Abstract
We report here the first integrated investigation of both ancient DNA and proteins in archaeobotanical samples: medieval grape (Vitis vinifera L.) seeds, preserved by anoxic waterlogging, from an early medieval (seventh–eighth century A.D.) Byzantine rural settlement in the Salento area (Lecce, Italy) and a late (fourteenth–fifteenth century A.D.) medieval site in York (England). Pyrolysis gas chromatography mass spectrometry documented good carbohydrate preservation, whilst amino acid analysis revealed approximately 90% loss of the original protein content. In the York sample, mass spectrometry-based sequencing identified several degraded ancient peptides. Nuclear microsatellite locus (VVS2, VVMD5, VVMD7, ZAG62 and ZAG79) analysis permitted a tentative comparison of the genetic profiles of both the ancient samples with the modern varieties. The ability to recover microsatellite DNA has potential to improve biomolecular analysis on ancient grape seeds from archaeological contexts. Although the investigation of five microsatellite loci cannot assign the ancient samples to any geographic region or modern cultivar, the results allow speculation that the material from York was not grown locally, whilst the remains from Supersano could represent a trace of contacts with the eastern Mediterranean.
Electronic supplementary material
The online version of this article (doi:10.1007/s00114-009-0629-3) contains supplementary material, which is available to authorized users.
Keywords: Archaeobotany, Grape, Ancient DNA, Ancient proteins, Mass spectrometry, Proteomics, Vitis vinifera
Introduction
Wine represents one of the most important products of plant domestication during the development of human civilisation in western Eurasia. It is intimately bound up with the expansion of agriculture, trade and commerce and is also important in socioreligious, cultural and political aspects of many societies, particularly around the Mediterranean. Winemaking enabled humans to produce a beverage which was the most widespread drug and medicine of antiquity, a drug upon which economies and communities were built (Bassermann-Jordan 1975; Olmo 1976).
Archaeobotanical evidence of Vitis sp. consists principally of waterlogged, mineralised and charred seeds. Morphological differences can only tentatively distinguish wild and cultivated subspecies (Mangafa and Kotsakis 1996). Furthermore, in many cases, this discrimination relies more on indirect evidence, such as the absence of wild Vitis in the geographic area, or on the archaeological context and the time period. Vitis seeds and in particular those that are waterlogged store biomolecular information, which can provide greater detail on viticulture diffusion and the wine trade.
Genetics have advanced our understanding of crop domestication, for example, studies of modern grapes have revealed changes brought about by human selection (Meredith 2001; This et al. 2006; Arroyo-García et al. 2006; Vouillamoz and Grando 2006). Ancient DNA (aDNA) recovered from archaeological plant residues has also begun to play a role by shaping our understanding of past grape diffusion (Manen et al. 2003). With their morphology and structure specifically designed to store genetic information, seeds are a promising target for studies on ancient biomolecules, in some cases germinating after centuries (Sallon et al. 2008). In grape seeds in particular, protection is further enhanced by the hard coat and the high concentrations of antioxidant molecules (Yilmaz and Toledo 2004). Remarkable preservation of lipids and nucleic acids has been reported in 1,400-year-old radish (Raphanus) seeds by O'Donoghue et al. (1996). Previous work on blackberry (Rubus fruticosus) seeds and crab apple (Malus silvestris) pips from York revealed that the preservation is good for lignin and cellulose but only limited for proteins (McCobb et al. 2001).
In this study, we have undertaken a combined DNA/carbohydrate/protein analysis of several ancient grape seeds. We explored the survival of polymerase chain reaction (PCR) amplifiable nuclear DNA (nuDNA) using five microsatellite loci in order to tentatively compare ancient samples with modern grape varieties. Such information can help to clarify ambiguities about indigenous cultivation and commercial trade of grapes and wine in two different archaeological contexts: a seventh–eighth century A.D., Byzantine rural settlement in the ‘heel’ of the Italian peninsula, near the town of Supersano (Salento area, Lecce, Italy; Arthur 1999, 2004; Arthur et al. 2008), and a late medieval, fourteenth–fifteenth century A.D., site on Low Petergate, in York city centre (northern England; Akeret et al. 2006; Reeves 2006). The present-day climate in York is unsuitable for outdoor grape cultivation, but the chronology of the site, between the Medieval Warm Period and the Little Ice Age (Lamb 1965; Bradley et al. 2003), does not completely exclude the possibility that viticulture was practised in the area at that time.
Considering that in ancient samples DNA survival may be related to general biomolecular preservation, we also analysed carbohydrates and proteins from the same samples of archaeological grape seeds, adopting multiple approaches. Pyrolysis gas chromatography mass spectrometry (py-GC/MS) was used to investigate carbohydrate preservation. Protein degradation was estimated observing amino acid composition and racemisation and by direct sequencing of preserved peptides.
The protein component represents an important indicator of seed and grain quality. Pioneering studies on archaeological cereal seeds (Derbyshire et al. 1977; Shewry et al. 1982) suggested that the investigation of proteins from ancient grains could aid in understanding of plant domestication, but this work was hampered by the prevailing technology of that time. However, the application of mass spectrometry to fossil bone samples has enabled the identification and sequencing of single ancient proteins, namely osteocalcin (Nielsen-Marsh et al. 2002, 2005; Humpula et al. 2007) and collagen (Asara et al. 2007; Schweitzer et al. 2009; but see also Buckley et al. 2008). Recently Solazzo et al. (2008) were able to identify protein remains in archaeological potsherds, whilst Hollemeyer et al. (2008) used matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry to analyse some fragments of the 5,300-year-old Tyrolean mummy's clothing.
The integration of a range of technical improvements has enabled rapid and sensitive identification of proteins present in small and complex biological samples. To test the potential of this ensemble of technologies for the analysis of archaeobotanical samples and to characterise the effects of long-term protein degradation processes, we have extracted and analysed the proteins from ancient grape seeds, using both electrospray ionisation (ESI) and MALDI.
Materials and methods1
Modern and ancient sample description
Vitis vinifera L. modern seeds from well-characterised grape cultivars ‘Cabernet Sauvignon’, ‘Cabernet Franc’, ‘Pinot Noir’, ‘Syrah’, ‘Cot’, ‘Chardonnay’ and ‘Croatina’ were collected in the Regional Agency for Services to Agriculture and Forests (ERSAF) experiment collection in Torrazza Coste, Pavia, Italy. Ancient grape seeds from the Byzantine rural settlement in ‘Località Scorpo’ to the north of the small town of Supersano (Lecce, Italy) were found on July 2007 at the bottom of a 6 m-deep well. Associated artefacts and radiocarbon dating (Arthur et al. 2008) suggest that the well was backfilled between the seventh and eighth centuries A.D. The lower part of the well was found to be waterlogged and contained large amounts of preserved organic remains, including hundreds of grape seeds (see S2a). Ancient grape seeds from York, S2b, were preserved by anoxic ‘waterlogging’ in the basal fill of a cobble-lined pit, dated by artefacts to the fourteenth/fifteenth century, identified during excavation of a series of medieval tenements at 62-8 Low Petergate in York city centre.
Ancient DNA analysis
DNA was extracted from, and PCR was attempted on, nine ancient grape seeds (three from York, six from Supersano). The aDNA extractions and PCR setup were performed in a dedicated ancient DNA laboratory at the University of Copenhagen, where stringent measures are taken to prevent contamination with modern sources of DNA. In addition to the ancient samples, modern grape seeds were extracted in a separate laboratory. To remove external contaminant sources of DNA, the seeds were briefly washed in dilute bleach solution (10% commercial strength) then rinsed in analytical grade H2O. Following this, the seeds were allowed to dry naturally, before being manually crushed. The crushed grape seeds were then digested overnight at 55°C with agitation in 400 μL of a proteinase K-containing digestion buffer (Gilbert et al. 2007). Post digestion, DNA was purified from the mixture using two phenol and one chloroform extractions. Subsequently, the final aqueous layer was purified further and concentrated using a Qiagen Qiaquick PCR cleanup column (Qiagen, Valencia, CA, USA) following the manufacturer's guidelines. In the final stage, the DNA was eluted from the silica filter in 50 μL elution buffer after a 10-min incubation at room temperature. In addition to the nine ancient samples, four extraction controls were performed to screen for contamination derived from the laboratory, reagents or between extracts.
The grape extracts were subjected to PCR amplification using five microsatellite markers (see S3). Initially, all extracts were screened with marker VVS2 (Thomas and Scott 1993) to assess for DNA survival. The results indicated that good quality DNA was present in three of the samples (two from York, one from Supersano). These were subsequently PCR-amplified for four additional microsatellites: VVMD5, VVMD7 (Bowers et al. 1996), ZAG62 and ZAG79 (Sefc et al. 1999). PCR amplification was performed in 25 μL reactions, using Amplitaq Gold (Applied Biosystems). Each reaction contained 1 μL DNA template, 2.5 mM MgCl2, 1 μL bovine serum albumin (20 mg/mL), 0.2 mM deoxyribonucleotide triphosphates (dNTPs), 0.2 μL Amplitaq Gold, 400 nM of each primer and 1× PCR buffer. PCR parameters were as follows: enzyme activation 95°C for 4 min, 40 cycles of 95°C for 15 s, 60°C for 30 s, 72°C for 30 s and final extension 72°C for 7 min. PCR reactions incorporated one PCR blank reaction for every three ancient samples. Following PCR, 5 μL of the products was visualised on 2% Tris–acetate–ethylenediaminetetraacetic acid agarose gels stained with ethidium bromide. The results gave no indication of any contamination, whether in the extraction or PCR controls. Successful amplicons were subsequently purified using Qiaquick kit (Qiagen) then cloned using the Topo TA cloning system (Invitrogen, Carlsbad, CA, USA), and up to 16 clones were sequenced by a commercial facility (Macrogen, Korea). In addition to the cloned amplicons, an additional amplicon was generated by PCR for each marker, from each of the three extracts. Following gel confirmation and Qiagen purification, using Qiaquick kit (Qiagen), these PCR amplicons were sent to Milan for capillary short tandem repeat (STR) analysis.
Capillary STR analysis was performed on a 1,000-fold dilution of PCR products received from Copenhagen. All PCR reactions were carried out in a 20-μL volume using a PTC-100 (MJ Research, Waltham, MA, USA) thermocycler. Each reaction contained 1 μL DNA template, 2.5 mM MgCl2, 0.2 mM dNTPs, 1 U Platinum Taq Polymerase (Invitrogen), 200 nM each primer and 1× PCR buffer (Invitrogen). PCR parameters were as follows: initial denaturation 95°C for 2 min, 35 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 30 s and final extension 72°C for 30 min. Capillary electrophoresis was performed on an ABI 310 Genetic Analyser (Applied Biosystems). Samples (1 μL in 12 μL highly deionised formamide) were injected at 15 kV for 5 s, and separation was performed at 15 kV and 60°C during 28 min. Data were collected with data collection software and analysed by GeneMapper 3.7 software (Applied Biosystems, Foster City, CA, USA). GeneScan-500 (250) standard fragments in the size range of 75–350 bp were used for the calculation of relative sizes of microsatellite alleles. It has been well documented that the adoption of different protocols in different laboratories can result in perceived size differences of as much as 5 bp for identical microsatellite alleles (This et al. 2004). To counter against this, for each capillary run, possible microsatellite size shift due to the experimental procedure adopted was corrected comparing the size of the microsatellites of the ancient samples with standards from the seven modern reference grape cultivars, reported above. Runs were repeated at least in triplicate for each sample until the best peak height and resolution were attained. The cloning procedure followed by amplicon sequencing, commonly adopted as a good practice procedure in ancient DNA investigation, also allowed confirmation of the size of the alleles. When allele sizes at one or more loci diverged from the odd-even profile shown by the reference accessions, both the original and the corrected values (allele size −1 base) were used for both the GeneClass2 and the genetic distance calculations.
Area-specific allele frequencies were used in a likelihood-based assignment test (Paetkau et al. 1995) that calculates the probability of each individual's assignment to a particular area as the cumulative products of each allele's frequency of occurrence in any of several European areas under examination. We generated significance levels for individuals that cross-assigned to a neighbouring area using the simulation routine within GeneClass 2.0 software (Paetkau et al. 2004; Piry et al. 2004). Significance levels were determined by comparing the individual genotypes of cross-assigned individuals with a simulated set of 10,000 genotypes that were generated using area-specific allele frequencies using a type I (alpha) error threshold set at 0.01.
In addition, genetic distance and cluster analyses were performed to compare the three ancient samples with a set of modern accessions representative of grapevines cultivated in Europe. The reference dataset consisted of 167 accessions described in the database of Sefc et al. (2000), also available at http://www.dagz.boku.ac.at/15271.html. Loci VVS2, VVMD5, VVMD7, ZAG62 and ZAG79 were included in the analysis. Distance matrices were calculated by the programme GenAlEx 6.1 (Peakall and Smouse 2006) using the ‘codominant’ and the ‘total distance only’ options. Cluster analysis was performed by the Neighbour programme of the PHYLIP package (Felsenstein 2005) using the neighbour-joining (Saitou and Nei 1987) algorithm. Dendrograms were plotted by the NJplot 2.2 programme (Perrière and Gouy 1996).
Amino acid analysis and Py-GC/MS
A 10.41 mg of powdered modern sample, 4.93 mg of ancient material from York and 1.18 mg from Supersano were analysed to evaluate amino acid composition and racemisation rate following the procedure previously reported (Penkman et al. 2008). The York sample was analysed using Curie point Py-GC/MS. The sample was ground and compressed onto a ferromagnetic wire using a hydraulic press. The temperature of the pyrolysis unit was set at 250°C. After heating to a temperature of 600°C for 5 s, the pyrolysis products were separated on a Fison Instruments GC8060 gas chromatograph fitted with a fused silica capillary column (25 m × 0.25 mm ID) with a CP-SIL 5CB-MS coating, film thickness 0.40 μm. The GC oven was set at 40°C and was heated to 320°C at a rate of 7°C/min; finally, it was kept isothermal for 15 min. The GC was connected to a Fison Instruments MD800 quadrupole mass spectrometer, operating at 70 eV and scanning the m/z range 45–650 at a cycle time of 1.5 s. Helium was used as the carrier gas with a constant flow rate of 0.7 mL/min.
Mass spectrometric protein analysis
Three pools, made of six modern grape seeds, six ancient seeds from Supersano and four ancient seeds from York were powdered in liquid nitrogen; 136.81 mg of the modern sample, 98.16 mg of the ancient one from York and 141.24 mg from Supersano were extracted adopting the phenol-based protocol number four reported by Vincent et al. (2006), with minor changes. Since it was not possible to recover a pellet from the Supersano sample, this pool was not processed further. The other pellets were resuspended in 50 mM ammonium bicarbonate pH 8.00 and were digested overnight at 37°C using Sequencing Grade Modified Trypsin (Promega Corporation, Madison, WI, USA).
Three microlitres of the digest solution was loaded onto an Ultimate micro-capillary high-performance liquid chromatography (HPLC) system; the eluate was on-line mixed with a matrix solution containing α-cyano-4-hydroxy cinnamic and then collected onto a MALDI sample plate. Plates were loaded into an AB 4700 Proteomics Analyser TOF/TOF (Applied Biosystems, Foster City, CA, USA), mass spectrometer operated in positive ion reflectron mode. The same amount of the digest solution was also loaded onto a micro-capillary HPLC system interfaced with a QSTAR API Pulsar LC-MS/MS System (Applied Biosystems) with a Micro Ion Spray source. Positive ESI-MS and MS/MS spectra were acquired using information dependent acquisition. Peak lists generated by the software associated with the mass spectrometers were searched using the Mascot 2.2 algorithm (Matrix Science Ltd., London, UK), in December 2007, after the grape genome was made publicly available (Jaillon et al. 2007; Velasco et al. 2007). Post-translational modifications, semi-tryptic peptides and single amino acid substitutions were searched for by selecting the Mascot automatic error tolerant database search option. Each tryptic digest solution was labelled using iTRAQ reagents (Applied Biosystems), reporter mass 114 for the modern and 116 for the ancient sample, and analysed on the MALDI mass spectrometer following the manufacturer’s instructions. The quantitative proteomic approach has been used very recently on modern grape samples (Lucker et al. 2009).
Results
Protein and carbohydrate preservation
Py-GC/MS analysis demonstrated that lignin- and cellulose-derived compounds are present in the coat of the York seeds (Fig. 1). The catechyl and guaiacyl compounds detected are pyrolytic breakdown products of the lignin structure of the seed coat. The data confirm that V. vinifera L. has no syringyl compounds in its lignin structure. Within the early-eluting compounds, there are some indicators of the presence of sugars, e.g. 3-hydroxy-2-methyl-2-cyclopenten-1-one, in the sample, as well as some compounds linked to the presence of proteins, e.g. toluene and phenol (Ralph and Hatfield 1991).
Fig. 1.
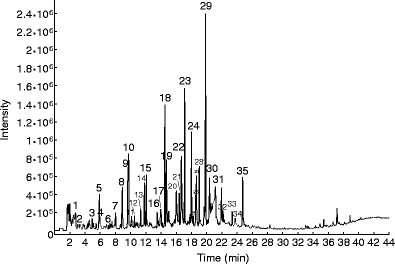
Total ion current of on-line flash pyrolysate of the seed coat of the York samples (for compound identification, see S4 in ESM)
Amino acid analysis documents a high level of biomolecular degradation for the Supersano samples. Compared to modern grape seeds, the samples from Supersano have lost 87% of their amino acids, slightly less than the York seeds (89%). The levels of aspartic acid and asparagine (Asx = Asp + Asn) racemisation, however, are higher in the Supersano sample, D/L Asx 0.17 (n = 3, SD = 0.01), than in the York sample, D/L Asx = 0.13 (n = 3, SD = 0.01).
A more detailed characterisation of the proteinaceous residues was achieved using a phenol-based protocol to extract proteins from three sample pools generated on powdering (a) six modern seeds, (b) six ancient seeds from Supersano and (c) four ancient seeds from York. It was possible to extract approximately 62.5 mg of proteins per gram of modern grape seed (6.25% by wt), as estimated by Bradford assay and confirmed by amino acid analysis. The medieval York sample contained an order of magnitude less protein; no protein pellet was recovered from the Supersano sample. The sodium dodecyl sulfate polyacrylamide gel electrophoretic (SDS-PAGE) profile of the protein pellet from the York sample revealed a 23-kDa band with other more diffuse bands between 6 and 14 kDa (Fig. 2), all emerging from a continuous background smear.
Fig. 2.

SDS-PAGE separation of archaeological grape seed proteins. The proteins were resolved in a 4–12% (w/v) gradient acrylamide gel and stained with colloidal Coomassie Blue. MW lane, molecular mass markers. The molecular mass (in kilodaltons) of each protein is shown on the left. Lane 1, ancient proteins isolated by phenolic extraction from medieval grape seeds excavated in York. Lane 2, modern grape seed proteins extracted under the same conditions
In the absence of a sedimented protein pellet from the Supersano sample, nano-LC MS/MS was only performed on the late medieval sample from York. Protein extracts of the modern and the York samples were digested using trypsin, isobarically labelled (iTRAQ), and the resulting peptide mixtures were resolved using a micro-capillary HPLC system for analysis on both MALDI TOF/TOF and ESI quadrupole TOF mass spectrometers. The MS/MS peak lists from the MALDI and ESI experiments were searched using the Mascot search engine against the National Center for Biotechnology Information protein database. Despite the age of the sample, the quality of the mass spectra from the ancient sample is, in some cases, remarkable. For the storage protein peptide, for example, DSGFEYVAIK (see Fig. 3b, c), it was possible to identify almost complete ‘y’ and ‘b’ ion series. However, due to the low protein recovery, as documented by amino acid analysis, a total of only six protein accessions, based on 30 unique peptide attributions, were identified in the ancient sample (Table 1; S5), representing a few percent of the total number of proteins typically identified from modern dry seeds (Rajjou et al. 2006; Chibani et al. 2006). 2S albumin was directly identified (Li and Gray 2005), whilst a Basic Local Alignment Search Tool (BLAST) search (see S1 on ESM) allowed confident identification of different seed storage and metabolic proteins: 7S and 11S globulins, peptidase A1 and a non-specific lipid transfer protein.
Fig. 3.

Example of nano-LC/tandem MS analysis results for the archaeological grape seed protein extract from York: a nano-LC chromatogram of the tryptic digest. The baseline in the ancient sample is raised, compared to the modern one reported in the inset, indicating the presence of degraded proteinaceous material. b Mass spectrum at the retention time of 14.3 min (m/z 650–4,400). c MS/MS spectrum of m/z 1,128.59, which identified the peptide DSGFEYVAIK, unique to the unnamed protein product gi|157350579 from V. vinifera showing high homology, BLAST score = 379, with a seed storage protein from Juglans regia (accession no. gi|56788031)
Table 1.
List of the ancient proteins extracted from medieval grape seeds excavated in York
| Modern sample | York medieval sample | Quantification | BLAST homology attributiond | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession number | No. of peptidesa | Coverage %b | Mowse scorec | No. of peptidesa | Coverage % b | Mowse scorec | Ancient/modern % | Accession number | Description | Score (bits) | E value |
| gi|157350579 | 12 | 54 | 723/627 | 10 | 37 | 442/262 | 29.8 | gi|56788031 | Seed storage protein (Juglans regia) | 379 | 3.00 E−103 |
| gi|157350582 | 11 | 57 | 684/488 | 8 | 33 | 299/149 | 57.0 | gi|56788031 | Seed storage protein (Juglans regia) | 294 | 5.00 E−78 |
| gi|157347968 | 16 | 42 | 709/357 | 6 | 11 | 585/59 | 7.6 | gi|13183177 | 7S globulin (Sesamum indicum) | 442 | 3.00 E−122 |
| gi|147821120 | 7 | 27 | 511/282 | 3 | 8 | 178/- | 18.9 | gi|87240526 | Peptidase A1, pepsin (Medicago truncatula) | 414 | 5.00 E−114 |
| gi|30421130 | 4 | 35 | 369/52 | 2 | 15 | 203/- | 0.5 | N.A. | 2S albumin precursor (Vitis vinifera) | N.A. | |
| gi|157344576 | 3 | 30 | 134/109 | 3 | 30 | 77/- | 9.0 | gi|128378 | Nonspecific lipid-transfer protein A (phospholipid transfer protein) | 126 | 3.00 E−28 |
Ancient proteins, extracted from medieval grape seeds excavated in York, identified by the Mascot search engine from the data produced by nano-LC/tandem MS
N.A. not applicable
aThe number of peptides reported is calculated as the sum of the unique peptides identified by ESI and MALDI together
bThe same approach was used to calculate the sequence coverage
cFor each protein, the Mowse scores from both MALDI, including quantitative data, and ESI experiments are reported
dThe proteins described as ‘unnamed protein product’ were identified by BLAST homology (see S1 on ESM for details). Quantitative evaluation was based on the data produced using stable isotope isobaric labelling—iTRAQ
Quantitative evaluation shows that the seed storage protein content in the ancient sample is 30–57% of the values observable from modern samples, significantly higher than for other proteins (6–20%). Error-tolerant searching of the data obtained from the medieval sample allowed the identification of a series of semi-tryptic peptides generated by hydrolytic processes. A clear example is represented by the detection of the fragments generated after cleavage of the peptidic backbone next to five of the eight consecutive glutamine (Q) residues in the tryptic peptide FYLAGNPQNEFQQQQQQQQGSEGQQQQQEGGGSEGR. The full-length peptide was detected unaltered in the modern sample and, in a very limited amount, in the ancient one (see S5 and S6 on ESM). It also appears that, for the ancient sample, the number of peptides matched is lower using ESI rather than MALDI.
Ancient DNA
DNA was successfully PCR amplified from two out of three York seeds (here referred to as ‘York 1’ and ‘York 2’) and one out of six Supersano seeds (here referred to as ‘Supersano’). The size of the microsatellites of the ancient samples, reported in Table 2, are consistent with the size of the amplicon sequences obtained after cloning the PCR products (see S7).
Table 2.
Microsatellite sizes as determined by capillary electrophoresis
| Sample | VVS2 (bp) | VVMD5 (bp) | VVMD7 (bp) | ZAG62 (bp) | ZAG79 (bp) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| York 1 | 132 | 140 | 230 | 232 | 240 | 246 | 201 | 201 | 244 | 244 |
| York 2 | 132 | 140 | 230 | 232 | 240 | 246 | 187 | 201 | 244 | 244 |
| Supersano | 130 | 142 | 228 | 228 | 246 | 248 | 197 | 197 | 246 | 248 |
The Supersano sample yielded a 228-bp allele at locus VVMD5 that is not represented in the reference set, whilst the two York samples showed the allele 201 bp at locus ZAG62 which is shared by reference samples ‘Agiorgitiko’, ‘Dermatas’, ‘Akominato’ (Greek) and ‘Cortese’ (Italian). For those loci with homozygous profiles, such as ZAG79 for both the York samples, it is not possible to exclude undetected heterozygosis due to allele drop after degradation of the ancient genetic material. However, a certain amount of homozygosis is likely considering grape hermaphroditism and its consequent propensity for selfing.
GeneClass2 (Piry et al. 2004) was used to statistically assign the archaeological specimens to European populations of V. vinifera cultivars using the database of Sefc et al. (2000). The input data list contains all possible genotype combinations per specimen as described in Manen et al. (2003). The profile of the two York samples is identical except for one of the two ZAG62 alleles. ‘York 2’ was placed, by likelihood-based assignment testing (Paetkau et al. 1995), in the Greek, Italian, Austrian/German and French groups with probabilities of 0.106, 0.023, 0.019 and 0.013, respectively. Sample ‘York 1’ was placed only in the Greek group with a probability of 0.020. No assignment was possible for the ‘Supersano’ sample. Cluster analysis placed the ancient samples next to each other, on particularly long branches. The closest group of modern cultivars includes the Spanish accessions ‘Ondarrabi Beltza’ and ‘Blanca Cayetana’, the Italian ‘Cortese’, the Portuguese ‘Boal Ratinho’ and the Greek ‘Dafnia’, ‘Kristalli’, ‘Syriki’, ‘Dermatas’ and ‘Agiorgitiko’ (see Fig. 4; S8 on the ESM).
Fig. 4.
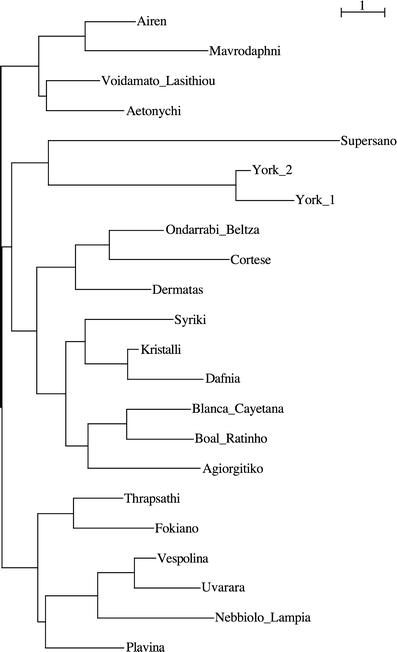
Detail of dendrogram, reported in full in S8, based on five microsatellites, showing the position of the three ancient samples: ‘York 1’, ‘York 2’ and ‘Supersano’ relative to the closest accessions from the database of Sefc et al. (2000). The bar corresponds to 1 U of genetic distance
Discussion
Biomolecular preservation
Py-GC/MS reveals the high abundance of both 4-methyl-catechol and 4-methyl-guaiacol, considered indicators of good preservation as the loss of methyl-side chains is commonly associated to lignin degradation (van Bergen et al. 1994). The abundance of the protein-derived compounds is lower than is commonly seen in waterlogged archaeological seeds (McCobb et al. 2001; Johan Kool, unpublished data).
Ancient protein degradation is also confirmed by the diffuse smear present in the electrophoretic profile (Fig. 2), as previously reported by Shewry et al. (1982), and by the ‘hump’ in the nano-LC chromatogram (cf. Fig. 3a with inset of modern sample with flat baseline; Moreno et al. 2005). Most of the proteins identified by mass spectrometry are storage proteins, which represent a source of carbon, nitrogen and sulfur for the shoot. They are usually present in large amounts in discrete deposits called protein bodies, and their high initial abundance in the seeds is probably the crucial factor for the persistence of residual fragments amenable to identification after centuries. Storage proteins are characterised by a high level of polymorphism, due to the presence of multigene families and to post-translational modifications (Shewry 1995; Shewry and Halford 2002). Consequently, a mass spectrometry-based approach, investigating storage protein isoform distribution, could be possibly developed for species or sub-species attribution of archaeological seeds, integrating conventional DNA-based methods. Preliminary results, documenting distinctive protein profiles between grape seeds from different modern varieties, have recently been reported (Pesavento et al. 2007).
The molecular investigation of this protein subset from archaeological seeds enabled, for the first time, a more detailed understanding of protein damage mechanisms over extended time periods. The role of asparagine and glutamine in protein ageing reactions clearly emerges. These amino acids are subject to deamidation (Ritz-Timme and Collins 2002), one of the most relevant hydrolytic protein degradation mechanisms (Groenen et al. 1994). The importance of deamidation/racemisation control for maintenance of seed viability is documented by the detection of active protein l-isoaspartyl methyltransferase in seeds that remained viable for centuries. This enzyme repairs l-isoaspartyl residues accumulating in ageing proteins as a consequence of deamidation/racemisation (Shen-Miller et al. 1995; Shen-Miller 2002; Clarke 2003). We found, in the ancient sample only, deamidation of Q3 in the peptide LHQGAMVLPYYNVNAH and of Q12 in SSVTGYDLPVLQK (see S5 and S9 in ESM). The estimated in vitro deamidation half-time life for glutamine in these contexts is more than 2 and 16 years, respectively (Robinson et al. 2004). Although it is possible that some diagenesis may occur as an artefact of laboratory preparation, the slow rates of these reactions convince us that they represent real products of the prolonged action of time.
Where multiple peptides from the same protein are detected, it is also instructive to identify those with atypically poor preservation. As an example, the quantitative analysis showed, for the peptide VVPVNPALLNR, a ‘survival level’ of approximately 6% in the ancient sample, two to four times lower than the other peptides from the same protein. Asn residues form a heterocyclic succinimide by attack on the amide of the C-terminally adjacent amino acid, but in the case of the imine Pro, Asn-Pro cyclisation leads to spontaneous cleavage of the peptide bond (Capasso et al. 1996), as previously reported in fossil proteins (McNulty et al. 2002). GNGYEETICSLR is another peptide characterised by atypically low recovery in the ancient sample. The Asn2-Gly3 peptide bond is particularly prone to cleavage, due to the limited steric hindrance of the glycine residue affording little protection (Bada 1985; Collins et al. 1998). Our results are consistent with the proposal that specific positions represent ‘hot spots’ for protein fragmentation (Dehart and Anderson 2007).
Racemisation and peptide bond cleavage are hydrolytic reactions (Dal Monte et al. 2002), enhanced in conditions, such as oxidative stress, that increase polypeptide chain flexibility (Ingrosso et al. 2002). Oxidative damage of ancient biomolecules is well documented (Höss et al. 1996) and carbonylation in particular is the predominant alteration recently detected in the Arabidopsis thaliana seed proteome, after ageing treatments (Rajjou et al. 2008). Carbonylation principally converts arginine and proline to glutamic semialdehyde and lysine to aminoadipic semialdehyde (Nyström 2005), removing from arginine and lysine side chains the basic motif necessary for trypsin substrate recognition. The limited tryptic peptide recovery can thus represent an indicator of oxidative damage, despite waterlogging and high levels of antioxidant compounds in grape seeds. The investigation of molecular degradation in relation to seed viability has practical application beyond the realms of archaeology, for example, in the forestry industry, and for the management of seed banks created to protect plant genetic diversity.
Different preservation of proteins and DNA from the two archaeological sites is to be expected, given the considerably warmer climate at Supersano compared with York, as anticipated by thermal age modelling (Smith et al. 2001). Supersano (40.02 N, 18.24 W) has a thermal age of 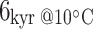 , an order of magnitude higher than that of the York specimens
, an order of magnitude higher than that of the York specimens 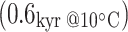 . The results from amino acid racemisation, protein degradation and DNA preservation, however, do not appear to be closely linked. The presence of aDNA—even nuclear microsatellites—in the Supersano sample seems inconsistent with our inability to obtain an insoluble protein extract and consequently peptide sequences. Further method development is probably required to improve peptide recovery from archaeological samples.
. The results from amino acid racemisation, protein degradation and DNA preservation, however, do not appear to be closely linked. The presence of aDNA—even nuclear microsatellites—in the Supersano sample seems inconsistent with our inability to obtain an insoluble protein extract and consequently peptide sequences. Further method development is probably required to improve peptide recovery from archaeological samples.
Ancient DNA
The recovery of five microsatellite loci from ancient grape seed samples demonstrates good nuclear DNA preservation and is quite encouraging for future work. The variation in cloned sequences, however, showed a certain degree of microsatellite stutter, as expected for aDNA (Burger et al. 1999). The data also indicate that it is highly unlikely that any of the results derived from contamination either between samples or from external sources. Specifically, all samples yielded identifiable grape sequences for all microsatellite loci, all samples differed at the markers and no samples or extraction and PCR controls showed any example of cross contamination. Although it has been popular to demand independent replication of aDNA results (Cooper and Poinar 2000), we subscribe to the view of Gilbert et al. (2005) that aDNA studies should be validated using a cognitive approach. As the controls showed no evidence of contamination, the samples yielded clearly grape DNA sequences, there was no evidence of any cross contamination in the sequences and the data were generated from multiple PCR reactions analysed in two different ways, i.e. cloning plus sequencing and capillary analysis; the results are unlikely to derive from contamination, thus do not require independent validation.
The microsatellite data, however, allow just tentative comparison of the ancient samples with the modern varieties. Conventionally at least six and possibly 20 microsatellites are required to fully resolve the cultivar for modern grape samples (Vouillamoz et al. 2003, 2006; This et al. 2004). We acknowledge that the investigation of five microsatellite loci is still not enough to clearly assign the ancient samples to a geographic region or to a cultivar. The low resolution resulting from so few microsatellites is clearly observed in the tree, as closely related cultivars (Bowers and Meredith 1997; Sefc et al. 1998; Vouillamoz and Grando 2006) do not group. The results of this analysis cannot be used to draw conclusions with regard to the attribution of the ancient samples to modern cultivars since clusters illustrate similarity rather than kinship or identity (Karatas et al. 2007). Despite both size calibration and correction for microsatellite shift, the allelic profiles of the ancient samples were quite distant from those of all the modern cultivars they were compared to. Ancient sample assignment to any geographic region is law, and their genetic distance from the modern varieties is high (Fig. 4), suggesting that they either represent unsequenced or extinct cultivars. Future analysis searching larger databases may lead to greater precision. Because V. vinifera is a hermaphrodite, seeds are mostly the result of selfing; nevertheless, crosses cannot be excluded, and consequently, analyses of ancient wood remains would also be useful to characterise old cultivars and compare their DNA profiles with those from modern ones (This et al. 2006). Although tentative, the data from the Supersano archaeological sample suggest shorter distance from the modern Greek cultivars ‘Dermatas’, ‘Syriki’, ‘Kristalli’, ‘Dafnia’ and ‘Agiorgitiko’. The affinity with Greek cultivars may suggest grape or wine trading between southern Italy and the eastern Mediterranean or perhaps cultivation of similar varieties in both the areas during the Byzantine period, or even earlier.
All the cultivars in the group closer to the York archaeological seeds are from regions in the south of Europe, with no particular resistance to cold environments. Despite the extreme latitude, viticulture in York cannot be excluded during the ‘Medieval Warm Period’ (about 1100–1300 A.D.; Lamb 1965; Seward 1979, p. 128; Goosse et al. 2006). However, assuming the York grape sample belongs to a variety with climatic requirements similar to those of the modern cultivars it clusters closer to, a more plausible explanation than local viticulture is that the seeds were imported from southern growing areas as dried raisins. Historical records document consistent import of raisins and wine from southern Europe into England for centuries starting from the twelfth century A.D. (Salzmann 1931, p. 410; Seward 1979, p. 135). Also, the lack in the York context of conventional archaeological evidence, such as tools or dedicated structures associated to winemaking activity, is consistent with this option. Further integration of aDNA and classical pollen data from Roman and medieval sites in southern areas of Britain could help to clarify the dynamics of viticulture and winemaking in this region (Unwin 1990; Brown et al. 2001).
Conclusions
In addition to observing good preservation of the lignin- and cellulose-derived compounds (as detected by Py-GC-MS), peptide sequencing allowed us to identify several ancient proteins and to characterise chemical changes. We determined that the highly abundant seed storage proteins were affected by hydrolytic damage and that specific motifs are particularly prone to cleavage. The finding also defines the basis for the development of a proteomic approach for species or sub-species attribution of archaeological seeds to integrate DNA-based methods.
Archaeological grape seeds have several traits that make them attractive for future studies on the domestication and diffusion processes of this important crop. The data suggest that the material from York was not grown locally, whilst the remains from Supersano evoke relations with the eastern Mediterranean. Successful genetic analysis of five microsatellite markers enabled a tentative comparison of the ancient profiles with the modern varieties, providing indications to corroborate archaeological hypotheses. This is a significant achievement with promise of much more with the integration of high-throughput sequencing (Poinar et al. 2006; Green et al. 2006; Gilbert et al. 2008) for both ancient proteins and DNA.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary methods (DOC 42 kb)
Photomicrographs of the archaeological grape seeds excavated in: a Supersano, Lecce, Italy, and b York, England. (DOC 1016 kb)
Primer sets used to amplify each microsatellite locus. (DOC 39 kb)
Compounds identified in the pyrolysate. (DOC 42 kb)
List of the peptides identified in protein extracts from modern and ancient grape seeds in ESI and MALDI MS experiments. (DOC 156 kb)
MALDI mass spectra (next page) of the protein digest from the medieval York seeds documenting a series of semi-tryptic peptides: FYLAGNPQNEFQQQQ (M+H+ m/z = 1,811.81; GRAVY = −1.340), FYLAGNPQNEFQQQ (M+H+ m/z = 1,683.75; GRAVY = −1.186), FYLAGNPQNEFQQ (M+H+ m/z = 1,555.68; GRAVY = −1.008) and FYLAGNPQNEFQ (M+H+ m/z = 1,427.62; GRAVY = −0.800) of the peptide FYLAGNPQNEFQQQQQQQQGSEGQQQQQEGGGSEGR. We believe that they represent diagenetic products rather than fragments generated by the MALDI laser from a single peptide presenting terminal polyglutamine (Sen and Pasha 2004), considering that they are detected in different spots on the plate—70, 71, 72; spectra a, b, and c, respectively—suggesting they existed at the stage of separation and thus before laser desorption. Elution is in agreement with their grand average of hydropathicity (GRAVY; Kyte and Doolittle 1982; Gasteiger et al. 2005). (DOC 605 kb)
Sequences of the clones of the microsatellite PCR products (in FASTA format) (DOC 58 kb)
Dendrogram (DOC 34 kb)
MS/MS spectra of peptides from the ancient sample presenting low velocity deamidations (DOC 25312 kb)
Acknowledgements
EC, JTO and MJC were supported by the Natural Environment Research Council ‘Palaeoproteomics’ grant (no. NE511148) allotted to MJC and JTO. EC and MJC would like to acknowledge Dr. Kirsty Penkman and Dr. Caroline Solazzo for technical support. PFC was supported by the ‘Genetime’ European Marie Curie Training Network. JK would like to acknowledge Eef Velthorst for help with Py-GC/MS analysis and Wageningen University for access to their equipment. The Centre of Excellence in Mass Spectrometry at York, chaired by JTO, is supported by Science City York and Yorkshire Forward, using funds from the Northern Way Initiative. The authors express their gratitude to Prof. Raffaele Testolin of the University of Udine-Italy for the helpful discussion about the genetic data.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Further details of the methods used in this work are provided in S1 as electronic supplemental material (ESM).
Data deposition note
All the mass spectrometric raw data associated with this manuscript may be downloaded from ProteomeCommons.org Tranche, http://tranche.proteomecommons.org, using the following hash: wY9mO2A9E/dcKFY7ZzWvX4Aa0BiMqU9G3BPyhvsMEUI9x3o7fls8thTmEts0CtbOrQrenfWw3vUPJwfzk0DtPJZHFxgAAAAAAA+RnA== or this link: http://www.proteomecommons.org/data-downloader.jsp?fileName= wY9mO2A9E/dcKFY7ZzWvX4Aa0BiMqU9G3BPyhvsMEUI9x3o7fls8thTmEts0CtbOrQrenfWw3vUPJwfzk0DtPJZHFxgAAAAAAA%2BRnA==. The hash may be used to prove exactly what files were published as part of this manuscript's dataset, and the hash may also be used to check that the data have not changed since publication.
References
- Akeret Ö, Carrott J, Mant J, Jaques D, Stewart G, Reeves B. Environmental samples. In: Reeves B, editor. 62–68 Low Petergate. York. Assessment report on an archaeological excavation. York: York Archaeological Trust; 2006. pp. 160–161. [Google Scholar]
- Arroyo-García R, Ruiz-García L, Bolling L, Ocete R, López MA, Arnold C, Ergul A, Söylemezoglu G, Uzun HI, Cabello F, Ibáñez J, Aradhya MK, Atanassov A, Atanassov I, Balint S, Cenis JL, Costantini L, Goris-Lavets S, Grando MS, Klein BY, McGovern PE, Merdinoglu D, Pejic I, Pelsy F, Primikirios N, Risovannaya V, Roubelakis-Angelakis KA, Snoussi H, Sotiri P, Tamhankar S, This P, Troshin L, Malpica JM, Lefort F, Martinez-Zapater JM. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol Ecol. 2006;15:3707–3714. doi: 10.1111/j.1365-294X.2006.03049.x. [DOI] [PubMed] [Google Scholar]
- Arthur P. Grubenhäuser nella Puglia bizantina. A proposito di recenti scavi a Supersano (LE) Archeol Mediev. 1999;XXVI:171–178. [Google Scholar]
- Arthur P. Il territorio di Supersano in età bizantina. In: Arthur P, Melissano V, editors. Supersano. Un paesaggio antico del Basso Salento. Galatina: Congedo; 2004. pp. 53–66. [Google Scholar]
- Arthur P, Fiorentino G, Imperiale ML (2008) L’insediamento in Loc. Scorpo (Supersano, LE) nel VII–VIII secolo. La scoperta di un paesaggio di età altomedievale. Archeol Mediev XXXV:365–380
- Asara JM, Schweitzer MH, Freimark LM, Phillips M, Cantley LC. Protein sequences from mastodon and Tyrannosaurus Rex revealed by mass spectrometry. Science. 2007;316:280–285. doi: 10.1126/science.1137614. [DOI] [PubMed] [Google Scholar]
- Bada JL. Amino acid racemization dating of fossil bones. Annu Rev Earth Planet Sci. 1985;13:241–268. doi: 10.1146/annurev.ea.13.050185.001325. [DOI] [Google Scholar]
- Bassermann-Jordan F (1975) Geschichte des Weinbaus, 3rd edn. Pfälzische Verlagsanstalt GmbH., Neustadt an der Weinstraße, reprint of the 2nd edn. Frankfurter Verlags-Anstalt A.G., Frankfurt am Main, 1923; vol II, pp 362–416
- Bowers JE, Meredith CP. The parentage of a classic wine grape, Cabernet Sauvignon. Nat Genet. 1997;16:84–87. doi: 10.1038/ng0597-84. [DOI] [PubMed] [Google Scholar]
- Bowers JE, Dangl GS, Vignani R, Meredith CP. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.) Genome. 1996;39:628–633. doi: 10.1139/g96-080. [DOI] [PubMed] [Google Scholar]
- Bradley RS, Hughes MK, Diaz HF. Climate change: climate in medieval time. Science. 2003;302:404–405. doi: 10.1126/science.1090372. [DOI] [PubMed] [Google Scholar]
- Brown A, Meadows I, Turner S, Mattingly D. Roman vineyards in Britain: stratigraphic and palynological data from Wollaston in the Nene Valley, England. Antiquity. 2001;75:745–757. [Google Scholar]
- Buckley M, Walker A, Ho SYW, Yang Y, Smith C, Ashton P, Oates JT, Cappellini E, Koon H, Penkman K, Elsworth B, Ashford D, Solazzo C, Andrews P, Strahler J, Shapiro B, Ostrom P, Gandhi H, Miller W, Raney B, Zylber MI, Gilbert MTP, Prigodich RV, Ryan M, Rijsdijk KF, Janoo A, Collins MJ. Comment on “protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry”. Science. 2008;319:33c. doi: 10.1126/science.1147046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger J, Hummel S, Herrmann B, Henke W. DNA preservation: a microsatellite-DNA study on ancient skeletal remains. Electrophoresis. 1999;20:1722–1728. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1722::AID-ELPS1722>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Capasso S, Mazzarella L, Sorrentino G, Balboni G, Kirby AJ. Kinetics and mechanism of the cleavage of the peptide bond next to asparagine. Peptides. 1996;17:1075–1077. doi: 10.1016/0196-9781(96)00153-2. [DOI] [PubMed] [Google Scholar]
- Chibani K, Ali-Rachedi S, Job C, Job D, Jullien M, Grappin P. Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiol. 2006;142:1493–1510. doi: 10.1104/pp.106.087452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. Aging as war between chemical and biochemical processes: protein methylation and the recognition of age-damaged proteins for repair. Ageing Res Rev. 2003;2:263–285. doi: 10.1016/S1568-1637(03)00011-4. [DOI] [PubMed] [Google Scholar]
- Collins MJ, Walton D, King A. The geochemical fate of proteins. In: Stankiewicz BA, van Bergen PF, editors. Nitrogen-containing macromolecules in the bio- and geosphere. American Chemical Society Symposium Series, 707. Washington, D.C.: American Chemical Society; 1998. pp. 74–87. [Google Scholar]
- Cooper A, Poinar HN. Ancient DNA: do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- Dal Monte PR, Edmond Rouan SK, Bam NB. Biotechnology-based pharmaceuticals. In: Banker GS, Rhodes CT, editors. Modern pharmaceutics, vol. 121. 4. New York: Dekker; 2002. pp. 1025–1066. [Google Scholar]
- Dehart MP, Anderson BD. The role of the cyclic imide in alternate degradation pathways for asparagine-containing peptides and proteins. J Pharm Sci. 2007;96:2667–2685. doi: 10.1002/jps.20905. [DOI] [PubMed] [Google Scholar]
- Derbyshire E, Harris N, Boulter D, Jope EM. The extraction, composition and intra-cellular distribution of protein in early maize grains from an archaeological site in N.E. Arizona. New Phytol. 1977;78:499–504. doi: 10.1111/j.1469-8137.1977.tb04856.x. [DOI] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package), version 3.6. Distributed by the author. Seattle: Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- Gilbert MTP, Bandelt H-J, Hofreiter M, Barnes I. Assessing ancient DNA studies. Trends Ecol Evol. 2005;20:541–544. doi: 10.1016/j.tree.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Gilbert MTP, Moore W, Melchior L, Worobey M. DNA extraction from dry museum beetles without conferring external morphological damage. PLoS ONE. 2007;2:e272. doi: 10.1371/journal.pone.0000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MTP, Kivisild T, Grønnow B, Andersen PK, Metspalu E, Reidla M, Tamm E, Axelsson E, Götherström A, Campos PF, Rasmussen M, Metspalu M, Higham TFG, Schwenninger J-L, Nathan R, De Hoog C-J, Koch A, Moller LN, Andreasen C, Meldgaard M, Villems R, Bendixen C, Willerslev E. Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science. 2008;320:1787–1789. doi: 10.1126/science.1159750. [DOI] [PubMed] [Google Scholar]
- Goosse H, Arzel O, Luterbacher J, Mann ME, Renssen H, Riedwyl N, Timmermann A, Xoplaki E, Wanner H. The origin of the European “Medieval Warm Period”. Clim Past. 2006;2:99–113. [Google Scholar]
- Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, Simons JF, Du L, Egholm M, Rothberg JM, Paunovic M, Pääbo S. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- Groenen PJTA, Merck KB, Jong WW, Bloemendal H. Structure and modifications of the junior chaperone α-crystallin. From lens transparency to molecular pathology. Eur J Biochem. 1994;225:1–19. doi: 10.1111/j.1432-1033.1994.00001.x. [DOI] [PubMed] [Google Scholar]
- Hollemeyer K, Altmeyer W, Heinzle E, Pitra C. Species identification of Oetzi's clothing with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry based on peptide pattern similarities of hair digests. Rapid Commun Mass Spectrom. 2008;22:2751–2767. doi: 10.1002/rcm.3679. [DOI] [PubMed] [Google Scholar]
- Höss M, Jaruga P, Zastawny TH, Dizdaroglu M, Pääbo S. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humpula JF, Ostrom PH, Gandhi H, Strahler JR, Walker AK, Stafford TW, Jr, Smith JJ, Voorhies MR, George Corner R, Andrews PC. Investigation of the protein osteocalcin of Camelops hesternus: sequence, structure and phylogenetic implications. Geochim Cosmochim Acta. 2007;71:5956–5967. doi: 10.1016/j.gca.2007.09.003. [DOI] [Google Scholar]
- Ingrosso D, Amelia C, D'Angelo S, Alfinito F, Zappia V, Galletti P. Protein methylation as a marker of aspartate damage in glucose-6-phosphate dehydrogenase-deficient erythrocytes. Eur J Biochem. 2002;269:2032–2039. doi: 10.1046/j.1432-1033.2002.02838.x. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyere C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pe ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quetier F, Wincker P. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Karatas Hs, Degirmenci D, Velasco R, Vezzulli S, Bodur A, Agaoglu YS. Microsatellite fingerprinting of homonymous grapevine (Vitis vinifera L.) varieties in neighboring regions of south-east Turkey. Sci Hortic. 2007;114:164–169. doi: 10.1016/j.scienta.2007.07.001. [DOI] [Google Scholar]
- Lamb HH. The early medieval warm epoch and its sequel. Palaeogeogr Palaeoclimatol Palaeoecol. 1965;1:13–37. doi: 10.1016/0031-0182(65)90004-0. [DOI] [Google Scholar]
- Li ZT, Gray DJ. Isolation by improved thermal asymmetric interlaced PCR and characterization of a seed-specific 2S albumin gene and its promoter from grape (Vitis vinifera L.) Genome. 2005;48:312–320. doi: 10.1139/g05-030. [DOI] [PubMed] [Google Scholar]
- Lucker J, Laszczak M, Smith D, Lund S. Generation of a predicted protein database from EST data and application to iTRAQ analyses in grape (Vitis vinifera cv. Cabernet Sauvignon) berries at ripening initiation. BMC Genomics. 2009;10:50. doi: 10.1186/1471-2164-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manen JF, Bouby L, Dalnoki O, Marinval P, Turgay M, Schlumbaum A. Microsatellites from archaeological Vitis vinifera seeds allow a tentative assignment of the geographical origin of ancient cultivars. J Archaeol Sci. 2003;30:721–729. doi: 10.1016/S0305-4403(02)00244-3. [DOI] [Google Scholar]
- Mangafa M, Kotsakis K. A new method for the identification of wild and cultivated charred grape seeds. J Archaeol Sci. 1996;23:409–418. doi: 10.1006/jasc.1996.0036. [DOI] [Google Scholar]
- McCobb LME, Briggs DEG, Evershed RP, Hall AR, Hall RA. Preservation of fossil seeds from a 10th century AD Cess Pit at Coppergate, York. J Archaeol Sci. 2001;28:929–940. doi: 10.1006/jasc.2000.0617. [DOI] [Google Scholar]
- McNulty T, Calkins A, Ostrom P, Gandhi H, Gottfried M, Martin L, Gage D. Stable isotope values of bone organic matter: artificial diagenesis experiments and paleoecology of Natural Trap Cave, Wyoming. Palaios. 2002;17:36–49. doi: 10.1669/0883-1351(2002)017<0036:SIVOBO>2.0.CO;2. [DOI] [Google Scholar]
- Meredith CP. Grapevine genetics: probing the past and facing the future. Agric Cons Scient. 2001;66:21–25. [Google Scholar]
- Moreno FJ, Maldonado BM, Wellner N, Mills ENC. Thermostability and in vitro digestibility of a purified major allergen 2S albumin (Ses i 1) from white sesame seeds (Sesamum indicum L.) Biochim Biophys Acta Proteins Proteomics. 2005;1752:142–153. doi: 10.1016/j.bbapap.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Nielsen-Marsh CM, Ostrom PH, Gandhi H, Shapiro B, Cooper A, Hauschka PV, Collins MJ. Sequence preservation of osteocalcin protein and mitochondrial DNA in bison bones older than 55 ka. Geology. 2002;30:1099–1102. doi: 10.1130/0091-7613(2002)030<1099:SPOOPA>2.0.CO;2. [DOI] [Google Scholar]
- Nielsen-Marsh CM, Richards MP, Hauschka PV, Thomas-Oates JE, Trinkaus E, Pettitt PB, Karavanic I, Poinar H, Collins MJ. Osteocalcin protein sequences of Neanderthals and modern primates. Proc Natl Acad Sci U S A. 2005;102:4409–4413. doi: 10.1073/pnas.0500450102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005;24:1311–1317. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue K, Clapham A, Evershed RP, Brown TA. Remarkable preservation of biomolecules in ancient radish seeds. Proc Biol Sci. 1996;263:541–547. doi: 10.1098/rspb.1996.0082. [DOI] [PubMed] [Google Scholar]
- Olmo HP. Grapes. In: Simmonds NW, editor. Evolution of crop plants. London: Longman; 1976. pp. 294–298. [Google Scholar]
- Paetkau D, Calvert W, Stirling I, Strobeck C. Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol. 1995;4:347–354. doi: 10.1111/j.1365-294X.1995.tb00227.x. [DOI] [PubMed] [Google Scholar]
- Paetkau D, Slade R, Burden M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol Ecol. 2004;13:55–65. doi: 10.1046/j.1365-294X.2004.02008.x. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. Genalex 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkman KEH, Kaufman DS, Maddy D, Collins MJ. Closed-system behaviour of the intra-crystalline fraction of amino acids in mollusc shells. Quat Geochronol. 2008;3:2–25. doi: 10.1016/j.quageo.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrière G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Pesavento IC, Bertazzo A, Flamini R, Dalla Vedova A, De Rosso M, Seraglia R, Traldi P. Differentiation of Vitis vinifera varieties by MALDI-MS analysis of the grape seed proteins. J Mass Spectrom. 2007;43:234–241. doi: 10.1002/jms.1295. [DOI] [PubMed] [Google Scholar]
- Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A. GENECLASS2: a software for genetic assignment and first-generation migrant detection. J Hered. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Poinar HN, Schwarz C, Qi J, Shapiro B, Macphee RD, Buigues B, Tikhonov A, Huson DH, Tomsho LP, Auch A, Rampp M, Miller W, Schuster SC. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- Rajjou L, Belghazi M, Huguet R, Robin C, Moreau A, Job C, Job D. Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol. 2006;141:910–923. doi: 10.1104/pp.106.082057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Lovigny Y, Groot SPC, Belghazi M, Job C, Job D. Proteome-wide characterization of seed aging in Arabidopsis. A comparison between artificial and natural aging protocols. Plant Physiol. 2008;148:620–641. doi: 10.1104/pp.108.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Hatfield RD. Pyrolysis-GC-MS characterization of forage materials. J Agric Food Chem. 1991;39:1426–1437. doi: 10.1021/jf00008a014. [DOI] [Google Scholar]
- Reeves B. 62–68 Low Petergate. York. Assessment report on an archaeological excavation. York: York Archaeological Trust; 2006. [Google Scholar]
- Ritz-Timme S, Collins MJ. Racemization of aspartic acid in human proteins. Ageing Res Rev. 2002;1:43–59. doi: 10.1016/S0047-6374(01)00363-3. [DOI] [PubMed] [Google Scholar]
- Robinson NE, Robinson ZW, Robinson BR, Robinson AL, Robinson JA, Robinson ML, Robinson AB. Structure-dependent nonenzymatic deamidation of glutaminyl and asparaginyl pentapeptides. J Pept Res. 2004;63:426–436. doi: 10.1111/j.1399-3011.2004.00151.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sallon S, Solowey E, Cohen Y, Korchinsky R, Egli M, Woodhatch I, Simchoni O, Kislev M. Germination, genetics, and growth of an ancient date seed. Science. 2008;320:1464. doi: 10.1126/science.1153600. [DOI] [PubMed] [Google Scholar]
- Salzmann LF. English trade in the middle ages. Oxford: Clarendon; 1931. [Google Scholar]
- Schweitzer MH, Zheng W, Organ CL, Avci R, Suo Z, Freimark LM, Lebleu VS, Duncan MB, Vander Heiden MG, Neveu JM, Lane WS, Cottrell JS, Horner JR, Cantley LC, Kalluri R, Asara JM. Biomolecular characterization and protein sequences of the Campanian Hadrosaur B. canadensis. Science. 2009;324:626–631. doi: 10.1126/science.1165069. [DOI] [PubMed] [Google Scholar]
- Sefc KM, Steinkellner H, Glössl J, Kampfer S, Regner F. Reconstruction of a grapevine pedigree by microsatellite analysis. Theor Appl Genet. 1998;97:227–231. doi: 10.1007/s001220050889. [DOI] [Google Scholar]
- Sefc KM, Regner F, Turetschek E, Glössl J, Steinkellner H. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome. 1999;42:367–373. doi: 10.1139/gen-42-3-367. [DOI] [PubMed] [Google Scholar]
- Sefc KM, Lopes MS, Lefort F, Botta R, Roubelakis-Angelakis KA, Ibáñez JIP, Wagner HW, Glössl J, Steinkellner H. Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theor Appl Genet. 2000;100:498–505. doi: 10.1007/s001220050065. [DOI] [Google Scholar]
- Seward D. Monks and wine. London: Mitchell Beazley; 1979. [Google Scholar]
- Shen-Miller J. Sacred lotus, the long-living fruits of China Antique. Seed Sci Res. 2002;12:131–143. doi: 10.1079/SSR2002112. [DOI] [Google Scholar]
- Shen-Miller J, Mudgett MB, Schopf JW, Clarke S, Berger R. Exceptional seed longevity and robust growth: ancient sacred lotus from China. Am J Bot. 1995;82:1367–1380. doi: 10.2307/2445863. [DOI] [Google Scholar]
- Shewry PR. Plant storage proteins. Biol Rev. 1995;70:375–426. doi: 10.1111/j.1469-185X.1995.tb01195.x. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Halford NG. Cereal seed storage proteins: structures, properties and role in grain utilization. J Exp Bot. 2002;53:947–958. doi: 10.1093/jexbot/53.370.947. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Kirkman MA, Burgess SR, Festenstein GN, Miflin BJ. A comparison of the protein and amino acid composition of old and recent barley grain. New Phytol. 1982;90:455–466. doi: 10.1111/j.1469-8137.1982.tb04478.x. [DOI] [Google Scholar]
- Smith CI, Chamberlain AT, Riley MS, Cooper A, Stringer CB, Collins MJ. Neanderthal DNA: not just old but old and cold? Nature. 2001;410:771–772. doi: 10.1038/35071177. [DOI] [PubMed] [Google Scholar]
- Solazzo C, Fitzhugh WW, Rolando C, Tokarski C. Identification of protein remains in archaeological potsherds by proteomics. Anal Chem. 2008;80:4590–4597. doi: 10.1021/ac800515v. [DOI] [PubMed] [Google Scholar]
- This P, Jung A, Boccacci P, Borrego J, Botta R, Costantini L, Crespan M, Dangl G, Eisenheld C, Ferreira-Monteiro F, Grando S, Ibáñez J, Lacombe T, Laucou V, Magalhães R, Meredith C, Milani N, Peterlunger E, Regner F, Zulini L, Maul E. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor Appl Genet. 2004;109:1448–1458. doi: 10.1007/s00122-004-1760-3. [DOI] [PubMed] [Google Scholar]
- This P, Lacombe T, Thomas MR. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006;22:511–519. doi: 10.1016/j.tig.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Thomas MR, Scott NS. Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as sequence-tagged sites (STSs) Theor Appl Genet. 1993;86:985–990. doi: 10.1007/BF00211051. [DOI] [PubMed] [Google Scholar]
- Unwin T. Saxon and early Norman viticulture in England. J Wine Res. 1990;1:61–75. doi: 10.1080/09571269008717857. [DOI] [Google Scholar]
- van Bergen PF, Collinson ME, Sinninghe Damsté JS, de Leeuw JW. Chemical and microscopical characterization of inner seed coats of fossil water plants. Geochim Cosmochim Acta. 1994;58:231–239. doi: 10.1016/0016-7037(94)90460-X. [DOI] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, FitzGerald LM, Vezzulli S, Reid J, Malacarne G, Iliev D, Coppola G, Wardell B, Micheletti D, Macalma T, Facci M, Mitchell JT, Perazzolli M, Eldredge G, Gatto P, Oyzerski R, Moretto M, Gutin N, Stefanini M, Chen Y, Segala C, Davenport C, Dematte L, Mraz A, Battilana J, Stormo K, Costa F, Tao Q, Si-Ammour A, Harkins T, Lackey A, Perbost C, Taillon B, Stella A, Solovyev V, Fawcett JA, Sterck L, Vandepoele K, Grando SM, Toppo S, Moser C, Lanchbury J, Bogden R, Skolnick M, Sgaramella V, Bhatnagar SK, Paolo F, Gutin A, Peer Y, Salamini F, Viola R. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PloS One. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent D, Wheatley MD, Cramer GR. Optimization of protein extraction and solubilization for mature grape berry clusters. Electrophoresis. 2006;27:1853–1865. doi: 10.1002/elps.200500698. [DOI] [PubMed] [Google Scholar]
- Vouillamoz JF, Grando MS. Genealogy of wine grape cultivars: ‘Pinot’ is related to ‘Syrah’. Heredity. 2006;97:102–110. doi: 10.1038/sj.hdy.6800842. [DOI] [PubMed] [Google Scholar]
- Vouillamoz J, Maigre D, Meredith CP. Microsatellite analysis of ancient alpine grape cultivars: pedigree reconstruction of Vitis vinifera L. ‘Cornalin du Valais’. Theor Appl Genet. 2003;107:448–454. doi: 10.1007/s00122-003-1265-5. [DOI] [PubMed] [Google Scholar]
- Vouillamoz JF, McGovern PE, Ergul A, Söylemezoğlu G, Tevzadze G, Meredith CP, Grado MS. Genetic characterization and relationships of traditional grape cultivars from Transcaucasia and Anatolia. Plant Genet Resour. 2006;4:144–158. doi: 10.1079/PGR2006114. [DOI] [Google Scholar]
- Yilmaz Y, Toledo RT. Major flavonoids in grape seeds and skins: antioxidant capacity of catechin, epicatechin, and gallic acid. J Agric Food Chem. 2004;52:255–260. doi: 10.1021/jf030117h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods (DOC 42 kb)
Photomicrographs of the archaeological grape seeds excavated in: a Supersano, Lecce, Italy, and b York, England. (DOC 1016 kb)
Primer sets used to amplify each microsatellite locus. (DOC 39 kb)
Compounds identified in the pyrolysate. (DOC 42 kb)
List of the peptides identified in protein extracts from modern and ancient grape seeds in ESI and MALDI MS experiments. (DOC 156 kb)
MALDI mass spectra (next page) of the protein digest from the medieval York seeds documenting a series of semi-tryptic peptides: FYLAGNPQNEFQQQQ (M+H+ m/z = 1,811.81; GRAVY = −1.340), FYLAGNPQNEFQQQ (M+H+ m/z = 1,683.75; GRAVY = −1.186), FYLAGNPQNEFQQ (M+H+ m/z = 1,555.68; GRAVY = −1.008) and FYLAGNPQNEFQ (M+H+ m/z = 1,427.62; GRAVY = −0.800) of the peptide FYLAGNPQNEFQQQQQQQQGSEGQQQQQEGGGSEGR. We believe that they represent diagenetic products rather than fragments generated by the MALDI laser from a single peptide presenting terminal polyglutamine (Sen and Pasha 2004), considering that they are detected in different spots on the plate—70, 71, 72; spectra a, b, and c, respectively—suggesting they existed at the stage of separation and thus before laser desorption. Elution is in agreement with their grand average of hydropathicity (GRAVY; Kyte and Doolittle 1982; Gasteiger et al. 2005). (DOC 605 kb)
Sequences of the clones of the microsatellite PCR products (in FASTA format) (DOC 58 kb)
Dendrogram (DOC 34 kb)
MS/MS spectra of peptides from the ancient sample presenting low velocity deamidations (DOC 25312 kb)


