Abstract
The genomes of herpes simplex virus type 1 (HSV-1) are regularly chromatinized during latency such that their digestion with micrococcal nuclease (MCN) releases nucleosome-sized DNA fragments. In lytically infected cells, in contrast, MCN releases HSV-1 DNA in primarily heterogeneously sized fragments. Consistently, only a small percentage of this HSV-1 DNA coimmunoprecipitates with histones. Most current models propose that histones associate with HSV-1 DNA during lytic infections at low occupancy. However, histone modification or occupation is also proposed to regulate HSV-1 transcription. It remains unclear how the histones associated with a small percentage of HSV-1 DNA may regulate transcription globally. Moreover, the physical properties of the complexes containing histones and HSV-1 DNA are unknown. We evaluated the HSV-1 DNA-containing complexes at 5 h after (lytic) infection by biochemical fractionations. Nuclear HSV-1 DNA did not fractionate as protein-free HSV-1 DNA but as DNA in cellular nucleosomes. Moreover, MCN released HSV-1 DNA in complexes that fractionate as cellular mono- and dinucleosomes by centrifugation followed by sucrose gradients and size-exclusion chromatography. The HSV-1 DNA in such complexes was protected to heterogeneous sizes and was more accessible to MCN than DNA in most cellular chromatin. Using a modified MCN digestion to trap unstable digestion intermediates, HSV-1 DNA was quantitatively recovered in discrete mono- to polynucleosome sizes in complexes fractionating as cellular mono- to polynucleosomes. The HSV-1 DNAs in complexes fractionating as mono- to dinucleosomes were stabilized by cross-linking. Therefore, most HSV-1 DNA forms particularly unstable nucleosome-like complexes at 5 h of lytic infection.
Herpes simplex virus type 1 (HSV-1) is a double-stranded DNA virus that transcribes and replicates in the nucleus. It establishes lytic infections in epithelial cells and latent infections in neurons in vivo. During lytic infections, all HSV-1 genes are expressed, and the genomes are replicated. During latent infections, HSV-1 gene expression is restricted, and the genomes are not replicated.
Nuclear DNA is typically packaged into a nucleoprotein complex, chromatin. Chromatin is a chain of nucleosomes, 146 bp of DNA wrapped 1.75 turns around a histone octamer composed of two copies each of H2A, H2B, H3, and H4 (44). Linker histone H1 binds at entry and exit points and to linker DNA in between nucleosomes, promoting the compaction of chromatin into higher-order structures (31, 60, 61).
The structure of chromatin is classically probed with nucleases (69) such as micrococcal (endo)nuclease (MCN). MCN preferentially cleaves exposed linker DNA between nucleosomes (6, 26). Mild MCN digestions first cleave linker DNA sparsely, releasing polynucleosome chains. Longer digestions eventually cleave the polynucleosomes into mononucleosomes. Under even more stringent digestion conditions, MCN eventually degrades even the DNA in mononucleosomes. Normal MCN digestions of regularly chromatinized DNA result in the protection of poly- and mononucleosome-sized DNA fragments.
Like cellular DNA, the genomes of most nuclear replicating DNA viruses also associate with histones albeit to various degrees (54). The association between histones and the genomes of small DNA-tumor viruses of the polyomavirus and papillomavirus families, for example, is maintained throughout the replicative cycle even in encapsidated virions (12, 27). As a result, MCN digestions of polyomavirus- or papillomavirus-infected cells release mostly nucleosome-sized viral DNA fragments (7, 17, 27). As a notable exception, the simian virus 40 (SV40) origin of replication, which is bound by the large T antigen, is free of nucleosomes (38).
Chromatinization also plays an important role in the life cycles of most herpesviruses (50, 54). Unlike the genomes of the small DNA viruses, however, those of the herpesviruses are not associated with histones in the nucleocapsids (15, 24, 30, 58, 63, 70, 71). Once in the nucleus, the genomes of the Gammaherpesvirinae Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) become regularly chromatinized, and histone modifications are thought to regulate their transcription (1, 8, 9, 18, 19, 39, 43, 59, 73). The roles of chromatin in transcription regulation for the members of the Alphaherpesvirinae and Betaherpesvirinae HSV-1 and human cytomegalovirus (HCMV) are more complex. HSV-1 genomes are regularly chromatinized during latency, and MCN digestions therefore release fragments of sizes characteristic of nucleosome DNA (20, 65). As evaluated by chromatin immunoprecipitation (ChIP), histones bearing posttranslational modifications (PTMs) characteristic of facultative heterochromatin are associated with the transcriptionally restricted latent genomes of HSV-1 and HCMV (2, 3, 13, 25, 35, 46, 51, 68, 74, 80). During reactivation from latency, however, there is a shift from PTMs associated with silencing to PTMs associated with transcription (2). These results strongly suggest a role for chromatin in the regulation of latency and reactivation.
In contrast to latent infections, MCN digestions of HSV-1 DNA in lytically infected cells result in DNA fragments of primarily heterogeneous sizes and only a minority of nucleosome sizes (20, 42, 52, 65). Moreover, infecting HSV-1 genomes localize to nuclear domains adjacent to ND10s, domains which are devoid of cellular chromatin and histones, and the HSV-1 replication compartments at late times postinfection, which contain the HSV-1 genomes, are also mostly free of histones (4, 36, 62, 64, 76). It was therefore classically concluded that most HSV-1 DNA is not associated with histones during lytic infections (52, 65). Data from more recent ChIP assays, however, indicated that core histones do interact with HSV-1 DNA during lytic infections (14, 33, 34, 42, 47-50, 72). These interactions occur as early as 1 h postinfection (hpi) and may correlate with HSV-1 transcription (14, 23, 49, 72). The mobilization of linker (16) and core (K. L. Conn et al., unpublished data) histones during infection likely allows the histones to reach the sites where the HSV-1 genomes localize, to then interact with them.
Despite the lack of experimental evidence of proper chromatinization of HSV-1 genomes during lytic infections, the HSV-1 transcriptional activators VP16, ICP4, and ICP0 and the VP16-interacting protein VP22 all have chromatin-disrupting activities. As evaluated by ChIP, for example, VP16 promotes the depletion of core histones from HSV-1 genomes (33, 49) and recruits histone-modifying proteins such as histone acetyltransferases (HATs) and ATP-dependent chromatin-remodeling complexes (33, 45, 66, 67, 78). However, neither the depletion of core histones nor the recruitment of HATs or chromatin-remodeling complexes to HSV-1 DNA appears to be required for HSV-1 transcription (47, 49). ICP0, an HSV-1 E3 ubiquitin ligase, promotes the degradation of the histone H3 variant CENP A (among many other proteins) (22, 55, 56) and causes a disruption of histone deacetylase (HDAC) complexes associated with transcriptional repression (28, 29, 57). Like VP16, ICP0 also promotes the depletion of histone H3 from HSV-1 DNA (14). Another HSV-1 transcriptional transactivator, ICP4, together with ICP0, disrupts the silencing of cellular genes (10). The tegument protein VP22, which strongly interacts with VP16, inhibits the TAF-1-dependent deposition of histones on DNA templates (79). The nontranscribed genomes of HSV-1 mutants in all immediate-early (IE) genes, or expressing only ICP0, are associated with increased levels of histone H3 (23). However, only a small percentage of HSV-1 DNA coimmunoprecipitates with histones in most studies (0.0069% to 11% of the amounts of coimmunoprecipitated cellular DNA in the same reaction mixtures) (42, 47, 49, 72). Only one paper reported equivalent coimmunoprecipitations of HSV-1 or cellular DNA with a particular histone, H3 (14). Consistent with the majority of the ChIP results, only a small percentage of HSV-1 DNA is protected from MCN in sizes corresponding to nucleosome DNA (20, 42, 65). It therefore remains unclear how the PTM of histones associated with only a minority of HSV-1 DNA may regulate HSV-1 transcription globally. Moreover, the experimental evidence does not support a model in which most HSV-1 DNA is in regular nucleosomes, although HSV-1 genomes do interact with all core histones (49). The physical properties of the complexes containing histones and HSV-1 DNA also remain unclear.
Using biochemical fractionation techniques, we systematically analyzed the biophysical properties of the complexes formed with HSV-1 DNA in lytically infected cells at 5 hpi. We found that at 5 hpi, most HSV-1 DNA is in complexes that fractionate as cellular mono- to dinucleosomes. Although these complexes share many of the biophysical properties of cellular nucleosomes, they also show many unique properties. Most prominently, the HSV-1 DNA nucleoprotein complexes are most unstable.
MATERIALS AND METHODS
Cells and viruses.
Vero African green monkey kidney fibroblast cells were maintained in Dulbecco's modified minimum Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS), 50 mU/ml penicillin, and 50 ng/ml streptomycin. Low-passage (passage 10) HSV-1 strain KOS was used throughout this study. Viral stocks were propagated and titrated on monolayers of Vero cells according to standard procedures (21).
HSV-1 infection.
Vero cells were infected with a multiplicity of infection (MOI) of 5 PFU of HSV-1 per cell in serum-free medium. After inoculation for 1 h at 37°C, inocula were removed, and cells were washed twice with cold phosphate-buffered saline (PBS) (1 mM KH2PO4, 154 mM NaCl, 3 mM Na2HPO4 [pH 7.4]). Fresh DMEM-5% FBS was then added to the infected cells, which were harvested at 5 h postinfection (hpi).
Isolation of nuclei.
Cells were rinsed with PBS at 4°C, trypsinized, and resuspended in 20 ml of DMEM-5% FBS. Cells were then pelleted by centrifugation, resuspended in hypotonic RSB buffer (10 mM Tris [pH 7.5], 10 mM NaCl, 5 mM MgCl2), and lysed with 0.5% (vol/vol) Nonidet P-40. Nuclei were then isolated by differential centrifugation (1,800 × g for 25 min at 4°C).
Nuclease digestion.
Nuclei were digested with either BamHI or micrococcal nuclease (MCN). For BamHI digestions, nuclei were rinsed twice with a solution containing 50 mM Tris (pH 7.5), 10 mM MgCl2, and 100 mM NaCl (React 3 buffer), pelleting the nuclei at 5,000 × g for 15 min between rinses. Nuclei were then resuspended in React 3 buffer containing 10 U of BamHI per 1 × 106 nuclei. Digestions were carried out at 37°C for 4 h. For MCN digestions, nuclei were resuspended in MCN digestion buffer (10 mM Tris [pH 8], 1 mM CaCl2) containing 0.005, 0.05, 0.5, or 5 U of MCN per 1 × 107 nuclei. Digestions were carried out at 39°C and stopped at the indicated times with the addition of EGTA to a final concentration of 5 mM. For MCN redigestion or serial MCN digestion experiments, MCN was used at a concentration of 0.05 U/ml. Serial digestions were performed at room temperature. DNA was isolated by digestion with proteinase K, phenol-chloroform extraction, and ethanol precipitation.
Chromatin fractionation.
MCN-digested nuclei were lysed by adding 1 volume of chromatin extraction buffer (CEB) (1 mM EGTA, 2% Triton X-100, 3 mM MgCl2, 2 mM Tris [pH 8]) and incubated with rotation for 10 min at 4°C. The so-called “soluble” and “insoluble” chromatin fractions were then separated by differential centrifugation (8,000 × g for 20 min).
Sucrose gradient ultracentrifugation.
Continuous 0 to 10% sucrose gradients were prepared by using a Gradient Master (Biocomp, NB, Canada) with sucrose gradient buffer (0.5 mM EGTA, 1.5 mM MgCl2, 10 mM Tris [pH 8]) and 0, 80, 225, or 450 mM NaCl. Soluble chromatin was loaded on top of preformed gradients and centrifuged at 40,000 rpm in an SW-40 rotor for 3 h at 4°C. Fractions were collected from the bottom of the tube. After the removal of the final fraction, the pellet was recovered in 1 ml of STE (10 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA). DNA was isolated from each fraction by standard proteinase K digestion and phenol-chloroform extraction.
Size-exclusion chromatography.
Size-exclusion chromatography was performed by using Econo-Pac disposable chromatography columns (Bio-Rad, Hercules, CA) packed with Sephadex G-100 (GE Healthcare, Piscataway, NJ). Relevant fractions from the sucrose gradients were loaded onto the columns. Columns were washed twice with 1 volume and once with 5 volumes of gel filtration buffer (0.5 mM EGTA, 1.5 mM MgCl2, 150 mM NaCl, 10 mM Tris [pH 8]). Fractions were collected and cross-linked with formaldehyde (final concentration of 0.1%) under rotation for 1 h at 4°C. Cross-linking was quenched by the addition of 1 M glycine to a final concentration of 125 mM. Cross-linked samples were then subjected to MCN redigestion.
Hybridization.
DNA was resolved on 2% agarose gels and Southern blotted onto positively charged nylon membranes according to standard protocols. Membranes were prehybridized with 10 ml rapid hybrid buffer (Amersham Biosciences, Piscataway, NJ) at 75°C for HSV-1 DNA or 60°C for cellular DNA. The probes used were an HSV-1 EcoRI library (except for fragment F) or the JK fragment or total Vero cell DNA. Plasmid DNA from the HSV-1 EcoRI library was isolated by using a GeneElute HP plasmid maxiprep kit (Sigma). Vero cell DNA was isolated from uninfected Vero cells by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation. Probes were random-prime labeled according to the manufacturer's instructions (Amersham Biosciences). The denatured probe was added to 5 ml of rapid hybrid buffer at 75°C for HSV-1 DNA or 60°C for cellular DNA and hybridized at the respective temperatures for 3 h. Membranes were washed twice for 15 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at room temperature. If needed, membranes were further washed for 10 min in 75 mM NaCl-7.5 mM sodium citrate (0.5× SSC)-0.5% SDS at 60°C (HSV) or 50°C (cell). Membranes were exposed to Kodak PhosphorImager screens and quantitated by using a Bio-Rad FX molecular imager.
RESULTS
At 5 h after infection, HSV-1 DNA is in complexes that do not fractionate as protein-free DNA.
As a first step in characterizing the biophysical properties of nuclear HSV-1 DNA in lytically infected cells, we used standard biochemical fractionation techniques to test whether most nuclear HSV-1 DNA had the biophysical properties of protein-free DNA. Briefly, nuclei from HSV- or mock-infected cells were harvested at 5 hpi, and protein-free DNA was resolved from DNA in larger complexes by differential centrifugation. However, replicating HSV-1 genomes form long, branched concatemers (75). Nuclei were therefore first digested with BamHI to cleave these long, branched concatemers into subgenomic linear fragments (ranging from 50 to 5,000 bp) and thus minimize mechanical trapping during fractionation. Nuclei were then lysed, and DNA-protein complexes were resolved by differential centrifugation into so-called “soluble” and “insoluble” fractions. The “soluble” fraction contains soluble proteins, protein-free DNA, and small complexes including mono-, di-, and some short polynucleosomes. The “insoluble” fraction contains larger complexes including large polynucleosome chains and nuclear matrix. As a control, protein-free HSV-1 DNA was added to nuclei of mock-infected cells before BamHI digestion.
As expected, 83% of the detected protein-free HSV-1 DNA added to mock-infected nuclei fractionated to the “soluble” fraction (Fig. 1A and Table 1). In contrast, and also as expected, only 19% of the detected chromatinized cellular DNA fractionated to the “soluble” fraction (Fig. 1A and Table 1). Likewise, only 27% of the detected HSV-1 DNA from the nuclei of infected cells fractionated to the “soluble” fraction, whereas 73% fractionated to the insoluble fraction (Fig. 1A and Table 1). The majority of nuclear HSV-1 DNA and chromatinized cellular DNA therefore fractionated to the insoluble fraction during the first differential centrifugation (73% and 81%, respectively) (Table 1). This HSV-1 DNA, however, was still digested to heterogeneously sized fragments and therefore was accessible to BamHI (Fig. 1A).
FIG. 1.
HSV-1 DNA in nuclei of lytically infected cells does not fractionate as protein-free DNA. Vero cells were mock infected or infected with HSV-1 (Nuclear) and harvested 5 h later. Half of the mock-infected nuclei were then spiked with protein-free HSV-1 DNA. Infected, mock-infected, and spiked mock-infected nuclei were digested with BamHI and lysed. Soluble and insoluble protein complexes were separated by differential centrifugation. The soluble DNA-protein complexes were then further resolved on sucrose gradients (Fraction). DNA in the pellet and each gradient fraction was analyzed by Southern blotting with HSV-1 or cellular probes. (A) Images of the hybridizations. Bottom fractions loaded to the left show different exposures under each condition. Exposures optimized for soluble fractions are also shown for insoluble fractions, which are therefore overexposed (Equal exposure). Lower exposures optimized for the insoluble fractions are also shown (Short Exposure). (B) Line graph presenting HSV-1 and cellular DNA in each fraction as a percentage of DNA in the gradient. The panels on the top are the same hybridizations shown in A, shown as a reference for the graph below and resized to fit the figure.
TABLE 1.
HSV-1 DNA in nuclei of lytically infected cells does not fractionate as protein-free DNAa
| DNA | % of DNA in fraction |
|
|---|---|---|
| Insoluble | Soluble | |
| HSV, protein free | 17 | 83 |
| HSV, nuclear | 73 | 27 |
| Cellular, nuclear | 81 | 19 |
Percentages of DNA from nuclei digested with BamHI that fractionate to soluble or insoluble fractions.
The DNA and DNA-protein complexes that fractionated in the “soluble” fraction were then further resolved by sucrose gradient fractionation. As expected for protein-free DNA, the detected protein-free HSV-1 DNA fractionated to the lowest-density fractions (fractions 9 to 12) (Fig. 1A and B). In contrast, the detected HSV-1 DNA from nuclei of infected cells fractionated to heavier fractions (fractions 6 to 10) (Fig. 1A and B). These were mostly the same fractions to which the detectable “soluble” cellular DNA in mono-, di-, and short polynucleosomes fractionated (Fig. 1A).
In summary, the majority of the nuclear HSV-1 DNA detected at 5 hpi in lytically infected cells does not fractionate as protein-free DNA. Instead, it fractionates to the same fractions as chromatinized cellular DNA (Fig. 1B).
MCN digestion releases HSV-1 DNA in complexes that fractionate as cellular nucleosomes.
HSV-1 DNA released as “soluble” complexes cofractionated with chromatinized cellular DNA. We then tested whether this nuclear HSV-1 DNA was in complexes with properties similar to those of nucleosomes.
Nuclei from infected cells were harvested at 5 hpi and digested with MCN. Digested nuclei were then lysed and resolved into “soluble” and “insoluble” fractions. The soluble fraction is often referred to as “soluble chromatin” and contains mono-, di-, and relatively short polynucleosome complexes. Complexes released as soluble chromatin were then further resolved by sucrose gradient centrifugation (Fig. 2A). Polynucleosomes resolve to heavier fractions, mono- and dinucleosomes resolve to lighter fractions, and protein-free DNA resolves to the lightest fractions. As expected, the cellular DNA released as soluble chromatin resolved as various sizes of polynucleosomes in fractions 2 to 9 and as mono-, di-, and trinucleosomes in fractions 10 and 11 (Fig. 2A). In contrast, all the HSV-1 DNA released under these conditions as soluble chromatin was in complexes that resolved to the same fractions as cellular mono- to trinucleosomes, and most of it was in the fractions containing mostly mono- to dinucleosomes (HSV fractions 10 and 11) (Fig. 2A and B). No HSV-1 DNA released with the soluble chromatin fractionated as polynucleosomes or to the lightest fraction.
FIG. 2.
MCN digestion releases HSV-1 DNA in complexes that fractionate as cellular mono- to dinucleosomes. Nuclei of infected cells were digested for 150 s with 0.05 U of MCN per 1 × 107 nuclei, and the soluble DNA-protein complexes were resolved on sucrose gradients (A and B). Following a similar experiment, fractions 10 and 11 from the gradients were further fractionated by size-exclusion chromatography (C). DNA from each fraction was analyzed by Southern blotting with HSV-1 or cellular probes. (A) Images of the membrane hybridized with HSV-1 or cellular probes. Bottom fractions are to the left. (B) Line graphs presenting HSV-1 and cellular DNA in each fraction as a percentage of DNA in the gradient. The panels on the top are the same as those shown in A, shown as reference for the graph below and resized to fit in the figure. (C) Images of the membranes hybridized with cellular or HSV-1-specific probes. Arrowheads indicate the migration of mono- or dinucleosomes.
The complexes in fractions 10 and 11 were then subjected to size-exclusion chromatography. Fractions were collected and analyzed by Southern blotting (Fig. 2C). For this experiment, we performed a slightly more stringent MCN digestion such that fractions 10 and 11 were more enriched for mono- and dinucleosomes over trinucleosomes. The HSV-1 DNA-containing complexes continued to fractionate as cellular mono- and dinucleosomes after chromatography (fraction 4) (Fig. 2C). Therefore, MCN digestion releases HSV-1 DNA in small complexes that fractionate as cellular mono- to dinucleosomes following differential centrifugation followed by sucrose gradient centrifugation and size-exclusion chromatography. However, unlike the DNA in cellular nucleosomes, which is protected to homogeneous sizes corresponding to mono- and dinucleosomes (146 and 292 bp, respectively), the HSV-1 DNA in such complexes was protected to a range of heterogeneous fragments from mono- to dinucleosome sizes (146 to 292 bp) (Fig. 2A).
Nuclear HSV-1 DNA is more accessible to MCN than DNA in most cellular chromatin.
Basically all HSV-1 DNA released as soluble chromatin by 150 s of MCN digestion (Fig. 2) was in small fragments in complexes that fractionated as mono- to dinucleosomes, whereas the DNA in cellular chromatin was primarily in large fragments in polynucleosome complexes. We next tested whether these differences directly reflected differences in MCN accessibility.
Nuclei of infected cells were harvested at 5 hpi and digested with different concentrations of MCN for increasing times to reach from almost no to almost total DNA digestion. Nuclear DNA was purified, resolved by agarose gel electrophoresis, and analyzed by ethidium bromide staining and Southern blotting. Cellular and HSV-1 hybridizations are shown in standard exposures to compare total DNA levels and also in overexposures to analyze the similarities and differences between the smaller cellular and HSV-1 DNA fragments (Fig. 3A).
FIG. 3.
Nuclear HSV-1 DNA is more accessible to MCN than DNA in most cellular chromatin. Purified nuclei of infected cells were digested for 0.5, 2.5, 5, 15, 30, and 60 min with 0.005, 0.05, 0.5, or 5 U MCN per 1 × 107 nuclei, and the DNA was analyzed by Southern blotting with HSV-1 or cellular probes. (A) Images of ethidium bromide-stained gels (Total) and membranes hybridized with cellular or HSV-1-specific probes. M, molecular mass marker (in basepairs). Normal exposures and overexposures (bottom), in which the nucleosome-sized and MCN-resistant HSV-1 DNAs are more clearly visible, are shown. To achieve comparable signal intensities, only 50% of the sample was loaded for 0.5 min at 0.005 to 5.0 U of MCN. (B and C) Line graphs presenting normalized levels of cellular (B) and HSV-1 (C) DNAs against digestion time (averages ± standard deviations) (n = 3).
As expected for chromatinized cellular DNA, low MCN concentrations digested cellular DNA to a typical nucleosome ladder of sizes corresponding to multiples of 160 bp (0.005 U) (Fig. 3A, lanes 4 to 6). In contrast, and as previously reported (42, 52, 53, 65), HSV-1 DNA was digested primarily to a smear presumably reflecting heterogeneously sized fragments (most apparent at 0.005 U) (Fig. 3A, lane 5). Also consistent with data from previous reports (42, 52, 53, 65), a minor population of HSV-1 DNA was protected to sizes consistent with mono- or dinucleosomes (most apparent at 0.05 U) (Fig. 3A, lanes 4 and 5).
As the concentration of MCN increased, eventually most (98%) cellular DNA was completely degraded (Fig. 3B, compare data for 0.005 and 5 U). Also consistent with data from previous reports (42, 52, 53, 65), a minor percentage (approximately 10%) of HSV-1 DNA was poorly accessible to even high MCN concentrations (0.5 U) (Fig. 3C). However, even this poorly accessible population was almost completely digested (up to 98%) when the concentration of MCN was increased by 100-fold (HSV, 5 U) (Fig. 3C).
In addition to differences in the size distribution of the DNA fragments released, the kinetics of MCN digestion were also different between HSV-1 DNA and DNA in cellular chromatin. HSV-1 DNA was digested approximately 3-fold-more rapidly than DNA in cellular chromatin (Table 2). For example, 50% of HSV-1 or cellular DNA was digested by 0.05 U MCN in 6.74 or 19.5 min, respectively (time required to digest 50% of the HSV-1 DNA [T50]) (Fig. 3B and C and Table 2). Most HSV-1 DNA is therefore more accessible to MCN than most DNA in cellular chromatin. However, the 10% of HSV-1 DNA that was poorly accessible was 5.3-fold-more resistant to MCN digestion than DNA in cellular chromatin (0.5 U) (Table 2). For example, 0.5 U MCN digested 90% of HSV-1 or cellular DNA in 27.4 or 5.2 min, respectively (T90) (Fig. 3B and C and Table 2).
TABLE 2.
Nuclear HSV-1 DNA is more accessible to MCN than DNA in most cellular chromatina
| Amt of MCN (U) |
T50 (min) |
T90 (min) |
||
|---|---|---|---|---|
| Cellular | HSV | Cellular | HSV | |
| 0.005 | >60 | 51.1 | >60 | >60 |
| 0.05 | 19.5 | 6.7 | >60 | 32.6 |
| 0.5 | 0.7 | 0.4 | 5.2 | 27.4 |
| 5 | 0.3 | 0.3 | 2.5 | 0.2 |
Digestion times required to degrade 50% (T50) or 90% (T90) of HSV-1 or cellular DNA were graphically calculated from the average digestion curves (n = 3).
In summary, most HSV-1 DNA was far more accessible to MCN at 5 hpi than DNA in cellular chromatin. However, a small percentage of HSV-1 DNA was far less accessible to MCN than DNA in cellular chromatin. Further contrasting with DNA in most cellular chromatin, most HSV-1 DNA was cleaved primarily to heterogeneous sizes, whereas the least accessible fraction was protected from digestion as long fragments.
HSV-1 DNA released as soluble chromatin is mostly in complexes that fractionate as mono- to dinucleosomes.
HSV-1 DNA is far more accessible to MCN than DNA in cellular chromatin and is digested in 150 s (2.5 min), primarily to heterogeneously sized fragments (Fig. 3). Nevertheless, only a small percentage of this digested HSV-1 DNA is released as soluble chromatin in complexes that fractionate as cellular mono- to dinucleosomes in sucrose gradients (Fig. 2). Instead, the vast majority is in heterogeneously sized complexes that fractionate as insoluble chromatin. Different MCN digestion times release cellular chromatin as progressively shorter polynucleosome complexes. We therefore tested whether different times of MCN digestion could also release HSV-1 DNA in differently sized nucleosome-like complexes. We selected conditions that result in digestions ranging from either most HSV-1 DNA not being digested (equivalent to HSV at 0.005 U) (Fig. 3, lanes 2 and 3) to most HSV-1 DNA being digested to heterogeneous sizes (equivalent to HSV at 0.05 U) (Fig. 3, lanes 2 and 3).
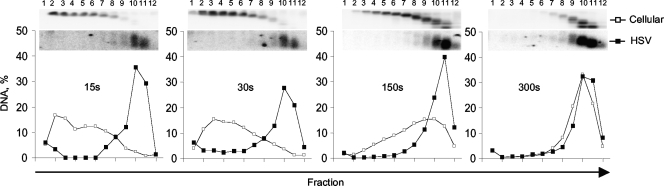
Briefly, nuclei of infected cells were harvested at 5 hpi and digested with MCN for 15, 30, 150, or 300 s before being lysed and resolved into “soluble” and “insoluble” chromatin. Soluble chromatin was further resolved by sucrose gradient centrifugation and analyzed by Southern blotting. As expected, brief digestions (15 or 30 s) released only large polynucleosomes from cellular chromatin (cellular fractions 1 to 8) (Fig. 4). Cellular mono- and dinucleosomes fractionating to fractions 10 and 11 were released only after 150 s. At 300 s, mono- and dinucleosomes accounted for 55% of the cellular DNA released as soluble chromatin (Fig. 4). In contrast, HSV-1 DNA was released already at 15 s only in complexes that resolved to fractions 10 and 11 (Fig. 4). In fact, mostly all HSV-1 DNA released into the soluble fraction at any MCN digestion time was in complexes that fractionate as mono- to dinucleosomes (Fig. 4) even at the mildest MCN digestions (which release >97% of cellular DNA in large polynucleosomes) (Fig. 4, compare fractions 10 and 11 for HSV and cellular polynucleosomes at 15 s). Consistent with the results presented in Fig. 2, the HSV-1 DNA in the released complexes was protected to heterogeneous sizes ranging from mono- to dinucleosome-sized DNA.
FIG. 4.
Most HSV-1 DNA released by MCN as soluble chromatin is in complexes that fractionate as mono- to dinucleosomes. Nuclei of infected cells were digested for 15, 30, 150, or 300 s with 0.05 U of MCN per 1 × 107 nuclei. The soluble DNA-protein complexes were resolved on sucrose gradients. DNA from each fraction was analyzed by Southern blotting with HSV-1- or cell-specific probes. Line graphs present cellular and HSV-1 DNAs in each fraction as a percentage of DNA in the gradient. Inserts on top show images of the membranes hybridized with HSV-1 or cellular probes, resized to fit the graph. Bottom fractions are loaded to the left.
Complexes containing HSV-1 or cellular DNA resolve to different fractions at low salt concentrations.
HSV-1 DNA is in complexes that fractionate as cellular mono- to dinucleosomes even after extensive fractionation. However, the HSV-1 DNA within these complexes is more accessible to MCN than the DNA in cellular nucleosomes (Fig. 2). The increased accessibility may result from an instability of the HSV-1 DNA-protein interactions. HSV-1 DNA may alternatively interact nonspecifically with cellular nucleosomes, thereby leading to cofractionation and partial protection from MCN. We therefore evaluated the stability of these interactions. If HSV-1 DNA interacts with cellular chromatin nonspecifically, it should cofractionate better in the absence of salt, whereas a disruption of such interactions with salt should result in less cofractionation. Neither inter- nor intranucleosome interactions are disrupted at NaCl concentrations below 225 mM NaCl (5, 11), whereas 450 mM NaCl disrupts inter- but not intranucleosome interactions (5, 11).
Nuclei from infected cells were harvested at 5 hpi and digested with MCN for 150 s. The soluble chromatin was resolved on sucrose gradients containing various NaCl concentrations. The fractions were collected and analyzed by Southern blotting. Consistent with the experiments presented in Fig. 4, cellular DNA was released primarily as large polynucleosomes. The pattern of fractionation of cellular DNA did not change much as the concentration of NaCl was increased from 0 to 225 mM, as expected (5, 11). Only ∼15% of the DNA in soluble cellular chromatin was released as mono- or dinucleosomes (cellular fractions 10 and 11 at 0 and 80 mM NaCl and fractions 11 and 12 at 225 mM NaCl) (Fig. 5). When the concentration of salt was increased to 450 mM, however, the cellular DNA that fractionated as mono- and dinucleosomes increased from 15% to 24% (fractions 11 and 12) (Fig. 5), as expected from the disruption of internucleosome interactions. Without such a disruption, some mononucleosomes fractionated in the pellet by interacting with larger polynucleosomes. Therefore, ∼9% of the cellular DNA released by 150 s of MCN digestion was in mono- or dinucleosomes bound tightly to larger polynucleosomes. In contrast, the HSV-1 DNA released as soluble chromatin was primarily in complexes that fractionated as mono- to dinucleosomes even in the absence of NaCl (fractions 9 to 11) (Fig. 5), when most cellular DNA fractionated as polynucleosomes (fraction 9) (Fig. 5).
FIG. 5.
HSV-1 DNA- or cellular DNA-containing complexes resolve to different fractions at low salt concentrations. Nuclei of infected cells were digested for 150 s with 0.05 U MCN per 1 × 107 nuclei and then lysed. The soluble DNA-protein complexes were resolved on sucrose gradients containing 0, 80, 225, or 450 mM NaCl. DNA from each fraction was analyzed by Southern blotting with HSV-1- or cell-specific probes. Line graphs present cellular and HSV-1 DNA in each fraction as a percentage of DNA in the gradient. Insets on the top show images of the membrane hybridized with HSV-1 or cellular probes, resized to fit the graphs. Bottom fractions are loaded to the left.
Although all the HSV-1 DNA that was released into the soluble chromatin at any NaCl concentration always migrated as mono- to dinucleosomes, the absolute amounts released into the soluble chromatin as mono- to dinucleosomes increased as the salt concentration increased. Therefore, a percentage of the HSV-1 DNA released as mono- to dinucleosomes still fractionated to the pellet, with the largest cellular and viral polynucleosomes at salt concentrations that do not disrupt internucleosome interactions, but was released into the soluble chromatin at concentrations that disrupt them. These results suggest that HSV-1 DNA is in nucleosome-like complexes that interact with nucleosome-like affinities with longer cellular (or viral) polynucleosomes.
At 5 hpi, therefore, HSV-1 DNA does not cofractionate with cellular nucleosomes through nonspecific interactions. Instead, it is in complexes with biophysical properties similar to those of cellular nucleosomes.
Nuclear HSV-1 DNA is in unstable nucleoprotein complexes.
Different MCN digestions released as little as 2% (15 s) or as much as 23% (150 s) of detectable HSV-1 DNA as soluble chromatin. Curiously, all this HSV-1 DNA was in complexes that fractionate as cellular mono- to dinucleosomes (Fig. 4). Therefore, only a small percentage of nuclear HSV-1 DNA may be in nucleosome-like complexes, whereas most may not be associated with proteins, and therefore quickly degraded by MCN (Fig. 6A and B). Alternatively, HSV-1 DNA may be in unstable nucleosome-like complexes, which, because of their instability, still allow MCN access to their DNA. Unstable HSV-1 DNA-containing complexes are expected to be rapidly released by MCN but then also rapidly degraded, resulting in only a small percentage of nucleosome-like complexes being detected at any given digestion time (Fig. 6C). To differentiate between these possibilities, we modified the MCN digestions to “trap” the potential digestion intermediates, preventing their degradation (Fig. 7A).
FIG. 6.
Potential types of HSV-1 nucleoprotein complexes. Cartoons represent three potential models of HSV-1 DNA nucleoprotein complexes in lytically infected cells. (A and B) A small percentage of HSV-1 genomes is either regularly chromatinized (A) or irregularly chromatinized with randomly positioned nucleosomes (B). Protein-free genomes are represented as straight lines without histones. Sites of MCN digestion are indicated by arrows. MCN first randomly cleaves the protein-free genomes and the linker region between nucleosomes. Protein-free genomes are completely digested by longer digestions, whereas chromatinized genomes are protected to mononucleosome sizes. As a result, only a small percentage of HSV-1 DNA is protected to nucleosome-sized fragments or coimmunoprecipitates with histones. (C) Most HSV-1 DNA is in unstable nucleosome-like complexes. Unstable nucleosomes are represented with dotted lines. MCN first cleaves the DNA within the unstable nucleosomes and the linker DNA. DNA within the unstable nucleosomes is then promptly degraded. As a result, only a minor fraction of HSV-1 is protected to nucleosome-sized fragments at any given time or coimmunoprecipitates with histones.
FIG. 7.
HSV-1 DNA fractionates as polynucleosomes after modified MCN digestion. (A) Cartoon representing the modified MCN digestion protocol designed to “trap” potential intermediate unstable HSV-1 nucleosome-like complexes. MCN digestion randomly cleaves the protein-free genomes and linker DNA as well as the DNA within the unstable nucleosomes. “Soluble chromatin” remains in the supernatant, unlike the “insoluble chromatin” pellets. Soluble chromatin is removed, and MCN is quenched. Insoluble chromatin is resuspended with fresh MCN digestion buffer. The process is repeated nine times. (B) Nuclei of infected cells were lysed, and soluble chromatin and insoluble chromatin were fractionated. Insoluble chromatin was resuspended in MCN digestion buffer (0.05 U MCN/ml) and subjected to modified MCN digestions. Supernatants were periodically removed and quenched, and the insoluble pellets were resuspended with fresh MCN. Soluble DNA-protein complexes were pooled and further resolved on sucrose gradients. DNA from each fraction was analyzed by Southern blotting with HSV-1- or cell-specific probes. Images of the membranes hybridized with cell- or HSV-1-specific probes are shown.
Briefly, cells were infected and nuclei were harvested at 5 hpi. So-called soluble and insoluble chromatins were fractionated without MCN digestion. The insoluble chromatin was then digested with MCN, and the released soluble chromatin was periodically removed. To do so, insoluble chromatin was resuspended in MCN digestion buffer (0.05 U MCN/ml) and digested during differential centrifugation. The insoluble chromatin was thus pelleted, whereas the soluble chromatin released by MCN digestion remained in the supernatant. The supernatant was removed after 5 min, and the MCN in it was immediately quenched to prevent further digestion of the released soluble chromatin. The insoluble chromatin pellet was resuspended with fresh MCN digestion buffer, and the entire procedure was repeated nine times. The soluble fractions were pooled, resolved together by sucrose gradients, and analyzed by Southern blotting (Fig. 7).
Under these conditions, the cellular DNA released as soluble chromatin fractionated as mono- and dinucleosome (fractions 10 and 11) (Fig. 7B) and larger polynucleosome complexes (fractions 5 to 9) (Fig. 7B), as expected from regularly chromatinized DNA. Unlike the standard continuous MCN digestion, however, the HSV-1 DNA released by our modified digestion was also in complexes that resolved to the same fractions as cellular polynucleosomes (HSV fractions 5 to 9) (Fig. 7B) as well as in complexes fractionating as mono- to dinucleosomes (HSV fractions 10 and 11) (Fig. 7B). Moreover, the HSV-1 DNA was protected to more discrete sizes under these conditions than under conditions of continuous MCN digestions (Fig. 2A and 7, compare fractions 10 and 11). Therefore, HSV-1 DNA fractionates as cellular polynucleosomes when MCN digestion of the soluble fraction is restricted (Fig. 7). Standardized by the recovery of cellular DNA, 78% of the total nuclear HSV-1 DNA fractionated as soluble chromatin, leaving only 22% in the insoluble chromatin pellet (Table 3), very much like the DNA in cellular chromatin (Table 3). These results indicate that the majority of nuclear HSV-1 DNA is in unstable complexes that fractionate as cellular nucleosomes and protect their DNA to similar sizes as cellular nucleosomes do.
TABLE 3.
HSV-1 DNA is quantitatively recovered as polynucleosomes after modified MCN digestiona
| DNA | % of DNA in fraction |
|
|---|---|---|
| Insoluble | Soluble | |
| Cellular | 26 | 74 |
| HSV | 22 | 78 |
Shown are percentages of HSV-1 or cellular DNA fractionating in soluble and insoluble fractions after being digested according to the modified MCN digestion protocol, standardized by the recovery of cellular DNA.
In summary, HSV-1 DNA can be isolated in complexes that fractionate as cellular polynucleosomes as long as MCN activity is rapidly quenched after the release of the complexes containing HSV-1 DNA into the soluble chromatin. These results suggest that the majority of nuclear HSV-1 DNA is in complexes that are highly unstable and, as such, are rapidly degraded by standard MCN digestions.
HSV-1 nucleosome-like complexes are stabilized by cross-linking.
Particularly unstable cellular nucleosomes are digested by MCN unless stabilized by cross-linking (40, 41). We therefore evaluated the stability of the HSV-1 DNA-containing complexes by subjecting them to MCN redigestion.
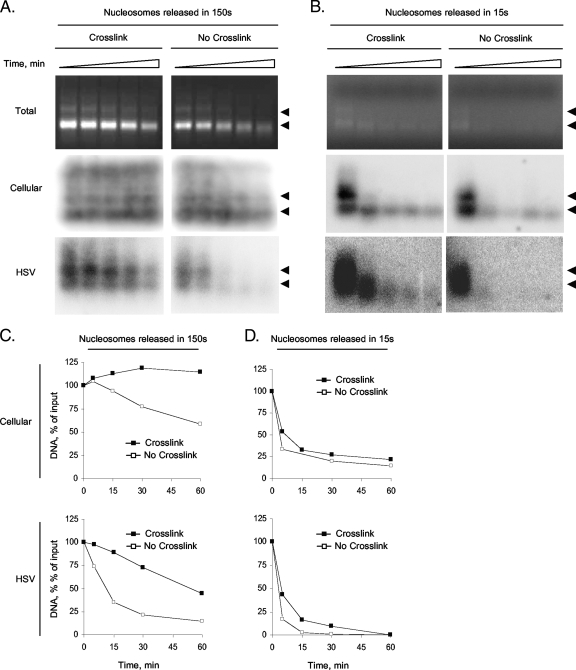
We first evaluated the stability of the complexes released by moderate (150-s) MCN digestions. These digestions release mostly mononucleosomes, with a small percentage of dinucleosomes (Fig. 8A). Cross-linking resulted in essentially complete protection from MCN redigestion (T50, >60 min) (Table 4 and Fig. 8A and C). Consistent with the stability of most cellular nucleosomes, moreover, even the non-cross-linked cellular nucleosomes were still resistant to MCN redigestion. More than 50% of cellular DNA in non-cross-linked nucleosomes was still protected from 60 min of MCN redigestion (T50, >60 min) (Table 4 and Fig. 8A and C). Cross-linking also stabilized the HSV-1 nucleosome-like complexes, resulting in a 6-fold increase in the time required to digest 50% of the HSV-1 DNA (T50, 9 and 54 min, respectively) (Table 4 and Fig. 8A and C). Consistent with the proposed instability of the HSV-1 nucleosome-like complexes, however, HSV-1 DNA in the non-cross-linked samples was digested >6-fold-more rapidly than cellular DNA (T50, 9 and >60 min, respectively; T90, 54 min and nondeterminable, respectively) (Table 4 and Fig. 8A and C). As in the experiments presented in Fig. 3, 4, and 7, therefore, HSV-1 DNA in non-cross-linked (or cross-linked) nucleosome-like complexes is far more accessible to MCN than DNA in cellular nucleosomes.
FIG. 8.
Unstable HSV-1 nucleosome-like complexes are partially stabilized by cross-linking. Nuclei of infected cells were digested with 0.05 U MCN per 1 × 107 nuclei for 150 s or 15 s and lysed. Soluble DNA-protein complexes were resolved on sucrose gradients, and the relevant fractions from the sucrose gradients were then further fractionated by size-exclusion chromatography. Relevant fractions from the size-exclusion columns were either cross-linked or not for 1 h at 4°C, quenched with 125 mM glycine for 10 min, and redigested with 0.05 U/ml MCN for 0, 5, 15, 30, and 60 min. DNA was analyzed by Southern blotting with HSV-1 or cellular probes. (A and B) Images of ethidium bromide-stained gels (Total) and membranes hybridized with cell- or HSV-1-specific probes. Different exposures are shown for each hybridization. Arrowheads indicate the migration of mono- or dinucleosomes. (C and D) Line graphs presenting cellular and HSV-1 DNAs against digestion time, expressed as a percentage of DNA prior to digestion.
TABLE 4.
Unstable HSV-1 nucleosomes are partially stabilized by cross-linkinga
| Nucleosome and presence of cross-link | T50 (min) | T90 (min) |
|---|---|---|
| In moderately accessible chromatin | ||
| Cell | ||
| No cross-link | >60 | >60 |
| Cross-link | >60 | >60 |
| HSV | ||
| No cross-link | 9 | >60 |
| Cross-link | 54 | >60 |
| In mostly accessible chromatin | ||
| Cell | ||
| No cross-link | 3.8 | >60 |
| Cross-link | 6.5 | >60 |
| HSV | ||
| No cross-link | 3 | 10 |
| Cross-link | 4.4 | 30 |
Shown are the digestion times required to degrade 50% (T50) and 90% (T90) of the total DNA from nucleosome fractions previously cross-linked with formaldehyde.
We next evaluated the stability of the complexes released by very mild (15-s) MCN digestions (Fig. 8B and D), which, by definition, release only the most accessible complexes. According to the proposed model, these complexes should be the most unstable. As expected, very little DNA was released as mononucleosomes by such mild digestions, as observed by ethidium bromide staining (Fig. 8B). Nonetheless, both cellular and HSV-1 DNAs were detected by Southern blotting.
Consistent with the release of the most unstable nucleosomes, the cellular nucleosomes in this soluble chromatin were >15-fold-more accessible to MCN redigestion than those released by moderate (150-s) MCN digestions (T50, 3.8 and >60 min, respectively) (Table 4 and Fig. 8A and C). The nucleosomes released by 15-s digestions were in fact so unstable that cross-linking only modestly (1.7-fold) stabilized them (T50, 3.8 and 6.5 min for non-cross-linked and cross-linked complexes, respectively) (Table 4 and Fig. 8B and D). Approximately 75% of the DNA in the highly accessible cellular nucleosomes was in these unstable nucleosomes and therefore digested during the first 15 min (Fig. 8D). In addition to these unstable nucleosomes, however, there was also a population of more stable ones. The DNA in these nucleosomes was much less accessible to MCN, and it was thus digested at a much lower rate. Only approximately 10% of the DNA in the latter complexes was digested between 15 and 60 min (Fig. 8B and D).
The HSV-1 DNA in the unstable nucleosomes released by the very brief MCN digestion was also rapidly degraded by MCN redigestion, resulting in a T50 very similar to that of the DNA in the most unstable cellular nucleosomes (3 and 3.8 min, respectively) (Table 4 and Fig. 8). Also, like the most unstable cellular nucleosomes, cross-linking resulted in only slight (1.5-fold) protection from redigestion (T50, 4.4 and 3 min, respectively) (Table 4 and Fig. 8). Cross-linking also resulted in a modest increase (3-fold) in the time required to digest 90% of DNA (T90) (Table 4 and Fig. 8). In contrast to the DNA in the most accessible cellular nucleosomes, however, there was no significant population of HSV-1 DNA that was resistant to MCN redigestion even after cross-linking (Table 4 and Fig. 8). These results indicate that most of the complexes released from cellular and HSV-1 genomes by a brief MCN digestion are similarly unstable. However, whereas the unstable cellular nucleosomes are only a minority, essentially all the HSV-1 DNA-containing complexes are unstable. In summary, HSV-1 DNA is in unstable complexes that are as accessible as the most unstable cellular nucleosomes.
DISCUSSION
Here we show that most nuclear HSV-1 DNA in lytically infected cells is in complexes that fractionate as DNA in cellular nucleosomes at 5 hpi. Furthermore, these complexes have many biophysical properties similar to those of cellular mono- to di- nucleosomes. However, they also have several unique properties. Most importantly, the nucleosome-like complexes containing HSV-1 DNA are more unstable than most cellular nucleosomes.
Classical MCN digestions showed that HSV-1 DNA in nuclei of lytically infected cells is digested to heterogeneously sized fragments (52, 53, 65). Such results were inconsistent with regular chromatinization. It was therefore concluded that the majority of HSV-1 DNA was not chromatinized during lytic infections (52, 53, 65). In recent years, however, ChIP assays have repeatedly shown that histones and chromatin-modifying proteins (such as CBP/P300, Brm-1, and Brg) interact with HSV-1 genomes in the nuclei of lytically infected cells and that such an association may correlate with transcription (14, 33, 34, 42, 47-49, 72). Most current models therefore propose that chromatin regulates HSV-1 transcription in lytically infected cells. However, only a small percentage of HSV-1 DNA was found to coimmunoprecipitate with histones in all previously published papers (33, 34, 42, 47-49, 72) but one (14) or to be protected from MCN in sizes corresponding to nucleosome DNA (52, 65). It therefore remains unclear how the chromatinization of only a small percentage of HSV-1 DNA may globally regulate HSV-1 transcription. Furthermore, the complexes containing HSV-1 DNA remain uncharacterized. Our objective was therefore to evaluate the HSV-1 DNA-containing complexes in lytically infected cells. Differing from previous work, however, we used classical chromatin purification techniques, an approach that led us to unexpected results.
To our knowledge, this is the first systematic characterization of the biophysical properties of HSV-1 DNA-containing complexes in lytically infected cells. For this initial characterization, we selected a single time point, 5 hpi. At this time, all classes of HSV-1 genes are being transcribed, HSV-1 DNA is being replicated, and histones interact with HSV-1 DNA (14, 33, 42, 47, 49, 72). The nucleosome instability detected in our experiments may occur only at specific times of infection. In ongoing experiments, therefore, we are analyzing the biophysical properties of the complexes formed by HSV-1 DNA in lytically infected cells at times other than 5 hpi.
Most current models, which propose that only a minor percentage of HSV-1 DNA is associated with histones, predict that most HSV-1 DNA should fractionate as protein-free DNA. However, only 28% of the nuclear HSV-1 DNA detected fractionated as such (Fig. 1), indicating that the majority of nuclear HSV-1 DNA is in some sort of complex. Further analyses of these complexes after partial MCN digestion revealed that they fractionate as cellular mono- to dinucleosomes by differential centrifugation followed by sucrose gradients and then size-exclusion chromatography. However, these complexes differ from most cellular nucleosomes in three major aspects. First, the HSV-1 DNA in these complexes is far more accessible to MCN than DNA in most cellular chromatin (Fig. 3, 7, and 8) (52, 53, 65). Second, the interactions between the HSV-1 DNA complexes or between these complexes and cellular polynucleosomes are more unstable than the interactions between or within cellular nucleosomes (Fig. 5 and 7). Such an instability leads to the prompt dissociation of the HSV-1 DNA-containing complexes released into the soluble chromatin, allowing MCN access to the DNA within them, which is thus degraded. If digestion of the soluble fraction was prevented, however, the unstable digestion intermediates were obvious (Fig. 7). Third, the unstable HSV-1 DNA-containing complexes could be only partially stabilized from MCN redigestion by cross-linking. These unstable HSV-1 complexes have the same instability as the most unstable cellular nucleosomes (Fig. 8).
The instability of the HSV-1 DNA-containing complexes could be the result of weak HSV-1 DNA-protein or weak protein-protein interactions. Particularly unstable cellular nucleosomes were characterized previously (32). Jin et al. previously showed that such unstable cellular nucleosomes required cross-linking for stabilization (40, 41), similar to the unstable HSV-1 nucleosome-like complexes that we characterize here (Fig. 8). Instability within the cellular nucleosomes was attributed to the presence of histone variants such as H3.3. H3.3 is the H3 variant synthesized in G1, S, G2, and G0 phases (81) and assembled into nucleosomes via replication-independent pathways (77). H3.3 would therefore be the most likely H3 variant available to be assembled in nucleosomes with infecting HSV-1 genomes. Consistently, Placek et al. showed by ChIP that H3.3 associates with infecting HSV-1 DNA (72). The association was observed from as early as 1 hpi to as late as 10 hpi (72). In contrast, the association between HSV-1 DNA and the canonical H3 variant H3.1 was not observed until 6 hpi (i.e., later than the times tested herein) and was dependent on HSV-1 DNA replication (72), consistent with the normal replication-coupled deposition of H3.1 (77). Unpublished results from our laboratory show that the available pool of free H3.3 increases during HSV-1 infection (Conn et al., unpublished), suggesting a potential source of H3.3 available for its incorporation into unstable nucleosome-like complexes containing HSV-1 DNA.
The proposed unstable nucleosome-like complexes are consistent with the heterogeneously sized HSV-1 DNA fragments released by MCN digestion, which we and many others have observed (Fig. 3) (42, 52, 53, 65). HSV-1 DNA in unstable complexes would still remain randomly accessible to MCN. Such unstable complexes would, by definition, be dynamic and therefore would not expected to be tightly confined to 160 bp of DNA. They would be expected to quickly change positions by either unbinding and rebinding to, or by sliding through, the HSV-1 DNA. In contrast to DNA in most cellular chromatin, therefore, MCN cleavage of HSV DNA would not be restricted by distinctly exposed (linker) or protected (core) sites.
If HSV-1 DNA was indeed in unstable nucleosomes, histones must continuously bind and unbind to HSV-1 DNA and consequently should be mobilized in infected cells. Consistently, linker (16) and core (Conn et al., unpublished) histones are mobilized in infected cells at 4 and 7 hpi (i.e., flanking the times tested in the current experiments).
Our proposed model is also consistent with the otherwise intriguing distribution of the sizes of HSV-1 DNA fragments released by standard MCN digestions (Fig. 3) (42, 52, 65). Besides the heterogeneously sized fragments, only mono-, di-, and, occasionally, trinucleosome-sized DNA fragments are visible, whereas tetra-, penta-, or other polynucleosome-sized DNA fragments are not. Each of the unstable nucleosome-like complexes has an equal and independent probability of dissociating in any given period. Therefore, the probability of detecting several adjacent complexes still together at any given time decreases exponentially. If each complex has a 75% chance of disassembling in a given period, for example, then the relative abundance of mono-, di-, tri-, and tetranucleosome-like complexes at any given digestion time would be 1:0.25:0.06:0.01, respectively, making the detection of tetranucleosome- or longer polynucleosome-sized DNA extremely difficult. Furthermore, consistent with the proposed model, the polynucleosome-sized HSV-1 DNA is obvious when the degradation of the unstable digestion intermediates is prevented by the modified MCN digestion protocol.
With only one exception (14), ChIP assays consistently showed that a small and variable percentage of HSV-1, in comparison to cellular DNA, coimmunoprecipitates with histones (33, 34, 42, 47-49, 72). This small percentage has often been interpreted as low histone occupancy (50). However, the proposed unstable HSV-1 nucleosomes are also entirely consistent with such small percentages. Interactions between HSV-1 DNA and histones in unstable nucleosomes are transient by definition. Histones in unstable complexes therefore spend less time within cross-linking distance from DNA. At any given time, the probability of a given number of histones being cross-linked to DNA is lower, resulting in a small percentage of HSV-1 DNA coimmunoprecipitating with them (41). Jin et al. recently characterized highly unstable cellular nucleosomes (40, 41). Under standard conditions, the histone variants H2A.Z and H3.3 could not be detected within the same nucleosomes. When the interactions within these complexes were stabilized by cross-linking, however, nucleosomes containing both variants were detected (41).
ChIP results also show that although the occupancy appears to be low, the coverage throughout the HSV-1 genome appears to be relatively even (14, 33, 42, 49, 72). The most common interpretation is that histones bind randomly through the entire HSV genome at low occupancy. Through the modification of the MCN digestion protocol, however, we quantitatively “trapped” most HSV-1 DNA in polynucleosome-like complexes, consistent with more or less regularly spaced nucleosomes (Fig. 7). These results suggest that the entire HSV-1 genome may instead be “regularly chromatinized” albeit in unstable nucleosomes.
We have extensively characterized the highly accessible HSV-1 DNA released as soluble chromatin. HSV-1 DNA also fractionates to the insoluble fraction during the first differential centrifugation (Fig. 1 and 2). The HSV-1 DNA in this insoluble fraction could be encapsidated. However, three pieces of evidence strongly suggest that most of it is not. First, the HSV-1 DNA fractionating to the insoluble fraction after BamHI digestion was partially digested by BamHI (Fig. 1). Therefore, the HSV-1 DNA in this fraction was still accessible to BamHI. Likewise, the HSV-1 DNA in the insoluble fraction after MCN digestion was also partially digested to heterogeneously sized fragments (Fig. 2). Second, most of the HSV-1 DNA migrating as a “resistant” band at the top of the agarose gel after standard MCN digestion was still digested by higher MCN concentrations, indicating again that this DNA is still accessible (Fig. 3). Third, the experiments were performed at 5 hpi (at a multiplicity of infection of 5 PFU per cell), when most HSV-1 DNA is in replication intermediates and therefore decapsidated (37).
In summary, the results presented here show that at 5 hpi, most HSV-1 DNA in lytically infected cells is in complexes with the biophysical properties of highly unstable nucleosomes. Our efforts are now directed at further characterizing these novel HSV-1 DNA-containing complexes at different times after infection and at testing their importance in the HSV-1 replication cycle.
Acknowledgments
This work was supported by the Canadian Institute for Health Research (CIHR) and the Burroughs Wellcome Fund (BWF).
L.M.S. is a BWF Investigator in the Pathogenesis of Infectious Disease.
Footnotes
Published ahead of print on 9 December 2009.
In memory of Priscilla A. Schaffer.
REFERENCES
- 1.Alazard, N., H. Gruffat, E. Hiriart, A. Sergeant, and E. Manet. 2003. Differential hyperacetylation of histones H3 and H4 upon promoter-specific recruitment of EBNA2 in Epstein-Barr virus chromatin. J. Virol. 77:8166-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amelio, A. L., N. V. Giordani, N. J. Kubat, J. E. O'Neil, and D. C. Bloom. 2006. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 80:2063-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amelio, A. L., P. K. McAnany, and D. C. Bloom. 2006. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J. Virol. 80:2358-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ascoli, C. A., and G. G. Maul. 1991. Identification of a novel nuclear domain. J. Cell Biol. 112:785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausio, J., F. Dong, and K. E. van Holde. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 206:451-463. [DOI] [PubMed] [Google Scholar]
- 6.Axel, R., W. Melchior, Jr., B. Sollner-Webb, and G. Felsenfeld. 1974. Specific sites of interaction between histones and DNA in chromatin. Proc. Natl. Acad. Sci. U. S. A. 71:4101-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellard, M., P. Oudet, J. E. Germond, and P. Chambon. 1976. Subunit structure of simian-virus-40 minichromosome. Eur. J. Biochem. 70:543-553. [DOI] [PubMed] [Google Scholar]
- 8.Chang, L. K., and S. T. Liu. 2000. Activation of the BRLF1 promoter and lytic cycle of Epstein-Barr virus by histone acetylation. Nucleic Acids Res. 28:3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chau, C. M., X. Y. Zhang, S. B. McMahon, and P. M. Lieberman. 2006. Regulation of Epstein-Barr virus latency type by the chromatin boundary factor CTCF. J. Virol. 80:5723-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, P., B. Panning, and J. R. Smiley. 1997. Herpes simplex virus immediate-early proteins ICP0 and ICP4 activate the endogenous human alpha-globin gene in nonerythroid cells. J. Virol. 71:1784-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christiansen, G., and J. Griffith. 1977. Salt and divalent cations affect the flexible nature of the natural beaded chromatin structure. Nucleic Acids Res. 4:1837-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christiansen, G., T. Landers, J. Griffith, and P. Berg. 1977. Characterization of components released by alkali disruption of simian virus 40. J. Virol. 21:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cliffe, A. R., D. A. Garber, and D. M. Knipe. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 83:8182-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cliffe, A. R., and D. M. Knipe. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82:12030-12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. J. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conn, K. L., M. J. Hendzel, and L. M. Schang. 2008. Linker histones are mobilized during infection with herpes simplex virus type 1. J. Virol. 82:8629-8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cremisi, C., P. F. Pignatti, O. Croissant, and M. Yaniv. 1975. Chromatin-like structures in polyoma virus and simian virus 10 lytic cycle. J. Virol. 17:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Day, L., C. M. Chau, M. Nebozhyn, A. J. Rennekamp, M. Showe, and P. M. Lieberman. 2007. Chromatin profiling of Epstein-Barr virus latency control region. J. Virol. 81:6389-6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng, Z., C. J. Chen, M. Chamberlin, F. Lu, G. A. Blobel, D. Speicher, L. A. Cirillo, K. S. Zaret, and P. M. Lieberman. 2003. The CBP bromodomain and nucleosome targeting are required for Zta-directed nucleosome acetylation and transcription activation. Mol. Cell. Biol. 23:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deshmane, S. L., and N. W. Fraser. 1989. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 63:943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diwan, P., J. J. Lacasse, and L. M. Schang. 2004. Roscovitine inhibits activation of promoters in herpes simplex virus type 1 genomes independently of promoter-specific factors. J. Virol. 78:9352-9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferenczy, M. W., and N. A. Deluca. 2009. Epigenetic modulation of gene expression from quiescent herpes simplex virus genomes. J. Virol. 83:8514-8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson, W., and B. Roizman. 1971. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc. Natl. Acad. Sci. U. S. A. 68:2818-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giordani, N. V., D. M. Neumann, D. L. Kwiatkowski, P. S. Bhattacharjee, P. K. McAnany, J. M. Hill, and D. C. Bloom. 2008. During herpes simplex virus type 1 infection of rabbits, the ability to express the latency-associated transcript increases latent-phase transcription of lytic genes. J. Virol. 82:6056-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greil, W., T. Igo-Kemenes, and H. G. Zachau. 1976. Nuclease digestion in between and within nucleosomes. Nucleic Acids Res. 3:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffith, J. D. 1975. Chromatin structure: deduced from a minichromosome. Science 187:1202-1203. [DOI] [PubMed] [Google Scholar]
- 28.Gu, H., Y. Liang, G. Mandel, and B. Roizman. 2005. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:7571-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu, H., and B. Roizman. 2007. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc. Natl. Acad. Sci. U. S. A. 104:17134-17139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall, M. R., N. Aghili, C. Hall, J. Martinez, and S. St. Jeor. 1982. Chromosomal organization of the herpes simplex virus type 2 genome. Virology 123:344-356. [DOI] [PubMed] [Google Scholar]
- 31.Hendzel, M. J., M. A. Lever, E. Crawford, and J. P. Th'ng. 2004. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 279:20028-20034. [DOI] [PubMed] [Google Scholar]
- 32.Henikoff, S. 2008. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9:15-26. [DOI] [PubMed] [Google Scholar]
- 33.Herrera, F. J., and S. J. Triezenberg. 2004. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 78:9689-9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, J., J. R. Kent, B. Placek, K. A. Whelan, C. M. Hollow, P. Y. Zeng, N. W. Fraser, and S. L. Berger. 2006. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J. Virol. 80:5740-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ioudinkova, E., M. C. Arcangeletti, A. Rynditch, F. De Conto, F. Motta, S. Covan, F. Pinardi, S. V. Razin, and C. Chezzi. 2006. Control of human cytomegalovirus gene expression by differential histone modifications during lytic and latent infection of a monocytic cell line. Gene 384:120-128. [DOI] [PubMed] [Google Scholar]
- 36.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacob, R. J., and B. Roizman. 1977. Anatomy of herpes simplex virus DNA. VIII. Properties of the replicating DNA. J. Virol. 23:394-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakobovits, E. B., S. Bratosin, and Y. Aloni. 1980. A nucleosome-free region in SV40 minichromosomes. Nature 285:263-265. [DOI] [PubMed] [Google Scholar]
- 39.Jenkins, P. J., U. K. Binne, and P. J. Farrell. 2000. Histone acetylation and reactivation of Epstein-Barr virus from latency. J. Virol. 74:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin, C., and G. Felsenfeld. 2007. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21:1519-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin, C., C. Zang, G. Wei, K. Cui, W. Peng, K. Zhao, and G. Felsenfeld. 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 41:941-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kent, J. R., P. Y. Zeng, D. Atanasiu, J. Gardner, N. W. Fraser, and S. L. Berger. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 78:10178-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knight, J. S., K. Lan, C. Subramanian, and E. S. Robertson. 2003. Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J. Virol. 77:4261-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kornberg, R. D. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184:868-871. [DOI] [PubMed] [Google Scholar]
- 45.Kristie, T. M., Y. Liang, and J. L. Vogel. 12 August 2009, posting date. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim. Biophys. Acta [Epub ahead of print.] doi: 10.1016/j.bbagrm.2009.08.003. [DOI] [PMC free article] [PubMed]
- 46.Kubat, N. J., A. L. Amelio, N. V. Giordani, and D. C. Bloom. 2004. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J. Virol. 78:12508-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kutluay, S. B., S. L. DeVos, J. E. Klomp, and S. J. Triezenberg. 2009. Transcriptional coactivators are not required for herpes simplex virus type 1 immediate-early gene expression in vitro. J. Virol. 83:3436-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kutluay, S. B., J. Doroghazi, M. E. Roemer, and S. J. Triezenberg. 2008. Curcumin inhibits herpes simplex virus immediate-early gene expression by a mechanism independent of p300/CBP histone acetyltransferase activity. Virology 373:239-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kutluay, S. B., and S. J. Triezenberg. 2009. Regulation of histone deposition on the herpes simplex virus type 1 genome during lytic infection. J. Virol. 83:5835-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kutluay, S. B., and S. J. Triezenberg. 2009. Role of chromatin during herpesvirus infections. Biochim. Biophys. Acta 1790:456-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwiatkowski, D. L., H. W. Thompson, and D. C. Bloom. 2009. Polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 83:8173-8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leinbach, S. S., and W. C. Summers. 1980. The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion. J. Gen. Virol. 51:45-59. [DOI] [PubMed] [Google Scholar]
- 53.Lentine, A. F., and S. L. Bachenheimer. 1990. Intracellular organization of herpes simplex virus type 1 DNA assayed by staphylococcal nuclease sensitivity. Virus Res. 16:275-292. [DOI] [PubMed] [Google Scholar]
- 54.Lieberman, P. M. 2006. Chromatin regulation of virus infection. Trends Microbiol. 14:132-140. [DOI] [PubMed] [Google Scholar]
- 55.Lomonte, P., and E. Morency. 2007. Centromeric protein CENP-B proteasomal degradation induced by the viral protein ICP0. FEBS Lett. 581:658-662. [DOI] [PubMed] [Google Scholar]
- 56.Lomonte, P., K. F. Sullivan, and R. D. Everett. 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276:5829-5835. [DOI] [PubMed] [Google Scholar]
- 57.Lomonte, P., J. Thomas, P. Texier, C. Caron, S. Khochbin, and A. L. Epstein. 2004. Functional interaction between class II histone deacetylases and ICP0 of herpes simplex virus type 1. J. Virol. 78:6744-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loret, S., G. Guay, and R. Lippe. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605-8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu, F., J. Zhou, A. Wiedmer, K. Madden, Y. Yuan, and P. M. Lieberman. 2003. Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J. Virol. 77:11425-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu, X., B. Hamkalo, M. H. Parseghian, and J. C. Hansen. 2009. Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry 48:164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu, X., and J. C. Hansen. 2004. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J. Biol. Chem. 279:8701-8707. [DOI] [PubMed] [Google Scholar]
- 62.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75. [DOI] [PubMed] [Google Scholar]
- 63.Maxwell, K. L., and L. Frappier. 2007. Viral proteomics. Microbiol. Mol. Biol. Rev. 71:398-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Monier, K., J. C. Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2:661-665. [DOI] [PubMed] [Google Scholar]
- 65.Muggeridge, M. I., and N. W. Fraser. 1986. Chromosomal organization of the herpes simplex virus genome during acute infection of the mouse central nervous system. J. Virol. 59:764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Narayanan, A., W. T. Ruyechan, and T. M. Kristie. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. U. S. A. 104:10835-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4:649-655. [DOI] [PubMed] [Google Scholar]
- 68.Neumann, D. M., P. S. Bhattacharjee, N. V. Giordani, D. C. Bloom, and J. M. Hill. 2007. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J. Virol. 81:13248-13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Noll, M. 1974. Subunit structure of chromatin. Nature 251:249-251. [DOI] [PubMed] [Google Scholar]
- 70.Oh, J., and N. W. Fraser. 2008. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 82:3530-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pignatti, P. F., and E. Cassai. 1980. Analysis of herpes simplex virus nucleoprotein complexes extracted from infected cells. J. Virol. 36:816-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Placek, B. J., J. Huang, J. R. Kent, J. Dorsey, L. Rice, N. W. Fraser, and S. L. Berger. 2009. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J. Virol. 83:1416-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reeves, M. B., P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J. Gen. Virol. 86:2949-2954. [DOI] [PubMed] [Google Scholar]
- 75.Severini, A., D. G. Scraba, and D. L. Tyrrell. 1996. Branched structures in the intracellular DNA of herpes simplex virus type 1. J. Virol. 70:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Simpson-Holley, M., R. C. Colgrove, G. Nalepa, J. W. Harper, and D. M. Knipe. 2005. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 79:12840-12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 78.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 79.van Leeuwen, H., M. Okuwaki, R. Hong, D. Chakravarti, K. Nagata, and P. O'Hare. 2003. Herpes simplex virus type 1 tegument protein VP22 interacts with TAF-I proteins and inhibits nucleosome assembly but not regulation of histone acetylation by INHAT. J. Gen. Virol. 84:2501-2510. [DOI] [PubMed] [Google Scholar]
- 80.Wright, E., M. Bain, L. Teague, J. Murphy, and J. Sinclair. 2005. Ets-2 repressor factor recruits histone deacetylase to silence human cytomegalovirus immediate-early gene expression in non-permissive cells. J. Gen. Virol. 86:535-544. [DOI] [PubMed] [Google Scholar]
- 81.Wu, R. S., S. Tsai, and W. M. Bonner. 1982. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell 31:367-374. [DOI] [PubMed] [Google Scholar]