Abstract
Circoviruses are known to infect birds and pigs and can cause a wide range of severe symptoms with significant economic impact. Using viral metagenomics, we identified circovirus-like DNA sequences and characterized 15 circular viral DNA genomes in stool samples from humans in Pakistan, Nigeria, Tunisia, and the United States and from wild chimpanzees. Distinct genomic features and phylogenetic analysis indicate that some viral genomes were part of a previously unrecognized genus in the Circoviridae family we tentatively named “Cyclovirus” whose genetic diversity is comparable to that of all the known species in the Circovirus genus. Circoviridae detection in the stools of U.S. adults was limited to porcine circoviruses which were also found in most U.S. pork products. To determine whether the divergent cycloviruses found in non-U.S. human stools were of dietary origin, we genetically compared them to the cycloviruses in muscle tissue samples of commonly eaten farm animals in Pakistan and Nigeria. Limited genetic overlap between cycloviruses in human stool samples and local cow, goat, sheep, camel, and chicken meat samples indicated that the majority of the 25 Cyclovirus species identified might be human viruses. We show that the genetic diversity of small circular DNA viral genomes in various mammals, including humans, is significantly larger than previously recognized, and frequent exposure through meat consumption and contact with animal or human feces provides ample opportunities for cyclovirus transmission. Determining the role of cycloviruses, found in 7 to 17% of non-U.S. human stools and 3 to 55% of non-U.S. meat samples tested, in both human and animal diseases is now facilitated by knowledge of their genomes.
Animal viruses with small, circular, single-stranded DNA (ssDNA) genomes comprise the Circoviridae family and the Anellovirus genus, while viruses in the Geminiviridae and Nanoviridae families infect plants (3, 25, 34, 37, 40). The genomes of these small viruses without a lipid envelope replicate through a rolling-circle mechanism, possibly sharing a common origin with bacterial plasmids (6), and show high recombination and nucleotide substitution rates (7, 19).
The Circoviridae family consists of the Circovirus genus whose member species are currently known to infect only birds and pigs, and the Gyrovirus genus, including a single species, Chicken anemia virus (CAV). Circoviruses infect several avian groups, including parrots, pigeons, gulls, anserids (ducks, geese, and swans), and numerous passerines (ravens, canaries, finches, and starlings) (12, 15, 16, 22, 26, 31, 35, 38, 39). Avian circoviruses have been associated with a variety of symptoms, including developmental abnormalities, lymphoid depletion, and immunosuppression (22, 26, 28, 35, 39). Mammalian circoviruses include only two closely related species, Porcine circovirus 1 and 2 (PCV1 and PCV2, respectively), infecting pigs (21). PCV2 has been associated with porcine circovirus-associated diseases, which can manifest as a systemic disease, respiratory disease complex, enteric disease, porcine dermatitis and nephropathy syndrome or as reproductive problems, causing great losses in the pork industry (1, 29, 32). Circovirus infections are thought to occur mainly through fecal-oral transmission (37).
We describe here highly diverse, circovirus-like, circular DNA viral genomes discovered in human and chimpanzee stool samples, and we propose their inclusion in a new genus of the Circoviridae family that we tentatively name “Cyclovirus” pending review by the International Committee on Taxonomy of Viruses (ICTV). Cycloviruses were also found to be prevalent in the muscle tissue of farm animals, such as chickens, cows, sheep, goats, and camels. The cyclovirus species found in human stool samples and in animal meat samples showed limited genetic overlap, suggesting that most of the cycloviruses found in human stool samples are not from consumed animal meat. Rather, these cycloviruses in human stools might cause human enteric infections. The presence of cycloviruses in human stool samples and in farm animal tissue also suggests the potential for frequent cross-species exposure and zoonotic transmissions.
MATERIALS AND METHODS
Detection of circoviruses using degenerate primers.
Nucleic acids were extracted from stool supernatants and plasma samples using the QIAamp viral RNA kit which extracts both RNA and DNA (Qiagen). DNA was extracted from animal tissue specimens using a QIAamp DNA minikit (Qiagen). Degenerate primers for nested PCR were as follows: CV-F1 (5′-GGIAYICCICAYYTICARGG), CV-R1 (5′-AWCCAICCRTARAARTCRTC), CV-F2 (5′-GGIAYICCICAYYTICARGGITT), and CV-R2 (5′-TGYTGYTCRTAICCRTCCCACCA). The degenerate primers were designed on the basis of the consensus sequence from an alignment of replicase (Rep) proteins from CyCV1-PK5006 and 12 representative Circovirus species. Multiple-sequence alignment of the Rep amino acid sequences was performed using ClustalW2, with default settings. PCRs with the degenerate Rep primers were performed with the following cycling profile: 5 min at 95°C; 40 cycles, with 1 cycle consisting of 1 min at 95°C, 1 min at 52°C (56°C for the 2nd PCR round), and 1 min at 72°C; and a final incubation for 10 min at 72°C. Products with a size of approximately 400 bp were purified and sequenced using primer CV-R2. Most of the products were sequenced directly. Amplicons with low concentrations or multiple bands were cloned to obtain high-quality sequence data.
Phylogenetic analysis.
Phylogenetic analyses based on aligned amino acid sequences from full-length or partial Rep proteins were generated by the neighbor-joining (NJ) method in MEGA 4.1 (18), using amino acid p-distances, with 1,000 bootstrap replicates. Other tree-building methods, maximum parsimony (MEGA) and maximum likelihood (PhyML [11]), were carried out to confirm the NJ tree. The GenBank accession numbers of the Rep sequences from plasmids, viruses, and protists used in the phylogenetic analyses are as follows (shown in brackets): Beak and feather disease virus (BFDV) [AF071878], Canary circovirus (CaCV) [AJ301633], Columbid circovirus (CoCV) [AF252610], Duck circovirus (DuCV) [DQ100076], Goose circovirus (GoCV) [AJ304456], Gull circovirus (GuCV) [DQ845074], Finch circovirus (FiCV) [DQ845075], Raven circovirus (RaCV) [DQ146997], Starling circovirus (StCV) [DQ1729062], Cygnus olor circovirus (SwCV) [EU056310], Porcine circovirus 1 (PCV1) [AY660574], Porcine circovirus 2 (PCV2) [AY424401], Chicken anemia virus (CAV) [M55918], Milk vetch dwarf virus [AB009047], Pepper golden mosaic virus [U57457], Canarypox virus [NC_005309], Giardia intestinalis [AF059664], Bifidobacterium pseudocatenulatum plasmid p4M [NC_003527], and Entamoeba histolytica [XM_643662].
Genome analyses.
Putative open reading frames (ORFs) with a coding capacity greater than 100 amino acids were predicted by Vector NTI Advance 10.3 (Invitrogen). The stem-loop structure was predicted using Mfold (version 3.2) (43).
Sample collections. (i) Stool samples from South Asian children.
A total of 107 fecal specimens were collected by the WHO Regional Reference Laboratory for polio eradication at the National Institute of Health in Islamabad, Pakistan, between December 2005 and May 2008: 57 samples from non-polio-infected children with acute flaccid paralysis (AFP), 9 from closely related but healthy contacts, and 41 from clinically healthy children living in the same geographic region. The median age of the children was 3 years (range, 1 month to 15 years).
(ii) Stool samples from Nigerian children.
Ninety-six stool samples from non-polio-infected children with AFP were collected by the WHO National Polio Laboratory at the University of Maiduguri Teaching Hospital in Maiduguri, Nigeria, during February to April 2007. The median age of these children was 2.5 years (range, 6 months to 12 years).
(iii) Stool samples from Tunisian children and adults.
A total of 192 stool samples were collected by the WHO Regional Reference Laboratory for Poliomyelitis and Measles, Institut Pasteur de Tunis, Tunis-Belvédère, Tunisia, from 2005 to 2008, including 94 stool samples from non-polio-infected children with AFP and 82 samples from closely related healthy contact children. Two stool samples from AFP cases and 14 stool samples from healthy contacts were from adults (>15 years). The median age of the cohort was 5 years (range, 6 months to 54 years).
(iv) Stool samples from Minnesota patients with gastroenteritis and healthy controls.
A total of 247 stool samples from the Minnesota Department of Health were collected from 2004 to 2006, including 107 specimens from clinically healthy donors and 140 specimens from patients with acute gastroenteritis.
(v) Stool samples from African chimpanzees.
Forty-four stool samples from individual wild chimpanzees were collected from Central Africa (Tanzania, Cameroon, Rwanda, Uganda, Central African Republic, Republic of the Congo, and Democratic Republic of the Congo). All of the samples were collected from the common chimpanzee (Pan troglodytes) between 2002 and 2007. The geographic sites where the chimpanzee stool samples were collected are shown in Table S1 in the supplemental material.
(vi) Plasma specimens from U.S. blood donors.
Ninety-six plasma specimens were collected from unremunerated blood donors in the United States.
(vii) Plasma specimens from African bush hunters.
A total of 113 plasma specimens were collected from nonsymptomatic bush-hunting African adults (95 specimens) or adults with a nonmalarial fever (18 specimens).
(viii) U.S. pork.
Thirteen specimens of pork products were purchased from markets in stores in the United States (San Francisco) in September 2008.
(ix) Meat products from Pakistan.
A total of 57 meat samples were collected from Islamabad, Rawalpindi, and Lahore in Pakistan between August 2008 and March 2009.
(x) Meat products from Nigeria.
A total of 147 meat product samples from markets in Maiduguri, Nigeria, were collected during March to April 2009.
All studies were reviewed and approved by the University of California in San Francisco Committee on Human Research.
Nucleotide sequence accession numbers.
The sequences of 15 genomes have been deposited in GenBank under accession numbers GQ404844 to GQ404858. Partial Rep gene sequences were deposited in GenBank under accession numbers GQ404858 to GQ404986.
RESULTS
A highly divergent circovirus in human stool samples.
Viral particles in human stool samples from Pakistani children were enriched by filtration, and contaminating host DNA and RNA were digested by nuclease treatment. Nucleic acids protected within viral capsids were then extracted, amplified using random PCR, and pyrosequenced (41). The resulting DNA sequences were assembled into contigs, translated, and analyzed by protein similarity search (BLASTx). A contig (1,164 bp) composed of eight sequence reads from the stool sample of a healthy South Asian child (PK5006) was found to have significant similarity to the replicase (Rep) protein of circoviruses (E-value < 1e−10) (41). Since species in the genus Circovirus have a circular genome, the full viral genome was then amplified by inverse nested PCR, and the amplicon was sequenced by primer walking. The virus of the assembled genome was tentatively named “Cyclovirus species 1 strain PK5006” (CyCV1-PK5006) (cyclo means circular in Greek). Sequence alignment of the putative Rep protein of CyCV1-PK5006 with that of known species in the genus Circovirus identified several highly conserved amino acids motifs (see Fig. S1 in the supplemental material) (15, 31, 38).
Frequent detection and analysis of circovirus-like Rep sequences in human and animal specimens.
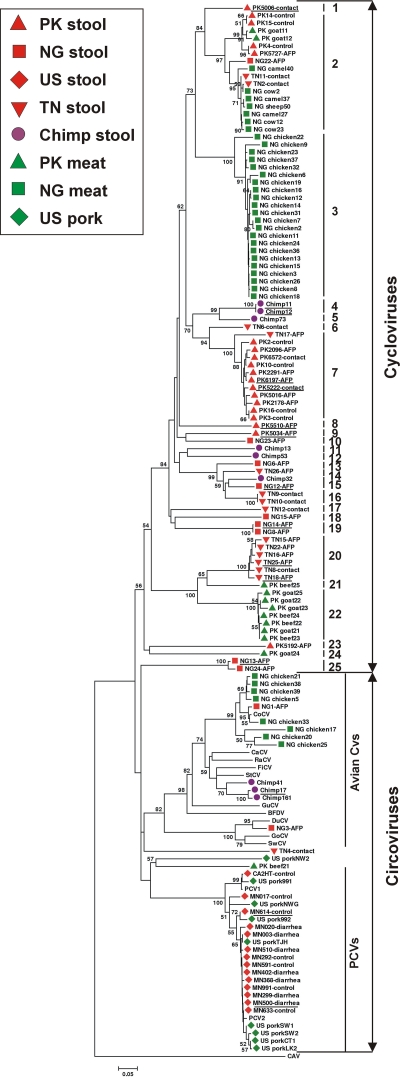
To screen for related viruses, we designed pan-Rep PCR primers to hybridize to the Rep genes of known avian and porcine circoviruses as well as to the Cyclovirus prototype CyCV1-PK5006. Ten specimen collections of 1,112 samples, including human stool and plasma samples and animal stool and muscle tissue samples were then screened with these primers (Table 1). Rep sequences were detected in 137 samples from all but the two human plasma collections. The approximately 400-bp amplicons were sequenced, and translated amino acids were aligned using the homologous region of the putative Rep-associated protein of CAV as the outgroup. The derived phylogenetic tree was consistent with prior analyses based on the complete Rep protein sequences and on the complete genome of animal circoviruses (12, 16, 22, 38, 39). A densely populated cluster of Rep sequences (including that of the Cyclovirus prototype genome) was labeled cycloviruses in Fig. 1. Some of the Rep sequences fell outside the Circovirus and Cyclovirus clades, together with the non-Circoviridae Rep proteins from Nanovirus, Geminivirus, Gyrovirus, Canarypox virus, Bifidobacterium pseudocatenulatum plasmid p4M, Giardia intestinalis, and Entamoeba histolytica (10) (see Fig. S2 in the supplemental material). The possibility that some outlier Rep sequences belong to ingested plant viruses distantly related to nanoviruses and geminiviruses cannot be discounted (42).
TABLE 1.
Sample collections tested by using the degenerate primers for replication-associated proteins
| Sample collection | Collection site(s) (country) | Specimen type | No. of specimens | No. of specimens (%) |
|||
|---|---|---|---|---|---|---|---|
| Pan-Rep PCR-positive | With cyclovirus | With circovirus | With other Rep proteins | ||||
| South Asian children with | Pakistan, Afghanistan | Human stool | 57 (diseased) | 12 (21.1) | 9 (15.8) | 0 (0.0) | 3 (5.3) |
| AFP and healthy | 9 (contact) | 3 (33.3) | 3 (33.3) | 0 (0.0) | 0 (0.0) | ||
| contact and control children | 41 (control) | 8 (19.5) | 7 (17.1) | 0 (0.0) | 1 (2.4) | ||
| Minnesotans with | United States | Human stool | 140 (diseased) | 7 (5.0) | 0 (0.0) | 7 (5.0) | 0 (0.0) |
| gastroenteritis and healthy controls | 107 (control) | 6 (5.6) | 0 (0.0) | 6 (5.6) | 0 (0.0) | ||
| Nigerian children with AFP | Nigeria | Human stool | 96 | 18 (18.8) | 9 (9.4) | 2 (2.1) | 7 (7.3) |
| Tunisian children with | Tunisia | Human stool | 96 (diseased) | 7 (7.3) | 7 (7.3) | 0 (0.0) | 0 (0.0) |
| AFP and healthy contact children | 96 (contact) | 9 (9.4) | 7 (7.3) | 1 (1.0) | 1 (1.0) | ||
| African chimpanzees | Middle African countriesa | Chimpanzee stool | 44 | 9 (20.5)a | 6 (13.6) | 3 (6.8) | 1 (2.3) |
| U.S. blood donors | United States | Human plasma | 96 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| African bush hunters | Africa | Human plasma | 113 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| U.S. pork products | United States | Pork | 13 | 9 (69.2) | 0 (0.0) | 9 (69.2) | 0 (0.0) |
| Pakistani meat | Pakistan | Chicken | 13 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Beef | 26 | 5 (19.2) | 4 (15.4) | 1 (3.8) | 0 (0.0) | ||
| Goat | 18 | 7 (38.9) | 7 (38.9) | 0 (0.0) | 0 (0.0) | ||
| Nigerian meat | Nigeria | Chicken | 40 | 30 (75.0) | 22 (55.0) | 8 (20.0) | 0 (0.0) |
| Beef | 25 | 3 (12.0) | 3 (12.0) | 0 (0.0) | 0 (0.0) | ||
| Camel | 27 | 3 (11.1) | 3 (11.1) | 0 (0.0) | 0 (0.0) | ||
| Goat | 26 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Sheep | 29 | 1 (3.4) | 1 (3.4) | 0 (0.0) | 0 (0.0) | ||
| Total | 1,112 | 137 | 88 | 37 | 13 | ||
Specimens were collected from Tanzania, Cameroon, Uganda, Rwanda, Central Africa Republic, Republic of the Congo, and the Democratic Republic of the Congo. Two different Rep sequences were obtained from one chimpanzee specimen by subcloning.
FIG. 1.
Phylogenetic analysis of the translated Rep sequences amplified by pan-Rep PCR. Sequences derived from human stool samples (red), chimpanzee stool samples (purple), and farm animal meat samples (green) are indicated. Cycloviruses in the same species are defined as having >85% identity in Rep region and are labeled by vertical bars 1 to 25. The scale bar labeled 0.05 at the bottom of the figure represents 5% estimated phylogenetic divergence. The countries of origin are Pakistan (PK), Nigeria (NG), Tunisia (TN), and the United States (US).
The prevalence of Cyclovirus in human stool samples ranged from 17% in Pakistani children to 0% in U.S. adult stools that exclusively contained PCV1 or PCV2 (Table 1). Rep sequences amplified from the stool samples from two Nigerian children (Fig. 1, NG1-AFP and NG3-AFP) grouped within the avian Circovirus clade, while sequence amplified from the stool sample of a Tunisian child formed a distinct lineage of circovirus-like sequence (Fig. 1, TN4-contact). Cycloviruses were found in 6 out of 44 stool samples from chimpanzees (14%), and avian circovirus-like sequences were amplified from another 3 chimpanzee stool samples (Fig. 1, purple circles). No statistical association was found between detection of cyclovirus or circovirus Rep sequences with the occurrence of disease (non-polio AFP in Pakistan or Tunisia or unexplained gastroenteritis in Minnesota).
Genome characteristics and phylogeny of cycloviruses.
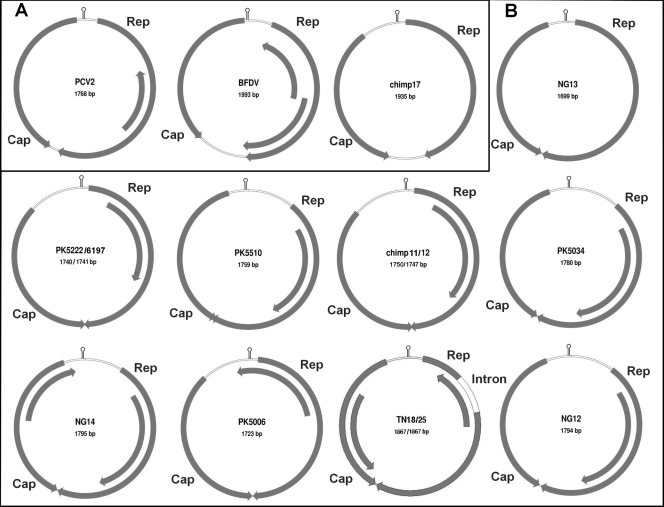
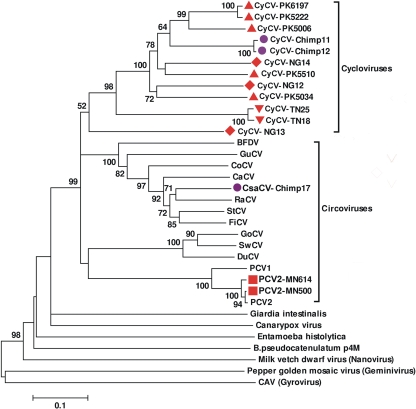
To confirm the presence of diverse cycloviruses and to characterize the genome of this novel group, inverse PCR was used to amplify and sequence complete viral genomes from human and chimpanzee stool samples. Each of the 15 sequenced circular genomes has two main open reading frames arranged in opposite directions, encoding the putative Rep and capsid (Cap) proteins, an arrangement typical of circoviruses (Fig. 2). The complete Rep proteins were used for phylogenetic analysis. The resulting tree confirms the presence of a new Cyclovirus clade within the Circoviridae, including now 12 genomes (Fig. 3). The ORFs of the Cyclovirus genomes were similar to those of circoviruses but with some distinctive features (Fig. 2). On average, cycloviruses have smaller genomes (average, 1,772 bp; range, 1,699 to 18,67 bp) than circoviruses do (average, 1,902 bp; range, 1,759 to 2,063 bp), encoding relatively smaller Rep and Cap proteins (Table 2). NG13 had the smallest genome size of any reported virus (1,699 bp).
FIG. 2.
Genomic organizations of circoviruses (A) and cycloviruses (B). The 2 major ORFs, encoding the putative replication-associated protein (Rep) and the putative capsid protein (Cap), and other ORFs with a coding capacity greater than 100 amino acids are shown. The locations of the stem-loop structures are marked.
FIG. 3.
Phylogenetic analysis of 15 Circoviridae replicase proteins from 12 human stool samples and 3 chimpanzee stool samples. Outlier taxa are non-Circoviridae Rep proteins. Sample designation is the same as in Fig. 1. CyCV, cyclovirus.
TABLE 2.
Genome organization of newly discovered cycloviruses and representative circoviruses
| Virus type and circular DNA virus species | No. of nt in the genome | No. of aa in protein |
Length (nt) of region (start-end)a |
||
|---|---|---|---|---|---|
| Rep | Cap | 5′ Intergenic region | 3′ Intergenic region | ||
| Cycloviruses | |||||
| PK5006 | 1,723 | 278 | 219 | 230 (1516-22) | |
| PK5222 | 1,740 | 279 | 218 | 247 (1516-22) | |
| PK5510 | 1,759 | 280 | 219 | 271 (1679-190) | |
| PK5034 | 1,780 | 277 | 218 | 293 (1691-203) | |
| PK6197 | 1,741 | 279 | 218 | 248 (1516-22) | |
| Chimp11 | 1,750 | 280 | 220 | 258 (1515-22) | |
| Chimp12 | 1,747 | 280 | 220 | 255 (1515-22) | |
| NG12 | 1,794 | 281 | 218 | 284 (1691-180) | 7 (1027-1033) |
| NG14 | 1,795 | 286 | 230 | 245 (1707-156) | |
| TN18 | 1,867 | 286 | 222 | 160 (1762-54) | 6 (1087-1092) |
| TN25 | 1,867 | 286 | 222 | 160 (1762-54) | 6 (1087-1092) |
| NG13 | 1,699 | 307 | 221 | 105 (1622-27) | 4 (952-955) |
| Circoviruses | |||||
| Chimp17 | 1,935 | 291 | 232 | 198 (1772-34) | 162 (911-1072) |
| MN614 | 1,767 | 314 | 233 | 83 (1735-50) | 37 (996-1032) |
| MN500 | 1,768 | 314 | 233 | 83 (1736-50) | 38 (996-1033) |
| PCV1 | 1,759 | 312 | 233 | 82 (1724-46) | 36 (986-1021) |
| PCV2 | 1,768 | 314 | 233 | 83 (1736-50) | 38 (996-1033) |
| DuCV | 1,991 | 292 | 257 | 110 (1929-47) | 228 (927-1154) |
| GoCV | 1,821 | 293 | 250 | 132 (1762-72) | 54 (955-1008) |
| CoCV | 2,037 | 317 | 273 | 90 (1988-40) | 171 (995-1165) |
| RaCV | 1,898 | 291 | 243 | 86 (1848-35) | 204 (912-1115) |
| SwCV | 1,785 | 293 | 251 | 107 (1726-47) | 40 (930-969) |
| BFDV | 1,993 | 299 | 244 | 126 (1975-107) | 232 (1008-1239) |
| GuCV | 2,035 | 305 | 245 | 207 (1928-99) | 172 (1018-1189) |
| FiCV | 1,962 | 291 | 249 | 29 (1962-28) | 307 (905-1211) |
| StCV | 2,063 | 289 | 276 | 79 (2021-36) | 283 (907-1189) |
| CaCV | 1,952 | 290 | 250 | 77 (1907-31) | 249 (905-1153) |
Fifteen genome sequences obtained in this study are shown in boldface type. Nucleotide position 1 was set at the residue A at position 8 of the nonamer sequence. nt, nucleotides; aa, amino acids.
The 3′ intergenic regions between the stop codons of the two major ORFs were either absent or only a few base pairs long in cycloviruses, while those of circoviruses were significantly larger. The 5′ intergenic regions between the start codons of the two major ORFs of cycloviruses were larger than those of circoviruses (Table 2). The Rep ORFs of the two closely related genomes, TN18 and TN25 (97% nucleotide similarity) were both interrupted by an apparent 171-bp intron with a typical splice donor site (GT) and splice acceptor site (AG) (Fig. 2).
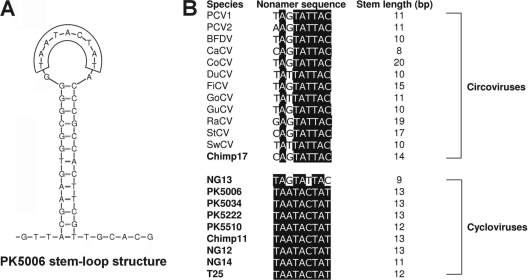
The stem-loop structure with a conserved nonanucleotide motif located at the 5′ intergenic region of circovirus genomes is thought to initiate rolling-cycle replication (37). A highly conserved stem-loop structure is also found in the 5′ intergenic regions of cycloviruses (Fig. 2 and Fig. 4A). The consensus sequence for the loop nonamer of the circoviruses is 5′-TAGTATTAC-3′, with slight variation among the sequenced genomes (21, 26, 31, 35, 38, 39) (Fig. 4B). A different and conserved loop nonamer sequence (5′-TAATACTAT-3′) was observed for all the cycloviruses except CyCV-NG13, which is a cyclovirus group outlier but carries a typical circovirus nonamer (Fig. 4B). The highly conserved nonamer atop the stem-loop structure is one of the distinct characteristics of the new Cyclovirus genus.
FIG. 4.
(A) Stem-loop of Cyclovirus prototype CyCV1-PK5006 and (B) nonamer sequences and stem lengths of the stem-loop structures for circoviruses and cycloviruses.
The two genomes derived from human stool samples in the United States (MN614 and MN500) shared 99% overall genome nucleotide similarity with PCV2. The Chimp17 genome, from a chimpanzee stool sample, grouped with the raven circovirus RaCV, sharing 79% amino acid similarity to its Rep protein. We have named this virus “Chimpanzee Stool avian-like circovirus-chimp17” (CsaCV-chimp17). No suitably located ATG was identified for either ORF of CsaCV-chimp17. Considering the common usage of alternative start codons in avian circoviruses, such as TCT (26, 31, 35), GTG (38), and ATA (39), CTG was considered the most likely candidate for a start codon in the genome, producing ORFs of expected lengths.
The average amino acid similarity among cyclovirus Rep proteins is 59% (range, 42 to 80%), and the value for circovirus Rep proteins is 56% (range, 40 to 87%), reflecting a comparable range of viral diversity within both genera (see Table S2 in the supplemental material). For the capsid protein, the average amino acid similarity is 29% (range, 11 to 56%) for cycloviruses and 34% (range, 18 to 76%) for circoviruses (see Table S2 in the supplemental material). An amino acid alignment shows that cycloviruses also possess some of the highly conserved Rep amino acid motifs typical of circoviruses, including WWDGY, DDFYGW, and DRYP. Motifs associated with rolling-circle replication (FTLNN, TPHLQG, and CSK) and deoxynucleoside triphosphate (dNTP) binding (G—GSK) were also identified, with some alterations (see Fig. S1 in the supplemental material) (15, 31, 38). The N-terminal region of the cyclovirus Cap proteins was highly basic and arginine-rich, as is typical for circoviruses (17, 35).
PCVs frequently detected in U.S. human stool samples and pork products.
All Rep sequences derived from human stool samples from the United States clustered closely with PCVs (Fig. 1, red diamonds). In order to test the possible dietary origin of these PCV sequences, pork specimens purchased from different U.S. stores were tested. Out of 13 U.S. pork products, 9 (70%) were Rep positive, 7 of which clustered with PCV2, 1 with PCV1, and 1 highly divergent sequence (US porkNW2) (Fig. 1, green diamonds). Pork sample US porkNW2 may represent a yet uncharacterized porcine circovirus species. Out of 23 Rep sequences from the U.S. samples, 22, including all 14 from human stool samples and 8 out of 9 from pork specimens, therefore belonged to PCV1 or PCV2.
Circovirus-like Rep sequences in consumed meats.
The frequent detection of PCVs in U.S. stool samples and U.S. pork products suggested that the cycloviruses found in non-U.S. human stool samples and wild chimpanzee stool samples might similarly originate from the consumption of meat contaminated with cycloviruses. To test this hypothesis, commonly eaten meat products were acquired from markets in Pakistan and Nigeria and analyzed by pan-Rep PCR (Table 1). Of 204 meat samples tested, 24% were positive, and all amplicons were sequenced. The Rep sequence detection rates differed substantially between countries for the same type of meat. None of 13 chicken samples from Pakistan was positive, while 30 out of 40 (75%) chicken samples from Nigeria were positive. Of the 30 Rep sequences from Nigerian chicken samples, 22 sequences clustered tightly within the Cyclovirus genus, and 8 sequences clustered together in a cluster with pigeon Circovirus (as did the Rep sequence NG1-AFP from the stool sample of a Nigerian child). Of the 26 goat samples from Nigeria, none was positive, while 7 out of 18 (38%) goat specimens from Pakistan were positive for cycloviruses. Of the total 19 Rep sequences obtained from farm mammals (cows, goats, sheep, and camels), only 1 (PK beef21) grouped deeply with the circovirus clade, while 18 fell within the cyclovirus clade. The majority (40 out of 49) of Rep sequences obtained from animal tissue therefore belonged to the cyclovirus clade. Some cyclovirus Rep sequences from different animal species (e.g., cows and goats, even-toed ungulates in the Bovidae family) were very closely related (Fig. 1, species 22).
The ICTV defines different circovirus species based on sequence similarity; genomic sequences having <75% nucleotide identity and <70% identity in the capsid protein qualify as different species (37). We adopted a criterion of <85% amino acid identity in the highly conserved Rep protein region amplified by pan-Rep PCR as the criterion for Cyclovirus species designation by comparing the amino acid identity of the same Rep region among known circovirus species. Using this criterion, 25 species of Cyclovirus were found in human and chimpanzee stool samples and meat samples from farm animals. Of these 25 species, only a single Cyclovirus species (Fig. 1, species 2), represented by 16 out of a total of 88 Rep sequences (18.2%), was found in both human stool and farm animal tissue samples. Four species were specific to chimpanzee stool samples, and another four species were specific to farm animals. Sixteen cyclovirus species were specific to non-U.S. human stool samples. The consumption of meat from infected animals is therefore unlikely to account for the majority of cycloviruses detected in non-U.S. human stool samples.
DISCUSSION
We report on the frequent detection of viral, circular DNA genomes related to porcine and avian circoviruses in human and chimpanzee stool samples and genetically characterize a previously unrecognized genus in the family Circoviridae. These viruses were both widely dispersed (Tunisia, Pakistan, and Nigeria) and highly prevalent (7 to 17% of children's stool samples).
Cycloviruses are not closely related phylogenetically to the recently described circular DNA viruses chimpanzee stool-associated circular viruses (ChiSCV) found in chimpanzee stool samples (5) or the circular ssDNA viruses in aquatic environments (20, 33), nor is their genome organization related to human or animal anelloviruses (e.g., torque teno virus [TTV]) (4, 33).
PCVs were frequently detected in stool samples from adults in the United States (5%), and store-bought pork products also frequently contain PCV sequences (70%). These results indicated that detection of PCV DNA in stool may reflect dietary consumption of PCV-infected pork.
Evidence for circovirus infection in mammals other than pigs is equivocal, and studies have been restricted to PCVs. PCV2 DNA in cows with respiratory symptoms and in aborted bovine fetuses has been reported only once (24). PCV2 was also reported in a colon biopsy specimen from a patient with ulcerative colitis, although contamination with PCV2 from stool is difficult to exclude in this case (2). No PCV DNA was found by PCR in screening more than 1,000 samples from various tissues of both healthy and immunosuppressed humans and plasma samples from 18 xenotransplantation recipients of pig islet cells (9, 13). In this study, the results of screening plasma samples from 96 U.S. blood donors and 113 Central African bush hunters via pan-Rep PCR were also negative (Table 1). A study showed that viral protein expression, cytopathic effect, and DNA persistence occurs in human cell lines infected with PCV2, but the virus could not be passed to new cultures (14). One study reported the presence of PCV-reactive antibodies, although with somewhat distinctive properties in sera of humans, cows, and mice (36), while another reported the lack of PCV antibodies in cows and horses (8). Whether PCVs simply pass through or are capable of replication in the human gut remains unknown.
Avian circovirus-like DNA was found in 3/44 wild chimpanzee stool samples and in 2/96 stool samples from Nigerian children (Table 1). This observation may reflect consumption of infected birds or conceivably contamination of food with bird droppings.
Cycloviruses were found in the muscle tissue of all the species of farm animals tested (goats, sheep, cows, camels, and chickens), suggesting that viral infection occurs in these species. In previous studies, different tissues have been shown to retain small DNA viruses (e.g., parvoviruses) long after primary infection viremia (23, 27). The detection of cycloviruses and circoviruses in muscle tissue could therefore reflect prior and/or ongoing infection. The detection of closely related cyclovirus Rep sequences in both cows and goats from Pakistan (Fig. 1, species 22) might reflect cross-species transmission.
A wide diversity of cycloviruses was identified in human stool samples collected from children in developing countries. In contrast, in the United States, all Rep sequences obtained from stool samples belonged to the PCV clade. An important distinction between U.S. and non-U.S. human stool samples was the younger age of the non-U.S. donors, which may have impacted host susceptibility to infections or the duration of viral shedding. Exposure to cyclovirus may therefore also occur in the United States but was not detected because of the older age of the subjects.
In total, 17 Cyclovirus species were identified in 395 human stool samples, and 5 Cyclovirus species were found in muscle tissue samples from 204 farm animals, with only a single species found in common in both groups of samples. The meat samples analyzed were acquired from three major cities in Pakistan and one major city in Nigeria, while the children from these countries shedding cycloviruses were geographically more dispersed. It is therefore conceivable that despite the large number of cyclovirus replicase sequences generated, more geographically dispersed sampling of farm animals would have shown greater overlap with human stool-derived cyclovirus sequences. Using the current sampling, the limited overlap between Cyclovirus species found in human stool samples and in meat from farm animals from the same countries does suggest that most of the cycloviruses found in the stool samples of children in Nigeria and Pakistan were not from consumed meat. Possibly, the 16 cycloviruses species found only in human stool samples were transmitted via a fecal-oral route from other infected children, a common pathway for many enteric viral infections. The detection of cycloviruses in 14% of stool samples from chimpanzees (who consume very limited amounts of meat) also argues in favor of transmission within this primate species rather than simply reflecting consumption of infected meats. It is not known whether the viral species found in both human stool samples and tissue samples from farm animals, such as PCVs in the United States and cyclovirus species 2 (Fig. 1, species 2) in Pakistan, Nigeria, and Tunisia, can replicate in their human host. Since transmission of PCV2 from one pig to another through consumption of meat was recently shown (30), the potential for zoonotic transfer also exists for other circoviruses and cycloviruses.
Given the high prevalence of cyclovirus infections in non-U.S. farm animals, the possibility of cross-species transmission (cyclovirus species 22 in different members of the family Bovidae), the high diversity of cycloviruses in human stool samples, the documented pathogenicity of closely related Circovirus species, and the high rate of mutation and recombination of some ssDNA viruses, the pathogenic potential of cycloviruses in both humans and farm animals merits further study.
Supplementary Material
Acknowledgments
This work was supported by Blood Systems Research Institute and NIH grant R01 HL083254 to E.D. The chimpanzee sample collection was supported by grants from National Institutes of Health to B.H.H. (R01 AI50529 and R01 AI58715), the Bristol Myers Freedom to Discover Program, and the Jane Goodall Institute. Additional support was provided by the Global Viral Forecasting Initiative, Google.org, and the Skoll Foundation.
We thank M. P. Busch for helpful discussions; Marycelin Mandu Baba and David Nadeba Bukbuk from the WHO National Polio Laboratory, University of Maiduguri Teaching Hospital, Maiduguri, Borno State, Nigeria, for assistance collecting stool samples from non-polio-infected children with AFP; and John McGee, Jason Reilly, and Mats Rynge from the Renaissance Computing Institute (RENCI) for assistance with computing analysis of the pyrosequencing data. We also thank Farbod Babrzadeh and Baback Gharizadeh at Stanford University for assistance with pyrosequencing. We thank the staff of Project PRESICA for chimpanzee sample collection in southern Cameroon and the Central African Republic; the staff of the Gombe Stream Research Centre for sample collection in Gombe National Park in Tanzania; Michael A. Huffman for sample collection in Mahale Mountain National Park in Tanzania; and Crickette Sanz and David Morgan for sample collection in the Goualougo Triangle in the Democratic Republic of Congo. We thank the Cameroonian Ministries of Health, Environment and Forestry, and Research for permission to collect samples in Cameroon; the Democratic Republic of Congo Ministry of Science and Technology and Ministry of Forest Economy for permission to collect samples in the Goualougo Triangle in the Democratic Republic of Congo; the Tanzania National Parks, the Tanzania Commission for Science and Technology, and the Tanzania Wildlife Research Institute for permission to conduct research in Gombe Stream and Mahale Mountain National Parks in Tanzania; the Uganda Wildlife Authority, the Uganda National Council for Science and Technology, and the Makerere University Biological Field Station for permission to conduct research in Kibale National Park and Kyambura Gorge in Uganda; the Rwandan Office of Tourism and National Parks for permission to collect samples in Nyungwe National Park in Rwanda; and the Department of Ecology and Management of Plant and Animal Resources (University of Kisangani) for authorization to collect samples in the Democratic Republic of Congo.
Footnotes
Published ahead of print on 9 December 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Allan, G. M., and J. A. Ellis. 2000. Porcine circoviruses: a review. J. Vet. Diagn. Investig. 12:3-14. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, C. N., G. Nayar, A. Hamel, and J. F. Blanchard. 2003. Study of animal-borne infections in the mucosas of patients with inflammatory bowel disease and population-based controls. J. Clin. Microbiol. 41:4986-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biagini, P. 2005. Anellovirus, p. 335-341. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 4.Biagini, P. 2009. Classification of TTV and related viruses (anelloviruses). Curr. Top. Microbiol. Immunol. 331:21-33. [DOI] [PubMed] [Google Scholar]
- 5.Blinkova, O., J. Victoria, Y. Li, B. Keele, C. Sanz, J. B. Ndjango, M. Peeters, D. Travis, E. Lonsdorf, M. Wilson, A. Pusey, B. Hahn, and E. Delwart. 2010. Novel circular DNA viruses in stool samples of wild-living chimpanzees. J. Gen. Virol. 91:74-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. K. 2006. Rolling-circle replication of an animal circovirus genome in a theta-replicating bacterial plasmid in Escherichia coli. J. Virol. 80:8686-8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffy, S., L. A. Shackelton, and E. C. Holmes. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9:267-276. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, J. A., C. Konoby, K. H. West, G. M. Allan, S. Krakowka, F. McNeilly, B. Meehan, and I. Walker. 2001. Lack of antibodies to porcine circovirus type 2 virus in beef and dairy cattle and horses in western Canada. Can. Vet. J. 42:461-464. [PMC free article] [PubMed] [Google Scholar]
- 9.Garkavenko, O., M. C. Croxson, M. Irgang, A. Karlas, J. Denner, and R. B. Elliott. 2004. Monitoring for presence of potentially xenotic viruses in recipients of pig islet xenotransplantation. J. Clin. Microbiol. 42:5353-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibbs, M. J., V. V. Smeianov, J. L. Steele, P. Upcroft, and B. A. Efimov. 2006. Two families of rep-like genes that probably originated by interspecies recombination are represented in viral, plasmid, bacterial, and parasitic protozoan genomes. Mol. Biol. Evol. 23:1097-1100. [DOI] [PubMed] [Google Scholar]
- 11.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 12.Halami, M. Y., H. Nieper, H. Muller, and R. Johne. 2008. Detection of a novel circovirus in mute swans (Cygnus olor) by using nested broad-spectrum PCR. Virus Res. 132:208-212. [DOI] [PubMed] [Google Scholar]
- 13.Hattermann, K., A. Maerz, H. Slanina, C. Schmitt, and A. Mankertz. 2004. Assessing the risk potential of porcine circoviruses for xenotransplantation: consensus primer-PCR-based search for a human circovirus. Xenotransplantation 11:547-550. [DOI] [PubMed] [Google Scholar]
- 14.Hattermann, K., C. Roedner, C. Schmitt, T. Finsterbusch, T. Steinfeldt, and A. Mankertz. 2004. Infection studies on human cell lines with porcine circovirus type 1 and porcine circovirus type 2. Xenotransplantation 11:284-294. [DOI] [PubMed] [Google Scholar]
- 15.Hattermann, K., C. Schmitt, D. Soike, and A. Mankertz. 2003. Cloning and sequencing of Duck circovirus (DuCV). Arch. Virol. 148:2471-2480. [DOI] [PubMed] [Google Scholar]
- 16.Johne, R., D. Fernandez-de-Luco, U. Hofle, and H. Muller. 2006. Genome of a novel circovirus of starlings, amplified by multiply primed rolling-circle amplification. J. Gen. Virol. 87:1189-1195. [DOI] [PubMed] [Google Scholar]
- 17.Johne, R., R. Raue, C. Grund, E. F. Kaleta, and H. Muller. 2004. Recombinant expression of a truncated capsid protein of beak and feather disease virus and its application in serological tests. Avian Pathol. 33:328-336. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., M. Nei, J. Dudley, and K. Tamura. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefeuvre, P., J. M. Lett, A. Varsani, and D. P. Martin. 2009. Widely conserved recombination patterns among single-stranded DNA viruses. J. Virol. 83:2697-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Bueno, A., J. Tamames, D. Velazquez, A. Moya, A. Quesada, and A. Alcami. 2009. High diversity of the viral community from an Antarctic lake. Science 326:858-861. [DOI] [PubMed] [Google Scholar]
- 21.Mankertz, A., R. Caliskan, K. Hattermann, B. Hillenbrand, P. Kurzendoerfer, B. Mueller, C. Schmitt, T. Steinfeldt, and T. Finsterbusch. 2004. Molecular biology of Porcine circovirus: analyses of gene expression and viral replication. Vet. Microbiol. 98:81-88. [DOI] [PubMed] [Google Scholar]
- 22.Mankertz, A., K. Hattermann, B. Ehlers, and D. Soike. 2000. Cloning and sequencing of columbid circovirus (coCV), a new circovirus from pigeons. Arch. Virol. 145:2469-2479. [DOI] [PubMed] [Google Scholar]
- 23.Manning, A., S. J. Willey, J. E. Bell, and P. Simmonds. 2007. Comparison of tissue distribution, persistence, and molecular epidemiology of parvovirus B19 and novel human parvoviruses PARV4 and human bocavirus. J. Infect. Dis. 195:1345-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nayar, G. P., A. L. Hamel, L. Lin, C. Sachvie, E. Grudeski, and G. Spearman. 1999. Evidence for circovirus in cattle with respiratory disease and from aborted bovine fetuses. Can. Vet. J. 40:277-278. [PMC free article] [PubMed] [Google Scholar]
- 25.Ng, T. F., C. Manire, K. Borrowman, T. Langer, L. Ehrhart, and M. Breitbart. 2009. Discovery of a novel single-stranded DNA virus from a sea turtle fibropapilloma by using viral metagenomics. J. Virol. 83:2500-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niagro, F. D., A. N. Forsthoefel, R. P. Lawther, L. Kamalanathan, B. W. Ritchie, K. S. Latimer, and P. D. Lukert. 1998. Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch. Virol. 143:1723-1744. [DOI] [PubMed] [Google Scholar]
- 27.Norja, P., K. Hokynar, L. M. Aaltonen, R. Chen, A. Ranki, E. K. Partio, O. Kiviluoto, I. Davidkin, T. Leivo, A. M. Eis-Hubinger, B. Schneider, H. P. Fischer, R. Tolba, O. Vapalahti, A. Vaheri, M. Soderlund-Venermo, and K. Hedman. 2006. Bioportfolio: lifelong persistence of variant and prototypic erythrovirus DNA genomes in human tissue. Proc. Natl. Acad. Sci. U. S. A. 103:7450-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noteborn, M. H., G. F. de Boer, D. J. van Roozelaar, C. Karreman, O. Kranenburg, J. G. Vos, S. H. Jeurissen, R. C. Hoeben, A. Zantema, G. Koch, et al. 1991. Characterization of cloned chicken anemia virus DNA that contains all elements for the infectious replication cycle. J. Virol. 65:3131-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Opriessnig, T., X. J. Meng, and P. G. Halbur. 2007. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J. Vet. Diagn. Investig. 19:591-615. [DOI] [PubMed] [Google Scholar]
- 30.Opriessnig, T., A. R. Patterson, X. J. Meng, and P. G. Halbur. 2009. Porcine circovirus type 2 in muscle and bone marrow is infectious and transmissible to naive pigs by oral consumption. Vet. Microbiol. 133:54-64. [DOI] [PubMed] [Google Scholar]
- 31.Phenix, K. V., J. H. Weston, I. Ypelaar, A. Lavazza, J. A. Smyth, D. Todd, G. E. Wilcox, and S. R. Raidal. 2001. Nucleotide sequence analysis of a novel circovirus of canaries and its relationship to other members of the genus Circovirus of the family Circoviridae. J. Gen. Virol. 82:2805-2809. [DOI] [PubMed] [Google Scholar]
- 32.Ramamoorthy, S., and X. J. Meng. 2009. Porcine circoviruses: a minuscule yet mammoth paradox. Anim. Health Res. Rev. 10:1-20. [DOI] [PubMed] [Google Scholar]
- 33.Rosario, K., S. Duffy, and M. Breitbart. 2009. Diverse circovirus-like genome architectures revealed by environmental metagenomics. J. Gen. Virol. 90:2418-2424. [DOI] [PubMed] [Google Scholar]
- 34.Stanley, J. 2005. Geminiviridae, p. 301-326. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 35.Stewart, M. E., R. Perry, and S. R. Raidal. 2006. Identification of a novel circovirus in Australian ravens (Corvus coronoides) with feather disease. Avian Pathol. 35:86-92. [DOI] [PubMed] [Google Scholar]
- 36.Tischer, I., L. Bode, J. Apodaca, H. Timm, D. Peters, R. Rasch, S. Pociuli, and E. Gerike. 1995. Presence of antibodies reacting with porcine circovirus in sera of humans, mice, and cattle. Arch. Virol. 140:1427-1439. [DOI] [PubMed] [Google Scholar]
- 37.Todd, D. 2005. Circoviridae, p. 326-334. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 38.Todd, D., A. N. Scott, E. Fringuelli, H. L. Shivraprasad, D. Gavier-Widen, and J. A. Smyth. 2007. Molecular characterization of novel circoviruses from finch and gull. Avian Pathol. 36:75-81. [DOI] [PubMed] [Google Scholar]
- 39.Todd, D., J. H. Weston, D. Soike, and J. A. Smyth. 2001. Genome sequence determinations and analyses of novel circoviruses from goose and pigeon. Virology 286:354-362. [DOI] [PubMed] [Google Scholar]
- 40.Vetten, H. J. 2005. Nanoviridae, p. 343-352. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 41.Victoria, J. G., A. Kapoor, L. Li, O. Blinkova, B. Slikas, C. Wang, A. Naeem, S. Zaidi, and E. Delwart. 2009. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol. 83:4642-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, T., M. Breitbart, W. H. Lee, J. Q. Run, C. L. Wei, S. W. Soh, M. L. Hibberd, E. T. Liu, F. Rohwer, and Y. Ruan. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.