Abstract
Recent studies show that the extrahepatic human UDP-glucurono-syltransferase (UGT)1A10 is capable of phase II glucuronidation of several major cytochrome P450 metabolites of warfarin (i.e., 6-, 7-, and 8-hydroxywarfarin). This study expands on this finding by testing the hypothesis that the UGT1A10 F90-M91-V92-F93 amino acid motif is important for proper recognition and conjugation of hydroxywarfarin derivatives. Site-directed mutagenesis studies demonstrate that F90 is critical for 6- and 7-hydroxywarfarin glucuronidation based on the complete loss of enzymatic activity toward these substrates. In contrast, V92A and F93A mutants lead to higher rates of substrate turnover, have minimum changes in Km values, and demonstrate substrate inhibition kinetics. A completely different activity profile is observed in the presence of 8-hydroxywarfarin. No change in either activity or affinity is observed with F90A when compared with wild type, whereas F93A and V92A mutants show increases in Vmax (3- and 10-fold, respectively) and minimum changes in Km. Liquid chromatography-tandem mass spectrometry studies show that enzymatic products produced by mutants are identical to wild-type products produced in the presence of 6-, 7-, and 8-hydroxywarfarin. Because F90 is not critical for the glucuronidation of 8-hydroxywarfarin, there is likely another, different amino acid responsible for binding this compound. In addition, an inhibitory binding site may be formed in the presence of 6- and 7-hydroxywarfarin. This new knowledge and continued characterization of the hydroxywarfarin binding site(s) for UGT1A10 will help elucidate the molecular mechanism of hydroxywarfarin glucuronidation and potentially result in more effective anticoagulant therapies.
Warfarin (Coumadin; Bristol-Myers Squibb, NY, NY) is an anticoagulant drug that is routinely administered worldwide to manage thromboembolic disease. Even though this drug has been administered and studied for many decades, there remain gaps in our understanding of potential adverse side effects and biological fate of this compound. Efforts have primarily focused on the role of phase I enzymes, specifically cytochromes P450 (P450 or CYP for a specific isoform), in the inactivation and elimination of warfarin and its metabolites [reviewed in Kaminsky and Zhang (1997)]. Administered as a racemic mixture of R and S forms, warfarin undergoes different phase I oxidations that depend on the chirality of substrate. CYP2C9 preferentially hydroxylates the more active S-isomer to 6- and 7-hydroxy-warfarin. In contrast, R-warfarin undergoes hydroxylation at the 6-position by CYP1A2, the 8 position by CYP2C19, and the 10 and 4′ positions by CYP3A4. The modification of the hydroxywarfarin ring at positions 6-, 7, and 8 inactivates warfarin anticoagulant activity; however, the impact of the other hydroxylations has not been investigated. Despite the contribution of multiple P450s to phase I metabolism of warfarin, CYP2C9 activity predominates in the inactivation of warfarin anticoagulant activity and thus is the focus of many pharmacogenetic approaches to managing warfarin treatment.
The introduction of hydroxyl groups by P450 activity transforms warfarin to molecules suitable for downstream phase II conjugative processes that facilitate removal of the metabolites. For example, glucuronidation is a phase II reaction catalyzed by the superfamily of UDP glucuronosyltransferase enzymes that conjugate the cosubstrate UDP glucuronic acid to the hydrophobic aglycons resulting in a metabolite with greater polarity and solubility. As a result of these chemical properties, glucuronides are readily excreted in the bile or urine. Removal of the hydroxylated warfarins would eliminate any residual activity (e.g., by 10- or 4′-hydroxywarfarin) and prevent product inhibition as observed for CYP2C9 by 8-hydroxywarfarin (Zielinska et al., 2008).
Despite the potential contribution of downstream processes to warfarin metabolism, only a few studies have explored the activity of phase II enzymes and the corresponding biological consequences in the form of excretable metabolites. Jansing et al. (1992) used rat hepatocytes to identify glucuronidation metabolic pathways for warfarin. The formation of 4′-hydroxywarfarin and subsequent transport from the cell occurred as a single glucuronide conjugate. Although 6-hydroxywarfarin was found mainly as a sulfate conjugate, the 7- and 8-hydroxywarfarin metabolites were converted to both glucuronide and sulfate products. In addition, the glucuronide products were found as both mono- and diglucuronide conjugates. The only evidence of human warfarin conjugation came from a single set of unpublished in vivo studies in which racemic warfarin was administered (Kaminsky and Zhang, 1997). These studies indicated that 6-, 8-, and 10-hydroxywarfarin were excreted as either sulfate or glucuronide conjugates; however, the specific human UGT isoforms were not characterized. In vitro studies using human recombinant UGTs characterized many of these UGTs and showed that several hydroxylated derivatives of warfarin generated by P450-oxidative metabolism, but not warfarin itself, are substrates for glucuronidation (Zielinska et al., 2008). Products were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS), and spectral analysis suggests formation of the O-glucuronide conjugate at the 6, 7, and 8 positions, respectively. Of special interest is the fact that a major extrahepatic isoform UGT1A10 located in the intestine shows significant activity toward 6-, 7-, and 8-hydroxywarfarin.
In this study, we have investigated the role of the UGT1A10 structure in the specificity toward hydroxywarfarins. We targeted a proposed substrate-binding motif, F90-M91-V92-F93, for site-directed mutagenesis and assessed the impact on catalytic activity. Specifically, we measured steady-state glucuronidation of 6-, 7-, and 8-hydroxywarfarin by single-site alanine mutants of F90, V92, and F93. Of these mutants, UGT1A10 F90A lacked activity toward 6- and 7-hydroxywarfarin but retained activity toward 8-hydroxywarfarin. The differences in activity could reflect either the loss in binding of 6- and 7-hydroxywarfarin or a change in binding to a nonproductive orientation for catalysis. To explore these possibilities, we performed inhibition studies for UGT1A10 F90A with 6- and 7-hydroxywarfarin. We included in that effort experiments with warfarin, which was shown previously not to be a substrate for UGT1A enzymes (Zielinska et al., 2008). Taken together, these studies complement recent findings associating genetic variations of cytochromes P450 with differential warfarin responses (Higashi et al., 2002), and it is anticipated that continued characterization of UGT active sites and potential UGT1A10 polymorphisms will lead to novel anticoagulant therapies having reduced adverse side effects.
Materials and Methods
Materials
All chemicals used for this study were of at least reagent grade. 6-Hydroxywarfarin, 7-hydroxywarfarin, 8-hydroxywarfarin, and UDP-glucuronic acid were purchased from Sigma-Aldrich (St. Louis, MO). Ethyl alcohol (95%) was purchased from AAPER (Shelbyville, KY). Unless otherwise specified, all other chemicals and reagents were of reagent grade and purchased from Sigma-Aldrich. Recombinant human UGT1A10 and it mutants were produced in baculovirus-infected insect cells as previously described (Kurkela et al., 2003; Kuuranne et al., 2003). Despite the inclusion of a histidine tag with the UGT constructs, we were not able to use the tag for affinity purification and retain activity, and thus membrane preparations were used for catalytic studies.
Recombinant UGT Isoform Incubations
For kinetic assays, UGT recombinant membrane protein (5 μg) was incubated in reaction buffer [100 μM Tris-HCl (pH 7.4)/5 mM MgCl2/5 mM saccharolactone] with 100 to 2000 μM substrate at a fixed concentration of the cosubstrate, UDP-GlcUA (4 mM), in a total volume of 30 μl. Substrates were added in dimethyl sulfoxide with a final concentration of 2%, and controls omitting substrates were run with each assay. No additional detergents or other activators were used in the incubations. Reactions were started by the addition of cosubstrate and incubated at 37°C for 90 min. The rate of glucuronidation of 8-hydroxywarfarin was shown to be linear for up to 3 h (data not shown). After 90 min, reactions were stopped by addition of 40 μl of ethanol. At least two sets of experiments were prepared in duplicate on different days. These data were compiled and analyzed to obtain the appropriate kinetic parameters.
HPLC-UV/Vis Analysis
For kinetic assays, each sample was centrifuged at 14,000 rpm for 8 min to spin down the protein, and 60 μl of the supernatant was transferred to an autosampler vial for analysis by high-performance liquid chromatography (HPLC). Analyses were performed using an HP 1050 HPLC system (Hewlett Packard, Palo Alto, CA) equipped with a UV-Vis diode array detector using conditions identical to those previously published (Zielinska et al., 2008). Primary standards for the glucuronidated monohydroxylated warfarin metabolites were not available; therefore, product concentrations were semiquantified using the responses for external standards of each warfarin substrate. It has been shown previously that the addition of the glucuronic acid moiety does not alter the extinction coefficients from that of the unreacted substrate (Doerge et al., 2000). Detection limits were the same as those previously published (Zielinska et al., 2008).
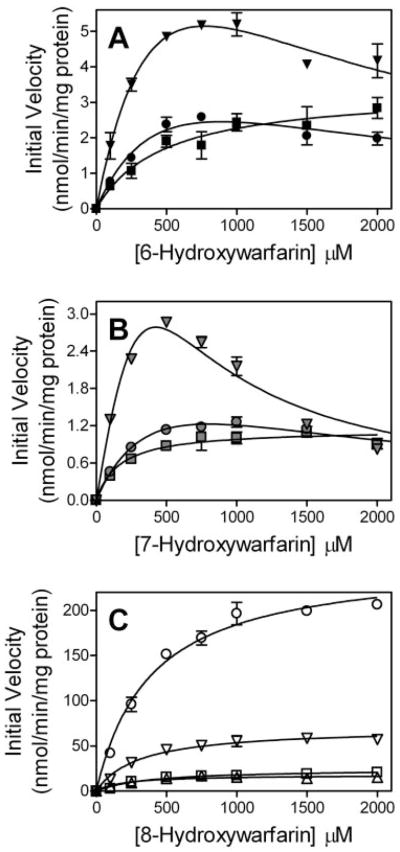
The kinetic parameters for UGT1A10 and the mutants were determined using two different mechanisms as described previously (Collom et al., 2007). For all hydroxywarfarins, wild-type UGT1A10 demonstrated hyperbolic kinetics, which is consistent with single-site reaction kinetics. These data were fit to the traditional Michaelis-Menten kinetic scheme yielding Vmax and Km (Fig. 1). This simple mechanism was also applicable for analyzing UGT1A10 mutant steady-state activity with the exception of 6- and 7-hydroxywarfarin glucuronidation by UGT1A10 V92A and F93A. In the latter studies, initial velocities increased as a function of substrate concentration and then decreased, indicating the mutations caused substrate inhibition. To analyze those data sets, a two-site substrate inhibition model in which a second substrate binding site becomes occupied at higher substrate concentration to generate an inactive ternary complex (ESS) was used (Fig. 1). This mechanism yielded Vmax, Km, and the equilibrium constant for the inhibitory site, Ks. The fitting of all data sets to the respective mechanisms was done through the use of DynaFit software (BioKin, Watertown, MA) (Kuzmic, 1996).
Fig. 1.
Possible reaction mechanisms for UGT1A10 activity toward hydroxywarfarins.
To assess the binding of nonsubstrates, the significance of inhibition and the corresponding IC50 value were determined by plotting the decrease in activity as a function of log of warfarin and hydroxywarfarin concentration and fitting to the standard IC50 equation. Specifically, under standard reaction conditions, glucuronidation of 250 μM 8-hydroxywarfarin was monitored in the presence of 0.5 to 2500 μM competitor. Two to four sets of experiments were performed in duplicate. Data were compiled, normalized to 100% activity, and fit globally to calculate the appropriate IC50 value using GraphPad Prism software (La Jolla, CA). For simplicity, we assumed that inhibition would be complete under saturating conditions, so that the minimal activity was set at 0%. IC50 values were then converted to the corresponding inhibition constant (Ki) based on the following relationship: IC50 = Ki(1 + [S]/Km), where S is the concentration of hydroxywarfarin substrate and Km is the corresponding kinetic constant. The hydroxywarfarin Km values for wild-type and mutant UGT1A10 enzymes are given in Tables 1 and 2.
TABLE 1.
Steady-state kinetics for 6-hydroxywarfarin glucuronidation by UGT1A10 and its mutants
| Enzyme | Kinetic model | Kinetic Parameters | ||
|---|---|---|---|---|
| Vmax | Km | Ks | ||
| nmol/min/mg protein | μM | μM | ||
| Wild type | Michaelis-Menten | 3.3 (2.7–4.2)a | 480 (200–760)a | —b |
| F90A | —b | —b | —b | —b |
| V92A | Substrate inhibition | 6.3 (3.6–78)c | 700 (250–12000)c | 1100 (61–4000)c |
| F93A | Substrate inhibition | 15 (8.7–66)a | 720 (310–4300)a | 820 (130–2200)a |
95% confidence intervals.
No activity was detected.
93% confidence intervals.
TABLE 2.
Steady-state kinetics for 7-hydroxywarfarin glucuronidation by UGT1A10 and its mutants
| Enzyme | Kinetic Model | Kinetic Parameters | ||
|---|---|---|---|---|
| Vmax | Km | Ks | ||
| nmol/min/mg protein | μM | μM | ||
| Wild type | Michaelis-Menten | 1.1 (1.0–1.3)a | 170 (77–250)a | —b |
| F90A | —b | —b | —b | —b |
| V92A | Substrate inhibition | 2.9 (1.9–7.5)a | 560 (270–1900)a | 1100 (320–2800)a |
| F93A | Substrate inhibition | N.D. | N.D. | N.D. |
N.D., not able to be determined (see Results).
95% confidence intervals.
No activity was detected.
LC-MS/MS Analysis
For product conformation, samples were centrifuged at 14,000 rpm for 8 min to spin down the protein, and 60 μl of the supernatant was transferred to an autosampler vial for analysis by LC-MS/MS. Analyses were performed using an Agilent 1100 HPLC system (Agilent Technologies, Santa Clara, CA), which was interfaced with an API 4000 triple quadrupole (MS/MS) mass spectrometer (Applied Biosystems, Foster City, CA). Conditions used were identical to those that have been previously published (Zielinska et al., 2008).
Results
Glucuronidation of Warfarin and Its Hydroxylated Derivatives, 6-, 7-, and 8-Hydroxywarfarin, by Recombinant UGT1A10, F90A, V92A, and F93A
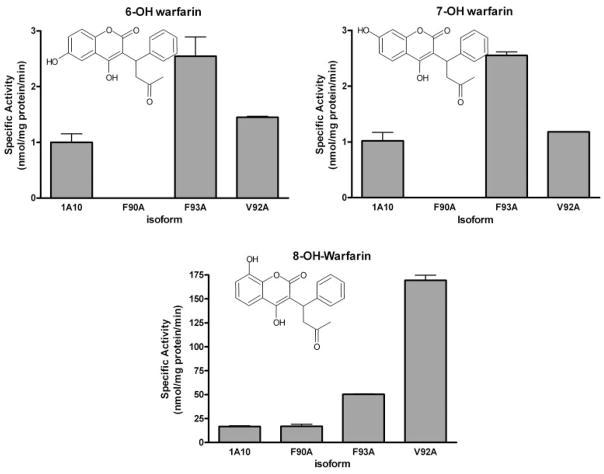
Human recombinant UGT1A10 and its mutants expressed as Histag proteins in baculovirus-infected Sf9 insect cells were evaluated for their ability to glucuronidate warfarin and racemic mixtures of 6-, 7-, and 8-hydroxywarfarin. Racemic incubations represented clinical dosing because warfarin is usually administered as a racemic mixture of its two enantiomeric forms. Screening data from the glucuronidation experiments are shown (Fig. 2). These preliminary studies were assessed by measuring the UDP-GlcUA-dependent formation of conjugates in the presence of 750 μM of each substrate.
Fig. 2.
Glucuronidation activities of UGT1A10, F90A, F93A, and V92A toward 6-, 7-, and 8-hydroxywarfarin. Glucuronidation activities of 1A10 and its mutants were measured using membrane fractions of recombinant UGTs expressed as Histag proteins in baculovirus-infected Sf9 insect cells. The substrate and cosubstrate (UDP-GlcUA) concentrations were 750 μM and 4 mM, respectively. Specific activities are expressed in nmol/mg protein/min and shown with S.E.M. based on two experiments.
Similar patterns of glucuronidation were seen with wild-type UGT1A10 and the V92A and F93A mutants. The 8-hydroxywarfarin glucuronide was by far the predominant product (20, 183, and 50 nmol/min/mg protein for wild type, V92A, F93A, respectively) followed by much lower biosynthesis of the 6- and 7-hydroxywarfarin glucuronides (6-hydroxywarfarin: 1.8, 2.5, and 4.5 nmol/min/mg protein for wild type, V92A, and F93A, respectively; 7-hydroxywarfarin: 1.0, 1.2, and 2.5 nmol/min/mg protein for wild type, V92A, and F93A, respectively). The F90A mutant was unique in that the 8-hydroxy-glucuronide was the only product detected (20 nmol/min/mg protein).
Kinetic Analysis of Recombinant UGT1A10, F90A, V92A, and F93A toward 6-, 7-, and 8-Hydroxywarfarin
The analysis of the steady-state kinetics for glucuronidation of the racemic hydroxywarfarins required fitting the data to mechanisms that are dependent on the particular mutation and substrate (Fig. 3; Tables 1–3). Consistent with traditional Michaelis-Menten kinetics (Fig. 1), the glucuronidation of all substrates by wild-type UGT1A10 yielded a hyperbolic correlation between substrate concentration and the initial velocity. A similar trend was observed for all UGT1A10 mutants toward 8-hydroxywarfarin (Fig. 3C). In contrast, the glucuronidation of 6- and 7-hydroxywarfarin by UGT1A10 V92A and F93A was best described by a substrate inhibition model (Fig. 1) whereby increasing concentrations of substrate led to an initial rise in the observed velocity followed by a decrease (Fig. 3, A and B). For simplicity, we assumed the ternary ESS complex was inactive. Otherwise, an active ESS complex introduces another variable to the mechanism, which renders the resulting equation unsolvable because of overparameterization. Despite activity toward 8-hydroxywarfarin, UGT1A10 F90A did not glucuronidate either 6- or 7-hydroxywarfarin.
Fig. 3.
Steady-state glucuronidation of 6-, 7-, and 8-hydroxywarfarin (A, B, and C, respectively) by UGT1A10 and its mutants. Initial velocities for glucuronidation were measured by incubating membrane fractions containing recombinant UGT1A10 and its mutants with increasing concentrations of substrate at a constant concentration of UDP-GlcUA (4 mM). Symbols are as follows: wild type, ■; F90A, ▲; V92A,●; F93A, ▼;. Shading for the symbols denotes the specific substrates used in the assays. The resulting data were fit to either a single-site Michaelis-Menten mechanism or substrate inhibition mechanism shown in Fig. 1 using DynaFit software (Kuzmic, 1996). The corresponding kinetic parameters are listed in Table 1.
TABLE 3.
Steady-state kinetics for 8-hydroxywarfarin glucuronidation by UGT1A10 and its mutants
| Enzyme | Kinetic model | Kinetic Parameters | |
|---|---|---|---|
| Vmax | Km | ||
| nmol/min/mg protein | μM | ||
| Wild type | Michaelis-Menten | 25 (22–28)a | 390 (240–530)a |
| F90A | Michaelis-Menten | 18 (15–20)a | 190 (73–310) |
| V92A | Michaelis-Menten | 260 (240–280)a | 390 (300–480) |
| F93A | Michaelis-Menten | 69 (63–75)a | 304 (220–400) |
95% confidence intervals.
Mutations at all three sites significantly altered kinetic parameters for glucuronidation of the hydroxywarfarins (Tables 1–3). Substitution of F90 with alanine led to a complete loss of activity toward 6-and 7-hydroxywarfarin, whereas this mutant retained activity toward 8-hydroxywarfarin. UGT1A10 F90A displayed a Vmax similar to wild type and a 2-fold lower Km.
The V92A mutation resulted in an increase in substrate turnover often at the expense of substrate recognition. For 6- and 7-hydroxywarfarin, UGT1A10 V92A demonstrated a 2-fold higher Vmax along with a 1.5- and 3-fold higher Km for these respective substrates. In addition, the kinetic mechanisms for 6- and 7-hydroxywarfarin glucuronidation by UGT1A10 required the formation of an inhibitory complex with a Ks value slightly greater than the corresponding Km. UGT1A10 V92A was the most effective at glucuronidating 8-warfarin; Vmax was 10-fold greater than wild type and demonstrated a comparable Km.
The catalytic parameters for UGT1A10 F93A were similar to the results observed for UGT1A10 V92A. For 6-hydroxywarfarin, Vmax increased 5-fold and Km increased 1.5-fold compared with wild type. The maximal rate of turnover was suppressed by the occupancy of a second substrate site with a Ks value 1.5-fold greater than Km. Unlike 6-hydroxywarfarin, higher concentrations of 7-hydroxywarfarin led to a rapid increase in turnover followed by a rapid loss in activity. The effect occurred over a narrow concentration range, which obviated the determination of parameters to describe the mechanism. Clearly, the mutation increased Vmax, Km, or both, but it also created a high affinity second site which, when occupied, severely inhibited turnover. 8-Hydroxywarfarin glucuronidation by UGT1A10 F93A resulted in a 3-fold higher Vmax and similar Km relative to wild type.
Competitive Inhibition Studies for Recombinant UGT1A10, F90A, V92A, and F93A toward Warfarin and Hydroxywarfarins
Because of the observed lack of activity toward 6- and 7-hydroxywarfarin, we assayed competition for UGT1A10 F90A between these nonsubstrates and 8-hydroxywarfarin and, thus, estimated the impact of the mutations on binding. The IC50 value for 6-hydroxywarfarin was 520 μM (430 to 650, 95% confidence interval), whereas the corresponding value for 7-hydroxywarfarin was approximately 2-fold higher at 950 μM (800 to 1100, 95% confidence interval). Based on the Km value for 8-hydroxywarfarin (Table 3), the corresponding Ki values are 300 μM for 6-hydroxywarfarin and 540 μM for 7-hydroxywarfarin, respectively.
Similarly, we measured the IC50 values for wild-type and mutant UGT1A10 toward warfarin, which was shown previously not to be a substrate for UGT1A enzymes (Zielinska et al., 2008). The IC50 value for wild-type UGT1A10 was at the limit of detection, 2500 μM (1600 to 3400, 95% confidence interval). The mutations displayed a very different effect on warfarin affinity relative to wild type. Loss of F90 actually increased binding almost 3-fold (IC50 = 920 with 720 to 1100, 95% confidence interval). UGT1A10 V92A displayed an IC50 value similar to wild type, that is, 3000 μM (2200 to 3700, 95% confidence interval). The F93 mutation increased the IC50 value above the highest concentration of competitor used in the assay, and thus the corresponding value is > 2500 μM. Based on the 8-hydroxywarfarin Km values (Table 3), the warfarin Ki values were 1500, 520, 1700, and > 1500 μM for wild type, F90A, V92A, and F93A, respectively.
Product Confirmations/MS Spectral Interpretation
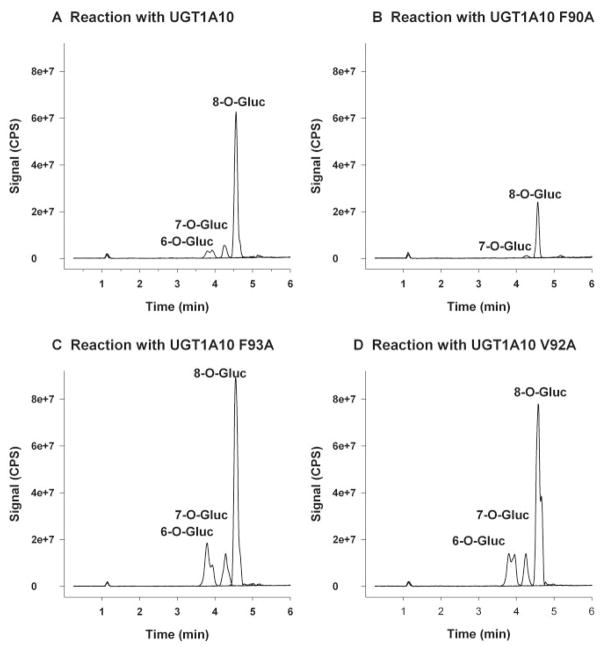
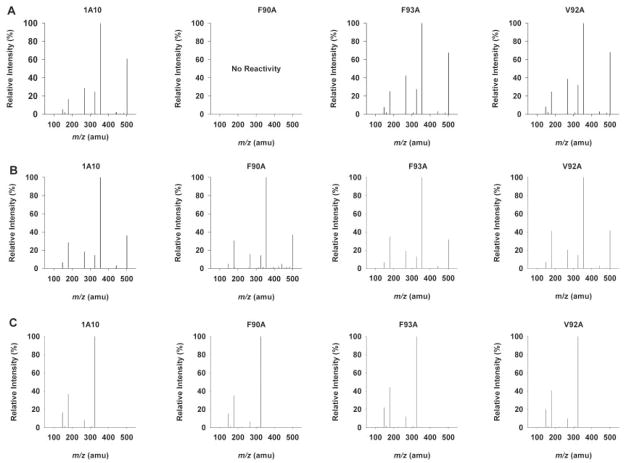
Glucuronidation of racemic hydroxywarfarins via UGT1A10 conjugation is thought to occur at the C6-, C7-, and C8-positions (Zielinska et al., 2008). It is possible that mutation in the active site could give rise to new products. Product mixtures were analyzed by LC-MS/MS in order to elucidate major product structure and to determine relative amounts of products formed. The overlaid MS2 chromatograms (m/z 501) (Fig. 4) show products formed from reactions of 6-hydroxywarfarin, 7-hydroxywarfarin, and 8-hydroxywarfarin with UGT1A10 wild-type recombinant protein (Fig. 4A), UGT1A10 F90A mutant (Fig. 4B), UGT1A10 F93A mutant (Fig. 4C), and UGT1A10 V92A mutant (Fig. 4D). Baculovirus negative controls for each substrate, as well as negative controls with no UGT1A10 or mutant protein, confirm that glucuronidation is seen only in the presence of UGT1A10 or its mutated isoforms (data not shown). No difference in retention times was observed in products produced by the various enzymes and substrates (Fig. 4, A–D), and comparisons of MS2 spectra obtained from each product show no major differences between the wild type and respective mutants (Fig. 5, A–D). As previously reported (Zielinska et al., 2008), the base peak for 8-hydroxywarfarin glucuronide (m/z 321) was different from the base peak formed with the 6- and 7-hydroxywarfarin glucuronides (m/z 355). This observation was consistent with each mutant and indicative that identical products were being formed. MS/MS fragmentation pathways have been proposed for all the products observed in Fig. 5, A–C (Zielinska et al., 2008).
Fig. 4.
Reversed phase-HPLC chromatograph of product ion (m/z 501) experiments. Tracings represent organic-soluble metabolites generated during incubation of UGT1A10 and mutants with UDP-glucuronic acid (4 mM) and 750 μM of each substrate [6-hydroxywarfarin (A), 7-hydroxywarfarin (B), and 8-hydroxywarfarin (C), 10-hydroxywarfarin (D)]. Each substrate was incubated individually for 180 min. All other incubation conditions are noted in Materials and Methods.
Fig. 5.
Product ion (m/z 501) spectra. Each spectrum reflects the organic-soluble metabolites generated during incubation of 750 μM 6-hydroxywarfarin (A), 7-hydroxywafarin (B), or 8-hydroxywarfarin (C) with UGT1A10 or UGT1A10 F90A, F93A, or V92A in the presence of UDP-glucuronic acid (4 mM). Reversed phase-HPLC chromatograph of product ion (m/z 501) experiments. Each substrate was incubated individually for 180 min. All other incubation conditions are noted in Materials and Methods.
Discussion
Glucuronidation of hydroxywarfarin derivatives is important for proper warfarin detoxification and excretion. However, specific UGT enzymatic mechanisms have yet to be characterized which give rise to glucuronidated products. Previous investigations of human hepatic and intestinal UGT glucuronidation of native warfarin and hydroxywarfarin derivatives identified UGT1A10 as a major isoform involved in the glucuronidation of the largest number of hydroxywarfarin compounds (Zielinska et al., 2008). Other studies have also identified phenolic and estrogen binding sites in UGT1A10 through site-directed mutagenesis, photoaffinity labeling, and MS analyses (Xiong et al., 2006; Starlard-Davenport et al., 2007). The current study extends our previous work by showing that the substrate-binding motif, F90-M91-V92-F93, is important for hydroxywarfarin metabolism. Mutation of F90, V92, and F93 with alanine led to substrate-dependent changes in the kinetic mechanism for glucuronidation and the corresponding parameters, Vmax, Km, and Ks (inhibitory constant). These differences indicate that the respective residues and position of the hydroxyl group play different roles in substrate binding and subsequent glucuronidation by UGT1A10.
The binding motif residue F90 appears to be the most significant contributor to glucuronidation of UGT1A10 hydroxywarfarin substrates. Mutation of this residue to alanine abolished activity toward 6-and 7-hydroxywarfarin, which was the same effect observed for other substrates [e.g., p-nitrophenol (Xiong et al., 2006), estrogens (Star-lard-Davenport et al., 2007), and hydroxyestrogens (Starlard-Davenport et al., 2007)]. These results were unexpected given the observed mutant activity toward 4-methylumbelliferone (4MU) and 7-hydroxy-4-methylcoumarin (Xiong et al., 2006). 4MU and 7-hydroxywarfarin share a common site for glucuronidation at the hydroxyl group located at the 7 position of the coumarin ring, yet 4MU is a substrate and 7-hydroxywarfarin is not. The determining factor in activity toward these molecules must then derive from the different location and size of alkyl groups present on the coumarin ring between 4MU and 7-hydroxywarfarin. The absence of phenylalanine must perturb binding contacts and packing of molecules within the active site. By contrast, glucuronidation of 8-hydroxywarfarin was not significantly affected by the mutation, indicating the location of the hydroxyl group determines whether these hydroxywarfarins serve as substrates (8-hydroxywarfarin) or not (6- and 7-hydroxywarfarin).
The magnitude of this subtle difference in substrate structure on UGT1A10 F90A activity may reflect the loss of binding and/or orientation of substrates, which is necessary for glucuronidation of 6- and 7-hydroxywarfarin but not 8-hydroxywarfarin. Based on our inhibition studies, the 6-hydroxywarfarin Ki value for UGT1A10 F90A (300 μM) is slightly less than the wild-type enzyme Km for the substrate (Table 1). In this case, the mutation likely does not impact the overall binding energy for the molecule. By contrast, the mutant Ki value for 7-hydroxy-warfarin is slightly greater than the Km exhibited by wild-type UGT1A10 (Table 2), suggesting a loss in binding and orientation occurs as a result of the mutation. Taken together, F90 seems to play a more important role in generating a catalytically competent complex between substrate and cofactor than binding substrate within the UGT1A10 active site.
The UGT1A10 valine residue at position 92 is not conserved across the UGT1A family and thus may play a role in the distinctive glucuronidation capacity of UGT1A10 toward hydroxywarfarins. UGT1A10 V92A displayed an increased Vmax and Km for hydroxywarfarins relative to wild type. Despite the changes in kinetic parameters, the overall efficiencies of UGT1A10 V92A toward 6- and 7-hydroxywarfarin were similar to wild type, indicating that V92 contributes to formation and turnover of the Michaelis complex equally. This was not the case for 8-hydroxywarfarin (Table 3). Although the binding interactions necessary to form the Michaelis complex (Km) were unchanged, the rate of the glucuronidation reaction (Vmax) was 10-fold higher, indicating that the mutation alters the active site environment and/or orientation of substrate and cofactor to improve coupling. Other significant alterations of the active site were evident at high 6- and 7-hydroxywarfarin concentrations, which led to substrate inhibition kinetics. A similar effect was observed for hydroxywarfarin glucuronidation by UGT1A8 and 1A9 (Zielinska et al., 2008). Like UGT1A10 V92A, these UGTs possess residues with small side chains at position 92, specifically aspartic acid for UGT1A8 and alanine for UGT1A9. For the mutant and wild-type UGTs, the absence of a bulky phenylalanine may create a binding pocket for a second substrate molecule, which, when occupied, compromises turnover of the Michaelis complex. Thus, V92 then controls the volume of the active site and thus influences the stoichiometry of bound substrates and the subsequent organization of substrate and cofactor in the active site.
The conservation of F93 among UGT1A family members (except UGT1A1) may reflect a significant common role for this residue in catalysis. Mutation of this residue to alanine weakened the Michaelis complex as evidenced by higher Km values for 6- and 7-hydroxywarfarin, but also led to a more significant increase in turnover rates (Vmax) for all substrates (Tables 1 and 2). Overall, the decrease in steric bulk of the side chain improved catalysis except at higher 6- and 7-hydroxywarfarin concentrations when substrate inhibition kinetics was observed. Like UGT1A10 V92A, the introduction of an alanine at this position may have created a second inhibitory binding site for those substrates. Nevertheless, the role of F93 in catalysis appears more complex than V92. Previous studies with UGT1A10 F93A revealed decreases in Vmax accompanied by increases and decreases in Km toward p-nitrophenol (Xiong et al., 2006), estrogens (Starlard-Davenport et al., 2007), and hydroxyestrogens (Starlard-Davenport et al., 2007). Although the significance of F93 in catalysis is clear, its specific role in substrate turnover appears dependent on the structure of substrate.
Despite high activity toward many hydroxywarfarins, UGT1A10 was not able to glucuronidate the parent warfarin compound. The addition of the hydroxyl group introduces steric bulk and hydrogen-bonding ability. Because warfarin lacks this group, the inability of UGT1A10 to glucuronidate this molecule may indicate low affinity or binding in a catalytically unproductive orientation as suggested by the variations in UGT1A10 F90A activity. The wild-type UGT1A10 Ki value for warfarin was very high (1500 μM) relative to the Km values for the hydroxywarfarins (Tables 1–3), thus the absence of activity toward this molecule reflects poor binding. In other words, the presence of the hydroxyl group enhances binding for warfarin derivatives and possibly favors orientation of the substrate for glucuronidation.
Inhibition studies with the UGT1A10 mutants indicated that the roles of specific residues within the binding motif were very different. The mutation of F90 to alanine actually increased the affinity for warfarin almost 3-fold, indicating more favorable binding interactions through the decrease in steric bulk at the active site. In contrast, a smaller residue at 92 (originally a valine) did not affect the warfarin binding. Interactions between F93 and warfarin are important, because the loss of the aromatic side chain resulted in the inability of the molecule to bind UGT1A10.
These studies add to our knowledge of the molecular features, which confer substrate specificity for UGT1A10, and the information generated will facilitate predictions as to its pharmaco- and toxicokinetic properties. These insights are ultimately exploitable in novel drug development and assessment of risk associated with exposure to environmental chemicals. Mutation of F90, V92, and F93 to alanine revealed the critical role these residues play in binding hydroxywarfarins and forming the overall active site. Several polymorphic variants of UGT1A10 have been identified (Nagar and Remmel, 2006); however, the effect of these nine single-nucleotide polymorphisms on the functional activity of UGT1A10’s glucuronidation of warfarins remains to be determined. Future studies on polymorphic variants of UGT1A10 would also provide valuable insight regarding the role of structure in the function of UGT1A10 in biological processes.
Acknowledgments
We thank Vi-Huyen Le, Neil Mitchell, and Johanna Mosorin for technical assistance and Joanna Little for expert editing of the manuscript. The authors would also like to thank Bob Kobelski (Centers for Disease Control) for contributions to this study.
This work was funded by National Institutes of Health Grants DK60109 and GM075893 (to A.R.-P.); a Bioterrorism Cooperative Agreement U90/CCU616974-07 (to J.H.M.), an Arkansas Public Health Laboratory Fellowship (to J.H.M.); a Center for Disease Control Contract 200-2007-21729 (to J.H.M.); and the Academy of Finland (Project 210933; to M.F.).
ABBREVIATIONS
- P450
cytochrome P450
- UDP-GlcUA
UDP-glucuronic acid
- UGT
UDP-glucuronosyltransferase
- GlcUA
glucuronic acid
- HPLC
high-performance liquid chromatography
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- 4MU
4-methylumbelliferone
- ESS
enzyme substrate complex containing one enzyme and two substrates
References
- Collom SL, Jamakhandi AP, Tackett AJ, Radominska-Pandya A, Miller GP. CYP2E1 active site residues in substrate recognition sequence 5 identified by photoaffinity labeling and homology modeling. Arch Biochem Biophys. 2007;459:59–69. doi: 10.1016/j.abb.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge DR, Chang HC, Churchwell MI, Holder CL. Analysis of soy isoflavone conjugation in vitro and in human blood using liquid chromatographymass spectrometry. Drug Metab Dispos. 2000;28:298–307. [PubMed] [Google Scholar]
- Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, Rettie AE. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- Jansing RL, Chao ES, Kaminsky LS. Phase II metabolism of warfarin in primary culture of adult rat hepatocytes. Mol Pharmacol. 1992;41:209–215. [PubMed] [Google Scholar]
- Kaminsky LS, Zhang ZY. Human P450 metabolism of warfarin. Pharmacol Ther. 1997;73:67–74. doi: 10.1016/s0163-7258(96)00140-4. [DOI] [PubMed] [Google Scholar]
- Kurkela M, García-Horsman JA, Luukkanen L, Mörsky S, Taskinen J, Baumann M, Kostiainen R, Hirvonen J, Finel M. Expression and characterization of recombinant human UDP-glucuronosyltransferases (UGTs). UGT1A9 is more resistant to detergent inhibition than other UGTs and was purified as an active dimeric enzyme. J Biol Chem. 2003;278:3536–3544. doi: 10.1074/jbc.M206136200. [DOI] [PubMed] [Google Scholar]
- Kuuranne T, Kurkela M, Thevis M, Schänzer W, Finel M, Kostiainen R. Glucuronidation of anabolic androgenic steroids by recombinant human UDP-glucuronosyltransferases. Drug Metab Dispos. 2003;31:1117–1124. doi: 10.1124/dmd.31.9.1117. [DOI] [PubMed] [Google Scholar]
- Kuzmic P. Program DYNAFIT for analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;239:260–276. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- Nagar S, Remmel RP. Uridine diphosphoglucroosyltransferase pharmacogenetics and cancer. Oncogene. 2006;25:1659–1672. doi: 10.1038/sj.onc.1209375. [DOI] [PubMed] [Google Scholar]
- Rettie AE, Korzekwa KR, Kunze KL, Lawrence RF, Eddy AC, Aoyama T, Gelboin HV, Gonzalez FJ, Trager WF. Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: a role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chem Res Toxicol. 1992;5:54–59. doi: 10.1021/tx00025a009. [DOI] [PubMed] [Google Scholar]
- Starlard-Davenport A, Xiong Y, Bratton S, Gallus-Zawada A, Finel M, Radominska-Pandya A. Phenylalanine(90) and phenylalanine(93) are crucial amino acids within the estrogen binding site of the human UDP-glucuronosyltransferase 1A10. Steroids. 2007;72:85–94. doi: 10.1016/j.steroids.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Bernardi D, Bratton S, Ward MD, Battaglia E, Finel M, Drake RR, Radominska-Pandya A. Phenylalanine 90 and 93 are localized within the phenol binding site of human UDP-glucuronosyltransferase 1A10 as determined by photoaffinity labeling, mass spectrometry, and site-directed mutagenesis. Biochemistry. 2006;45:2322–2332. doi: 10.1021/bi0519001. [DOI] [PubMed] [Google Scholar]
- Zielinska A, Lichti CF, Bratton S, Mitchell NC, Gallus-Zawada A, Le VH, Finel M, Miller GP, Radominska-Pandya A, Moran JH. Glucuronidation of monohydroxylated warfarin metabolites by human liver microsomes and human recombinant UDP-glucuronosyltransferases. J Pharmacol Exp Ther. 2008;324:139–148. doi: 10.1124/jpet.107.129858. [DOI] [PMC free article] [PubMed] [Google Scholar]