enami (Kanazawa University) for generously providing plasmids containing cDNAs to the influenza A virus M and NS genes. We also gratefully thank R. Sho (Department of Public Health, Yamagata University Faculty of Medicine) for statistical analysis.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan; by the Takeda Science Foundation; by the Terumo Life Science Foundation; and by a Grant-in-Aid from the Global COE program of the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 9 December 2009.
REFERENCES
- 1.Alamgir, A. S. M., Y. Matsuzaki, S. Hongo, E. Tsuchiya, K. Sugawara, Y. Muraki, and K. Nakamura. 2000. Phylogenetic analysis of influenza C virus nonstructural (NS) protein genes and identification of the NS2 protein. J. Gen. Virol. 81:1933-1940. [DOI] [PubMed] [Google Scholar]
- 2.Betakova, T., and A. J. Hay. 2007. Evidence that the CM2 protein of influenza C virus can modify the pH of the exocytic pathway of transfected cells. J. Gen. Virol. 88:2291-2296. [DOI] [PubMed] [Google Scholar]
- 3.Bukrinskaya, A. G., N. K. Vorunova, G. V. Kornilayeva, R. A. Narmanbetova, and A. J. Hay. 1982. Influenza virus uncoating in infected cells and effect of rimantadine. J. Gen. Virol. 60:49-59. [DOI] [PubMed] [Google Scholar]
- 4.Ciampor, F., C. A. Thompson, S. Grambas, and A. J. Hay. 1992. Regulation of pH by the M2 protein of influenza A viruses. Virus Res. 22:247-258. [DOI] [PubMed] [Google Scholar]
- 5.Enami, M., G. Sharma, C. Benham, and P. Palese. 1991. An influenza virus containing nine different RNA segments. Virology 185:291-298. [DOI] [PubMed] [Google Scholar]
- 6.Fortes, P., A. Beloso, and J. Ortin. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 13:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garaigorta, U., and J. Ortin. 2007. Mutation analysis of a recombinant NS replicon shows that influenza virus NS1 protein blocks the splicing and nucleo-cytoplasmic transport of its own viral mRNA. Nucleic Acids Res. 35:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hongo, S., F. Kitame, K. Sugawara, H. Nishimura, and K. Nakamura. 1992. Cloning and sequencing of influenza C/Yamagata/1/88 virus NS gene. Arch. Virol. 126:343-349. [DOI] [PubMed] [Google Scholar]
- 9.Hongo, S., K. Sugawara, H. Nishimura, Y. Muraki, F. Kitame, and K. Nakamura. 1994. Identification of a second protein encoded by influenza C virus RNA segment 6. J. Gen. Virol. 75:3503-3510. [DOI] [PubMed] [Google Scholar]
- 10.Hongo, S., K. Sugawara, Y. Muraki, F. Kitame, and K. Nakamura. 1997. Characterization of a second protein (CM2) encoded by RNA segment 6 of influenza C virus. J. Virol. 71:2786-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hongo, S., P. Gao, K. Sugawara, Y. Muraki, Y. Matsuzaki, Y. Tada, F. Kitame, and K. Nakamura. 1998. Identification of a 374 amino acid protein encoded by RNA segment 6 of influenza C virus. J. Gen. Virol. 79:2207-2213. [DOI] [PubMed] [Google Scholar]
- 12.Hongo, S., K. Sugawara, Y. Muraki, Y. Matsuzaki, E. Takashita, F. Kitame, and K. Nakamura. 1999. Influenza C virus CM2 protein is produced from a 374-amino-acid protein (P42) by signal peptidase cleavage. J. Virol. 73:46-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hongo, S., K. Ishii, K. Mori, E. Takashita, Y. Muraki, Y. Matsuzaki, and K. Sugawara. 2004. Detection of ion channel activity in Xenopus laevis oocytes expressing influenza C virus CM2 protein. Arch. Virol. 149:35-50. [DOI] [PubMed] [Google Scholar]
- 14.Kohno, Y., Y. Muraki, Y. Matsuzaki, E. Takashita, K. Sugawara, and S. Hongo. 2009. Intracellular localization of influenza C virus NS2 protein (NEP) in infected cells and its incorporation into virions. Arch. Virol. 154:235-243. [DOI] [PubMed] [Google Scholar]
- 15.Lamb, R. A., P. W. Choppin, R. M. Chanock, and C. J. Lai. 1980. Mapping of the two overlapping genes for polypeptides NS1 and NS1 on RNA segment 8 of influenza virus genome. Proc. Natl. Acad. Sci. U. S. A. 77:1857-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb, R. A., C. J. Lai, and P. W. Choppin. 1981. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc. Natl. Acad. Sci. U. S. A. 78:4170-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, Y., X. Y. Qian, and R. M. Krug. 1994. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 8:1817-1828. [DOI] [PubMed] [Google Scholar]
- 18.Marschall, M., A. Helten, A. Hechtfischer, A. Zach, C. Banaschewski, W. Hell, and H. Meier-Ewert. 1999. The ORF, regulated synthesis, and persistence-specific variation of influenza C viral NS1 protein. Virology 253:208-218. [DOI] [PubMed] [Google Scholar]
- 19.Martin, K., and A. Helenius. 1991. Transport of incoming influenza virus nucleocapsids into the nucleus. J. Virol. 65:232-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muraki, Y., H. Washioka, K. Sugawara, Y. Matsuzaki, E. Takashita, and S. Hongo. 2004. Identification of an amino acid residue on influenza C virus M1 protein responsible for formation of the cord-like structures of the virus. J. Gen. Virol. 85:1885-1893. [DOI] [PubMed] [Google Scholar]
- 21.Muraki, Y., T. Murata, E. Takashita, Y. Matsuzaki, K. Sugawara, and S. Hongo. 2007. A mutation on influenza C virus M1 protein affects virion morphology by altering the membrane affinity of the protein. J. Virol. 81:8766-8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemeroff, M. E., U. Utans, A. Kramer, and R. M. Krug. 1992. Identification of cis-acting intron and exon regions in influenza virus NS1 mRNA that inhibit splicing and cause the formation of aberrantly sedimenting presplicing complexes. Mol. Cell. Biol. 12:962-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimura, H., S. Hongo, K. Sugawara, Y. Muraki, F. Kitame, H. Washioka, A. Tonosaki, and K. Nakamura. 1994. The ability of influenza C virus to generate cord-like structures is influenced by the gene coding for M protein. Virology 200:140-147. [DOI] [PubMed] [Google Scholar]
- 24.Ohuchi, M., A. Cramer, M. Vey, R. Ohuchi, W. Garten, and H. D. Klenk. 1994. Rescue of vector-expressed fowl plague virus hemagglutinin in biologically active form by acidotropic agents and coexpressed M2 protein. J. Virol. 68:920-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paragas, J., J. Talon, R. E. O'Neill, D. K. Anderson, A. Garcia-Sastre, and P. Palese. 2001. Influenza B and C virus NEP (NS2) proteins possess nuclear export activities. J. Virol. 75:7375-7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pekosz, A., and R. A. Lamb. 1998. Influenza C virus CM2 integral membrane glycoprotein is produced from a polypeptide precursor by cleavage of an internal signal sequence. Proc. Natl. Acad. Sci. U. S. A. 95:13233-13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakaguchi, T., G. P. Leser, and R. A. Lamb. 1996. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J. Cell Biol. 133:733-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shih, S. R., M. E. Nemeroff, and R. M. Krug. 1995. The choice of alternative 5′ splice sites in influenza virus M1 mRNA is regulated by the viral polymerase complex. Proc. Natl. Acad. Sci. U. S. A. 92:6324-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih, S. R., and R. M. Krug. 1996. Novel exploitation of a nuclear function by influenza virus: the cellular SF2/ASF splicing factor controls the amount of the essential viral M2 ion channel protein in infected cells. EMBO J. 15:5415-5427. [PMC free article] [PubMed] [Google Scholar]
- 30.Sugawara, K., H. Nishimura, F. Kitame, and K. Nakamura. 1986. Antigenic variation among human strains of influenza C virus detected with monoclonal antibodies to gp88 glycoprotein. Virus Res. 6:27-32. [DOI] [PubMed] [Google Scholar]
- 31.Sugawara, K., H. Nishimura, S. Hongo, F. Kitame, and K. Nakamura. 1991. Antigenic characterization of the nucleoprotein and matrix protein of influenza C virus with monoclonal antibodies. J. Gen. Virol. 72:103-109. [DOI] [PubMed] [Google Scholar]
- 32.Takebe, Y., S. Motoharu, J. Fujisawa, P. Hoy, K. Yokota, K. Arai, M. Yoshida, and N. Arai. 1988. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol. Cell. Biol. 8:466-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi, K., and R. A. Lamb. 1994. Influenza virus M2 protein ion channel activity stabilizes the native form of fowl plague virus hemagglutinin during intracellular transport. J. Virol. 68:911-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valcárcel, J., P. Fortes, and J. Ortin. 1993. Splicing of influenza virus matrix protein mRNA expressed from a simian virus 40 recombinant. J. Gen. Virol. 74:1317-1326. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita, M., M. Krystal, and P. Palese. 1988. Evidence that the matrix protein of influenza C virus is coded for by a spliced mRNA. J. Virol. 62:3348-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokota, M., K. Nakamura, K. Sugawara, and M. Homma. 1983. The synthesis of polypeptides in influenza C virus-infected cells. Virology 130:105-117. [DOI] [PubMed] [Google Scholar]