Summary
Chromosomal translocations are a hallmark of leukemia/lymphoma and also appear in solid tumors, but the underlying mechanism remains elusive. By establishing a cellular model that mimics the relative frequency of authentic translocation events without proliferation selection, we report mechanisms of nuclear receptor-dependent tumor translocations. Intronic binding of liganded-AR first juxtaposes translocation loci by triggering intra- and interchromosomal interactions. AR then promotes site-specific DNA double-stranded breaks (DSBs) at translocation loci by recruiting two types of enzymatic machinery induced by genotoxic stress and liganded-AR, including Activation-Induced Cytidine Deaminase (AID) and the LINE-1 repeat-encoded ORF2 endonuclease. These enzymatic machineries synergistically generate site-selective DSBs at juxtaposed translocation loci that are ligated by Non-Homologous Ending Joining (NHEJ) pathway for specific translocations. Our data suggest that the confluence of two parallel pathways initiated by liganded-nuclear receptor and genotoxic stress underlie non-random tumor translocations, which may function in many types of tumors and pathological processes.
Keywords: Tumor Translocations, Prostate Cancer, Androgen Receptor, AID, PIWIL1, Nuclear Architecture, LINE-1 Repeats, ORF2, DNA Damage/Repair, LNCaP Cells
Introduction
Chromosomal translocations, caused by rearrangement of non-homologous chromosomes (Aplan, 2006), have been well described in leukemia and lymphomas (Greaves and Wiemels, 2003), but their occurrence in solid tumors is increasingly recognized, particularly in prostate cancer (Tomlins et al., 2005). It has been recognized that prevalent tumor translocations may be responsible for certain aggressive behaviors of prostate cancer (Wang et al., 2006). Androgen and its derivatives, which act via the androgen receptor (AR), are not only essential for development of the prostate gland, but also instrumental to prostate carcinogenesis (Heinlein and Chang, 2004). Recently, some high frequency gene fusion events have been discovered in prostate cancers, which involve translocation of the 5′ untranslated region of the AR target gene TMPRSS2 to two members of the ETS family of genes ERG and ETV1 (Tomlins et al., 2005). These gene fusion events, which may be present in 50-70% of prostate cancers, render specific members of the ETS family of genes under the control of androgens; such acquired androgen-dependent expression or overexpression of the ETS genes has been proposed to provide a key driving force to the development or aggressiveness of prostate cancers (Shaffer and Pandolfi, 2006).
While the linkage between chromosomal translocations and various forms of cancer has founded the theoretical grounds for cancer diagnosis and therapeutics, particularly for leukemia and lymphomas (Corral et al., 1996; Krivtsov and Armstrong, 2007), the underlying molecular mechanisms have remained incompletely understood. Although it is well established that transcriptionally active regions, such as promoters, can be particularly susceptible to DNA damage (Aguilera and Gomez-Gonzalez, 2008; Thomas and Rothstein, 1989), a prevalent view has been that tumor translocations may initially result from random chromosome rearrangement events, which are ultimately selected based on the proliferative and/or anti-apoptotic advantage provided by specific fusion gene products. However, precedents such as Gross Chromosomal Rearrangements (GCR) in yeast (Myung et al., 2001), V(D)J recombination, and Class Switch Recombination (CSR) during T and B cell development (Chaudhuri and Alt, 2004) argue for a role of genetically-based and cell lineage-specific juxtaposition of translocation loci, which may facilitate specific chromosomal translocations (Jhunjhunwala et al., 2008; Neves et al., 1999; Nikiforova et al., 2000; Roix et al., 2003). Because many types of cancer occur in tissues in which specific transcription factors may exert critical roles in tumor development, a potential mechanistic relationship between regulated transcription and the strategies that underlie tumor translocations, if any, remain an intriguing question.
Here, we present evidence that tumor translocations involving TMPRSS2, ERG and ETV1 in prostate cancer are non-random events, which require two critical roles of AR: (i) ligand-dependent binding of AR to intronic binding sites near the tumor translocation sites, causing chromosomal movements that result in specific intra- and interchromosomal interactions to create the spatial proximity for tumor translocation partners, and (ii) the actions of intron-bound AR to both alter local chromatin architecture and recruit the ligand and genotoxic stress-induced enzymes, including the Activation-Induced cytidine Deaminase (AID) and LINE-1 repeat-encoded ORF2 endonuclease to these specific regions for facilitating DNA double-stranded breaks (DSBs) generation. The generated DSBs are subsequently ligated by the Non-Homologous End Joining (NHEJ) machinery. These findings elucidate several unexpected general principles for non-random chromosomal translocations in tumors.
Results
Androgens and Genotoxic Stress Synergistically Induce Prostate Cancer-Specific Chromosomal Translocations
Based on the critical roles of AR in prostate development and tumor progression, and the observation that genotoxic stress is able to rapidly induce chromosomal translocations (Deininger et al., 1998), we first investigated whether androgen treatment and genotoxic stress, either alone or in combination, might induce chromosomal translocations of TMPRSS2:ERG and TMPRSS2:ETV1. To establish a cellular model, we utilized androgen-responsive LNCaP prostate cancer cells in which these chromosomal translocations had not occurred (Tomlins et al., 2005).
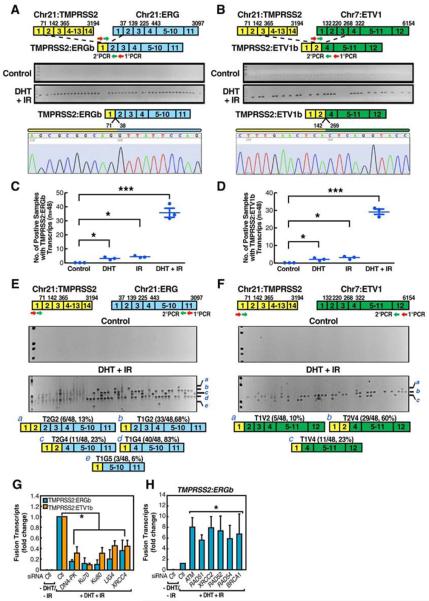
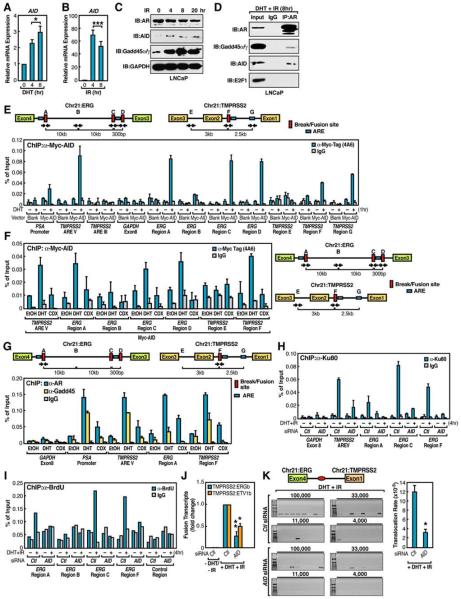
Surprisingly, the treatment of LNCaP cells with both DHT (10−7 M) and irradiation (IR) (50 Gy) dramatically induced both TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts in 24hr, with similar effects reproducibly observed at lowered levels of DHT (10−9 M) and IR (10 Gy). (Figures 1A and 1B and data not shown). Sequencing of induced fusion transcripts confirmed that they represented the authentic translocation fusion junctions (Figures 1A and 1B). The cell viability did not differ significantly after 24hr treatment. Actin expression was equivalent among samples (Figure S1). Other modalities that cause genotoxic stress, including Etoposide and Doxorubicin, when combined with the DHT treatment, also induced tumor translocation (Figures S3A and S3B).
Figure 1. Liganded-AR and Genotoxic Stress Synergistically Induce Chromosomal Translocations in Prostate Cancer Cells.
(A and B) Identification and characterization of induced TMPRSS2:ERGb and TMPRSS2:ETV1b translocations in LNCaP cells. Top: schematic structures for the TMPRSS2, ERG, and ETV1 mRNA indicating exon positions. Middle: RT-PCR amplification of TMPRSS2:ERGb (A) or TMPRSS2:ETV1b (B) fusion transcripts from 48 individual cell samples. Bottom: confirmation of position and fusion sites by automated DNA sequencing. (C and D) Statistical analysis of DHT and IR induced TMPRSS2:ERG (C) and TMPRSS2:ETV1 (D) translocations (n=3, ±SEM). (E and F) Identification and characterization of induced TMPRSS2:ERG (E) and TMPRSS2:ETV1 (F) translocation isoforms in LNCaP cells. Bottom: summary of distinct fusion types. (G and H) Involvement of DNA repair machinery in induced TMPRSS2:ERGb and TMPRSS2:ETV1b translocations by QPCR with indicated siRNAs (n=3, ±SEM).
To quantitate the frequency at which each type of fusion transcript was formed, 48 individual sets of cells were exposed (24hr) to either DHT alone, IR alone, or both. RT-PCR analyses of TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts revealed that either treatment gave rise to a reproducible, but minimal, induction of the fusion transcripts, but the combined treatment exhibited an unequivocal, striking synergy (Figures 1C and 1D). The frequency of these chromosomal translocations in LNCaP cells was estimated by dilution experiments to be ~1 event/10,000 cells (vide infra).
The finding that eight isoforms of TMPRSS2:ERG fusion transcripts have been identified in prostate cancer (Wang et al., 2006) led us to detect all potential induced fusion isoforms using primers targeting the first exon of TMPRSS2 and the last exon of ERG or ETV1, respectively (Figures 1E and 1F). This analysis revealed that the frequency of each induced TMPRSS2:ERG fusion isoform in LNCaP cells was very similar to that identified in prostate cancer tissues (Wang et al., 2006) (Figures 1E and S2). Together these data demonstrated that our cellular model and treatment conditions authentically recapitulate the in vivo chromosomal translocation events, and surprisingly, these specific patterns are established without proliferation selection.
To determine whether the generation of chromosomal translocation has cell type specificity, we analyzed IR-induced BCR:ABL1 translocation in KG-1 cells (Deininger et al., 1998) (Figure S4A) and LNCaP cells. Although BCR and ABL1 are expressed in LNCaP cells at a detectable level (Figures S4B and S4C), IR (50 Gy) induced BCR:ABL1 fusion in KG-1 cells at an efficiency ~8-fold higher than that in LNCaP cells (Figure S4D). Conversely, irradiation of LNCaP cells induced TMPRSS2:ERGb or TMPRSS2:ETV1b fusion >10 fold more efficiently than in KG-1 cells under the same conditions (Figures S4E and S4F). These observations demonstrated that general genotoxic stress signals preferentially induce gene fusions in a non-random, cell type-specific fashion.
Using validated siRNAs to target specific components of the Homologous Recombination (HR), NHEJ, or Microhomology-Mediated End Joining (MMEJ) pathways (Figure S8A), we found that knockdown of individual components of the NHEJ pathway generally attenuated the induction of both TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts, suggesting the error-prone NHEJ as the major repair mechanism to generate fusion genes (Figure 1G). Removal of the major components of the HR pathway actually enhanced the induction of TMPRSS2:ERGb translocations (Figure 1H). Knockdown of individual components of several other DNA damage repair pathways did not show any significant effect on the chromosomal translocation types we tested (Figure S5 and data not shown).
Liganded-AR Induces Intra- and Interchromosomal Interactions Required for Translocations
Based on the dynamic re-organization of chromosomes during development or in response to specific signals (Cremer et al., 2006), we investigated whether one role of AR might be to mediate androgen-dependent spatial proximity of the corresponding chromosomal translocating regions. ERG and TMPRSS2 are both located on Chromosome 21 (Chr21), ~3 megabases apart, while the ETV1 gene is located on Chromosome 7 (Chr7). As TMPRSS2 is a well-established direct AR target gene (Lucas et al., 2008), we tested whether DHT induced specific intra- and interchromosomal interactions between these regions.
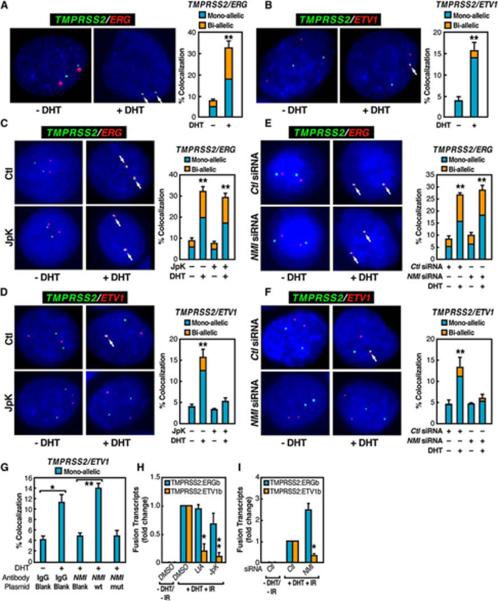
To avoid potential complications associated with aneuploidy of cancer cells, we focused on normal prostate epithelial cells (PrEC). While TMPRSS2 and ERG genes were independently localized in the nuclei of mock-treated PrEC cells, DHT stimulation (1hr) induced the apparent colocalization of these two genes in ~25% of cells, about half exhibiting mono-allelic interactions and the other half bi-allelic interactions as revealed by FISH analysis (Figure 2A). We performed a similar analysis for the TMPRSS2 and ETV1 genes in PrEC cells, which exhibited DHT-induced colocalization in ~10-15% of cells, the majority being mono-allelic interactions (Figure 2B). We observed similar induced colocalization in LNCaP cells (Figure S6A and S6B). In contrast, the BCR and ABL1 gene loci did not show any DHT-dependent colocalization, suggesting the specificity of DHT-dependent chromosomal interactions (Figure S6C).
Figure 2. AR-Induced and Motor-Dependent Chromosomal Interactions of TMPRSS2 and ERG or ETV1 Loci.
(A and B) Interphase FISH analysis on PrEC cells with TMPRSS2 (green), ERG (red) (A), or ETV1 (red) (B) probes. (C and D) Actin polymerization-dependent interchromosomal interactions. (E and F) Nuclear myosin-dependent interchromosomal interactions. (G) ATPase activity of NMI is required for DHT induced interchromosomal interactions. (H and I) Requirement of nuclear myosin/actin motor system for induced TMPRSS2:ERGb and TMPRSS2:ETV1b translocations in LNCaP cells pretreated with Latrunculin (LtA) or Jasplakinolide (Jpk) (H) or transfected with NMI siRNA (I) (n=3, ±SEM).
Based on the reported role of nuclear actin in transcriptional activation (Hofmann and de Lanerolle, 2006), we examined whether a nuclear myosin I (NMI)/actin-dependent mechanism might be involved in mediating intra- and interchromosomal interactions in PrEC cells, as recently documented for breast epithelial cells (Hu et al., 2008). Treatment of DHT-stimulated PrEC cells with Jasplakinolide (Jpk), which specifically inhibits depolymerization of actin networks (Holzinger, 2001), caused almost a complete loss of DHT-induced interchromosomal (TMPRSS2:ETV1) interactions, but did not affect intrachromosomal (TMPRSS2:ERG) interactions (Figures 2C and 2D). Likewise, single-cell nuclear microinjection of siRNA that specifically targeted NMI (Grummt, 2006) had little effect on intrachromosomal (TMPRSS2:ERG) interactions, but caused a complete loss of the DHT-induced TMPRSS2:ETV1 interchromosomal interactions (Figures 2E and 2F). Similar results were also obtained by single-cell nuclear microinjection of a neutralizing antibody against NMI, which could be functionally rescued by co-injecting a plasmid expressing wild-type NMI, but not mutant NMI that fails to bind to ATP (Wang et al., 2003)(Figures 2G and S7). LNCaP cells treated with Latrunculin (LtA), which blocks actin polymerization (Rizk and Walczak, 2005), or Jpk exhibited inhibition of induced TMPRSS2:ETV1 translocations, as was observed using an siRNA that specifically targeted NMI (Figures 2H and 2I). Together, these observations reveal the mechanistic link between ligand-induced, nuclear myosin/actin motor system-dependent interchromosomal proximity and tumor translocations.
Mapping TMPRSS2:ERG and TMPRSS2:ETV1 Translocation Breakpoints
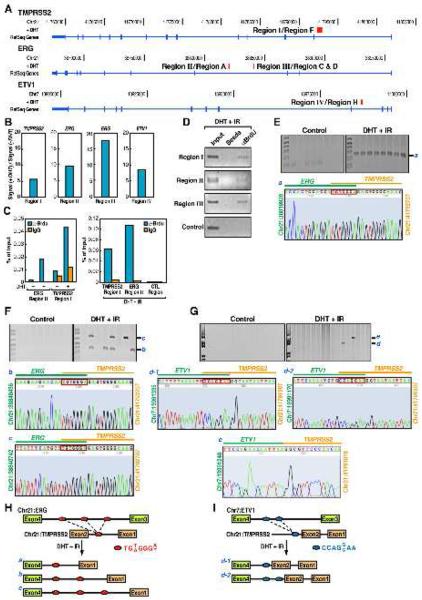
To identify the potential DSBs in the corresponding introns of the two translocation partners, we adapted a protocol of BrdU labeling by terminal deoxynucleotide transferase (TdT) (Ju et al., 2006) coupled with ChIP-seq analysis to detect potential DNA breaks in response to DHT, genotoxic stress, or both. To test the potential efficacy of this protocol for high-throughput screening experiments, we took advantage of the inducible 4-OHT-I-PpoI systems to generate specific cleavage sites on Chromosome 1 (Berkovich et al., 2008) and labeled these sites with BrdU. Indeed, anti-BrdU ChIP assay showed that after induction with 4-OHT, both the 5′ and 3′ chromatin fragments at the cleavage site, but not the flanking region, could be labeled by BrdU (Figure S9). By applying this protocol to LNCaP cells, our data revealed one potential breakpoint located within or nearby intron 1 of the TMPRSS2 gene (Region I), two potential breakpoints located within intron 3 of ERG gene (Regions II and III), and one potential breakpoint in the ETV1 gene intron 3 (Region IV) (Figure 3A). Compared to mock treatment, DHT induced an ~ 6-10-fold change at each of these putative DSBs (Figure 3B). These putative sites were further confirmed by conventional ChIP analysis, in DHT or DHT+IR treated LNCaP cells (Figures 3C and 3D).
Figure 3. Identification of Breakpoints for TMPRSS2:ERG and TMPRSS2:ETV1 Translocations.
(A) The tracks display the human genome coordinates (hg18 assembly), red band: predicted potential DSBs based on ChIP-seq using anti-BrdU antibodies (see Experimental Procedures). Boxes: exons; lines: introns; Region I to IV: DNA break points within corresponding loci. (B) The fold change of the tag density in the double-strand break regions I, II, III and IV after DHT treatment. (C and D) Conventional ChIP analysis with anti-BrdU antibodies on ERG and TMPRSS2 intronic break regions identified by ChIP-seq. (E-G) Identification and characterization of induced TMPRSS2:ERG and TMPRSS2:ETV1 translocation breakpoints. Top: genomic DNA extracted from LNCaP cells either non-transfected (E) or co-transfected with MeCP2 siRNA and FLAG-DOT1L expression vector (F and G) was subjected to PCR amplification using primers flanking Region II and I (E), Region III and I (F), or Region IV and I (G). Bottom: automated DNA sequencing aligned to ERG or ETV1 (green) and TMPRSS2 (orange) with genomic position of starting and ending nucleotides shown. Red box: common sequence shared by TMPRSS2 and ERG or ETV1. (H and I) Graphic illustration of translocation patterns corresponding to induced TMPRSS2:ERG (H) and TMPRSS2:ETV1 (I) translocations. Potential break/fusion sites are shown as Red oval: TMPRSS2:ERG; blue oval: TMPRSS2:ETV1; dot line: distinct fusion patterns.
As illustrated in Figure 3A, amplification with primer pairs flanking Region II and I resulted in a single PCR product (Figure 3E, band a) containing a breakpoint between nucleotides Chr21: 38819940-38819946 on ERG gene and nucleotide Chr21: 41792689-41792695 on TMPRSS2 gene (Figure 3E), which confirms that the DNA breaks identified in these pilot experiments corresponded to actual translocation breakpoints in these genes. Taking advantage of a finding that simultaneous cotransfection of FLAG-DOT1L (encoding an H3K79 methyltransferase) expression plasmid and MeCP2 siRNA into LNCaP cells resulted in a 60-fold induction of fusion transcript (Figure S10), we amplified two additional PCR products (Figure 3F, band b and c) using primer pair flanking Region III and I, finding that band b contains breakpoints between Chr21: 38840448-38840454 and Chr21: 41792689-41792695, and band c contains breakpoints between Chr21: 38840754-38840760 and Chr21: 41792689-41792695 (Figure 3F). Interestingly, both ERG and TMPRSS2 utilize a common motif (TGT/AGGGA/T) for break/ligation (Figure 3H).
For TMPRSS2:ETV1 translocation, two breakpoints between Chr7: 13991147-13991153 or Chr7: 13991182-13991188 and Chr21: 41798889-41798895 were revealed by sequencing PCR product d (Figure 3G, middle panel), containing a common CCAGG/CAA motif as their break/ligation sites (Figure 3I). An additional breakpoint between ETV1 and TMPRSS2 at Chr7: 13991254 and Chr21: 41791805 were identified from PCR product e (Figure 3G, lower panel). Together, these mapping results identified unique heptad repeats serving as breakpoints for non-random ligation sites of tumor translocation.
Sensitization of Intronic AR Binding Sites to Genotoxic Stress
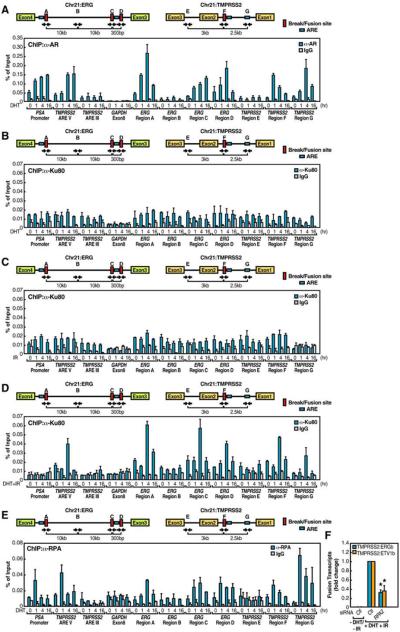
Since AR agonists induced both intra- and interchromosomal movements that involved the AR target gene TMPRSS2, we hypothesized that the ERG gene might also harbor some previously unsuspected AR binding sites, even though ERG expression is not induced by AR agonists (Tomlins et al., 2005). Because the identified “break” sites in ERG and TMPRSS2 are adjacent to potential Androgen Response Elements (AREs) or ARE half sites (Figure 4A, upper panel), we conducted conventional ChIP analysis for AR recruitment across the intronic translocation regions within the ERG and TMPRSS2 genes. As controls, we observed the binding of AR to PSA promoter and to the recently identified ARE located 13.5kb upstream of the TMPRSS2 gene (ARE V) (Wang et al., 2007), but no significant binding to either an AR negative binding region in the TMPRSS2 gene (ARE III) (Wang et al., 2007) or the GAPDH exon8 region (Figure 4A). Interestingly, upon DHT treatment, we found that AR was clearly recruited to the ERG “break” site Region A (corresponding to Region II in Figure 3A), C and D (corresponding to Region III in Figure 3A), TMPRSS2 “break” site Region F (corresponding to Region I in Figure 3A) and ETV1 “break” site Region H (corresponding to Region IV in Figure 3A) with temporal kinetics similar to these observed at known positive AR binding sites (PSA promoter and TMPRSS2 ARE V) (Figures 4A and S15A). AR was also recruited to a potential ARE (Region G) 2.5 kb upstream of the TMPRSS2 “break” site (Figure 4A). In contrast, the ERG Region B and the TMPRSS2 Region E, neither of which harbor any apparent ARE nor serve as break sites, showed no significant AR recruitment (Figure 4A).
Figure 4. AR-Dependent Local Chromatin Structural Alteration Sensitizes to Site-Specific Genotoxic Stress-Induced DSBs.
(A-E) Top: schematic diagram showing the relative positions of break/fusion sites and potential AREs located on ERG and TMPRSS2 loci. Blue boxes, potential AREs; red boxes, break/fusion sites; black arrows, relative positions of PCR primers. Bottom: LNCaP cells were treated with DHT (10−7 M) (A), DHT (10−7 M) (B), IR (50 Gy) (C) or both (D) and DHT (10−7 M) (E) for time courses as indicated. ChIP analyses were performed with indicated antibodies on indicated regions (n=2, ±SEM). (F) Examination of TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts with RPA2 siRNAs (n=3, ±SEM).
To understand the synergistic effect of liganded-AR and genotoxic stress on chromosomal translocation, we first examined the DNA damage response marked by γH2AX in DHT-, IR- or DHT+IR-treated LNCaP cells. No significant enrichment of γH2AX were detected at all the regions we tested with DHT alone; and a moderate recruitment was observed upon IR treatment alone with characteristic spreading to regions adjacent to DSBs. Surprisingly, there was a striking enrichment of γH2AX to the translocation regions (ERG Region A, C, D, the TMPRSS2 Region F and the ETV1 Region H) after combined treatment with DHT+IR (Figures S11 and S15B). Since Ku80 protein binds to DNA DSBs, nicks, gaps and hairpins in a sequence-independent manner and serves as a key component of the NHEJ repair machinery (Critchlow and Jackson, 1998), we next applied Ku80 as a mark to validate site-specific DSBs at translocation regions. Consistently, we observed that Ku80 bound to the TMPRSS2 and ERG intronic AR binding sites exclusively under the condition of combined treatment with DHT and IR, but not to control regions within the ERG gene (Figures 4B-4D). These results demonstrated a site-specific sensitization of AR-bound intronic sites to genotoxic stress-induced DSBs.
Because binding of nuclear receptors during transcriptional activation is characteristically accompanied by covalent histone modifications that mark structural changes of chromatin (Metivier et al., 2006), we next tested whether intronic AR binding causes histone modifications that facilitate sensitization of these regions to genotoxic stress. Enhanced H3K79 methylation, a mark of altered chromatin structure (Huyen et al., 2004) has been observed to correlate with V(D)J recombination in B cells (Ng et al., 2003). Histone H4K16 acetylation is also considered as a mark for chromatin relaxation (Shogren-Knaak et al., 2006) and is involved in DNA repair. We observed that DHT+IR provoked robust accumulation of di-methyl H3K79 and H4K16 acetylation markers in the ERG regions A, C, D and the TMPRSS2 site F (Figures S12A and S12B). TMPRSS2 ARE III and GAPDH exon 8 regions, at which no breaks occurred, provided negative controls. These data provide evidence that the androgen receptor acts to cause regional histone modifications at the intronic translocation sites.
As a 3′-5′ DNA helicase in yeast is required for GCR by enhancing binding of the replication protein A (RPA) (Banerjee et al., 2008), we tested whether binding of AR might induce the recruitment of RPA, which is known to bind to and stabilize single-stranded non-template DNA (Bochkarev and Bochkareva, 2004) by performing RPA ChIP analysis. Upon DHT stimulation for 1hr, RPA was rapidly recruited to the AR binding sites, persisting even after 16hr of DHT stimulation (Figure 4E). Consistent with a key role for RPA in yeast GCR, treatment of LNCaP cells with RPA2 siRNA inhibited both intra- and interchromosomal translocations induced by DHT+IR (Figure 4F).
Genotoxic Stress-Induced Enzymatic Machinery Contributes to Chromosomal Breaks at Intronic AR Binding Sites
Although irradiation may cause random DSBs in the genome, we next investigate the roles of irradiation in generating site-specific DSBs at translocation regions. Recently, Gadd45, which is a well-known irradiation-induced protein, was shown to play an important role in mediating AID actions by forming a functional complex with AID (Rai et al., 2008). Gadd45 is also known to function as a transcriptional co-activator for a number of nuclear receptors (Yi et al., 2000). Normally, AID is expressed at a very low level in LNCaP cells; however, we observed that AR agonists and genotoxic stress both induced AID expression by ~3-fold and >60-fold, respectively (Figures 5A and 5B). Correspondingly, the AID protein was highly induced in IR-treated cells as early as 4hr (Figure 5C).
Figure 5. Mechanisms that Initiate Extended DNA Breaks in AR-Dependent Tumor Translocations.
(A-C) Induction of AID expression by AR agonist and genotoxic stress. (n=3, ±SEM). (D) DHT-dependent interaction between AR and AID. Immunoprecipitates of anti-AR were subjected to immunoblotting analysis with indicated antibodies. Immunoblotting of E2F1 was included as negative control. (E) Ligand-dependent Myc-AID recruitment to AR-binding sites (n=2, ±SEM). (F and G) The recruitment of AID and Gadd45 to AR binding sites is mediated by liganded receptor. LNCaP cells were treated with ethanol (EtOH), DHT (10−7 M), or Bicalutamide (CDX, 10 μM) for 1hr followed by ChIP analyses with anti-Myc (F) or anti-AR and anti-Gadd45 (G) antibodies on indicated regions (n=2, ±SEM). (H and I) AID contributes to DSBs generation. ChIP analyses were performed on control siRNA or AID siRNA transfected, DHT+IR treated (4hr) LNCaP cells with anti-Ku80 antibodies (n=2, ±SEM) (H) or anti-BrdU antibody following BrdU labeling by TdT (I) on indicated regions. (J) Examination of TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts in LNCaP cells transfected with AID siRNA (n=3, ±SEM). (K) Left: representative agarose gels with PCR products corresponding to TMPRESS2:ERGa translocations (as illustrated in Figure 3H band a). The genomic DNA of control or AID siRNA transfected LNCaP cells were subjected to PCR using primers flanking ligation site (red oval). Right: Statistical analysis (n=3, ±SEM).
We therefore tested the potential roles of DHT-dependent recruitment of AID/Gadd45 to AR, observing that AR interacted with both AID and Gadd45 upon DHT+IR treatment (Figure 5D). This interaction was DHT-dependent, because IR alone did not induce such interactions (Figure S13). These data suggested that the DHT- and IR-induced AID might be recruited to the intronic translocation sites via direct binding of the Gadd45/AID complex to liganded-AR. Indeed, ChIP analysis of anti-Myc-tagged AID (Figure S14A) revealed that AID was recruited to the intronic AR-binding sites on ERG, TMPRSS2 and ETV1 translocation regions, as well as to known AR targets such as PSA promoter and TMPRSS2 enhancer, but not to non-AR binding sites (Figures 5E and S15C). To further confirm a functional role for AID and Gadd45 in AR dependent tumor translocations, we treated LNCaP cells with either DHT or Bicalutamide (Casodex, CDX) and observed only minimal recruitment of AID or Gadd45 to intronic AR binding sites in CDX-treated cells (Figures 5F and 5G).
Because AID mediates CSR and Somatic Hypermutation (SHM) by initiating deamination of bases of single-stranded non-template DNA during transcription (Chaudhuri et al., 2003), we investigated whether recruitment of AID is essential for the generation of DSBs by monitoring the enrichment of Ku80 at the translocation regions under the condition of AID knockdown by siRNA (Figure S8B). Knockdown of AID resulted in a dramatic decrease in the Ku80 enrichment as shown by ChIP analysis (Figure 5H), indicating an inhibition of DSBs generation in these translocation regions. To further validate this finding, we also used the BrdU/TdT assay to confirm that DSBs generated at these intronic sites are mediated by AID (Figure 5I). Consistent with these observations, siRNA knockdown of AID blocked the induction of both intra- and interchromosomal translocations (Figures 5J). The actual frequency of chromosomal translocation events was quantitated by PCR analysis in serial dilution experiments, finding that the rate of translocation induced by DHT+IR within 24hr fell from ~12×10−5 in control siRNA samples to ~3×10−5 in AID siRNA knockdown samples (Figure 5K).
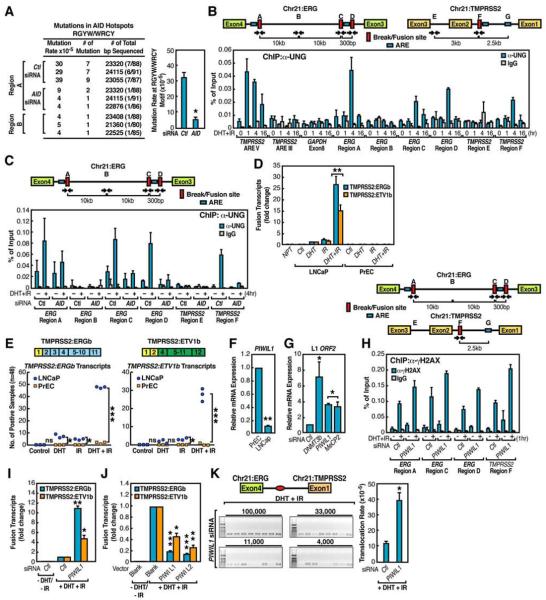
One hallmark of AID enzymatic activity is the induction of SHM (Honjo et al., 2002). PCR amplification and sequencing of the break region (ERG region A) following DHT+IR treatment revealed evidence of the C to U and G to A mutations characteristic of SHM (Figure 6A). These mutations were inhibited by knockdown of AID prior to the treatment, suggesting that AID is actively involved in the observed DSB generation at translocations sites. In adjacent control regions (ERG region B), only basal mutation rate was observed (Figure 6A). To examine the possibility that AID may also play an obligatory role in mediating AR-induced chromosomal movements, AID knockdown was performed in PrEC and LNCaP cells, finding that AID did not exert any effects on DHT-dependent interchromosomal movement, as revealed by FISH analysis (Figure S16).
Figure 6. Protective Effects of PIWIs on Chromosomal Translocation.
(A) Left: summary of identified RGYW/WRCY motif related mutation. The fusion chromatins of ERG region A in control siRNA and AID siRNA samples or ERG region B were amplified as in Figure 3E. Right: statistics analysis (n=3, ±SEM). (B) Ligand-dependent UNG recruitment to intronic regions of TMPRSS2 and ERG loci. (C) Recruitment of UNG to translocation regions is AID dependent. D) Quantitation of induced TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts in LNCaP or PrEC cells. NPT: normal prostate tissue. (E) Statistical comparison of induced TMPRSS2:ERGb (left) and TMPRSS2:ETV1b (right) fusion transcripts LNCaP and PrEC cells (48 samples per group and n=3, ±SEM). (F) The relative expression level of PIWIL1 in LNCaP and PrEC cells. (G) The expression level of LINE-1 ORF2 was examined in PrEC cells transfected with indicated siRNAs. (H) PIWIL1 knockdown enhances γH2AX enrichment at intronic ERG and TMPRSS2 break/fusion sites. (I and J) Examination of TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts in LNCaP cells electroporated with PIWIL1 siRNA (I) or indicated plasmids (J) (n=3, ±SEM). (K) Left: representative agarose gels with PCR products corresponding to TMPRESS2:ERGa translocations (as illustrated in Figure 3H band a). Right: Statistical analysis (n=3, ±SEM).
We also observed that the Uracil-DNA Glycosylase (UNG), which recognizes the U•G mismatch generated upon deamination of cytidine by AID and generate abasic sites in the C1 regions during CSR (Rajewsky and von Boehmer, 2008) localized to both ERG and TMPRSS2 intronic AR binding sites in DHT+IR-treated LNCaP cells (Figure 6B). The recruitment of UNG was dependent on AID, indicated by reduced UNG recruitment following the knockdown of AID (Figure 6C).
To determine whether AID also plays a role in SLC45A3:ETV1 chromosomal translocation identified in prostate cancer (Tomlins et al., 2007), we assessed these translocation events in DHT+IR-treated LNCaP cells following AID knockdown by specific siRNAs. With combined treatment of DHT and IR, we detected significant induction of SLC45A3:ETV1 translocation in LNCaP cells, but not in PrEC cells, even though the expression of SLC45A3 was similarly induced by DHT between these two cell lines (Figures S20 A-C). However, knockdown of AID abolished the induced SLC45A3:ETV1 translocation (Figure S20D). Similarly, in B cell leukemia KG-1 cell line, the BCR:ABL1 translocation is induced by high-dose irradiation, and knockdown of AID caused a decrease in irradiation-induced BCR:ABL1 translocation (Figures S8D, S17A and S17B), consistent with the previous report that AID is required for c-Myc:IgH translocation (Robbiani et al., 2008). While these findings strongly implicate the actions of AID to facilitate the DNA breaks at the intronic AR binding sites, they also raised the question whether other key enzymatic machinery was combinatorially required for induced chromosomal breaks.
Contribution of PIWI-Regulated LINE-1 Repeat/ ORF2 Endonuclease to Tumor Translocations
Intriguingly, DHT+IR was much less effective in inducing chromosomal translocations in PrEC cells than in LNCaP cells (Figures 6D, 6E and Figure S20B), even though the expression of the TMPRSS2 and SLC45A3 genes were induced similarly by DHT in both cell types (Figures S18 and S20C). This observation is consistent with the possibility that cancer cells express additional factors that predispose to genome instability (Peng and Karpen, 2008). We therefore surveyed the mRNA expression levels of a series of “epigenetic” modulators that have been implicated in genome instability (Goldberg et al., 2007). Whereas the expression of most of these modulators did not change, a few (e.g. MOF1) did exhibit ~2-3-fold higher expression level in PrEC cells compared to LNCaP cells (data not shown), and MOF knock-down noticeably enhanced the efficiency of both intra- and interchromosomal translocations (Figure S19).
Strikingly, the most dramatic discrepant target we tested between LNCaP and PrEC cells was PIWIL1 (~9-10 fold) (Figure 6F). PIWIs have been reported to suppress retrotransposition, including LINE-1 element during spermatogenesis (Aravin et al., 2007), but more recent studies have also implicated its role in somatic organs (Malone et al., 2009). Treatment of PrEC cells with PIWIL1 siRNA caused induction of LINE-1 ORF2 expression (Figures 6G and S8C). This finding raised the possibility that PIWIs might serve as an unexpected but key component of machinery that silences the expression of the LINE-1 encoded ORF2 in normal prostate epithelial cells. Indeed, knockdown of PIWIL1 enhanced the enrichment of γH2AX at ERG and TMPRSS2 intronic translocation regions in PrEC cells (Figure 6H). Despite its already low levels in LNCaP cells, further knockdown of PIWIL1 elevated the DHT+IR-induced intra- and interchromosomal translocations (Figure 6I), whereas overexpression of PIWIs in LNCaP cells largely inhibited the induced chromosomal translocations (Figure 6J). Serial dilution experiment confirmed that the actual translocation frequency rose to ~39×10−5 in LNCaP cell following PIWIL1 knockdown (Figure 6K).
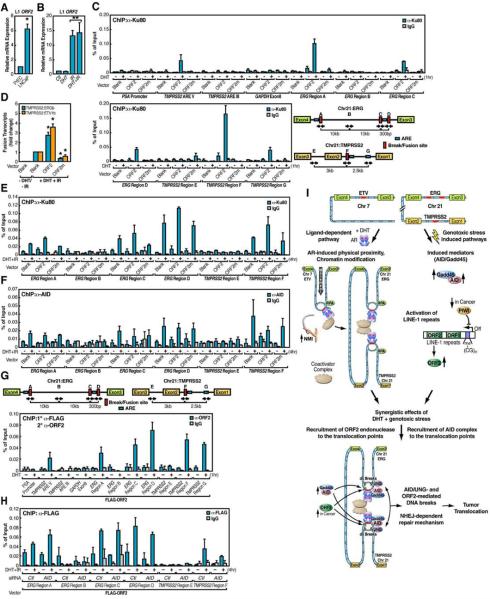
A series of observations including the close relationship between LINE-1 element and genome instability (Prak and Haoudi, 2006), the potent endonuclease activity of LINE-1-encoded ORF2 (Goodier et al., 2004), and the activation of LINE-1 retrotransposition by irradiation (Farkash et al., 2006), led us to address the possibility that expression of the LINE-1-encoded ORF2 endonuclease might itself exert a direct role in tumor translocation. Indeed, the relative level of LINE-1 ORF2 transcripts exhibited a significant (~6-fold) increase in LNCaP cells compared to PrEC cells, and ORF2 expression was robustly induced by genotoxic stress (Figures 7A and 7B). These data suggested that ORF2 might function as an additional irradiation/genotoxic stress-induced enzymatic machinery to mediate the chromosomal breaks involved in tumor translocations.
Figure 7. PIWI Regulated LINE-1 ORF2 Endonuclease Contributes to Chromosomal Translocations.
(A) The relative expression level of LINE-1 ORF2 in LNCaP and PrEC cells. (B) IR-dependent induction of LINE-1 ORF2 expression (n=3, ±SEM). (C) Overexpression of LINE-1 ORF2 enhances Ku80 enrichment at intronic ERG and TMPRSS2 break/fusion sites. (D) Examination of TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts in LNCaP cells electroporated with indicated plasmids (n=3, ±SEM). (E and F) ORF2 contributes to DSBs generation independent of AID. ChIP analyses with anti-Ku80 (E) or anti-AID (F) antibodies were performed in LNCaP cells electroporated with indicated plasmids (n=2, ±SEM). (G) Recruitment of LINE-1 ORF2 to translocation regions. ChIP analyses with anti-FLAG followed by anti-ORF2 antibodies were performed in LNCaP cells electroporated with FLAG-ORF2 plasmid (n=2, ±SEM). (H) The recruitment of ORF2 is independent of AID. ChIP analyses with anti-FLAG antibodies were performed in LNCaP cells electroporated with AID siRNA and FLAG-ORF2 plasmid (n=2, ±SEM). (I) Schematic illustration of molecular mechanisms of nuclear receptor-dependent non-random chromosomal translocations.
Consistent with this possibility, exogenous overexpression of LINE-1 ORF2 (Figure S14B) was sufficient to cause DHT-dependent DNA breaks at the established translocation sites, as revealed by specific recruitment of Ku80 to ERG and TMPRSS2 intronic translocation regions, but not at other AR binding regions, such as TMPRSS2 ARE III (Figure 7C). However, expression of an endonuclease-inactive mutant of ORF2 (Goodier et al., 2004) diminished the enrichment of Ku80 (Figure 7C). Furthermore, overexpression of the LINE-1 ORF2 induced a marked increase in both intra- and interchromosomal translocations whereas the endonuclease-inactive mutant partially blocked DHT+IR-induced translocations (Figure 7D). Interestingly, overexpression of the endonuclease-inactive mutant of ORF2 reduced the DSBs generation marked by Ku80 recruitment at ERG and TMPRSS2 intronic translocation regions in DHT+IR treated LNCaP cells, but with minimal effects on the recruitment of AID to the same regions (Figures 7E and 7F). These data suggest that the LINE-1 ORF2 contributes to DSBs generation, in combination with the parallel actions of AID.
Indeed, ChIP analysis revealed that LINE-1 ORF2 was selectively recruited in a DHT-dependent fashion to the translocation regions, as well as to the enhancer region of TMPRSS2, but not to the promoter of PSA (Figure 7G). However, we could not detect any direct physical interaction between AR and LINE-1 ORF2, suggesting the possibility that the recruitment of LINE-1 ORF2 may result from other events, including the AR-induced alterations in local chromatin architecture, and the exposed A/T-rich ssDNA that might then attract LINE-1 ORF2 to these chromatin regions. Importantly, the recruitment of ORF2 to translocation regions was not affected by removal of AID (Figure 7H), supporting the model that irradiation-induced LINE-1 ORF2 and AID act as independent mechanisms that combinatorially contribute to DSBs generation and tumor translocation. Thus, we conclude that activation of LINE-1 ORF2 expression in tumor cells appears to provide an additional endonuclease activity that makes an important quantitative contribution to ligand/genotoxic stress-induced tumor translocations. The functional role of LINE-1 ORF2 in chromosomal translocation has been further extended based on our findings that overexpression of LINE-1 ORF2 endonuclease-inactive mutant abolished DHT+IR-induced SLC45A3:ETV1 translocation (Figure S20E). Therefore, it is highly possible that LINE-1 ORF2 has a general function in facilitating chromosomal breaks and global genome instability in cancer cells.
Discussion
A long-standing concept in tumor translocations has been that genotoxic stress causes direct, random DSBs that lead to random translocations, with selection of those conferring growth advantage. Here, by devising and investigating a model of tumor translocations that fully mimics the frequency of in vivo events without proliferative selection, we suggested that, rather, there is a site-selective immediate pattern of DSBs that ultimately dictate the pattern of tumor translocations. This system has permitted the identification of several unexpected mechanisms, some sequential and others combinatorial, by which the androgen receptor acts in concert with genotoxic stress as parallel pathways to direct rapid, site-specific and cell type-specific tumor translocations in the absence of proliferative selection (Figure 7I). Translocation breakpoint preferences are dictated both by (i) site-specific binding of liganded-AR at intronic translocation regions that trigger rapid intra- and interchromosomal interactions, which results in spatial proximity required for translocations, and (ii) induced alterations of local chromatin architecture are permissive for sensitizing these regions to genotoxic stress. These transcription-related events, in combination with activation and induction of specific enzymatic machineries by ligand and genotoxic stress, mechanistically underlie the extended DNA breaks at the critical intronic AR binding sites. Thus, rather than being entirely the consequence of random damage-induced DSBs, with selection for the resulting tumor translocations that confer putative growth advantages as is commonly hypothesized, there appears to have an initial selective sensitization in a site-specific manner that presages the observed tumor translocations in prostate cancer. Therefore our findings suggest the provocative concept that liganded-receptor acts, in a sense, as a mutagen, licensed by the enzymatic machinery activated by genotoxic stress.
One initial puzzle was how the various TMPRSS2 fusion partners were brought together for DNA recombination. We found that AR bound to multiple intronic regions near break sites in TMPRSS2, ERG and ETV1 genes, juxtaposing DNA breaks for subsequent recombination. This spatial proximity induced by a specific transcription program is reminiscent of the DNA rearrangement in developing B cells, thus suggesting that a general strategy is exploited for DNA rearrangement in tumorigenesis as well as during normal development. The requirements of a nuclear motor for the induced proximity permitted our demonstration that the induction of this spatial proximity is actually prerequisite for the induced translocation events.
Our data indicate that AR acts in concert with a number of key enzymes induced by androgens and genotoxic stress, including AID, previously considered to be a B cell-specific factor (Chaudhuri et al., 2003), Gadd45 and quite unexpectedly, the LINE-1-encoded ORF2, to license extended DSBs. AID is recruited to the intronic AR binding sites as part of the genotoxic stress-induced Gadd45/AID complex. These recruitments are postulated to be permissive for potentiating the occurrence of specific translocation breakpoints that we have mapped within the TMPRSS2, ERG and ETV1 genes. Our data further suggest that only ~20% of intronic AR binding sites exhibiting induced DSBs. We have further documented that these breaks are marked by the recruitment of Ku80 and by direct incorporation of BrdU at these sites. The presence of micro-homology in multiple TMPRSS2:ERG and TMPRSS2:ETV1 fusion sites lends further support for the model that there are staggered sites of DNA breakage as might be expected from the sequential actions of AID and UNG, analogous to their predicted functions in CSR (Stavnezer et al., 2008). Further, we observed the mutations characteristic of somatic hypermutation within the short region of AID binding sites, reflecting actions of AID at these intronic break sites. Consistent with our findings, AID has been recently reported to be required for c-myc:IgH translocations in B cells (Robbiani et al., 2008).
A key challenge was to identify specific machineries altered in tumors that might sensitize the cell to tumor translocations. The observation that the DHT+IR induced chromosomal translocations observed in prostate cancer cells were not detected in normal prostate epithelial cells led to the discovery of the critical, independent role of the PIWI-regulated LINE-1 encoded-ORF2 endonuclease in translocation events. Thus, in addition to the induction of AID in prostate cancer cells, we have uncovered a second, unexpected component of this sensitization mechanism based on the diminished expression of PIWIs, which serves as a dedicated protective strategy to block retrotransposition of LINE-1 elements to ensure genome stability in germ cells (O'Donnell and Boeke, 2007). Surprisingly, we found that the level of PIWIL1 was dramatically lower (>9-fold) in LNCaP cells than in normal prostate epithelial cells. Even at this low level in LNCaP cells, knockdown with specific siRNA caused a further deprotection of the genome, resulting in increased chromosomal translocations.
To our surprise, even in the absence of genotoxic stress, the ORF2 endonuclease appears capable of targeting to the intronic AR bound sites to license DNA breakage, which emphasizes its independent, parallel function to AID in generating DSBs at the translocation sites. Our findings are consistent with the reported elevation of the LINE-1 ORF2 endonuclease activity in prostate cancer and the inducibility of this enzyme by irradiation (Farkash et al., 2006; Santourlidis et al., 1999), and also with the documented role of PIWIs in silencing LINE-1 repeat movement during neurogenesis (Muotri et al., 2005).
Because androgens exert important developmental roles in the prostate, we suggest that similar events might occur in other cancer types in which a selective, regulated DNA binding transcription factor(s) serves to promote DNA rearrangement. Finally, understanding the molecular mechanisms that underlie tumor translocations and specific “epigenetic” strategies used by normal cells to protect the genome against such deleterious DNA rearrangements have the promise to provide insights into the etiology of cancer and to facilitate the development of new diagnostic/therapeutic approaches.
Experimental Procedures
Cell Lines and Treatments
The prostate cancer cell line LNCaP was obtained from the American Type Culture Collection (ATCC). Human Prostate Epithelial Cells (PrEC) were obtained from Lonza and maintained in PrEGM media. Transfection of LNCaP cells with siRNA and plasmid DNA were performed using Lipofectamine2000™ (Invitrogen) and Nucleofector® Kit R (Lonza), respectively. Transfection of PrEC cells with siRNA was performed using DeliverX Plus siRNA transfection reagent (Panomics). MVA/T7RP-mediated expression of LINE-1 proteins was performed as previous described (Goodier et al., 2004). For induction of chromosomal translocation, LNCaP or PrEC cells were grown in charcoal-stripped serum containing media for 48 hr followed by mock, DHT (10−7 M), γ-irradiation (50 Gy) treatment, or both. After treatments, the cells were re-incubated for 24hr before being harvested for appropriate assays. Cell viability in cultures before and after irradiation was assessed by trypan blue exclusion.
Fluorescence in situ Hybridization
PrEC or LNCaP cells were stimulated with vehicle or 10−7 M of DHT for 1hr. Cell nuclei isolation and DNA-FISH was carried out according to a method previously described (Hu et al., 2008), and the probes were synthesized by Integrated DNA Technologies (IDT) and listed in Supplemental Table 1.
DNA-break Labeling and Chromatin Immunoprecipitation (ChIP)
The labeling of transient DNA break(s) by BrdU was performed as described (Ju et al., 2006). ChIP and Re-ChIP were performed as described previously (Shang et al., 2000). The enrichment of the DNA template was analyzed by QPCR using primers listed in Supplemental Table 1.
Single-Cell Nuclear Microinjection Assays
The single cell nuclear microinjection assays were performed as described (Perissi et al., 2004).
Data Analysis and Statistics
Relative quantities of TMPRSS2:ERG and TMPRSS2:ETV1 fusion transcript were normalized to Actin and further to the expression level of TMPRSS2. The relative amount of each fusion transcript was then calibrated to DHT alone or DHT+IR treated control sample as appropriately. Calibrated quantities are indicated and samples without detectable fusion transcript after 40 cycles of amplification are indicated by 0 (Tomlins et al., 2007). Results are reported as mean ± SEM of three independent experiments. Comparisons were performed using two-tailed paired Student t-test. *p<0.05, **p<0.01, ***p<0.001.
Supplementary Material
Acknowledgements
We thank C. Nelson for various cell culture assistance; J. Hightower for artwork; D. Benson for assistance with the manuscript; Dr. V. Lunyak and Dr. X. Zhu for critical comments; Dr. M. Schwartz and E. Nunez for assistance with fluorescence microscopy and FISH technology; Dr. Y. Zhang for providing the FLAG-DOT1L construct, Dr. M. B. Kastan for providing HA-ER-I-PpoI construct, and Dr. J. L. Goodier for providing LINE-1 ORF1, ORF2 constructs, Ankara vaccinia virus that expresses T7 polymerase and polyclonal antibody against ORF2. Dr. S. Heinz for providing PIWIL1 and PIWIL2 expression constructs, and AstraZeneca for providing Casodex®. M.G.R. is a Howard Hughes Medical Institute Investigator. This study was funded by grants from NIH/NCI , DoD and Prostate Cancer Foundation to M.G.R., X.D.F. and C.K.G. C. Lin is a Fellow of Susan G. Komen for the Cure (KG080247), and L. Yang is an awardee of Era of Hope Postdoctoral Award supported by DoD (GRANT00325108).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- Aplan PD. Causes of oncogenic chromosomal translocation. Trends Genet. 2006;22:46–55. doi: 10.1016/j.tig.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Science. Vol. 316. New York, NY: 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control; pp. 744–747. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Smith S, Oum JH, Liaw HJ, Hwang JY, Sikdar N, Motegi A, Lee SE, Myung K. Mph1p promotes gross chromosomal rearrangement through partial inhibition of homologous recombination. The Journal of cell biology. 2008;181:1083–1093. doi: 10.1083/jcb.200711146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Jr., Kastan MB. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nature protocols. 2008;3:915–922. doi: 10.1038/nprot.2008.54. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Current opinion in structural biology. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Corral J, Lavenir I, Impey H, Warren AJ, Forster A, Larson TA, Bell S, McKenzie AN, King G, Rabbitts TH. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Current opinion in cell biology. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends in biochemical sciences. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- Deininger MW, Bose S, Gora-Tybor J, Yan XH, Goldman JM, Melo JV. Selective induction of leukemia-associated fusion genes by high-dose ionizing radiation. Cancer research. 1998;58:421–425. [PubMed] [Google Scholar]
- Farkash EA, Kao GD, Horman SR, Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic acids research. 2006;34:1196–1204. doi: 10.1093/nar/gkj522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Engleka KA, Seleme MC, Kazazian HH., Jr. A potential role for the nucleolus in L1 retrotransposition. Human molecular genetics. 2004;13:1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nature reviews. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- Grummt I. Actin and myosin as transcription factors. Current opinion in genetics & development. 2006;16:191–196. doi: 10.1016/j.gde.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocrine reviews. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- Hofmann WA, de Lanerolle P. Nuclear actin: to polymerize or not to polymerize. The Journal of cell biology. 2006;172:495–496. doi: 10.1083/jcb.200601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A. Methods in molecular biology. Vol. 161. Clifton, NJ: 2001. Jasplakinolide. An actin-specific reagent that promotes actin polymerization; pp. 109–120. [DOI] [PubMed] [Google Scholar]
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annual review of immunology. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA, Jr., Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. Science. Vol. 312. New York, NY: 2006. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription; pp. 1798–1802. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nature reviews. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Lucas JM, True L, Hawley S, Matsumura M, Morrissey C, Vessella R, Nelson PS. The androgen-regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. The Journal of pathology. 2008;215:118–125. doi: 10.1002/path.2330. [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Reid G, Gannon F. Transcription in four dimensions: nuclear receptor-directed initiation of gene expression. EMBO reports. 2006;7:161–167. doi: 10.1038/sj.embor.7400626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- Neves H, Ramos C, da Silva MG, Parreira A, Parreira L. The nuclear topography of ABL, BCR, PML, and RARalpha genes: evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood. 1999;93:1197–1207. [PubMed] [Google Scholar]
- Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova MN, Stringer JR, Blough R, Medvedovic M, Fagin JA, Nikiforov YE. Science. Vol. 290. New York, NY: 2000. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells; pp. 138–141. [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Current opinion in genetics & development. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Prak NL, Haoudi A. LINE-1 Retrotransposition: Impact on Genome Stability and Diversity and Human Disease. Journal of biomedicine & biotechnology. 2006;2006:37049. doi: 10.1155/JBB/2006/37049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K, von Boehmer H. Lymphocyte development: overview. Current opinion in immunology. 2008;20:127–130. doi: 10.1016/j.coi.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk RS, Walczak CE. Chromosome dynamics: actin's gone fishing. Curr Biol. 2005;15:R841–842. doi: 10.1016/j.cub.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nature genetics. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. The Prostate. 1999;39:166–174. doi: 10.1002/(sici)1097-0045(19990515)39:3<166::aid-pros4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Shaffer DR, Pandolfi PP. Breaking the rules of cancer. Nature medicine. 2006;12:14–15. doi: 10.1038/nm0106-14. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Science. Vol. 311. New York, NY: 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions; pp. 844–847. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annual review of immunology. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Science. Vol. 310. New York, NY: 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer; pp. 644–648. [DOI] [PubMed] [Google Scholar]
- Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer research. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Molecular cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Moncman CL, Winkelmann DA. Mutations in the motor domain modulate myosin activity and myofibril organization. Journal of cell science. 2003;116:4227–4238. doi: 10.1242/jcs.00709. [DOI] [PubMed] [Google Scholar]
- Yi YW, Kim D, Jung N, Hong SS, Lee HS, Bae I. Gadd45 family proteins are coactivators of nuclear hormone receptors. Biochemical and biophysical research communications. 2000;272:193–198. doi: 10.1006/bbrc.2000.2760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.