Abstract
The purpose of this study was to determine the influence of age, sex, and strength training (ST) on large-scale gene expression patterns in vastus lateralis muscle biopsies using high-density cDNA microarrays and quantitative PCR. Muscle samples from sedentary young (20–30 yr) and older (65–75 yr) men and women (5 per group) were obtained before and after a 9-wk unilateral heavy resistance ST program. RNA was hybridized to cDNA filter microarrays representing ~4,000 known human genes and comparisons were made among arrays to determine differential gene expression as a result of age and sex differences, and/or response to ST. Sex had the strongest influence on muscle gene expression, with differential expression (>1.7-fold) observed for ~200 genes between men and women (~75% with higher expression in men). Age contributed to differential expression as well, as ~50 genes were identified as differentially expressed (>1.7-fold) in relation to age, representing structural, metabolic, and regulatory gene classes. Sixty-nine genes were identified as being differentially expressed (>1.7-fold) in all groups in response to ST, and the majority of these were downregulated. Quantitative PCR was employed to validate expression levels for caldesmon, SWI/SNF (BAF60b), and four-and-a-half LIM domains 1. These significant differences suggest that in the analysis of skeletal muscle gene expression issues of sex, age, and habitual physical activity must be addressed, with sex being the most critical variable.
Keywords: aging, exercise, gender, transcription, transcriptome
THE LOSS OF SKELETAL MUSCLE mass and strength with advancing age is associated with frailty, loss of function, and the deterioration of health status in the elderly (13, 33), and these changes may be influenced by sex differences (18, 20, 28, 35). The potential of strength training (ST) to reverse age-associated losses of muscle mass and strength in both men and women is well established (9, 17, 34); however, little is known about why muscle mass is lost with age or how muscle responds to ST, at the molecular level.
Recent evidence has indicated that changes in gene expression with advancing age may contribute to a deterioration in muscle function (11, 16, 19, 21, 25). Similarly, limited evidence supports the importance of differences in gene expression in explaining muscle phenotype differences between men and women (7, 31, 37). Changes in gene expression have been well-characterized for a number of muscle-specific genes in response to acute resistance-type exercise (3). A general limitation to previous gene expression research is the reliance on genes previously characterized in muscle or known a priori to be important for muscle structure and function. For this reason, several investigators have recently begun to use “exploratory” gene expression analyses to identify genes not previously known to be important to muscle function (15, 16, 43–45). For example, Lee et al. (16) reported that of 6,347 genes expressed in mouse soleus muscle, 58 genes displayed a greater than twofold increase in expression in old compared with young animals, whereas 55 genes exhibited greater than a twofold decrease in expression in old compared with young adult animals, although only a fraction of these corresponded with changes in human muscle (44, 45). In the comparison study of human muscle, Welle et al. (44) compared global gene expression patterns among young and older men and found that 89 of 702 genes were differentially expressed in relation to age, many of which were related to metabolic enzymes. Jozsi et al. (15) performed a medium-density (500 genes) cDNA microarray experiment in young and older men at baseline and in response to acute resistance exercise (24 h after exercise) and observed differential expression in several stress-related genes in the older men compared with the young men at baseline and differential responses in gene expression in some of these same genes in response to resistance exercise in the two groups.
To expand the limited work completed to date, the purpose of the present experiments was to identify genes whose expression in skeletal muscle is influenced by age and sex differences, and/or responses to ST using microarray and quantitative PCR (qPCR) techniques. To achieve this goal, we used a previously validated high-density cDNA microarray (22, 36, 38) representing over 4,000 human genes in a design that included 12 total microarray hybridizations providing ~32,000 experimental data points. This report outlines our analysis of these data, including general characteristics of gene expression patterns, as well as specific candidate genes indicated as being important for skeletal muscle function in relation to age, sex, and/or ST.
METHODS
Subjects
The subjects for these experiments were selected from a pool of over 40 sedentary young (20–30 yr) and older (65–75 yr) men and women who have been described previously (17, 34). Five individuals were selected from each group at random for the microarray experiments based on two criteria that were designed to minimize interindividual variability: 1) the subject was white (the most prominent racial group in the entire cohort) and 2) biopsy tissue was available for both time points (before and after ST). Five individuals were pooled from each group to minimize the impact of interindividual variation and improve our ability to identify genes differentially expressed in relation to age, sex, and/or ST (2). For the qPCR validation studies, samples were obtained from different subjects from the larger cohort to provide an additional level of validation in relation to the microarray data. Four subjects each for young men and women, and older women, and three older men were available for the validation experiments. All subjects were nonsmokers, free of significant cardiovascular, metabolic, or musculoskeletal disorders. Individuals enrolled in the study had not participated in a regular exercise program for at least 6 mo before their recruitment. After all methods and procedures were explained, the subjects read and signed a written consent form for the project that had been approved by the Institutional Review Boards at the Baltimore Veterans Affairs Medical Center and the University of Maryland, College Park. All subjects were reminded throughout the study not to alter their regular activity levels or dietary habits for the duration of the investigation.
ST program
The details of the study design and methods, as well as additional phenotype data (strength, mass, etc.) have been described previously (14, 17). Here, we briefly describe the ST intervention performed by the subjects (32). The intervention consisted of unilateral ST of the dominant leg's knee extensors with the nondominant leg serving as a control. The training program consisted of four sets of high-volume, heavy-resistance knee extension exercise performed 3 days per week. Subjects completed a set of five repetitions of knee extension at the 5RM resistance (following a warm-up). The resistance of subsequent sets was initially set at the 5RM, with the resistance incrementally decreased by the subject in a way that elicited near maximal effort on every repetition to perform a total of 10, 15, and 20 repetitions per set, respectively. Specified rest periods were allowed between sets. An exercise specialist directly supervised all sessions to verify compliance. Progressive increases in resistance occurred throughout the 9-wk ST program.
Biopsies
Biopsies were obtained from the vastus lateralis muscle of the trained leg before ST (2 wk prior to ST familiarization and baseline testing) and after ST (48–72 h following last training session) for each subject as outlined previously (32). Tissue samples were immediately dissected of fat, blood, and connective tissue, enclosed in cryovials, snap-frozen in liquid nitrogen, and stored at −80°C until analysis.
Microarray molecular biology
Total RNA was extracted using the SV RNA Isolation Kit (Promega) according to manufacturer's instructions (which included DNase I treatment) and quantitated by determining absorbance at 260 nm in triplicate, with the values averaged. For each microarray experiment, a total of 1 μg of total RNA was used for each hybridization, thus 200 ng of total RNA was taken from each sample and pooled for each group. Arrays were hybridized according to the manufacturer's instructions, once for each experimental condition (baseline, ST) within a single group. Thus four total microarrays, one for each of the four groups, were hybridized twice each (baseline and after ST). Total RNA was hybridized to GF211 Human Named Genes Gene-Filters (Release I; ResGen, Invitrogen) according to the manufacturer's instructions. Briefly, 1 μg of total pooled RNA was reverse transcribed using 12- to 18-mer oligo dT primers and SuperScript II (GIBCO-BRL). RNA was labeled with an [α-33P]dCTP probe during reverse transcription. Labeled cDNA was hybridized to the array for 18 h, washed, and exposed to a phosphor storage screen (Molecular Dynamics) for 92 h. The phosphor image was obtained using a Storm 860 PhosphorImaging system (Molecular Dynamics) at a 50 μm resolution. Image files were then imported into Pathways v3.0 microarray software (ResGen, Invitrogen) for analysis. Following image acquisition, arrays were stripped according to the manufacturer's instructions. Stripping efficiency was determined by assessing radioactive emissions, as well as by imaging stripped arrays with the PhosphorImaging system. In all cases, the baseline sample was hybridized first, followed by the after-ST sample. In addition to the experimental samples, these same microarrays were hybridized (final hybridization) as outlined above with cDNA generated from a commercially available skeletal muscle total RNA source (Ambion). Thus all four arrays were hybridized with an identical “control” sample, allowing for an analysis of inter-array variation (see RESULTS).
Walker and Rigley (38) and others (22, 36) have performed validation studies of these same microarrays (GF211) using quantitative PCR and Northern blot techniques for several differentially expressed genes. Based on this work, Walker and Rigley (38) recommended criteria of greater than 10-fold above background with at least 1.5-fold differential expression. In the present study, we relied on the more stringent criterion of >1.7-fold for defining differential expression to further decrease the chance of false-positive detection.
Quantitative PCR
qPCR gene expression studies were performed for caldesmon, SWI/SNF (BAF60b), and four-and-a-half LIM domains 1 (FHL1) as validation of the microarray results. Different subjects from those studied in the microarray experiments were selected for the qPCR experiments to provide an independent sample for the validation studies (see Subjects, above). These subjects (within their respective groups) did not differ significantly with regard to age, physical characteristics, or ST response from the subjects analyzed for the microarray experiments.
Total RNA was extracted from the before- and after-ST muscle samples using a standard phenol-based extraction method (Ambion), quantified by determining absorbance at 260 nm in triplicate, with the values averaged. Total RNA was treated with DNase I (Ambion) and reverse transcribed using the Reverse Transcription Reagents kit (Applied Biosystems) according to the manufacturer's instructions using random hexamers, with ~350 ng of total RNA reverse transcribed in a 20-μl reaction. qPCR was performed using the ABI 7700 DNA Sequence Detection System (TaqMan; Applied Biosystems) using standard fluorescent chemistries and thermal cycling conditions. Primer and probe sequences were designed for each experimental gene's mRNA sequence using Primer Express software (Applied Biosystems) as shown in Table 1. 18S rRNA was used as an internal expression control and was amplified using the Ribosomal RNA Control Reagents kit (Applied Biosystems). Primer and probe concentrations were optimized using the TaqMan Universal PCR Master Mix (Applied Biosystems). For each reaction, 14 ng of cDNA was added to optimized primer and probe concentrations, with 25 μl of PCR Master Mix. A corresponding well contained 7 ng of cDNA with reaction reagents for the qPCR of 18S rRNA. All reactions were performed in duplicate. Thermal cycling conditions were as specified by the manufacturer: 50°C for 2 min, 95°C for 10 min, and 40 cycles as follows: 95°C for 15 s, ramp to 60°C for 1 min. Known concentration standards were developed using cDNA (produced as outlined above) from a commercially available skeletal muscle total RNA source (Ambion). Standard curves were generated from five concentrations of total RNA (2, 4, 8, 16, and 32 ng) performed in duplicate for each experimental gene and 18S rRNA. Optimization reactions were performed to ensure that all experimental samples fell within the range of the standard curves for each gene.
Table 1.
Primer and probe sequences used for the qPCR validation experiments
| Gene | Forward Primer | Reverse Primer | Probe |
|---|---|---|---|
| Caldesmon | CACTAAGGTTTGAGACAGTTCCAGAA | GCGAATTAGCCCTCTACAACTGA | AACCCAAGCTCAAGACGCAGGACG |
| BAF60b | GGCACGAGCGGGAGTACA | CGGAGTCGGCCACAACTG | AACTGCAACCGTTACTTCCGCCAGATCT |
| FHL1 | CCAGTGTGGTGGCCTATGAAG | ACAGTCGGGACAATACACTTGCT | CAATCCTGGCACGACTACTGCTTCCAC |
All sequences are listed 5′ to 3′ and the probes were labeled in the standard manner with a 5′ 6-FAM reporter dye and a 3′ TAMRA quencher dye. FHL1; four-and-a-half LIM domains 1; BAF60b, SWI/SNF; qPCR, quantitative PCR; 6-FAM, 6-carboxyfluorescein; TAMRA, 6-carboxy-N,N,N′,N′-tetramethylrhodamine.
Relative quantitation of gene expression was performed according to the manufacturer's instructions. Briefly, the threshold cycle was determined for both the experimental gene (FAM reporter) and 18S rRNA (VIC reporter). Standard curves were then generated for each, and linear regression was used to determine the R-squared value and standard curve equation (all R2 > 0.993). The CT value for each sample was then used to calculate relative expression based on the respective standard curve equation, and the expression level of each experimental gene was normalized to the 18S rRNA control gene. Normalized expression values (with duplicate assays averaged) were used in all analyses. Although the standard curve estimates are based on nanogram values (from the standards), we report all data as arbitrary units (“U”), to reflect relative rather than absolute expression levels.
Data analysis
The GF211 microarray contains probe spots for over 4,000 known human genes. After each array image was imported into the Pathways v3.0 software, comparison images were normalized using the data point method according to the manufacturer's instructions. The data point method normalizes the data for each particular probe to the background intensity of the entire array, thereby controlling for hybridization variability. Following normalization, microarray comparisons were performed between age, sex, and ST conditions. In general, we performed two microarray comparisons (e.g., A vs. B and C vs. D) and then identified those genes differentially expressed in both comparisons to more specifically target genes related to a particular condition (e.g., age, sex, or ST). To decrease the prevalence of false-positive results, only genes expressed at 10-fold above background with a minimum of 1.7-fold differential expression between arrays were considered truly differentially expressed. To further decrease the chance of false-positive identification, comparisons were performed among the control microarrays to determine “differentially expressed” genes, which are not predicted. Any genes identified as differentially expressed on the control arrays were excluded in the experimental analysis. For brevity, some tables include only those genes with >2.0-fold differential expression. All Supplementary Tables1 (labeled Tables S1–S5) are available online at the Physiological Genomics web site and at http://www.inform.umd.edu/knes/research/genomics/data.
For the analysis of the qPCR results, averaged normalized data for each experimental gene was compared between and among groups using either independent or paired-samples t-tests, or one-way or repeated-measures ANOVA with LSD post hoc. Data are presented as means with standard deviation (SD) or means ± standard error (SE) as indicated, with P < 0.05 accepted as statistically significant.
RESULTS
General characteristics
Subject characteristics are shown in Table 2. The physical characteristics, including responses to ST, for the entire cohort from which these individuals were selected are described in detail elsewhere (14, 17). Physical characteristics and responses to ST did not differ significantly among the subjects used for the microarray vs. qPCR experiments (data not shown). Across all microarray analyses, ~1,000 of the probe spots on any particular array demonstrated an intensity >10-fold above background, or roughly one-fourth of the specific genes represented on the GF211 array.
Table 2.
Physical characteristics of the subjects used in the microarray experiments
| Older Men | Older Women | Young Men | Young Women | |
|---|---|---|---|---|
| n | 5 | 5 | 5 | 5 |
| Age, yr | 69.8 (1.6) | 66.0 (1.0) | 25.2 (2.6) | 25.8 (0.5) |
| Ht, cm | 170 (6) | 158 (3) | 182 (8) | 166 (5) |
| Wt, kg | 81.3 (11.6) | 72.6 (7.1) | 95.0 (18.2) | 68.0 (6.7) |
| Body fat, % | 29.7 (6.3) | 40.7 (6.5) | 24.8 (9.6) | 29.7 (4.6) |
Data are means, with SD in parentheses.
Control microarrays
All four arrays were hybridized with the same “control” RNA and compared to address interarray variability. Comparisons among the four arrays revealed strong correlations for all pairwise comparisons, with all correlations significant (all Pearson r = 0.962–0.979; all P < 0.001). Thirteen genes that were shown as differentially expressed at >2.0 fold in any comparison or at >1.7 fold in more than one comparison among all four arrays (six total comparisons) were rejected in all subsequent analyses of the experimental data to reduce false-positive identification.
Influence of sex
To identify genes differentially expressed in relation to sex, we first compared men vs. women both before and after ST to identify genes differentially expressed regardless of the influence of ST. A total of 210 genes were identified, with 175 of these with higher expression in men (see Supplementary Table S1 for all genes differentially expressed at >2.0-fold levels). We then determined sex differences regardless of the influence of age by comparing before-ST samples both between older men and women and between young men and women. That analysis revealed 179 differentially expressed genes (>1.7-fold), with 136 genes expressed higher in men (Supplementary Table S2). Of the 179 genes shown in Table S2 (sex differences regardless of age), only 28 of the genes (16%) are also represented in Table S1 (sex differences regardless of ST), with all but 5 of these being more highly expressed in men. Table 3 shows the majority of those 28 genes that were identified in both comparisons (i.e., present in both Supplementary Tables S1 and S2).
Table 3.
Genes differentially expressed between men and women regardless of age or ST (i.e., only those genes present in both Table S1 and S2)
| Older Men Before ST | Older Women Before ST | Young Men Before ST | Young Women Before ST | Accession No. | Gene |
|---|---|---|---|---|---|
| Higher expression in men regardless of age or ST | |||||
| 3.069 | 0.615 | 3.206 | 0.835 | W86860 | Nuclear VCP-like |
| 2.985 | 0.634 | 2.260 | 0.622 | R48320 | EphB1 |
| 2.091 | 0.462 | 2.404 | 0.418 | AA449361 | Ring finger protein 13 |
| 2.432 | 0.639 | 2.256 | 0.526 | N25204 | Leukemia associated gene 2 |
| 2.624 | 0.732 | 2.476 | 0.750 | N94713 | Intersectin 2 |
| 2.358 | 0.694 | 2.338 | 0.614 | AA496800 | Human transposon-like element mRNA |
| 1.784 | 0.559 | 2.110 | 0.493 | H15431 | Polymerase (RNA) II polypeptide D |
| 2.260 | 0.725 | 2.578 | 0.595 | H78385 | Huntingtin-interacting protein 2 |
| 2.480 | 0.801 | 1.660 | 0.621 | AA459401 | Kallikrein 10 |
| 1.895 | 0.677 | 1.703 | 0.563 | AA293192 | Putative nucleic acid binding protein RY-1 |
| 1.708 | 0.628 | 1.741 | 0.629 | N62245 | CDC7 (cell division cycle 7)-like 1 |
| 1.735 | 0.639 | 1.553 | 0.546 | AA088258 | Human protein immuno-reactive with anti-PTH polyclonal antibodies |
| 1.515 | 0.598 | 2.052 | 0.535 | AA460251 | NADH-ubiquinone, oxidoreductase 1, subcomplex 1 |
| 1.727 | 0.685 | 2.356 | 0.473 | AA046430 | Lung type-1 membrane-associated glycoprotein |
| 3.232 | 1.326 | 3.167 | 1.142 | AA418564 | EST with similarity to human cadherin 12 |
| 1.875 | 0.787 | 1.877 | 0.519 | N94385 | Cartilage oligomeric matrix protein |
| 3.185 | 1.361 | 3.699 | 1.194 | AA150301 | Adrenal gland protein AD-004 |
| 3.508 | 1.622 | 3.336 | 1.158 | AA450062 | Prostate differentiation factor |
| 3.053 | 1.432 | 2.888 | 1.135 | AA262504 | Eyes absent (Drosophila) homolog 3 |
| 1.273 | 0.622 | 1.302 | 0.471 | AA428196 | POU domain, class 4, transcription factor 1 |
| Higher expression in women regardless of age or ST | |||||
| 9.387 | 17.844 | 6.171 | 16.058 | AA455272 | ITBA1 gene |
| 0.486 | 1.891 | 0.763 | 1.906 | T94169 | Mitogen-activated protein kinase 8 |
| 3.293 | 13.784 | 2.587 | 14.735 | AA076063 | Caldesmon 1* |
| 0.501 | 2.163 | 0.522 | 1.143 | AA455925 | Four and a half LIM domains 1* |
| 0.716 | 3.589 | 1.348 | 3.063 | AA478436 | SWI/SNF complex (BAF60b)* |
Intensity values are shown within age comparisons of the before-ST condition (ST, strength training), along with the accession number and gene title or description. Intensity values represent the extent of radioactivity for each particular probe spot on the microarray, which is directly related to the extent of hybridization for that spot. All genes with >1.7-fold higher expression in women identified in both comparisons (across age and before and after ST) are shown, whereas only genes with >2.0-fold higher expression in men are shown for brevity. Complete lists for each comparison are available in Supplementary Tables S1 and S2 (available online at the Physiological Genomics web site).
Gene targeted for qPCR validation.
Influence of age
To assess age-related differences in gene expression, we identified those genes differentially expressed between young and older groups (sexes combined) separately at baseline and after ST, thus targeting genes with altered expression in the context of age regardless of the influence of ST. Those genes with >2.0-fold differential expression (n = 19) are shown in Table 4, with the entire list of genes (n = 54; >1.7-fold) shown in Supplementary Table S3.
Table 4.
Genes differentially expressed (>2-fold) between young and older groups both before ST and after ST
| Young Baseline | Older Baseline | Young After ST | Older After ST | Accession No. | Gene |
|---|---|---|---|---|---|
| 0.095 | 0.045 | 0.089 | 0.050 | AA670422 | ADP-ribosylation factor 3 |
| 0.176 | 0.456 | 0.206 | 0.368 | AA664077 | ATPase, vacuolar, 14 kDa |
| 0.127 | 0.063 | 0.119 | 0.060 | AA234671 | ATP-binding cassette, subfamily D, member 3 |
| 0.528 | 1.243 | 0.208 | 0.411 | AA481780 | Carbonic anhydrase III, muscle specific |
| 0.128 | 0.049 | 0.111 | 0.057 | W72697 | Cisplatin resistance associated |
| 0.424 | 0.732 | 0.353 | 0.734 | AA434144 | Claudin 3 |
| 0.269 | 0.125 | 0.506 | 0.154 | AA464748 | Collagen, type VI, alpha 2 |
| 0.069 | 0.145 | 0.088 | 0.164 | AA463452 | DiGeorge syndrome critical region gene DGSI |
| 0.514 | 0.242 | 0.432 | 0.192 | R60317 | Dihydrolipoamide dehydrogenase |
| 0.089 | 0.041 | 0.110 | 0.042 | H38839 | EST |
| 0.231 | 0.115 | 0.118 | 0.051 | N62761 | Fragile X mental retardation |
| 1.289 | 2.421 | 0.388 | 0.742 | H16958 | Glyceraldehyde-3-phosphate dehydrogenase* |
| 0.085 | 0.043 | 0.126 | 0.041 | H49592 | G protein, alpha activating activity polypeptide |
| 0.342 | 0.161 | 0.310 | 0.141 | N55480 | HMT1 (hnRNP methyltransferase)-like 2 |
| 0.175 | 0.322 | 0.215 | 0.388 | AA496360 | Protein kinase C, delta |
| 0.161 | 0.378 | 0.236 | 0.491 | R51209 | Protein phosphatase 2A, regulatory subunit B′ |
| 0.351 | 0.139 | 0.251 | 0.123 | N90273 | Ras homolog gene family, member H |
| 0.070 | 0.033 | 0.100 | 0.033 | H50344 | Tight junction protein 1 (zona occludens 1) |
| 0.251 | 0.100 | 0.223 | 0.070 | AA443634 | Ubiquitin-conjugating enzyme E2G 2 |
These genes exhibited different expression levels between young and older groups regardless of the influence of ST. Intensity values are shown for each of the groups, along with the accession number and gene title or description. Intensity values represent the extent of radioactivity for each particular probe spot on the microarray, which is directly related to the extent of hybridization for that spot. Only those genes with >2-fold differential expression are shown here, with the complete list of differentially expressed genes shown in Supplementary Table S3.
Two probe spots on the array for this gene were both identified in this analysis, with the averaged intensity values for both spots shown here.
We also identified those genes differentially expressed between young and older groups regardless of sex by comparing baseline samples between young and older men, and between young and older women, and then identifying those genes with differential expression in both comparisons. Table 5 outlines all of the genes (n = 8) with lower expression in older subjects regardless of sex (>1.7-fold) and all genes with >2.0-fold higher expression (n = 6) in the older subjects. The complete list of genes (n = 35) in this analysis is presented in Supplementary Table S4.
Table 5.
Genes differentially expressed (>1.7-fold) between young and older groups in both sexes
| Young Men | Older Men | Young Women | Older Women | Accession No. | Gene |
|---|---|---|---|---|---|
| Lower expression in older groups (all >1.7-fold) regardless of sex | |||||
| 0.143 | 0.068 | 0.150 | 0.076 | T62060 | Antithrombin III |
| 0.552 | 0.245 | 1.566 | 0.311 | R42609 | Capping protein (actin filament), muscle Z-line, alpha 2 |
| 0.329 | 0.185 | 0.614 | 0.230 | H80712 | Caspase 10, apoptosis-related cysteine protease |
| 0.301 | 0.177 | 0.198 | 0.086 | AA598974 | Cell division cycle 2, G1 to S and G2 to M |
| 0.131 | 0.071 | 0.270 | 0.139 | R54424 | Glutamate dehydrogenase I |
| 0.209 | 0.120 | 0.170 | 0.064 | AA877845 | LIM domain kinase 2 |
| 0.516 | 0.297 | 0.565 | 0.260 | AA451716 | Nuclear factor kappa B, subunit 1 (p105) |
| 0.881 | 0.459 | 0.487 | 0.164 | AA490538 | Zinc finger protein homologous to Zfp161 in mouse |
| Higher expression in older groups (all >2.0-fold) regardless of sex | |||||
| 0.113 | 0.238 | 0.234 | 0.674 | R88267 | Glutamate receptor, ionotropic, N-methyl-D-aspartate 1 |
| 0.240 | 0.591 | 0.215 | 0.596 | H02243 | Homeo box B5 |
| 0.048 | 0.110 | 0.078 | 0.183 | AA459308 | Human elastin gene |
| 0.322 | 0.701 | 0.230 | 0.605 | AA477400 | Tropomyosin 2 (beta) |
| 0.217 | 0.570 | 0.220 | 0.847 | H37774 | Tuberous sclerosis 2 |
| 0.092 | 0.252 | 0.110 | 0.373 | N64628 | Ubiquitin-like 4 |
These genes exhibited different expression levels between young and older groups regardless of the influence of sex. Intensity values are shown for each of the groups, along with the accession number and gene title or description. Intensity values represent the extent of radioactivity for each particular probe spot on the microarray, which is directly related to the extent of hybridization for that spot. All genes with >1.7-fold higher expression in the young group for both sexes are shown, whereas only genes with >2.0-fold higher expression in the older groups are shown for brevity. All genes >1.7-fold are shown in Supplementary Table S4.
Influence of ST
To target specifically those genes influenced by ST, we identified differentially expressed genes between before-ST and after-ST samples in both young and older subjects (men and women combined), as well as those genes differentially expressed in response to ST in both men and women (across age groups). Thus genes with altered expression in response to ST regardless of both age and sex were identified. As shown in Table 6, 14 genes were identified as differentially expressed in response to ST in both the age and sex comparisons (i.e., differentially expressed regardless of either age or sex). Nine and two genes were identified specifically in the context of age or sex, respectively. Finally, five genes (bold in Table 6) were identified with interaction effects, such that expression was significantly altered in response to ST, but the direction of change (increase or decrease) differed depending on the specific age or sex group. Our intent with this analysis was to maximize our ability to identify genes important in the response to ST, thereby minimizing the identification of false positives and narrowing the list of important candidates. All genes identified in both the age and sex comparisons are shown in Table 6. An analysis of differential expression for all groups (data combined) before (baseline) vs. after-ST revealed 69 differentially expressed genes, with 46 of those genes exhibiting decreased expression in the after-ST condition compared with baseline (Supplementary Table S5).
Table 6.
Genes differentially expressed (>1.7-fold) between before-ST and after-ST samples in both a comparison of men and women (age groups combined) and a comparison of young and older groups (men and women combined)
| Men Pre | Men Post | Women Pre | Women Post | Accession No. | Gene | Young Pre | Young Post | Older Pre | Older Post |
|---|---|---|---|---|---|---|---|---|---|
| Genes identified in both the age and sex comparisons | |||||||||
| 0.647 | 0.233 | 0.736 | 0.328 | AA634006 | Actin, alpha 2 | 0.510 | 0.226 | 0.874 | 0.335 |
| 0.269 | 0.133 | 0.238 | 0.127 | R40850 | Actin-related protein 1, centractin alpha | 0.231 | 0.133 | 0.276 | 0.127 |
| 0.180 | 0.104 | 0.288 | 0.120 | AA708298 | ATP synthase, mitochondrial F1 complex, beta polypeptide | 0.236 | 0.117 | 0.233 | 0.120 |
| 0.585 | 0.214 | 1.388 | 0.308 | AA455300 | Cold shock domain protein A | 0.723 | 0.215 | 1.250 | 0.308 |
| 0.541 | 0.274 | 0.703 | 0.324 | R44290 | Dynactin | 0.535 | 0.249 | 0.804 | 0.349 |
| 0.213 | 0.104 | 0.207 | 0.099 | R43973 | Eukaryotic translation elongation factor 1 gamma | 0.169 | 0.094 | 0.251 | 0.109 |
| 0.511 | 0.216 | 1.653 | 0.320 | AA455925 | Four-and-a-half LIM domains 1* | 0.832 | 0.169 | 1.332 | 0.367 |
| 1.627 | 0.534 | 1.969 | 0.535 | H16958 | Glyceraldehyde-3-phosphate dehydrogenase | 1.294 | 0.372 | 2.302 | 0.697 |
| 1.186 | 0.438 | 2.106 | 0.425 | N78927 | Myosin, light polypeptide 2 | 1.325 | 0.341 | 1.967 | 0.522 |
| 0.178 | 0.096 | 0.282 | 0.107 | AA192166 | Myosin, light polypeptide 3 | 0.220 | 0.118 | 0.241 | 0.086 |
| 0.133 | 0.309 | 0.101 | 0.190 | AA464601 | Tetraspan 5 | 0.109 | 0.190 | 0.125 | 0.309 |
| 0.051 | 0.089 | 0.036 | 0.074 | AA456295 | TNF receptor-associated factor 6 | 0.047 | 0.089 | 0.041 | 0.074 |
| 0.684 | 0.388 | 0.780 | 0.413 | R60160 | Topoisomerase (DNA) I | 0.900 | 0.479 | 0.565 | 0.322 |
| 0.395 | 0.210 | 0.446 | 0.218 | AA182848 | Troponin I | 0.404 | 0.207 | 0.437 | 0.220 |
| Genes only identified in within-sex comparison | |||||||||
| 0.112 | 0.065 | 0.159 | 0.092 | H85355 | ATPase, Ca2+ transporting, cardiac muscle, slow twitch 2 | ||||
| 0.242 | 0.527 | 0.099 | 0.216 | N92901 | Fatty acid binding protein 4, adipocyte | ||||
| Genes only identified in within-age comparison | |||||||||
| AA126265 | Calnexin | 0.241 | 0.124 | 0.205 | 0.112 | ||||
| AA481780 | Carbonic anhydrase III, muscle specific | 0.528 | 0.208 | 1.243 | 0.411 | ||||
| H61188 | EST | 0.044 | 0.093 | 0.036 | 0.062 | ||||
| T61948 | FBJ murine osteosarcoma viral oncogene homolog B | 0.322 | 0.141 | 0.528 | 0.270 | ||||
| AA112660 | Forkhead box F1 | 0.277 | 0.162 | 0.353 | 0.125 | ||||
| N62761 | Fragile X mental retardation, autosomal homolog 1 | 0.231 | 0.118 | 0.115 | 0.051 | ||||
| AA196393 | Human alkali myosin light chain 3 | 1.088 | 0.604 | 1.986 | 0.793 | ||||
| AA496359 | Immediate early protein | 0.106 | 0.216 | 0.084 | 0.148 | ||||
| W58092 | Tropomyosin 1 (alpha) | 0.466 | 0.261 | 0.928 | 0.269 | ||||
| Genes with age or sex interactions in response to ST | |||||||||
| 0.206 | 0.111 | 0.093 | 0.168 | AA782337 | Ankyrin 2, neuronal | ||||
| 0.135 | 0.072 | 0.058 | 0.105 | AA053674 | Mitogen-activated protein kinase kinase kinase 12 | ||||
| 0.310 | 0.162 | 0.209 | 0.404 | AA485347 | Protein phosphatase 1, regulatory (inhibitor) subunit 11 | ||||
| 0.363 | 0.121 | 0.127 | 0.252 | N90273 | Ras homolog gene family, member H | ||||
| H05919 | Eukaryotic translation initiation factor 4A, isoform 2 | 0.348 | 0.186 | 0.209 | 0.364 | ||||
Those genes identified in only one of the comparisons are shown separately. Intensity values are shown for each of the comparisons, along with the accession number and gene title or description. Intensity values represent the extent of radioactivity for each particular probe spot on the microarray, which is directly related to the extent of hybridization for that spot. Genes demonstrating an interaction effect (changes in response to ST that differ in direction among groups) are highlighted in bold at the bottom. Complete results for all groups combined are shown in Supplementary Table S5.
Gene targeted for qPCR validation. Pre, before-ST samples; Post, after-ST samples.
Quantitative PCR
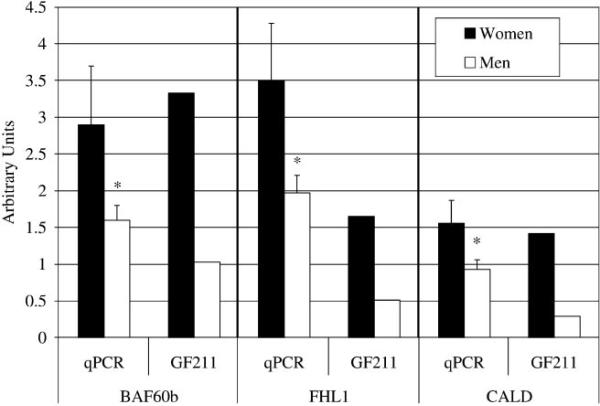
qPCR was performed for caldesmon, SWI/SNF (BAF60b), and FHL1 (all identified as differentially expressed in the microarray experiments) to provide validation of the microarray results. The GF211 has been similarly validated by others (22, 36, 38). We selected these genes based on both the microarray results and their importance to skeletal muscle as indicated by available background literature (see DISCUSSION). For caldesmon, 68% higher expression was observed in women than men, consistent with the microarray findings (P < 0.05; Fig. 1). Similarly for SWI/SNF, women exhibited 86% higher expression than men (P = 0.06; Fig. 1), validating the microarray results (Tables 3, S1, and S2). Finally, for FHL1, the qPCR results revealed significantly higher FHL1 expression in women than men (P < 0.05; Fig. 1), and a decrease in expression in response to ST was also observed (2.82 ± 0.74 vs. 1.89 ± 0.17 U; P = 0.06; see Table 6).
Fig. 1.
Microarray (GF211) and quantitative PCR (qPCR) expression data are presented for three genes used in the microarray validation experiments. BAF60b, SWI/SNF; FHL1, four-and-a-half LIM domains 1; CALD, caldesmon; ST, strength training. For the microarray results, data are intensity values averaged for men and women baseline (before-ST) samples as used in Tables 3–6. For the qPCR results, data are means with SE for relative expression levels (arbitrary units). Caldesmon data are shown at 1/10 actual values for the microarray intensity data. For the microarray data, women demonstrated greater than twofold higher expression than men for all three genes. *Significantly lower expression in men compared with women for the qPCR data (P ≤ 0.06; see RESULTS).
DISCUSSION
The purpose of this study was to identify candidate genes related to skeletal muscle function within the contexts of age, sex, and ST using microarray and qPCR gene expression techniques. The results indicate several important and novel findings. Perhaps most remarkable were the substantial differences in gene expression between men and women, independent of the influences of age or ST, with the majority of these identified genes (~75%) expressed at higher levels in men. The biological basis for these differences is unclear. Sex differences in muscle gene expression patterns were more extensive than age-related differences in gene expression, although a number of genes were identified as differentially expressed between young and older individuals independent of sex or ST. Despite the substantial changes in muscle phenotype in response to ST, only a moderate number of genes (n = 69) were demonstrated to be differentially expressed in response to ST, likely reflecting the diminished influence of altered transcription at the time of the after-ST biopsy rather than an indication of little influence of ST on transcription per se (29). In general, these results provide an initial attempt to define the “transcriptome” of skeletal muscle in the contexts of age, sex, and ST, and provide evidence of the importance of considering sex in genomic studies of gene expression in general, and for skeletal muscle in particular.
We performed the microarray experiments in the present study using pooled samples of five individuals per group. Our intent with this strategy was to define general patterns of gene expression for each group by diminishing the influence of interindividual variation in expression, which has been noted previously (2, 15). This strategy has been used successfully by other groups (15, 44) and has recently been recommended for the analysis of skeletal muscle gene expression using microarrays (2). Specifically, Bakay et al. (2) reported that high interindividual variability is common in skeletal muscle array experiments, which can obscure general patterns of expression. The authors further showed that pooling of patient samples was an effective way to normalize much of the intersubject “noise” while still identifying the large majority of differentially expressed genes. Bakay et al. (2) concluded that stringent yet robust data can be generated by pooling individuals (n = 5), with the caveat that follow-up experiments will be required to define interindividual variability.
The present results represent the first large-scale skeletal muscle gene expression study to address sex-related differences and indicate greater differential expression between the sexes than any other comparison, with ~200 genes differentially expressed between men and women regardless of either age or ST. Within these identified genes, which represent several functional classes, no specific patterns of genes emerged as being consistently differentially expressed between men and women, and only a fraction of the genes in either Supplementary Table S1 or S2 has been studied previously in the context of skeletal muscle. The basis for these extensive sex-related differences is unclear. Whether common transcription factor binding sites (e.g., estrogen and androgen response elements) within these genes might influence the sex-related differences in gene expression, as a result of hormonal differences between the sexes, is uncertain, although a remarkable 75% of the identified genes were more highly expressed in men than women. Another possibility is that these differences are related to the significant differences in relative body fat proportions between men and women, although we expect that hormonal differences are the predominant factor.
Previous work characterizing large-scale patterns of gene expression in human skeletal muscle is limited. In a smaller-scale gene expression study, Welle et al. (41) reported no age-related differences in muscle mRNA content of several muscle-specific contractile protein genes. Comparison with the present study is difficult, since α-actin is the only gene reported by Welle et al. (41) that is also represented on the GF211 array used here. No age-related differences in α-actin were observed in the present study, which supports their conclusion (41). More recently, Welle et al. (43, 44) used serial analysis of gene expression (SAGE) to generate transcript libraries of skeletal muscle from young and older men, the results of which indicated that ~12% of genes were differentially expressed as a result of age. In the present study, only ~5% of the ~1,000 genes expressed above background were differentially expressed in relation to age; however, our results were distilled from a conservative multiple comparison process. Of the genes identified by Welle et al. (44), seven were available for direct comparison on the GF211 microarray used in the present study (GAPDH, glycogen phosphorylase, triosephosphate isomerase, pyruvate kinase, glycogen synthase, phosphoglycerate kinase, and cytochrome c oxidase). Of those seven genes, all but one (phosphoglycerate kinase) similarly demonstrated higher expression in the young compared with older males in the present study, although similar differences were not observed in women, thus explaining their exclusion from the final lists of genes (Tables 4 and 5). This highlights the possibility that the molecular mechanisms of muscle aging may differ between men and women, consistent with the finding of broad sex-related differences in the present study. Finally, Jozsi et al. (15) performed a medium-density microarray investigation of skeletal muscle gene expression in young and older men both at baseline and 24 h following an acute bout of resistance exercise. They reported elevated expression of stress-related genes (e.g., HSP27 and XRCC1) in the older men at baseline, with increased expression of several stress-related genes (e.g., HSP27, MAP kinase kinase 3, and VEGF) in response to exercise, with differential expression indicated for some genes between young and older men. Of the genes identified in Jozsi et al. (15), five were available for comparison with the present study (XRCC1, IL-1β, RANTES, VEGF, and EGR-1). In relation to aging, only XRCC1 could be compared with the present study, and we observed higher XRCC1 levels in young compared with older men, consistent with Jozsi et al. (15), but this relationship was not found in women. Comparisons between acute exercise (24 h post) and chronic exercise training (48–72 h post; present study) require caution, as activity-induced transcription is likely transient (29). In that context, we did not observe changes in VEGF, XRCC1, RANTES, or EGR-1 levels in response to ST. We did observe small increases in IL-1β with ST in young men and women, but not in older men and women, which is consistent with the acute exercise response reported by Jozsi et al. (15). In summary, our data compare favorably to existing reports for comparable genes and extend the existing literature by providing the first investigation to examine skeletal muscle gene expression in relation to sex and chronic ST.
Several genes that might be hypothesized to be differentially expressed as a result of age (11, 16, 19, 21, 25) could not be verified in the present study because they were not represented on the GF211 microarray (e.g., muscle transcription factors). Few of the genes shown in Supplementary Tables S3 and S4, genes differentially expressed in relation to age, are immediately recognizable as previously studied muscle-related genes, with GAPDH, carbonic anhydrase III, and tropomyosin-β being the most studied in skeletal muscle. The genes most differentially expressed with age (Tables 4 and 5) represent several classes, including structural, metabolic, and regulatory genes. Similar to the issue raised with regard to sex-related differences in gene expression, whether shared transcription factor binding sites within these genes (with corresponding age-related changes in transcription factor levels) explain their differential expression or whether these genes are independently regulated remains to be seen.
With regard to ST-induced changes in gene expression, only a moderate number of genes (n = 69) were found to be differentially expressed as a result of ST across all groups (Table S5). Our aim with the present exploratory investigation was to characterize “trained” skeletal muscle, thus we chose our after-ST biopsy time point for 48–72 h after the last training session to minimize the residual effects of the last bout of acute exercise (29). Although translation of existing mRNA has been shown to be important to myofibrillar protein synthesis after resistive exercise (42), the hours following an acute ST bout are likely associated with substantial changes in transcription for several genes, which then return to near baseline by 48–72 h after exercise (4, 29). Twenty of the 69 genes in Supplementary Table S5, genes differentially expressed in response to ST, are immediately recognizable as previously studied muscle-related genes, and include several structural and metabolic genes such as carbonic anhydrase III, GAPDH, nebulin, and troponin I. The specific roles of the other identified genes in skeletal muscle adaptation remain to be determined. Multiple myosin light chains (MLC; no myosin heavy chains are represented on the GF211 array) were also identified as differentially expressed in response to ST. Although only limited work has been done in relation to MLC expression and ST (46), the decreases in expression of the MLCs are not predicted; however, these results must be viewed with caution. An inherent limitation in cDNA microarrays is cross-hybridization of genes within the same gene family (8). Moreover, the GF211 was not designed to distinguish between MLC isoforms, information that is critical for determining functional significance. Although Jozsi et al. (15) reported altered expression of stress-related genes between young and older men in relation to ST, signal transduction, stress, repair, and growth factor genes were specifically represented on the commercial array used in their investigation; the GF211 microarray used here does not represent specific classes of genes, but rather contains a broad spectrum of known genes selected without regard to functional class. Thus the GF211 microarray is well-suited for identification of novel genes (22, 36, 38), which was the purpose of our work. For example, caldesmon was highly expressed on the arrays in all groups (i.e., highly expressed in muscle) but was more highly expressed in women compared with men in all analyses. Caldesmon has been most studied with regard to the regulation of smooth muscle contraction, as it inhibits myosin ATPase activity by binding to actin and tropomyosin (5, 27, 40), especially when dephosphorylated (10). To our knowledge, its role in skeletal muscle contraction is unclear, although Heubach et al. (12) have determined that caldesmon does inhibit skeletal muscle force production in vitro.
The qPCR results confirmed a reduction in the expression of FHL1 following ST, as well as higher expression in women than men. FHL1 is a LIM protein family member, with three primary isoforms, FHL1, 2, and 3 (23, 24, 26). FHL1 and FHL3 are highly expressed in skeletal muscle, whereas FHL2 is expressed predominantly in cardiac muscle (24). Expression of FHL1 was associated with muscle cell differentiation in C2C12 cell culture studies (24), but no further work has been reported as of this writing. FHL1 differs from a related protein, muscle LIM protein (1).
Finally, SWI/SNF, or BAF60b, expression was higher in women than men. BAF60b is one of a series of proteins that are involved in the remodeling of chromatin during development (30) and which appear to be necessary for transcriptional activation of many genes, as well as cell cycle control (6, 39). BAF60b is one of three family members, a, b, and c, of which BAF60b and BAF60c are highly expressed in skeletal muscle (39). Recently, de la Serna et al. (6) reported that during myoD-mediated induction of muscle differentiation, the myogenic phenotype was completely absent when mutated forms of BAF60b and BAF60c genes were expressed. Moreover, the BAF60 enzymes appear to promote myoD-mediated differentiation by altering chromatin structure in promoter regions of endogenous, muscle-specific loci, including myogenin and myosin heavy chain (6), demonstrating the importance of this protein to muscle.
The three genes outlined above were chosen for “real-time” or qPCR validation experiments based on their patterns of expression, available background literature, and other unpublished microarray data. All three genes demonstrated similar expression patterns in the qPCR experiments compared with the microarray results, despite the fact that the subjects used in the qPCR experiments were different from those of the microarray experiments, thus providing an independent validation.
Perspective
The current investigation was an exploratory analysis designed to identify candidate genes related to differences in skeletal muscle gene expression in relation to age and sex, as well as changes in muscle in response to ST. Despite our validation experiments and the extensive validation of these particular GF211 microarrays by others (22, 36, 38), these results should be viewed as preliminary and not as a definitive view of the skeletal muscle “transcriptome” in these conditions. While the use of microarray technology is within the grasp of most laboratories, the analysis of the resulting data is complex and often specific to a particular type of array, with few established techniques for in-depth statistical analysis of the accompanying large datasets. Moreover, in practical terms it is impossible to verify each identified gene using qPCR or similar single gene techniques. Thus we refrain from in-depth speculation about the “interpretive meaning” of the results and rather present the data as an important initial step in understanding the underlying biology of skeletal muscle. From these data, specific genes can be targeted for individual study (e.g., caldesmon, FHL1, and BAF60b). The results of this and other large-scale gene expression studies will need to be combined and compared, with the ultimate goal of developing a model that describes the global patterns of gene expression in skeletal muscle.
Supplementary Material
Acknowledgments
We thank Drs. Greg Martel, Fred Ivey, Jeff Lemmer, and Brian Tracy for help with strength training, biopsies, and study coordination. Elisa Heidrich O'Hare was critical for technical support.
This work was supported by National Institutes of Health Grants AG-42148 and DK-46204 and by the Competitive Medical Research Fund of the Univ. Pittsburgh Medical Center Health System. S. M. Roth was supported by National Research Service Award Grant AG-05893.
Footnotes
Supplementary materials (Tables S1–S5) to this article are avail-online at http://physiolgenomics.physiology.org/cgi/content/full/10/3/181/DC1.
REFERENCES
- 1.Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 2.Bakay M, Chen YW, Borup R, Zhao P, Nagaraju K, Hoffman EP. Sources of variability and effect of experimental approach on expression profiling data interpretation. BMC Bioinform. 2002;3:4. doi: 10.1186/1471-2105-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booth FW, Baldwin KM. Handbook of Physiology. Exercise: Regulation and Integration of Multiple Systems. Am Physiol Soc; Bethesda, MD: 1996. Muscle plasticity: energy demanding and supply processes; pp. 1075–1123. sect. 12, part 3, chapt. 24. [Google Scholar]
- 4.Booth FW, Tseng BS, Fluck M, Carson JA. Molecular and cellular adaptation of muscle in response to physical training. Acta Physiol Scand. 1998;162:343–350. doi: 10.1046/j.1365-201X.1998.0326e.x. [DOI] [PubMed] [Google Scholar]
- 5.Chalovich JM, Cornelius P, Benson CE. Caldesmon inhibits skeletal muscle actomyosin subfragment-1 ATPase activity and the binding of myosin subfragment-1 to actin. J Biol Chem. 1987;262:5711–5716. [PubMed] [Google Scholar]
- 6.de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 7.Eason JM, Schwartz GA, Pavlath GV, English AW. Sexually dimorphic expression of myosin heavy chains in the adult mouse masseter. J Appl Physiol. 2000;89:251–258. doi: 10.1152/jappl.2000.89.1.251. [DOI] [PubMed] [Google Scholar]
- 8.Evertsz EM, Au-Young J, Ruvolo MV, Lim AC, Reynolds MA. Hybridization cross-reactivity within homologous gene families on glass cDNA microarrays. Biotechniques. 2001;31:1182–1186. doi: 10.2144/01315dd03. [DOI] [PubMed] [Google Scholar]
- 9.Fiatarone MA, Marks EC, Ryan ND, Meredith CN, Lipsitz LA, Evans WJ. High-intensity strength training in nonagenarians. JAMA. 1990;263:3029–3034. [PubMed] [Google Scholar]
- 10.Foster DB, Shen LH, Kelly J, Thibault P, Van Eyk JE, Mak AS. Phosphorylation of caldesmon by p21-activated kinase. J Biol Chem. 2000;275:1959–1965. doi: 10.1074/jbc.275.3.1959. [DOI] [PubMed] [Google Scholar]
- 11.Gomes RR, Booth FW. Expression of acetylcholine receptor mRNAs in atrophying and nonatrophying skeletal muscles of old rats. J Appl Physiol. 1998;85:1903–1908. doi: 10.1152/jappl.1998.85.5.1903. [DOI] [PubMed] [Google Scholar]
- 12.Heubach JF, Hartwell R, Ledwon M, Kraft T, Brenner B, Chalovich JM. Inhibition of cross-bridge finding to actin by caldesmon fragments in skinned skeletal muscle fibers. Biophys J. 1997;72:1287–1294. doi: 10.1016/S0006-3495(97)78775-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurley BF, Roth SM. Strength training in the elderly: effects on risk factors for age-related diseases. Sports Med. 2000;30:249–268. doi: 10.2165/00007256-200030040-00002. [DOI] [PubMed] [Google Scholar]
- 14.Ivey FM, Roth SM, Ferrell RE, Tracy BL, Lemmer JT, Hurlbut DE, Martel GF, Siegel EL, Fozard JL, Metter EJ, Fleg JL, Hurley BF. Effects of age, gender and myostatin genotype on the hypertrophic response to heavy resistance strength training. J Gerontol Med Sci. 2000;55A:M641–M648. doi: 10.1093/gerona/55.11.m641. [DOI] [PubMed] [Google Scholar]
- 15.Jozsi AC, Dupont-Versteegden EE, Taylor-Jones JM, Evans WJ, Trappe TA, Campbell WW, Peterson CA. Aged human muscle demonstrates an altered gene expression profile consistent with an impaired response to exercise. Mech Ageing Dev. 2000;120:45–56. doi: 10.1016/s0047-6374(00)00178-0. [DOI] [PubMed] [Google Scholar]
- 16.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 17.Lemmer JT, Hurlbut DE, Martel GF, Tracy BL, Ivey FM, Metter EJ, Fozard JL, Fleg JL, Hurley BF. Age and gender responses to strength training and detraining. Med Sci Sports Exerc. 2000;32:1505–1512. doi: 10.1097/00005768-200008000-00021. [DOI] [PubMed] [Google Scholar]
- 18.Lindle RS, Metter EJ, Lynch NA, Fleg JL, Fozard JL, Tobin JD, Roy TA, Hurley BF. Age and gender comparisons of muscle strength in 654 women and men aged 20–93 yr. J Appl Physiol. 1997;83:1581–1587. doi: 10.1152/jappl.1997.83.5.1581. [DOI] [PubMed] [Google Scholar]
- 19.Lowe DA, Lund T, Alway SE. Hypertrophy-stimulated myogenic regulatory factor mRNA increases are attenuated in fast muscle of aged quails. Am J Physiol Cell Physiol. 1998;275:C155–C162. doi: 10.1152/ajpcell.1998.275.1.C155. [DOI] [PubMed] [Google Scholar]
- 20.Lynch NA, Metter EJ, Lindle RS, Fozard JL, Tobin JD, Roy TA, Fleg JL, Hurley BF. Muscle quality. I. Age-associated differences between arm and leg muscle groups. J Appl Physiol. 1999;86:188–194. doi: 10.1152/jappl.1999.86.1.188. [DOI] [PubMed] [Google Scholar]
- 21.Marsh DR, Criswell DS, Carson JA, Booth FW. Myogenic regulatory factors during regeneration of skeletal muscle in young, adult and old rats. J Appl Physiol. 1997;83:1270–1275. doi: 10.1152/jappl.1997.83.4.1270. [DOI] [PubMed] [Google Scholar]
- 22.McCormick SM, Eskin SG, McIntire LV, Teng CL, Lu CM, Russell CG, Chittur KK. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc Natl Acad Sci USA. 2001;98:8955–8960. doi: 10.1073/pnas.171259298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan MJ, Madgwick AJA. Slim defines a novel family of LIM-proteins expressed in skeletal muscle. Biochem Biophys Res Commun. 1996;225:632–638. doi: 10.1006/bbrc.1996.1222. [DOI] [PubMed] [Google Scholar]
- 24.Morgan MJ, Madgwick AJA. The LIM proteins FHL1 and FHL3 are expressed differently in skeletal muscle. Biochem Biophys Res Commun. 1999;255:245–250. doi: 10.1006/bbrc.1999.0179. [DOI] [PubMed] [Google Scholar]
- 25.Musaro A, Cusella De Angelis MG, Germani A, Ciccarelli C, Molinaro M, Zani BM. Enhanced expression of myogenic regulatory genes in aging skeletal muscle. Exp Cell Res. 1995;221:241–248. doi: 10.1006/excr.1995.1372. [DOI] [PubMed] [Google Scholar]
- 26.Ng EKO, Lee SMY, Li HY, Ngai SM, Tsui SKW, Waye MMY, Lee CY, Fung KP. Characterization of tissue-specific LIM domain protein (FHL1C) which is an alternately spliced isoform of a human LIM-only protein (FHL1) J Cell Biochem. 2001;82:1–10. doi: 10.1002/jcb.1110. [DOI] [PubMed] [Google Scholar]
- 27.Ngai PK, Walsh MP. Inhibition of smooth muscle actin-activated myosin Mg2+-ATPase activity by caldesmon. J Biol Chem. 1984;259:13656–13659. [PubMed] [Google Scholar]
- 28.Pavol MJ, Owings TM, Foley KT, Grabiner MD. The sex and age of older adults influence the outcome of induced trips. J Gerontol Med Sci. 1999;54A:M103–M108. doi: 10.1093/gerona/54.2.m103. [DOI] [PubMed] [Google Scholar]
- 29.Pilegaard H, Ordway GA, Saltin B, Neufer PD. Transcriptional regulation of gene expression in human skeletal muscle during recovery from exercise. Am J Physiol Endocrinol Metab. 2000;279:E806–E814. doi: 10.1152/ajpendo.2000.279.4.E806. [DOI] [PubMed] [Google Scholar]
- 30.Ring HZ, Vameghi-Meyers V, Wang W, Crabtree GR, Francke U. Five SWI/SNF-related, matrix associated, actin-dependent regulator of chromatin (SMARC) genes are dispersed in the human genome. Genomics. 1998;51:140–143. doi: 10.1006/geno.1998.5343. [DOI] [PubMed] [Google Scholar]
- 31.Rosenkranz-Weiss P, Tomek RJ, Mathew J, Ehgbali M. Gender-specific differences in expression of mRNAs for functional and structural proteins in rat ventricular myocardium. J Mol Cell Cardiol. 1994;26:261–270. doi: 10.1006/jmcc.1994.1029. [DOI] [PubMed] [Google Scholar]
- 32.Roth SM, Martel GF, Ivey FM, Lemmer JT, Tracy BL, Hurlbut DE, Metter EJ, Hurley BF, Rogers MA. Ultra-structural muscle damage in young vs. older men after high-volume, heavy-resistance strength training. J Appl Physiol. 1999;86:1833–1840. doi: 10.1152/jappl.1999.86.6.1833. [DOI] [PubMed] [Google Scholar]
- 33.Roth SM, Ferrell RE, Hurley BF. Strength training for the prevention and treatment of sarcopenia. J Nutr Health Aging. 2000;4:143–155. [PubMed] [Google Scholar]
- 34.Roth SM, Ivey FM, Martel GF, Lemmer JT, Hurlbut DE, Siegel EL, Metter EJ, Fleg JL, Fozard JL, Kostek MC, Wernick DM, Hurley BF. Muscle size responses to strength training in young and older men and women. J Am Geriatr Soc. 2001;49:1428–1433. doi: 10.1046/j.1532-5415.2001.4911233.x. [DOI] [PubMed] [Google Scholar]
- 35.Schultz AB, Ashton-Miller JA, Alexander NB. What leads to age and gender differences in balance maintenance and recovery? Muscle Nerve. 1997;20:S60–S64. [PubMed] [Google Scholar]
- 36.Sgroi DC, Teng S, Robinson G, LeVangie R, Hudson JR, Elkahloun AG. In vivo gene expression profile analysis of human breast cancer progression. Cancer Res. 1999;59:5656–5661. [PubMed] [Google Scholar]
- 37.te Pas MFW, de Jong PJ, Verburg FJ, Duin M, Henning RH. Gender related and dexamethasone induced differences in the mRNA levels of the MRF gene in rat anterior tibial skeletal muscle. Mol Biol Rep. 1999;26:277–284. doi: 10.1023/a:1007042414993. [DOI] [PubMed] [Google Scholar]
- 38.Walker J, Rigley K. Gene expression profiling in human peripheral blood mononuclear cells using high-density filter-based cDNA microarrays. J Immunol Methods. 2000;239:167–179. doi: 10.1016/s0022-1759(00)00181-2. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 40.Watson MH, Kuhn AE, Novy RE, Lin JJ, Mak AS. Caldesmon-binding sites on tropomyosin. J Biol Chem. 1990;265:18860–18866. [PubMed] [Google Scholar]
- 41.Welle S, Bhatt K, Thornton C. Polyadenylated RNA, actin mRNA, and myosin heavy chain mRNA in young and old human skeletal muscle. Am J Physiol Endocrinol Metab. 1996;270:E224–E229. doi: 10.1152/ajpendo.1996.270.2.E224. [DOI] [PubMed] [Google Scholar]
- 42.Welle S, Bhatt K, Thornton CA. Stimulation of myofibrillar synthesis by exercise is mediated by more efficient translation of mRNA. J Appl Physiol. 1999;86:1220–1225. doi: 10.1152/jappl.1999.86.4.1220. [DOI] [PubMed] [Google Scholar]
- 43.Welle S, Bhatt K, Thornton CA. Inventory of high-abundance mRNAs in skeletal muscle of normal men. Genome Res. 1999;9:506–513. [PMC free article] [PubMed] [Google Scholar]
- 44.Welle S, Bhatt K, Thornton CA. High-abundance mRNAs in human muscle: comparison between young and old. J Appl Physiol. 2000;89:297–304. doi: 10.1152/jappl.2000.89.1.297. [DOI] [PubMed] [Google Scholar]
- 45.Welle S, Brooks A, Thornton CA. Senescence-related changes in gene expression in muscle: similarities and differences between mice and men. Physiol Genomics. 2001;5:67–73. doi: 10.1152/physiolgenomics.2001.5.2.67. [DOI] [PubMed] [Google Scholar]
- 46.Williamson DL, Godard MP, Porter DA, Costill DL, Trappe SW. Progressive resistance training reduced myosin heavy chain coexpression in single muscle fibers from older men. J Appl Physiol. 2000;88:627–633. doi: 10.1152/jappl.2000.88.2.627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.