Abstract
Hepatitis C virus (HCV) is a liver-tropic pathogen with severe health consequences for infected individuals. Chronic HCV infection can progress to cirrhosis and hepatocellular carcinoma and is a leading indicator for liver transplantation. The HCV core protein is an essential component of the infectious virus particle, but many aspects of its role remain undefined. The C-terminal region of the core protein acts as a signal sequence for the E1 glycoprotein and undergoes dual processing events during infectious virus assembly. The exact C terminus of the mature, virion-associated core protein is not known. Here, we performed genetic analyses to map the essential determinants of the HCV core C-terminal region, as well as to define the minimal length of the protein that can function for infectious virus production in trans.
Hepatitis C virus (HCV) is a major contributor to the development of human liver diseases, infecting approximately 2% of the population, or 130 million people, worldwide (2). Up to 80% of HCV infections progress to chronic hepatitis and can lead to cirrhosis and hepatocellular carcinoma (38). No vaccine exists to prevent HCV infection, and current treatments are frequently inadequate.
HCV is an enveloped virus of the genus Hepacivirus in the family Flaviviridae (30). The single-stranded, positive-sense RNA genome encodes a polyprotein of about 3,000 amino acids, which is processed by viral and host proteases into three structural proteins (the core protein, E1, and E2) and seven presumed nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). The core protein is thought to encapsidate the RNA genome within the virion, forming a complex that is surrounded by a host cell-derived lipid bilayer displaying the envelope glycoproteins, E1 and E2. Although not thought to be components of the virion, p7 and NS2 have recently been implicated in the production of infectious virus (5, 12, 29, 33, 39). The remaining nonstructural proteins, NS3 to NS5B, are essential for genome replication and have additional emerging roles in virus assembly. NS3 possesses RNA helicase/NTPase activities and together with its cofactor, NS4A, forms the major viral protease. NS5B is the RNA-dependent RNA polymerase (reviewed in references 16 and 27).
The core protein is the first protein produced during translation of the incoming viral genome. A signal sequence in its C-terminal region targets the nascent E1 glycoprotein to the endoplasmic reticulum (ER) membrane and is the substrate for processing by two host proteases. Cleavage by signal peptidase (SP) following core amino acid 191 (31) is thought to precede processing by signal peptide peptidase (SPP) (20, 26), an integral membrane aspartyl protease that cleaves within transmembrane segments (37). The C terminus of the mature, infectious-virion-associated core protein has not been determined, but it is speculated to lie between amino acids 173 and 182 (24, 31). SPP processing has been shown to mobilize the core from the ER membrane and enable it to traffic to lipid droplets (20). These triglyceride-rich storage organelles have recently been shown to be the sites of HCV particle assembly (21). Consistent with this finding, impaired SPP activity leads to decreased HCV infectious titers (34). Dual processing of the core proteins is a common feature of the Flaviviridae family. GB virus B, a hepacivirus, and classical swine fever virus, a related pestivirus, encode core proteins that undergo SP and SPP processing during maturation (8, 35). In the genus Flavivirus, the capsid protein undergoes regulated cleavage by the viral NS2B-3 protease and SP; this stepwise processing has been shown to be essential for proper encapsidation of genomes into infectious particles (3).
The development of an infectious cell culture system for HCV has been a major breakthrough in the field (7). Many details of virus morphogenesis and infectivity, however, are still unknown. In this study, we examined the role of the C-terminal portion of the HCV core protein and identified individual amino acids that are essential for infectious virus assembly and core protein stability. Findings from alanine-scanning and transcomplementation studies suggest that at least 177 residues of the core protein are needed to produce infectious particles.
MATERIALS AND METHODS
Cell culture.
Huh-7.5 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and 0.1 mM nonessential amino acids (complete medium). Cells were grown at 37°C in a humidified 5% CO2 atmosphere.
Plasmid construction.
Plasmids were constructed by standard methods, and constructs were verified by restriction enzyme digestion and sequencing of PCR-amplified segments. Descriptions of the cloning strategies are provided below; plasmid and primer sequences are available upon request.
Core mutant genomes.
Mutant genomes were based on the genome of J6/JFH(p7-Rluc2A) (12), a luciferase reporter virus derived from the J6/JFH genotype 2a chimera (15). To facilitate mutagenesis, the 1,290-bp EcoRI/KpnI fragment containing the complete coding sequence for the J6/JFH(p7-Rluc2A) core protein was subcloned into pSL1180 (Amersham, Piscataway, NJ) digested with the same enzymes. The resulting plasmid, pSL1180-EcoRI/KpnI, was used as a template for oligonucleotide-directed mutagenesis in core residues 170 to 191. Engineered mutations were verified by sequencing, and the appropriate EcoRI/KpnI fragments were ligated to the 12,073-bp fragment of J6/JFH(p7-Rluc2A) digested with the same enzymes.
Venezuelan equine encephalitis (VEE) virus vector-based core expression constructs.
A noncytopathic VEE virus green fluorescent protein replicon has been described previously and kindly provided by Ilya Frolov (28). The green fluorescent protein sequence was removed and replaced with a SapI-PstI-SapI multiple-cloning cassette. The full-length core protein coding sequence amplified from J6/JFH was then introduced between the SapI sites, and a stop codon and NotI site were engineered to follow core amino acid 191, yielding vector VEE-C191. To create core truncation mutants, the desired segments of the core sequence were amplified from VEE-C191, with a stop codon and NotI site engineered after the appropriate terminal codon in the reverse primer. PCR products digested with SwaI/NotI were then ligated to the 9,950-bp fragment of VEE-C191 digested with the same enzymes.
In vitro transcription of viral RNA.
Generation of HCV RNA was performed as described previously (15). Briefly, RNA was synthesized from 1 μg XbaI-linearized template using the T7 MEGAscript kit according to the protocol of the manufacturer (Ambion, Austin, TX). Reaction mixtures were incubated for 3 h at 37°C and then subjected to DNase treatment for 15 min at 37°C. Capped VEE virus RNA was similarly synthesized in vitro using the mMESSAGE mMACHINE SP6 kit with 1 μg MluI-linearized template according to the protocol of the kit manufacturer (Ambion). Synthesized viral RNA was purified using the RNeasy kit (Qiagen, Valencia, CA) and eluted in RNA storage buffer (Ambion). RNA was quantified by absorbance at 260 nm, and its quality was verified by agarose gel electrophoresis.
Transfection with RNA.
The introduction of in vitro-transcribed RNA into Huh-7.5 cells by electroporation was performed as described previously (15). Briefly, cells were trypsinized and washed twice before resuspension at 1.5 × 107 cells/ml in ice-cold phosphate-buffered saline (PBS). RNA (2 μg for HCV growth curves or 5 μg of each RNA for transcomplementation analyses) was combined with 6 × 106 cells, and the mixture was immediately subjected to five pulses at 820 V for 99 μs by using an ElectroSquare Porator ECM 830 system (BTX, Holliston, MA). Electroporated cells were incubated at room temperature for 10 min before resuspension in 30 ml of complete medium and plating.
HCV growth curves.
HCV replication and infectivity were measured at 6, 24, 48, and 72 h posttransfection. For analysis of replication, electroporated cells in 24-well plates were washed twice with PBS and lysed with 100 μl of Renilla lysis buffer (Promega, Madison, WI). To assay infectious virus production, culture supernatants from P100 dishes were harvested and replaced with fresh complete medium at each time point. Supernatants were clarified (with a 0.22-μm-pore-size filter) and used to infect naïve Huh-7.5 cells for 48 h before lysis in Renilla lysis buffer (Promega), as described above. Luciferase activity in 10 μl of cell lysate was quantified using a Renilla luciferase substrate (Promega) and a Centro LB960 luminometer (Berthold Technologies, Oak Ridge, TN) according to the manufacturers' protocols.
Transcomplementation assay.
Transcomplementation of the core protein was analyzed by cotransfection of a core deletion mutant lacking amino acids 57 to 160 [J6/JFH(p7-Rluc2A)/Δ57-160] (22) with core protein-expressing VEE virus replicon RNA. Transfected Huh-7.5 cells were seeded into six-well plates, and supernatants were harvested at 24, 48, and 72 h postelectroporation. Naïve Huh-7.5 cells seeded into 24-well plates were infected in triplicate with the clarified supernatants and incubated for 48 h at 37°C. Infected cells were then washed with PBS and lysed in Renilla lysis buffer (Promega) for the luciferase assay as described above.
Virus titration.
HCV infectious titers were determined by a limiting dilution assay as described previously (15). Briefly, Huh-7.5 cells seeded into 96-well plates (6 × 103 cells/well) were infected with serially diluted cell culture supernatants for 72 h at 37°C. Infected cells were detected by immunohistochemical staining for NS5A, and the 50% tissue culture infectious dose (TCID50) was calculated as described (15).
Isolation and quantification of released HCV RNA.
To quantify HCV genome release, total RNA was isolated from 1 ml clarified cell culture supernatant using the QiaAmp UltraSens kit (Qiagen). Quantitative reverse transcription-PCR (qRT-PCR) mixtures (20-μl volumes) were assembled using the LightCycler amplification kit with 2 μl extracted total RNA according to the instructions of the kit manufacturer (Roche, Basel, Switzerland). Primers are directed against the viral 5′ untranslated region. qRT-PCRs were performed using a LightCycler 480 (Roche).
Core protein ELISA.
Release of HCV core protein was quantified using an HCV antigen enzyme-linked immunosorbent assay (ELISA) system according to the instructions of the manufacturer (Ortho Clinical Diagnostics, Raritan, NJ). Briefly, clarified cell culture supernatants at appropriate dilutions were applied to plates coated with a mixture of mouse anti-core monoclonal antibodies (MAb). Antigen was detected by addition of a second MAb cocktail conjugated to horseradish peroxidase (HRP). After washing and development of the plates, the absorbance at 490 nm was measured and core protein was quantified by comparison to a standard curve using SoftMax software.
Intracellular infectivity assay.
To measure intracellular infectious HCV, electroporated cells were trypsinized and collected by centrifugation at 513 × g for 3 min and then resuspended in 500 μl of complete medium. Cells were then lysed by four freeze-thaw cycles and pelleted twice at 3,650 × g to remove cellular debris. Huh-7.5 cells inoculated with clarified supernatants were lysed and assayed for luciferase activity at 48 h postinfection as described above. Infections were carried out in the presence of 10 μg/ml anti-CD81 antibody (JS81; Pharmingen, San Diego, CA) or isotype control antiserum (anti-mouse IgG1; Pharmingen).
Western blot analysis.
Cells were lysed with Renilla lysis buffer (Promega) and homogenized using QIAshredder columns according to the instructions of the manufacturer (Qiagen). Lysates were separated on a NuPAGE Novex 4 to 12% bis-Tris minigel (Invitrogen) and transferred onto Hybond ECL membranes (Amersham). After being blocked with TBS-T (0.02 M Tris [pH 7.4], 0.2 M NaCl, 0.1% Tween 20) containing 6% skim milk, membranes were incubated for 1 h at room temperature or overnight at 4°C in TBS-T containing primary antibodies against the core protein (HCM-071-05 [1:1,000; Austral Biologicals, San Ramon, CA]), NS5A (9E10; 1:2,000) (15), E2 (HCM-091a-5 [1:500; Austral Biologicals]), or β-actin (1:10,000; Sigma; St. Louis, MO). After extensive washing with TBS-T, membranes were probed with HRP-conjugated rabbit anti-mouse IgG (diluted 1:10,000 in TBS-T; Pierce, Rockford, IL) for 1 h at room temperature. Membranes were repeatedly washed with TBS-T and developed with SuperSignal West Femto chemiluminescent substrate (Pierce).
Indirect immunofluorescence microscopy and lipid droplet staining.
Electroporated Huh-7.5 cells were seeded onto glass coverslips, and the coverslips were incubated for 48 h at 37°C. Cells were then fixed with 1% paraformaldehyde in PBS, permeabilized (using 0.1% Triton X-100 in PBS), and blocked for 30 min (using PBS containing 1% FBS). Fixed cells were incubated with primary antibody against the core protein (HCM-071-05 [Austral Biologicals] diluted 1:1,000 in PBS-1% FBS) at 4°C overnight and evaluated with Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes, Eugene, OR). After the cells were washed, lipid droplets were stained with Bodipy 493/503 (1 μg/ml; Invitrogen) for 10 min at room temperature. Coverslips were washed twice with PBS and water and mounted onto microscope slides using ProLong Gold antifade reagent (Molecular Probes).
RESULTS
Identification of residues in the C terminus of the core protein that are critical for infectious virus production.
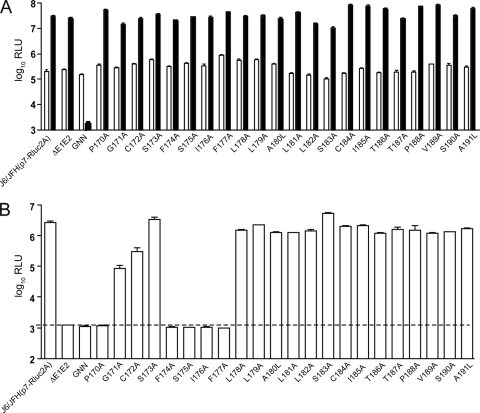
The C-terminal 20 to 22 amino acids of the HCV core protein are thought to function as a signal peptide for E1 and are processed by the cellular enzyme SPP, in addition to being cleaved by SP after amino acid 191. Comprehensive alanine-scanning mutagenesis of core amino acids 170 to 191 was performed to identify residues that play important roles in the C-terminal processing of the protein and/or in the production of infectious virus particles. Single alanine substitutions were created in the context of a J6/JFH reporter virus, J6/JFH(p7-Rluc2A) (12); alanine residues in the wild-type sequence (amino acids 180 and 191) were mutated to leucine. For each construct, in vitro-transcribed RNA was introduced into Huh-7.5 cells by electroporation and RNA replication was quantified by a luciferase assay at various time points; luciferase levels at 6 and 48 h postelectroporation are shown in Fig. 1A. All mutants exhibited replication kinetics comparable to those of the wild-type virus, confirming that the C-terminal region of the HCV core protein is not involved in RNA replication. As expected, a polymerase-defective mutant, J6/JFH(p7-Rluc2A)/GNN, did not replicate.
FIG. 1.
Core mutant replication and infectious virus production. (A) RNA replication, as quantified by a Renilla luciferase assay, at 6 h (white) and 48 h (black) postelectroporation. (B) Infectious virus production at 48 h postelectroporation was measured by assay of Renilla luciferase activity in infected Huh-7.5 cell lysates. The dashed horizontal line represents background levels of luciferase activity. J6/JFH(p7-Rluc2A) residues changed to alanine or leucine are identified. ΔE1E2, J6/JFH(p7-Rluc2A)/ΔE1E2; GNN, J6/JFH(p7-Rluc2A)/GNN; RLU, relative light units. Means and standard errors of the means (SEM) of results from at least duplicate electroporations are shown.
To analyze the production of infectious particles, naïve Huh-7.5 cells were inoculated with filtered supernatants harvested 48 h postelectroporation and Renilla luciferase activity in the infected cell lysates was quantified (Fig. 1B). Single alanine substitutions in the last 14 amino acids of the core protein did not affect infectious titers, as demonstrated by luciferase levels similar to those for the parental J6/JFH(p7-Rluc2A) virus. In contrast, mutation of core residues 170 and 174 to 177 resulted in undetectable infectious virus production, despite wild-type levels of replication. Mutations in residues 171 and 172, while not completely blocking progeny production, significantly decreased this process (Fig. 1B). No infectious virus was seen for a mutant lacking envelope proteins, J6/JFH(p7-Rluc2A)/ΔE1E2, or for the replication-deficient mutant J6/JFH(p7-Rluc2A)/GNN. To confirm the infectivity results for mutants showing a phenotype different from that of J6/JFH(p7-Rluc2A), a limiting dilution assay based on immunostaining for NS5A was performed. The TCID50 titers in supernatants harvested 48 h postelectroporation correlated well with the luciferase signals (Table 1). No NS5A-positive cells could be detected after infection with genomes encoding an alanine substitution at residue 170, 174, 175, 176, or 177, while the titers of mutants J6/JFH(p7-Rluc2A)/G171A and J6/JFH(p7-Rluc2A)/C172A were less than 10% that of the parent. As expected, no NS5A-positive cells were observed after infection with the supernatants from cells electroporated with J6/JFH(p7-Rluc2A)/ΔE1E2 or J6/JFH(p7-Rluc2A)/GNN. These results indicate that several residues in the core C-terminal region are essential for infectivity, at least in the J6/JFH(p7-Rluc2A) context.
TABLE 1.
Infectious virus production measured by a limiting dilution assaya
| Mutant | TCID50/ml |
|---|---|
| J6/JFH(p7-Rluc2A) | 5.5 × 103 |
| J6/JFH(p7-Rluc2A)/ΔE1E2 | 0 |
| J6/JFH(p7-Rluc2A)/GNN | 0 |
| J6/JFH(p7-Rluc2A)/P170A | 0 |
| J6/JFH(p7-Rluc2A)/G171A | 2.0 × 102 |
| J6/JFH(p7-Rluc2A)/C172A | 3.4 × 102 |
| J6/JFH(p7-Rluc2A)/F174A | 0 |
| J6/JFH(p7-Rluc2A)/S175A | 0 |
| J6/JFH(p7-Rluc2A)/I176A | 0 |
| J6/JFH(p7-Rluc2A)/F177A | 0 |
Infectious titers in culture supernatants collected at 48 h postelectroporation with the mutant genomes are indicated. Only titers of mutants with a phenotype different from that of J6/JFH(p7-Rluc2A) were quantified.
Core stability after alanine mutation.
The inability to produce infectious virus may result from instability of the mutant core proteins. To examine protein expression levels, cell lysates were harvested 48 h postelectroporation, separated on SDS-4 to 12% PAGE gels, and analyzed for the expression of the core protein, E2, NS5A, and β-actin by Western blotting (Fig. 2). All mutant genomes produced detectable levels of core protein. Of the five mutants that did not produce infectious virus, however, mutants P170A, F174A, and F177A showed lower levels of core protein than the parental virus (Fig. 2). Mutation of residue 171 or 172 did not drastically affect core protein levels, consistent with the ability of the corresponding genomes to produce detectable infectious virus. None of the tested mutants showed differences in core protein migration indicative of unprocessed forms, although it is not clear if our protein gel system would resolve SPP-processed versus uncleaved core protein. Unaltered migration of core protein with an alanine-to-leucine change at position 191 suggested that SP cleavage was not affected (data not shown). Consistent with the ability of the mutant genomes to replicate, NS5A was detectable in all cases (Fig. 2). These data indicate that the defects of P170A and F174A to F177A core mutants do not result from an absence of core protein, although the instability and/or low levels of P170A, F174A, and F177A mutant proteins may have contributed to the absence of released infectivity.
FIG. 2.
Intracellular protein expression. Lysates harvested 48 h postelectroporation were analyzed by Western blotting. Residues mutated to alanine are identified. WT, wild type [J6/JFH(p7-Rluc2A)]. Blots are representative of results of at least duplicate analyses from independent electroporations.
Core mutants do not release noninfectious particles.
Core mutations might result in the release of nonenveloped RNA-containing nucleocapsids, inefficient genome uncoating, or other defects affecting virion infectivity. To investigate if noninfectious particles were produced, we analyzed the release of HCV RNA and core protein into the supernatants of electroporated cells. Extracellular HCV RNA levels at 48 h postelectroporation were assessed by qRT-PCR (Fig. 3A). Consistent with previous reports (36), J6/JFH(p7-Rluc2A)/ΔE1E2 released significant amounts of HCV RNA into the culture supernatant. RNA levels released by noninfectious P170A and F174A to F177A core mutants did not exceed the levels detected for the assembly-defective control. Mutants previously observed to produce low levels of infectious virus, J6/JFH(p7-Rluc2A)/G171A and J6/JFH(p7-Rluc2A)/C172A, released RNA in proportion to the infectious titers.
FIG. 3.
Noninfectious particles are not released by core mutant genomes. (A) Viral RNA release into the culture supernatant at 48 h postelectroporation. Means and SEM of results from at least duplicate electroporations are shown. (B) HCV core protein release into the culture supernatant at 48 h postelectroporation. Residues mutated to alanine are identified. Horizontal lines indicate background levels based on that of negative controls.
To investigate the release of defective particles containing core protein, protein secretion was analyzed by ELISA of samples harvested 48 h postelectroporation (Fig. 3B). None of the mutants released substantially larger amounts of core protein than J6/JFH(p7-Rluc2A)/ΔE1E2. P170A and F177A mutants, which had decreased intracellular core protein levels, also showed reduced extracellular core protein levels. Taken together, these data indicate that impaired infectious virus production results from a defect after the intracellular accumulation of the mutant core proteins and before the release of infectious or noninfectious core protein- and RNA-containing particles.
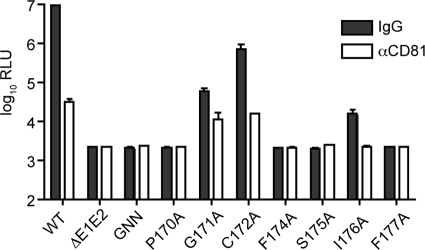
Intracellular infectivity of alanine mutants.
We next investigated whether the mutations in the core protein led to defects in infectious virus egress. To measure intracellular infectious virus production, washed cells were lysed by multiple freeze-thaw cycles at 72 h postelectroporation and the clarified supernatants were used to infect naïve Huh-7.5 cells. To confirm that luciferase transduction was the result of authentic viral entry pathways, infections were carried out in the presence of an anti-CD81 antibody (JS81) or isotype control antiserum (anti-mouse IgG1); luciferase activity was assayed 48 h postinfection. Intracellular infectious virus could be detected in J6/JFH(p7-Rluc2A)-infected cells, with over 99% of the signal neutralized by an anti-CD81 antibody (Fig. 4). Mutants containing a substitution in core residue 171 or 172 also produced infectious intracellular particles, but at levels significantly reduced compared to those produced by the wild type. These results mirrored the diminished extracellular virus titers. No intracellular infectivity could be observed for genomes encoding mutations at residues 170, 174, 175, and 177, indicating that the corresponding mutants are blocked at an earlier step in infectious virus assembly. Surprisingly, intracellular infectious virus was detected for the mutant J6/JFH(p7-Rluc2A)/I176A, despite the absence of any extracellular infectivity (Fig. 4).
FIG. 4.
Intracellular infectious virus production by core mutant genomes. Cell lysates generated by multiple rounds of freeze-thaw treatment at 48 h postelectroporation were used to infect naïve Huh-7.5 cells in the presence of isotype control antibody (anti-IgG1) or the HCV-blocking antibody (anti-CD81). Residues mutated to alanine are identified. Means and SEM of results from at least duplicate electroporations are shown.
Intracellular localization of core proteins and colocalization with lipid droplets.
Core protein has been reported to traffic from the ER membrane to lipid droplets after processing by SP and SPP (20), and several groups have shown the importance of lipid droplets as the site of HCV particle assembly (4, 21). To investigate whether the mutant core proteins localized to lipid droplets, staining for intracellular core protein at 48 h postelectroporation was conducted. In J6/JFH(p7-Rluc2A)-infected cells, core protein localized to distinct punctuate structures in the cytoplasm. Staining with Bodipy showed colocalization of the wild-type core protein with lipid droplets (Fig. 5, top). In cells harboring genomes expressing P170A, G171A, S175A, and I176A mutant proteins, patterns of core localization and colocalization with lipid droplets were indistinguishable from wild-type patterns (Fig. 5 and data not shown). Consistent with the Western blot analyses, J6/JFH(p7-Rluc2A)/P170A, J6/JFH(p7-Rluc2A)/F174A, and J6/JFH(p7-Rluc2A)/F177A showed weak core protein staining (data not shown). No core-specific staining was seen in Huh-7.5 cells transfected with J6/JFH(p7-Rluc2A)/GNN (data not shown).
FIG. 5.
Intracellular localization of mutant core proteins. Huh-7.5 cells transfected with J6/JFH(p7-Rluc2A) and mutant genomes encoding an alanine substitution at residue 175 or 176 were analyzed by immunofluorescence staining and confocal microscopy. Cells were fixed at 72 h postelectroporation and stained for core protein (core) and lipid droplets (Bodipy).
Transcomplementation of minimal core protein sequences.
Our mutational analyses suggested that core residues following amino acid 177 do not play a critical role in J6/JFH(p7-Rluc2A) infectivity. To investigate the minimal length of the core protein required to produce infectious virus particles, a transcomplementation assay was established. Full-length or truncated HCV core protein was expressed using a noncytopathic VEE virus vector (28). Capped transcripts of core protein-expressing VEE subgenomic replicons were produced in vitro and electroporated into Huh-7.5 cells together with a J6/JFH(p7-Rluc2A) mutant containing a large deletion in the core protein coding sequence, J6/JFH(p7-Rluc2A)/Δ57-160 (22) (Fig. 6A). Supernatants harvested 72 h postelectroporation were used to infect naïve Huh-7.5 cells for 48 h before the cells were assayed for luciferase activity. Coelectroporation of J6/JFH(p7-Rluc2A)/Δ57-160 and a VEE virus vector expressing the full-length HCV core protein (VEE-C191), led to significant levels of infectious virus release. Similar results were observed when J6/JFH(p7-Rluc2A)/Δ57-160 was coelectroporated with replicons VEE-C184, VEE-C180, VEE-C179, or VEE-C177 (Fig. 6A). No transcomplementation could be detected, however, when the core protein was truncated to amino acid 178 or to residue 176 and beyond. Western blot analysis indicated that each of the truncated core proteins were expressed (Fig. 6B). These data suggest that the C terminus of the HCV core protein must extend to at least amino acid 177 in order for infectious virus to be produced through transcomplementation.
FIG. 6.
Transcomplementation of a core deletion mutant by truncated core proteins. (A) Huh-7.5 cells were electroporated with a core deletion mutant, J6/JFH(p7-Rluc2A)/Δ57-160, alone (−) or together with a VEE virus replicon expressing truncated HCV core protein (VEE-C vectors). Core C-terminal amino acids are indicated. Infectious virus production at 72 h postelectroporation was measured by assay of Renilla luciferase activity in infected Huh-7.5 cells. Means and SEM from at least duplicate electroporations are shown. (B) Western blot analyses of cell lysates electroporated with VEE virus plasmids carrying truncations of core protein. The truncated residues of core protein are indicated.
DISCUSSION
The hydrophobic C-terminal region of the HCV core protein acts as a signal sequence to direct the nascent polyprotein to the ER membrane and undergoes processing by host proteases prior to initiation of infectious virus assembly (20, 34). Despite the importance of these activities for the viral life cycle, the critical determinants of this sequence have remained enigmatic. Furthermore, the exact C terminus of the mature core protein is unknown, as single amino acid substitutions capable of abolishing SPP cleavage have not been found. Here, we used a genetic approach to map the functional determinants of the core C-terminal domain and identified two subregions within this sequence with various levels of tolerance for alanine substitutions (Fig. 7).
FIG. 7.
Alignment of core protein C-terminal regions. Core amino acid sequences from H77 (genotype 1a), Con1 (genotype 1b), J6 and JFH-1 (genotype 2a), S52 (genotype 3a), ED43 (genotype 4a), SA13 (genotype 5a), and EUHK (genotype 6a) were aligned using ClustalW. The shaded bar at the bottom represents the infectivity phenotypes of the mutant viruses. Black, no infectious virus production; gray, reduced titers; white, wild-type titers. Numbers indicate amino acid positions, and dots represent residues identical to those in the consensus sequence.
The core protein is the most highly conserved HCV protein, and its C-terminal region includes a number of residues with close to 100% identity among genotypes (6). Surprisingly, we found that no individual residue among the last 14 amino acids of the core protein is absolutely required for infectious virus production, as individual substitutions resulted in wild-type titers and a core protein fragment comprising residues 1 to 177 efficiently complemented assembly in trans. In addition, engineered changes in the C-terminal 14 residues did not appear to disrupt processing of the core-E1 region, as no differences in gel mobility patterns were observed. These results support previous observations that multiple mutations in this region are necessary to inhibit cleavage by SPP and infectious virus production (34). Given this functional flexibility, it is unclear why several amino acids, such as serine 183 and cysteine 184, are very highly conserved (Fig. 7). Mutation of these residues together to leucine and valine decreases SPP cleavage in some (10, 14, 25) but not all (25, 34) contexts, and mutation to alanine individually (Fig. 1B) or in combination (22) is tolerated. Similarly, amino acid 188 is a highly conserved proline that might be expected to be important for signal peptide function (19), but when residue 188 in J6/JFH(p7-Rluc2A) was mutated to alanine, the mutant showed a wild-type phenotype. While our results indicate significant functional redundancy in cell culture, conserved residues may play important roles during virus infection in vivo. Additional functions for viral and cellular signal sequences have been proposed previously (18). Residues from the signal peptide of the lymphocytic choriomeningitis virus glycoprotein are presented by major histocompatibility complex class I as an immunodominant epitope (9), and the liberated HIV-1 gp160 leader sequence has been shown to associate with calmodulin (18, 19).
In contrast, several single amino acid substitutions in the region of residues 170 to 177 were found to completely abolish infectious virus production. Mutation of residue 170, 174, or 177 decreased intracellular core protein levels, which may account for the observed reduction in progeny release. The dual mutation I176A/F177L has previously been shown to impair the stability of core protein expressed in BHK cells (10) and to decrease SPP processing and membrane association of the core protein from strain H77 (24). These defects may reflect altered folding of the mutant proteins. In contrast, J6/JFH(p7-Rluc2A)/S175A showed unaltered levels of core and wild-type localization of the protein but no intracellular infectious virus assembly. We suspect that interactions with other proteins, such as NS5A, the glycoproteins, or host factors, or additional functions, such as roles in envelopment (1), may be disrupted for this mutant. Surprisingly, replacement of isoleucine 176 with alanine resulted in detectable levels of intracellular infectivity, despite the absence of extracellular virus. This outcome may reflect inefficient assembly, with infectious titers too low to appear in the culture supernatants. Alternatively, the I176A change may result in an egress defect, perhaps caused by instability or malformation of the particles.
Interestingly, amino acids 171, 172, and 173 are not essential for infectious virus assembly. Amino acids C172 and S173 are highly conserved across all genotypes, but the amino acids at the corresponding positions in strains JFH-1 (F172/P173) and JFH-2 (C172/P173) are different from those in any other reported sequence (13). The C172F change has been reported to enhance SPP processing (13), although engineering F172C/P173S into the JFH-1 background did not affect infectious virus production (32). Recently, palmitoylation of C172 has been demonstrated to control the association of core protein with ER membranes after processing by SPP (17). Mutation of C172 to serine significantly reduces the palmitoylation of core protein and leads to a block of infectious virus production (17). We found that alanine substitution for C172 or S173 had moderate to no effect on infectious titers of J6/JFH(p7-Rluc2A), suggesting that palmitoylation of this region may not be absolutely required.
Collectively, our results suggest that the mature core protein must extend to at least amino acid 177 to mediate infectious virus assembly in the J6/JFH context. In the transcomplementation system, proteins longer than 177 amino acids are expected to be substrates for SPP cleavage, and it is possible that truncation to amino acid 178 interferes with this process. SPP processing has been shown to be required for trafficking of core protein to lipid droplets and for infectious virus production (20, 34). Our results suggest, however, that SPP cleavage per se is not critically coupled to core localization or function. Furthermore, the ability to supply a functional truncated form of core protein in trans indicates that coordinated dual processing by SP and SPP is likely not integral to infectivity. Studies of core protein expressed in insect cells have indicated SPP cleavage after residue 177 (23) or 179 or 182 (11), and recent studies with human (293) cells have supported termination of the mature protein at residue 177 (24). Although resolution of the core C terminus awaits isolation of the mature protein from infectious particles, our results suggest important determinants relevant to the role of this region in virion production.
Acknowledgments
We thank Megan Holz, Maryline Panis, and Anesta Webson for technical support. We are grateful to Ilya Frolov for providing the noncytopathic VEE virus replicon.
This work was funded by The Greenberg Medical Research Institute and a grant from the National Institutes of Health (no. R01 AI075099). M.K. was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG). C.T.J. was supported by a Ruth L. Kirschstein National Research Service Award (NRSA; no. DK081193) from the NIDDK.
Footnotes
Published ahead of print on 9 December 2009.
REFERENCES
- 1.Ai, L.-S., Y.-W. Lee, and S. S.-L. Chen. 2009. Characterization of hepatitis C virus core protein multimerization and membrane envelopment: revelation of a cascade of core-membrane interactions. J. Virol. 83:9923-9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter, M. J. 2006. Epidemiology of viral hepatitis and HIV co-infection. J. Hepatol. 44:S6-S9. [DOI] [PubMed] [Google Scholar]
- 3.Amberg, S. M., A. Nestorowicz, D. W. McCourt, and C. M. Rice. 1994. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J. Virol. 68:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulant, S., M. W. Douglas, L. Moody, A. Budkowska, P. Targett-Adams, and J. McLauchlan. 2008. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic 9:1268-1282. [DOI] [PubMed] [Google Scholar]
- 5.Brohm, C., E. Steinmann, M. Friesland, I. C. Lorenz, A. Patel, F. Penin, R. Bartenschlager, and T. Pietschmann. 2009. Characterization of determinants important for hepatitis C virus p7 function in morphogenesis by using trans-complementation. J. Virol. 83:11682-11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukh, J., R. H. Purcell, and R. H. Miller. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. U. S. A. 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottwein, J. M., and J. Bukh. 2008. Cutting the Gordian knot—development and biological relevance of hepatitis C virus cell culture systems. Adv. Virus Res. 71:51-133. [DOI] [PubMed] [Google Scholar]
- 8.Heimann, M., G. Roman-Sosa, B. Martoglio, H. J. Thiel, and T. Rumenapf. 2006. Core protein of pestiviruses is processed at the C terminus by signal peptide peptidase. J. Virol. 80:1915-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hombach, J., H. Pircher, S. Tonegawa, and R. M. Zinkernagel. 1995. Strictly transporter of antigen presentation (TAP)-dependent presentation of an immunodominant cytotoxic T lymphocyte epitope in the signal sequence of a virus protein. J. Exp. Med. 182:1615-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hope, R. G., M. J. McElwee, and J. McLauchlan. 2006. Efficient cleavage by signal peptide peptidase requires residues within the signal peptide between the core and E1 proteins of hepatitis C virus strain J1. J. Gen. Virol. 87:623-627. [DOI] [PubMed] [Google Scholar]
- 11.Hussy, P., H. Faust, J. C. Wagner, G. Schmid, J. Mous, and H. Jacobsen. 1997. Evaluation of hepatitis C virus envelope proteins expressed in E. coli and insect cells for use as tools for antibody screening. J. Hepatol. 26:1179-1186. [DOI] [PubMed] [Google Scholar]
- 12.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato, T., M. Miyamoto, A. Furusaka, T. Date, K. Yasui, J. Kato, S. Matsushima, T. Komatsu, and T. Wakita. 2003. Processing of hepatitis C virus core protein is regulated by its C-terminal sequence. J. Med. Virol. 69:357-366. [DOI] [PubMed] [Google Scholar]
- 14.Lemberg, M. K., and B. Martoglio. 2002. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10:735-744. [DOI] [PubMed] [Google Scholar]
- 15.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 16.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 17.Majeau, N., R. Fromentin, C. Savard, M. Duval, M. J. Tremblay, and D. Leclerc. 2009. Palmitoylation of hepatitis C virus core protein is important for virion production. J. Biol. Chem. 284:33915-33925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martoglio, B. 2003. Intramembrane proteolysis and post-targeting functions of signal peptides. Biochem. Soc. Trans. 31:1243-1247. [DOI] [PubMed] [Google Scholar]
- 19.Martoglio, B., and B. Dobberstein. 1998. Signal sequences: more than just greasy peptides. Trends Cell Biol. 8:410-415. [DOI] [PubMed] [Google Scholar]
- 20.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 22.Murray, C. L., C. T. Jones, J. Tassello, and C. M. Rice. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogino, T., H. Fukuda, S. Imajoh-Ohmi, M. Kohara, and A. Nomoto. 2004. Membrane binding properties and terminal residues of the mature hepatitis C virus capsid protein in insect cells. J. Virol. 78:11766-11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto, K., Y. Mori, Y. Komoda, T. Okamoto, M. Okochi, M. Takeda, T. Suzuki, K. Moriishi, and Y. Matsuura. 2008. Intramembrane processing by signal peptide peptidase regulates the membrane localization of hepatitis C virus core protein and viral propagation. J. Virol. 82:8349-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto, K., K. Moriishi, T. Miyamura, and Y. Matsuura. 2004. Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein. J. Virol. 78:6370-6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pene, V., C. Hernandez, C. Vauloup-Fellous, J. Garaud-Aunis, and A. R. Rosenberg. 2009. Sequential processing of hepatitis C virus core protein by host cell signal peptidase and signal peptide peptidase: a reassessment. J. Viral Hepat. 16:705-715. [DOI] [PubMed] [Google Scholar]
- 27.Penin, F., J. Dubuisson, F. A. Rey, D. Moradpour, and J. M. Pawlotsky. 2004. Structural biology of hepatitis C virus. Hepatology 39:5-19. [DOI] [PubMed] [Google Scholar]
- 28.Petrakova, O., E. Volkova, R. Gorchakov, S. Paessler, R. M. Kinney, and I. Frolov. 2005. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in mammalian cells. J. Virol. 79:7597-7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phan, T., R. K. Beran, C. Peters, I. C. Lorenz, and B. D. Lindenbach. 2009. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 83:8379-8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson, B., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shin-i, P. Simmonds, D. Smith, L. Stuyver, and A. Weiner. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 31.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shavinskaya, A., S. Boulant, F. Penin, J. McLauchlan, and R. Bartenschlager. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158-37169. [DOI] [PubMed] [Google Scholar]
- 33.Steinmann, E., F. Penin, S. Kallis, A. H. Patel, R. Bartenschlager, and T. Pietschmann. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Targett-Adams, P., G. Hope, S. Boulant, and J. McLauchlan. 2008. Maturation of hepatitis C virus core protein by signal peptide peptidase is required for virus production. J. Biol. Chem. 283:16850-16859. [DOI] [PubMed] [Google Scholar]
- 35.Targett-Adams, P., T. Schaller, G. Hope, R. E. Lanford, S. M. Lemon, A. Martin, and J. McLauchlan. 2006. Signal peptide peptidase cleavage of GB virus B core protein is required for productive infection in vivo. J. Biol. Chem. 281:29221-29227. [DOI] [PubMed] [Google Scholar]
- 36.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weihofen, A., K. Binns, M. K. Lemberg, K. Ashman, and B. Martoglio. 2002. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296:2215-2218. [DOI] [PubMed] [Google Scholar]
- 38.WHO. 1999. Global surveillance and control of hepatitis C. Report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J. Viral Hepat. 6:35-47. [PubMed] [Google Scholar]
- 39.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2009. Trans-complementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog. 5:e1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]