Abstract
Clostridium difficile is the major cause of infectious diarrhea and a major burden to health care services. The ability of this organism to form endospores plays a pivotal role in infection and disease transmission. Spores are highly resistant to many forms of disinfection and thus are able to persist on hospital surfaces and disseminate infection. In order to cause disease, the spores must germinate and the organism must grow vegetatively. Spore germination in Bacillus is well understood, and genes important for this process have recently been identified in Clostridium perfringens; however, little is known about C. difficile. Apparent homologues of the spore cortex lytic enzyme genes cwlJ and sleB (Bacillus subtilis) and sleC (C. perfringens) are present in the C. difficile genome, and we describe inactivation of these homologues in C. difficile 630Δerm and a B1/NAP1/027 clinical isolate. Spores of a sleC mutant were unable to form colonies when germination was induced with taurocholate, although decoated sleC spores formed the same number of heat-resistant colonies as the parental control, even in the absence of germinants. This suggests that sleC is absolutely required for conversion of spores to vegetative cells, in contrast to CD3563 (a cwlJ/sleB homologue), inactivation of which had no effect on germination and outgrowth of C. difficile spores under the same conditions. The B1/NAP1/027 strain R20291 was found to sporulate more slowly and produce fewer spores than 630Δerm. Furthermore, fewer R20291 spores germinated, indicating that there are differences in both sporulation and germination between these epidemic and nonepidemic C. difficile isolates.
The Gram-positive anaerobe Clostridium difficile causes diarrheal diseases ranging from asymptomatic carriage to a fulminant, relapsing, and potentially fatal colitis (8, 30). This organism is resistant to various broad-spectrum antibiotics and capitalizes on disruption of the normal intestinal flora to colonize and cause disease symptoms through the action of toxins A and B (16, 40). While these toxins are the principal virulence factors, the ability of the organism to produce endospores is necessary for disease transmission.
Clostridial spores are extremely resistant to all kinds of chemical and physical agents and provide the mechanism by which C. difficile can evade the potentially fatal consequences of exposure to heat, oxygen, alcohol, and certain disinfectants (35). Thus, the spores shed in fecal matter are very difficult to eradicate and can persist on contaminated surfaces in health care facilities for extended periods of time (35). This leads to infection or reinfection of cohabitating individuals through inadvertent ingestion of infected material (10, 32). Once in the anaerobic environment of the gut, spores presumably germinate to form toxin-producing vegetative cells and, in susceptible individuals, diarrheal disease.
Spore germination is defined as the events that result in the irreversible loss of spore characteristics. However, current mechanistic knowledge of the germination process is based principally on data derived from studying Bacillus subtilis. Little is known about spore germination in clostridia and, in particular, in C. difficile. Germination is initiated when the bacterial spore senses specific effectors, termed germinants. These effectors can include nutrients, cationic surfactants, peptidoglycan, and a 1:1 chelate of pyridine-2,6-dicarboxylic acid (dipicolinic acid) and Ca2+ (CaDPA) (23, 34, 36). Spores of B. subtilis can germinate in response to nutrients through the participation of three sensory receptors located in the spore inner membrane, GerA, GerB, and GerK (23). After activation, the events include the release of monovalent cations (H+, K+, and Na+) and CaDPA (accounting for approximately 10% of the spore dry weight) (36). The third major step in germination involves hydrolysis of the spore peptidoglycan (PG) cortex. It is during this hydrolysis that the previously low water content of the spore is restored to the water content of a normal vegetative cell and the core is able to expand, which in turn allows enzyme activity, metabolism, and spore outgrowth (36).
CwlJ and SleB are two specific spore cortex-lytic enzymes (SCLEs) involved in Bacillus cortex hydrolysis, which break down PG containing muramic-δ-lactam (28). SleB has been shown to localize in both the inner and outer layers of B. subtilis spores through interaction of the enzyme peptidoglycan-binding motif and the δ-lactam structure of the cortex (7, 19) and in association with YpeB, which is required for sleB expression during sporulation (4, 7). SleB is a lytic transglycosylase muramidase, but so far its mode of activation is unknown (21). CwlJ is localized to the spore coat during sporulation (3) and is required for CaDPA-induced germination in B. subtilis. Activation can be due to either CaDPA released from the spore core at the onset of germination or exogenous CaDPA (22). Neither enzyme is individually essential for complete cortex hydrolysis during nutrient germination, although inactivation of both cwlJ and sleB in B. subtilis results in a spore unable to complete this process (15). The role of SleL has recently been studied in Bacillus anthracis. Mutants unable to produce this enzyme are still able to germinate, but the process is retarded (18).
The SCLEs of Clostridium are less well studied than those of Bacillus. The SCLEs SleC (20) and SleM (6) have been identified in Clostridium perfringens, and a recent study demonstrated that SleC is required during germination for complete cortex hydrolysis (26). Although SleM can degrade spore cortex peptidoglycan and inactivation of both sleC and sleM decreased the ability of spores to germinate more than inactivation of sleC alone did, SleM was not essential (26). It has also been shown that the germination-specific serine protease CspB is essential for cortex hydrolysis and converts the inactive pro-SleC found in dormant spores to an active enzyme (24). So far, there has been no detailed study of any gene responsible for spore germination in C. difficile, although genes showing homology to cwlJ and sleB of B. subtilis (CD3563) and sleC of C. perfringens (CD0551) have now been identified in the C. difficile 630 genome (33).
With germinant receptors in C. difficile yet to be identified, the mechanism by which the spores sense a suitable environment for germination is unclear. Recent studies have suggested that this process may involve the interaction of C. difficile with bile. Taurocholate has been shown to enhance recovery of C. difficile spores in nutrient-rich medium (42), and it has been proposed that glycine and taurocholate act as cogerminants (38), while chenodeoxycholate inhibits C. difficile spore germination (39).
The emergence of C. difficile B1/NAP1/027 strains has increased the burden on health care services worldwide. Such strains have been shown to produce higher levels of toxin in the laboratory than many other types of strains (41), although the mechanism behind this production is not fully understood. However, while the observed higher levels of toxin production is doubtless important, perhaps the recent attention given to B1/NAP1/027 strains has focused too much on toxins. As spores represent the infectious stage of C. difficile, processes such as spore germination may also contribute to the greater virulence of these strains. In this study we evaluated the sporulation and germination efficiencies of an “epidemic” B1/NAP1/027 C. difficile strain (R20291, isolated from the Stoke Mandeville outbreak in 2004 and 2005) and the “nonepidemic” strain 630Δerm (14). We then constructed strains with mutations in CD3563 (a cwlJ/sleB homologue) and a sleC homologue to analyze the role of these genes in the germination of C. difficile spores.
MATERIALS AND METHODS
Strains and growth conditions.
Unless indicated otherwise, all C. difficile strains were grown at 37°C in an anaerobic workstation (Don Whitley, United Kingdom) in BHIS (brain heart infusion supplemented with l-cysteine [0.1%; Sigma, United Kingdom] and yeast extract [5 mg/ml; Oxoid]) broth or agar, which has been shown to aid C. difficile sporulation (38). All Escherichia coli strains were grown using Luria-Bertani broth or agar at 37°C. Plasmid DNA was transferred into C. difficile 630Δerm and R20291 by conjugation from an E. coli donor, as previously described (13, 31). Table 1 shows the strains used in this study.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TOP10 | Invitrogen | |
| E. coli CA434 | Conjugation donor | 31 |
| CRG789 | C. difficile 630Δerm spo0A::intron ermB | 12 |
| C. difficile 630Δerm | 14 | |
| C. difficile R20291 | Stoke Mandeville (2004-2005) isolate | Anaerobe Reference Laboratory, Cardiff, Wales |
| CRG878 | C. difficile 630Δerm CD3563::intron ermB | This study |
| CRG1115 | C. difficile 630Δerm sleC::intron ermB | This study |
| CRG1166 | C. difficile R20291 sleC::intron ermB | This study |
| CRG1375 | C. difficile R20291 spo0A::intron ermB | S. A. Kuehne and N. P. Minton, unpublished data |
| CRG1555 | CRG1115 containing pMTL-DB1 (sleC complementation plasmid) | This study |
| CRG1556 | CRG1115 containing pMTL84151 | This study |
| CRG1628 | CRG1166 containing pMTL84151 | This study |
| CRG1634 | CRG1166 containing pMTL-DB1 (sleC complementation plasmid) | This study |
| CRG1651 | C. difficile 630Δerm containing pMTL84151 | This study |
| CRG1652 | C. difficile R20291 containing pMTL84151 | This study |
| Plasmids | ||
| pMTL007C-E2 | ClosTron plasmid containing catP and intron containing ermB RAM | 11 |
| pMTL84151 | Clostridium modular plasmid containing catP | 13 |
| pMTL-DB1 | pMTL84151 containing 1,272-bp SleC coding region and 244-bp upstream promoter region | This study |
Sporulation of C. difficile was achieved by incubating cultures anaerobically in BHIS broth for 5 days at 37°C. To ensure that no spores were present when the sporulation medium was inoculated, a starter culture was prepared in BHIS broth using a 1% inoculum of a C. difficile culture and incubated until the optical density at 600 nm (OD600) was between 0.2 and 0.5. The sporulation medium was then inoculated with 0.01 volume of this exponential starter culture. To measure the growth rates of C. difficile strains, 1-ml samples were removed from the sporulation medium at different time points and the OD600 was measured (Biomate 3; Thermo Scientific).
Measurement of C. difficile heat-resistant colony formation.
Sporulating cultures of C. difficile were prepared as described above. At different times, samples (500 μl) were removed from the anaerobic chamber and heated at 60°C for 25 min to kill the vegetative cells but not the spores. To control for any effects of oxygen exposure during heat treatment, a nonheated sample was also removed from the anaerobic chamber for 25 min. Samples were then returned to the anaerobic chamber, serially diluted in phosphate-buffered saline (PBS), and plated onto BHIS agar supplemented with 0.1% taurocholate (Sigma, United Kingdom). C. difficile spo0A mutants of both 630Δerm and R20291 were used as sporulation-negative controls (12). Plates were incubated for 24 h before CFU were enumerated. Samples were analyzed in the same way every 24 h for 5 days. The total sporulation after 5 days was measured by counting spores by phase-contrast microscopy using a Bright-Line hemocytometer (Sigma, United Kingdom).
Assaying the viability of C. difficile spores by decoating.
Sporulation cultures were set up as described above. To prepare pure C. difficile spores following 5 days of incubation, cultures were repeatedly washed with ice-cold distilled H2O (dH2O) until they contained no cell debris or vegetative cells as observed by phase-contrast microscopy. Spores were stored at −20°C in dH2O.
Purified spores were decoated by resuspending a spore pellet in 1 ml of 50 mM Tris-HCl (pH 8.0)-8 M urea-1% (wt/vol) sodium dodecyl-sulfate-50 mM dithiothreitol and incubating the preparation at 37°C for 90 min as described by Popham and coworkers (29). Following incubation, decoated spores were washed three times in PBS. Samples were then heat treated as described above and plated onto BHIS medium supplemented with lysozyme (1 μg/ml).
Strain construction.
Target genes were insertionally inactivated with the ClosTron system as previously described (12), using modular ClosTron plasmid pMTL007C-E2 (11). Table 2 lists the oligonucleotides used for construction of insertional mutants. Erythromycin (2.5 μg/ml) was used to select C. difficile 630Δerm integrants, while lincomycin (20 μg/ml) was used for selection of C. difficile R20291 integrants (Table 3). Mutants were confirmed by PCR and sequencing.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| Intron retargeting | |
| Cdi-CD3563-226s-IBS | AAAAAAGCTTATAATTATCCTTAATGAGCGACAGGGTGCGCCCAGATAGGGTG |
| Cdi-CD3563-226s-EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCGACAGGTTTAACTTACCTTTCTTTGT |
| Cdi-CD3563-226s-EBS2 | TGAACGCAAGTTTCTAATTTCGGTTCTCATCCGATAGAGGAAAGTGTCT |
| Cdi-sleC-493s-IBS | AAAAAAGCTTATAATTATCCTTAGTAGTCCCTGAAGTGCGCCCAGATAGGGTG |
| Cdi-sleC-493s-EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCCCTGAATTTAACTTACCTTTCTTTGT |
| Cdi-sleC-493s-EBS2 | TGAACGCAAGTTTCTAATTTCGATTACTACTCGATAGAGGAAAGTGTCT |
| Cdi-sleC-128a-IBS | AAAAAAGCTTATAATTATCCTTACATTACTTCTTAGTGCGCCCAGATAGGGTG |
| Cdi-sleC-128a-EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCTTCTTAGGTAACTTACCTTTCTTTGT |
| Cdi-sleC-128a-EBS2 | TGAACGCAAGTTTCTAATTTCGGTTTAATGTCGATAGAGGAAAGTGTCT |
| EBS Universal | CGAAATTAGAAACTTGCGTTCAGTAAAC |
| ClosTron sequencing | |
| Spofdx_F1 | GATGTAGATAGGATAATAGAATCCATAGAAAATATAGG |
| pMTL007_R1 | AGGGTATCCCCAGTTAGTGTTAAGTCTTGG |
| Mutant screening | |
| CD3563_F1 | CTTTTAGAACTGTTAATCCACCTAATCCCG |
| CD3563_R1 | CTTTACATTTTTGTGTTTAACAACAACTTATTTATCGC |
| sleC_F1 | GGGAACTAAATTCATTTAAAGAAAGGGTG |
| sleC_R1 (630Δerm) | GGCTGTTATGAACTAATATATACCATAAGTATTAC |
| sleC_R1 (R20291) | GTATTTTTACTTATAAGTATTGTAGTCTTAACAGCC |
| Complementation | |
| pSleC_F1 | TAAAGAATGCGGCCGCAGATTATTTTCCTTTCAAAATTTTTGATTTATTTATGATTTATATCATCTAC |
| pSleC_R1 | TAAAGAATCATATGATCACCCTTTCTTTAAATGAATTTAGTTCCC |
| SleC_F2 | TAAAGAATCATATGCAAGATGGTTTCTTAACAGTAAGCATAATTGATGC |
| SleC_R2 | TAAAGAATCTCGAGATCTCCATGGTTAAATTAAAGGATTTAAAGAAGCTATTCTAGTTGTAGC |
TABLE 3.
ClosTron insertion frequencies with erythromycin or lincomycin selection
| Strain | Target sitea | Frequency of desired mutant among clones screenedb |
|
|---|---|---|---|
| % | No. positive/no. screened | ||
| C. difficile 630Δerm CD3563 | 226s | 100 | 8/8 |
| C. difficile 630Δerm sleC | 493s | 0 | 0/20c |
| C. difficile 630Δerm sleC | 128a | 100 | 5/5 |
| C. difficile R20291 sleC | 128a | 100 | 4/4 |
Introns were inserted after the indicated number of bases from the start of the open reading frame in either the sense (s) or antisense (a) orientation.
Genomic DNA was extracted from erythromycin-resistant (630Δerm) or lincomycin-resistant (R20291) clones at random and used in a PCR to amplify the intron-exon junction. One clone of each desired mutant was selected, and the intron insertion site was verified by sequencing.
Further screening of a pool of genomic DNA from >100 erythromycin-resistant clones indicated that no desired mutants were present using the base 493 target site. The intron or target site was therefore judged to be inefficient (as occasionally occurs using group II intron technology), so another target (base 128 with the antisense orientation) was chosen for sleC.
For complementation studies, a 1,516-bp fragment encompassing the sleC structural gene and 5′ noncoding region was cloned into plasmid pMTL84151 (13) to generate plasmid pMTL-DB1. The 244-bp 5′ noncoding region likely encompassing the sleC promoter and the 1,272-bp region containing the sleC structural gene were independently amplified by PCR using oligonucleotide primer pairs pSleC_F1/pSleC_R1 and SleC_F2/SleC_R2, respectively. The primers were designed to allow subsequent cleavage of the two fragments generated with NotI/NdeI and NdeI/XhoI, respectively, where the ATG of the NdeI site was synonymous with the translational start codon of sleC. The two cleaved fragments were subsequently ligated with plasmid pMTL84151 cut with NotI and XhoI, which yielded plasmid pMTL-DB1, in which the two fragments were inserted contiguously. The strategy of separating the two regions into two independent fragments was adopted to allow subsequent use of a heterologous promoter if the presence of the 5′ noncoding region did not result in expression of sleC. As the protein encoded by sleC is the same in C. difficile 630Δerm and R20291, only one plasmid (pMTL-DB1) was constructed for use with both mutants.
Both the pMTL-DB1 complementation plasmid and a pMTL84151 empty vector control were transferred into C. difficile by conjugation, using additional lincomycin (20 μg/ml) selection in sleC mutant strains.
RESULTS
Comparison of sporulation and germination of C. difficile 630Δerm and C. difficile R20291.
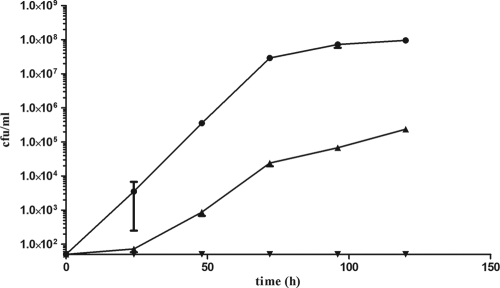
It has been suggested that B1/NAP1/027 “epidemic” strains of C. difficile may have different sporulation characteristics than nonepidemic strains (1, 9), although the evidence for this is currently limited. As sporulation is important in the study of spore germination, we decided to study C. difficile germination both in a B1/NAP1/027 strain, R20291, and in a nonepidemic strain, 630Δerm. First, we compared the differences in sporulation and germination between these two strains using a series of assays. In this analysis, the development of heat-resistant CFU was observed over a 5-day period (Fig. 1). At different times, samples were heated at 60°C for 25 min before they were plated onto BHIS agar supplemented with 0.1% taurocholate. The resultant colony formation clearly showed that C. difficile 630Δerm developed heat-resistant spores more rapidly than R20291. 630Δerm formed countable heat-resistant CFU after 24 h of incubation, while it was only after 48 h that this was the case for R20291. In addition, 630Δerm cultures contained approximately 400-fold more heat-resistant CFU than R20291 cultures after 5 days.
FIG. 1.
Development of C. difficile heat-resistant CFU over a 5-day period. Cultures of C. difficile 630Δerm (•), C. difficile R20291 (▴), and CRG789 (spo0A) (▾) were incubated anaerobically at 37°C in BHIS broth. At 24-h intervals, samples were heat treated at 60°C for 25 min before they were plated onto BHIS agar supplemented with 0.1% taurocholate and incubated at 37°C for 24 h. The numbers of heat-resistant CFU per milliliter were determined. The symbols indicate the averages of three independent experiments, and the error bars indicate the standard errors of the means. The detection limit for the assay was 50 CFU/ml.
It is possible that the observed difference in heat resistance was not due to sporulation or germination and instead was a result of growth differences between the two strains. The data for non-heat-treated CFU recovered on BHIS agar supplemented with taurocholate after 5 days were also compared, and as the colony formation was found to be 30-fold lower for R20291 than for 630Δerm (Fig. 2), the changes in OD600 for both 630Δerm and R20291 were analyzed over a 5-day period. No difference was seen between the two strains (data not shown), which ruled out an elementary growth difference between 630Δerm and R20291 and suggested that the observed difference in the number of non-heat-treated CFU for R20291 was due to the death of nonsporulating cells.
FIG. 2.
Numbers of untreated CFU, spores, and heat-resistant CFU of C. difficile 630Δerm and C. difficile R20291 after 5 days of incubation. Following 5 days of anaerobic incubation at 37°C in BHIS broth, the numbers of heat-resistant CFU were determined. Spore titers were determined by counting spores by phase-contrast microscopy using a Bright-Line hemocytometer (Sigma, United Kingdom). The bars indicate the averages of three independent experiments, and the error bars indicate standard errors of the means. The detection limit for colony counts was 50 CFU/ml, and the detection limit for spore counts was 1.6 × 103 spores.
To pinpoint any difference in sporulation between the two strains, the cultures used to measure development of heat resistance were analyzed by phase-contrast microscopy, and the numbers of spores per milliliter were determined. C. difficile 630Δerm was found to produce approximately 100-fold more spores per milliliter than R20291 (Fig. 2), suggesting that the previously observed difference in heat resistance between the two strains was due in part to sporulation. However, as 630Δerm produced 400-fold more heat-resistant CFU than R20291, we also hypothesized that fewer R20291 spores than 630Δerm spores germinated under the conditions used for our assay. To test the hypothesized differences in germination between 630Δerm and R20291, the observed spore titers were compared to the numbers of heat-resistant CFU on BHIS agar supplemented with taurocholate. It was found that a greater proportion of 630Δerm spores (97%) than of R20291 spores (30%) formed heat-resistant colonies (Fig. 2). Thus, our data suggest not only that the nonepidemic strain C. difficile 630Δerm sporulates earlier and to a greater degree than the B1/NAP1/027 isolate R20291 under the growth conditions employed but also that a higher proportion of 630Δerm spores than of R20291 spores form colonies in association with the bile salt taurocholate.
Heat-resistant colony formation by C. difficile mutant cultures supplemented with taurocholate.
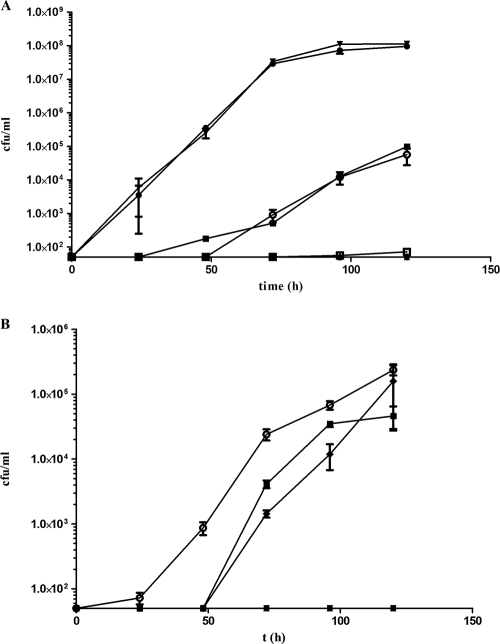
Previous studies have identified CwlJ and SleB as proteins that are important for cortex hydrolysis in B. subtilis (15), while SleC is essential for the same process in C. perfringens (26). Homologues are present in C. difficile 630 (33); the product of CD0551 (annotated as sleC) shows 53% amino acid identity to the previously characterized SleC of C. perfringens (17, 26), and the product of CD3563 shows 30% and 45% amino acid identity to B. subtilis CwlJ and SleB, respectively. Examination of an incomplete genome sequence available at http://www.sanger.ac.uk indicated that equivalent homologues are also present in C. difficile R20291. To assess the importance of these genes in C. difficile germination, the ClosTron system (12) was used to create independent insertional mutants of C. difficile 630Δerm in which either the CD3563 or sleC gene was inactivated, yielding strains CRG878 and CRG1115, respectively (Table 3 shows intron insertion sites and the frequencies of the desired mutants obtained). These strains were then tested to determine their abilities to form heat-resistant colonies over a 5-day period on BHIS agar supplemented with 0.1% taurocholate (Fig. 3A). Although inactivation of CD3563 had no effect on colony formation, a sleC mutant was unable to form heat-resistant CFU at any point over the 5-day period. The data suggest that sleC is absolutely required for sporulation and/or taurocholate-induced germination in C. difficile 630Δerm, while CD3563 plays no obvious role in either process.
FIG. 3.
Development of heat-resistant CFU of C. difficile mutants over 5 days. Cultures of C. difficile were incubated anaerobically at 37°C in BHIS broth. Every 24 h, samples were heat treated at 60°C for 25 min before they were plated onto BHIS agar supplemented with 0.1% taurocholate and incubated at 37°C for 24 h. The numbers of heat-resistant CFU per milliliter were determined. The symbols indicate the averages of three independent experiments, and the error bars indicate standard errors of the means. The detection limit for the assay was 50 CFU/ml. (A) •, C. difficile 630Δerm; ▴, CRG1115 (sleC); ○, CRG1555 (CRG1115 complemented with parental sleC gene); □, CRG1556 (CRG1115 harboring pMTL84151 empty vector control); ▪, CRG1651 (C. difficile 630Δerm harboring pMTL84151); ▾, CRG878 (CD3563); ⧫, CRG789 (spo0A). (B) (○), C. difficile R20291; •, CRG1166 (sleC); ▪, CRG1634 (CRG1166 complemented with parental sleC); ▾, CRG1628 (CRG1166 harboring pMTL84151); ⧫, CRG1652 (C. difficile R20291 harboring pMTL84151); ▴, CRG1375 (spo0A). All data points for CRG1115 and CRG789 in panel A and for CRG1166, CRG1628, and CRG1375 in panel B are overlaid because there was no heat-resistant colony formation.
In light of our findings of sporulation and germination differences between C. difficile 630Δerm and R20291, we analyzed the effect of sleC inactivation in both strains. In this analysis, an equivalent ClosTron-derived sleC mutant of C. difficile R20291 (CRG1166) was constructed (Table 3 shows the intron insertion site and frequency of mutants obtained), and the effect on colony formation was determined using the conditions employed for strain 630Δerm. This mutant exhibited the same phenotype as its 630Δerm counterpart (Fig. 3B), suggesting that the role of sleC in C. difficile is the same in both strains.
To ensure that the observed phenotypes of the sleC mutants of 630Δerm and R20291 did not result from growth deficiencies, the change in OD600 was used to monitor growth over 5 days. The growth of both mutants was indistinguishable from the growth of their parental strains (data not shown), suggesting that there were no obvious growth defects.
Complementation of sleC mutant with parental SleC.
In order to show that the observed phenotypes of CRG1115 and CRG1166 were a specific consequence of sleC inactivation, we performed complementation studies through construction of plasmid pMTL-DB1 carrying the parental sleC structural gene and the 244-bp region immediately upstream of the open reading frame presumed to contain its promoter. Strains CRG1555 and CRG1634 were created by introducing this plasmid into sleC mutants of C. difficile 630Δerm and C. difficile R20291, respectively. Empty vector control strains CRG1556 (630Δerm) and CRG1628 (R20291) were also created through introduction of pMTL84151 into the respective sleC mutant strains. Successful plasmid transfer was confirmed by recovery of the plasmid, followed by PCR and/or restriction analysis (data not shown). To control for the thiamphenicol selection needed to maintain the plasmids in culture, strains CRG1651 and CRG1652 were created by introducing pMTL84151 into parental strains 630Δerm and R20291, respectively. The development of heat-resistant CFU was then measured, and the data were compared to data obtained previously. The levels of heat-resistant colony formation associated with the taurocholate supplement were fully restored in the sleC derivatives of both C. difficile 630Δerm (Fig. 3A) and R20291 (Fig. 3B) carrying pMTL-DB1 to the levels in the parental strains containing the control vector pMTL84151. In the case of 630Δerm, there was a difference between the level obtained for the strain with the plasmid and the level obtained for the plasmid-free parental strain, presumably as a result of growth under thiamphenicol selection conditions. The sleC mutant controls containing the empty vector performed like the original mutant. Thus, it was possible to complement the heat-resistant CFU defect, indicating that the observed phenotype was due solely to inactivation of sleC.
Effect of sleC mutation on C. difficile sporulation.
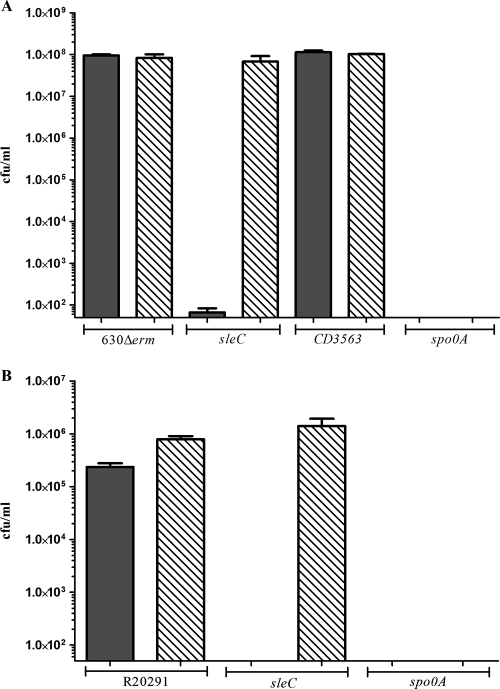
An inability to develop heat-resistant colonies could be a consequence of a number of possible defects. By definition, a heat-resistant colony represents a successfully sporulated vegetative cell that was able to survive heat treatment, germinate, and grow vegetatively. A reduction in the number of observed CFU following heat shock would suggest that a strain is unable to complete one or more of these processes. To examine the sporulation efficiencies of the mutant strains, spores of all C. difficile 630Δerm and R20291 strains were counted after 5 days by using phase-contrast microscopy, and the results were compared to the observed development of heat-resistant CFU (Fig. 4). Except for a sporulation-negative control, in which the master regulator of sporulation spo0A was insertionally inactivated (12), the sporulation frequencies of all mutant strains were found to be equivalent to those of the corresponding parental strains. This suggests that sleC mutants of both C. difficile 630Δerm and C. difficile R20291 sporulate at parental levels but are unable to form colonies even in the presence of taurocholate.
FIG. 4.
Numbers of heat-resistant CFU and spore titers after 5 days. Spore counts (bars with diagonal lines) and numbers of heat-resistant CFU (filled bars) were determined for (A) C. difficile 630Δerm, CRG1115 (sleC), CRG878 (CD3563), and CRG789 (spo0A) and (B) C. difficile R20291, CRG1166 (sleC), and CRG1375 (spo0A) after 5 days of anaerobic incubation at 37°C in BHIS broth. Following this incubation, numbers of heat-resistant CFU were determined as described in the legend to Fig. 2. Spore titers were obtained by counting spores by phase-contrast microscopy using a Bright-Line hemocytometer (Sigma, United Kingdom). The bars indicate the averages of three independent experiments, and the error bars indicate standard errors of the means. The detection limit for colony counts was 50 CFU/ml, and the detection limit for spore counts was 1.6 × 103 spores.
Effect of sleC mutation on C. difficile spore viability.
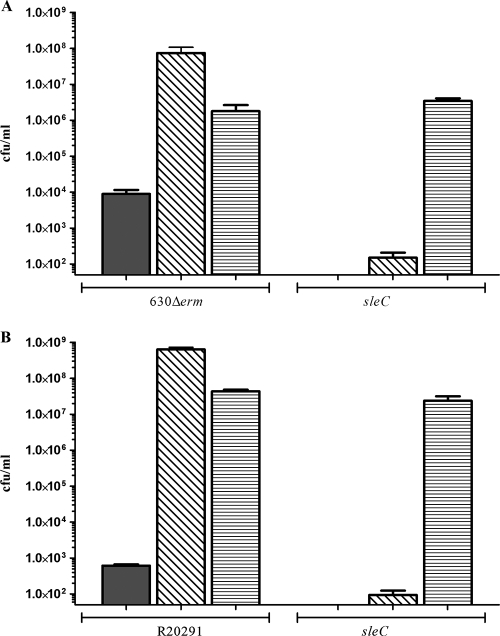
Having shown that the rate of sporulation of an sleC mutant was not affected compared to the rate of sporulation of the parental strain for both C. difficile 630Δerm and R20291, we next demonstrated that the previously noted reduction in heat-resistant colony formation was not a result of altered spore heat resistance properties. Decoating of spores has previously been used to distinguish between spore viability and germination phenotypes (26, 29) as decoating and plating with supplemental lysozyme allow a germination-deficient spore to form heat-resistant colonies. Purified spores of both C. difficile 630Δerm and R20291 plated without taurocholate formed few heat-resistant CFU compared to spores plated with taurocholate (Fig. 5). Decoating spores of the parental strains resulted in much higher numbers of heat-resistant CFU even in the absence of taurocholate. Purified spores of sleC mutants of both C. difficile 630Δerm and C. difficile R20291 were unable to form heat-resistant CFU on BHIS agar supplemented with taurocholate (Fig. 4), but decoated sleC spores formed parental levels of heat-resistant CFU (Fig. 5), confirming that they were viable.
FIG. 5.
Viability of C. difficile sleC mutant spores. Numbers of heat-resistant CFU were determined for (A) C. difficile 630Δerm and CRG1115 (sleC) spores and (B) C. difficile R20291 and CRG1166 (C. difficile R20291 sleC) spores. Spore cultures were heated at 60°C for 25 min before they were plated onto BHIS agar (filled bars) or BHIS agar supplemented with 0.1% taurocholate (bars with diagonal lines). Spores were also decoated as described in Materials and Methods before they were plated onto BHIS agar supplemented with 1 μg/ml lysozyme (bars with horizontal lines). The bars indicate the averages of three independent experiments, and the error bars indicate standard errors of the means. The detection limit for the assay was 50 CFU/ml.
Finally, we observed C. difficile spores by phase-contrast microscopy after heat treatment and incubation in BHIS broth supplemented with taurocholate. Spores of sleC mutants of both 630Δerm and R20291 appeared to lose their “phase-bright” characteristics to the same degree as parental spores after 2 h of incubation. This is consistent with the characterization of sleC in C. perfringens (26) and suggests that C. difficile sleC mutant spores undergo the initial stages of germination in association with taurocholate but do not complete the process and grow as vegetative cells.
DISCUSSION
In this study we identified a homologue of sleC from C. perfringens that is essential for complete germination (in the presence of taurocholate) of C. difficile spores in both the nonepidemic strain 630Δerm and the B1/NAP1/027 isolate R20291. Loss of SleC eliminated the development of heat-resistant CFU over 5 days despite spore formation by the parental strain, and the viability of sleC mutant spores was proven as their colony-forming ability after heat shock was restored by decoating. It is interesting that SleC is essential for C. difficile spore germination and vegetative cell outgrowth. While Bacillus has traditionally been thought of as the model genus for spore germination studies and previous work has identified three SCLEs, the cwlJ, sleB, and sleL products (4, 15, 18), these enzymes are not individually essential for complete cortex lysis (15, 22). On the other hand, recent work with C. perfringens suggests that the mechanisms of germination are somewhat different in Clostridium (26, 27). The identification of SleC as an essential C. perfringens SCLE underlined the difference between this clostridial species and the Bacillus paradigm (26), and our findings support the hypothesis that there is a generic difference between the germination of Bacillus spores and the germination of Clostridium spores. Furthermore, while germination-specific serine proteases have been characterized in C. perfringens (24, 37) and homologues have been identified in C. difficile 630 (33), these proteases are not known to play any role in Bacillus germination.
It has been shown that in B. subtilis CwlJ is important for CaDPA-induced germination (22), providing evidence that there is a CaDPA-mediated germination signaling pathway in Bacillus. In contrast, we found that a B. subtilis cwlJ homologue, CD3563, plays no obvious role in taurocholate-induced C. difficile spore germination. Indeed, recent studies have suggested that SCLEs in C. perfringens are not triggered by CaDPA (26), although further analysis of CD3563 is required to understand its importance, if any, to C. difficile spore germination. As CD3563 shares homology with both B. subtilis cwlJ and sleB, it is worthwhile to consider the possibility that a variety of functions are possible, although under our assay conditions CD3563 does not seem to play any role. This becomes more important with the knowledge that CD0552 has been annotated as sleB in the C. difficile 630 genome (33), although the 238-residue protein shares no similarity with any known SCLE.
While we can argue that our characterization of SleC and early studies of a cwlJ/sleB homologue suggest similarities between the germination of C. difficile spores and the germination of C. perfringens spores, it must be noted that there are obvious differences when germination characteristics as a whole are compared. Early characterization of C. perfringens germinant receptors indicated that monocistronic gerA and gerKB operons and a bicistronic gerKA-gerKC operon play a role in the early stages of germination (25, 27), and similar mechanisms have been described for other spore formers. B. subtilis has three tricistronic operons, gerA, gerB, and gerK (36), while a tricistronic gerA operon has also been identified and studied in both Clostridium botulinum and Clostridium sporogenes (2, 5). In contrast, none of these systems are present in C. difficile. Spore cortex lysis may proceed in similar ways in C. difficile and C. perfringens, but the circumstances under which the spores germinate likely differ. As a result, identification of C. difficile germinant receptors is crucial to understanding C. difficile spore germination.
The emergence of C. difficile B1/NAP1/027 strains (8) has strengthened the need to understand C. difficile spore germination, and our report is the first report of gene characterization by mutagenesis in a B1/NAP1/027 isolate of C. difficile. These strains are associated with more severe disease and greater virulence, and although it is still unclear how sporulation and germination in epidemic and nonepidemic strains compare, it has been suggested that the rate of sporulation is higher in particular B1/NAP1/027 isolates (1, 9). Therefore, the data that we present showing that R20291 sporulates and germinates less efficiently than 630Δerm is perhaps surprising. We showed that C. difficile R20291 sporulates slower (Fig. 1) and forms fewer spores than C. difficile 630Δerm over a 5-day period (Fig. 2). Furthermore, a lower number of the observed spores of R20291 than of the observed spores of 630Δerm formed heat-resistant colonies, indicating that fewer R20291 spores germinate under our assay conditions. These findings indicate that caution should be used when general conclusions are drawn about the properties of types of C. difficile strains without an adequate sample size. Studies encompassing a large number of C. difficile isolates are necessary to define what role sporulation and germination play in disease.
The absolute requirement for SleC for C. difficile to complete bile-induced spore germination could represent a therapeutic target in the health care environment. Spore germination in the patient could be prevented through inhibition of SleC, and this could reduce the ability of C. difficile to cause disease. Conversely, activating SleC could force spores to germinate on surfaces. As vegetative cells are far more susceptible to traditional decontamination procedures, this may allow a second, or combined, disinfection step. Thus, dissemination of C. difficile infection may be reduced, and control of disease in the health care environment could be a more realistic target.
Acknowledgments
This work was supported by the BBSRC (grant BBS/S/L/2003/10242), the MRC (grant G0601176), and Morvus Technology Ltd.
We thank Jon Brazier for donation of C. difficile R20291.
Footnotes
Published ahead of print on 20 November 2009.
REFERENCES
- 1.Akerlund, T., I. Persson, M. Unemo, T. Noren, B. Svenungsson, M. Wullt, and L. G. Burman. 2008. Increased sporulation rate of epidemic Clostridium difficile type 027/NAP1. J. Clin. Microbiol. 46:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberto, F., L. Botella, F. Carlin, C. Nguyen-The, and V. Broussolle. 2005. The Clostridium botulinum GerAB germination protein is located in the inner membrane of spores. FEMS Microbiol. Lett. 253:231-235. [DOI] [PubMed] [Google Scholar]
- 3.Bagyan, I., and P. Setlow. 2002. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J. Bacteriol. 184:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boland, F. M., A. Atrih, H. Chirakkal, S. J. Foster, and A. Moir. 2000. Complete spore-cortex hydrolysis during germination of Bacillus subtilis 168 requires SleB and YpeB. Microbiology 146:57-64. [DOI] [PubMed] [Google Scholar]
- 5.Broussolle, V., F. Alberto, C. A. Shearman, D. R. Mason, L. Botella, C. Nguyen-The, M. W. Peck, and F. Carlin. 2002. Molecular and physiological characterisation of spore germination in Clostridium botulinum and C. sporogenes. Anaerobe 8:89-100. [Google Scholar]
- 6.Chen, Y., S. Miyata, S. Makino, and R. Moriyama. 1997. Molecular characterization of a germination-specific muramidase from Clostridium perfringens S40 spores and nucleotide sequence of the corresponding gene. J. Bacteriol. 179:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chirakkal, H., M. O'Rourke, A. Atrih, S. J. Foster, and A. Moir. 2002. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 148:2383-2392. [DOI] [PubMed] [Google Scholar]
- 8.Dawson, L. F., E. Valiente, and B. W. Wren. 16 June 2009, posting date. Clostridium difficile—a continually evolving and problematic pathogen. Infect. Genet. Evol. doi: 10.1016/j.meegid.2009.06.005. [DOI] [PubMed]
- 9.Fawley, W. N., S. Underwood, J. Freeman, S. D. Baines, K. Saxton, K. Stephenson, R. C. Owens, Jr., and M. H. Wilcox. 2007. Efficacy of hospital cleaning agents and germicides against epidemic Clostridium difficile strains. Infect. Control Hosp. Epidemiol. 28:920-925. [DOI] [PubMed] [Google Scholar]
- 10.Gerding, D. N., C. A. Muto, and R. C. Owens, Jr. 2008. Measures to control and prevent Clostridium difficile infection. Clin. Infect. Dis. 46:S43-S49. [DOI] [PubMed] [Google Scholar]
- 11.Heap, J. T., S. A. Kuehne, M. Ehsaan, S. T. Cartman, C. M. Cooksley, J. C. Scott, and N. P. Minton. 3 November 2009, posting date. The ClosTron: mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods. doi: 10.1016/j.mimet.2009.10.018. [DOI] [PubMed]
- 12.Heap, J. T., O. J. Pennington, S. T. Cartman, G. P. Carter, and N. P. Minton. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452-464. [DOI] [PubMed] [Google Scholar]
- 13.Heap, J. T., O. J. Pennington, S. T. Cartman, and N. P. Minton. 2009. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78:79-85. [DOI] [PubMed] [Google Scholar]
- 14.Hussain, H. A., A. P. Roberts, and P. Mullany. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Δerm) and demonstration that the conjugative transposon Tn916ΔE enters the genome of this strain at multiple sites. J. Med. Microbiol. 54:137-141. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa, S., K. Yamane, and J. Sekiguchi. 1998. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J. Bacteriol. 180:1375-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, S., M. H. Samore, K. A. Farrow, G. E. Killgore, F. C. Tenover, D. Lyras, J. I. Rood, P. DeGirolami, A. L. Baltch, M. E. Rafferty, S. M. Pear, and D. N. Gerding. 1999. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N. Engl. J. Med. 341:1645-1651. [DOI] [PubMed] [Google Scholar]
- 17.Kumazawa, T., A. Masayama, S. Fukuoka, S. Makino, T. Yoshimura, and R. Moriyama. 2007. Mode of action of a germination-specific cortex-lytic enzyme, SleC, of Clostridium perfringens S40. Biosci. Biotechnol. Biochem. 71:884-892. [DOI] [PubMed] [Google Scholar]
- 18.Lambert, E. A., and D. L. Popham. 2008. The Bacillus anthracis SleL (YaaH) protein is an N-acetylglucosaminidase involved in spore cortex depolymerization. J. Bacteriol. 190:7601-7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masayama, A., H. Fukuoka, S. Kato, T. Yoshimura, M. Moriyama, and R. Moriyama. 2006. Subcellular localization of a germiantion-specific cortex-lytic enzyme, SleB, of bacilli during sporulation. Genes Genet. Syst. 81:163-169. [DOI] [PubMed] [Google Scholar]
- 20.Miyata, S., R. Moriyama, N. Miyahara, and S. Makino. 1995. A gene (sleC) encoding a spore-cortex-lytic enzyme from Clostridium perfringens S40 spores; cloning, sequence analysis and molecular characterization. Microbiology 141:2643-2650. [DOI] [PubMed] [Google Scholar]
- 21.Moir, A. 2006. How do spores germinate? J. Appl. Microbiol. 101:526-530. [DOI] [PubMed] [Google Scholar]
- 22.Paidhungat, M., K. Ragkousi, and P. Setlow. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J. Bacteriol. 183:4886-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paidhungat, M., and P. Setlow. 2000. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paredes-Sabja, D., P. Setlow, and M. R. Sarker. 23 July 2009, posting date. The protease CspB is essential for initiation of cortex hydrolysis and dipicolinic acid (DPA) release during germination of spores of Clostridium perfringens type A food poisoning isolates. Microbiology. doi: 10.1099/mic.0.030965-0. [DOI] [PubMed]
- 25.Paredes-Sabja, D., P. Setlow, and M. R. Sarker. 2009. Role of GerKB in germination and outgrowth of Clostridium perfringens spores. Appl. Environ. Microbiol. 75:3813-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paredes-Sabja, D., P. Setlow, and M. R. Sarker. 2009. SleC is essential for cortex peptidoglycan hydrolysis during germination of spores of the pathogenic bacterium Clostridium perfringens. J. Bacteriol. 191:2711-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paredes-Sabja, D., J. A. Torres, P. Setlow, and M. R. Sarker. 2008. Clostridium perfringens spore germination: characterization of germinants and their receptors. J. Bacteriol. 190:1190-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popham, D., J. Helin, C. Costello, and P. Setlow. 1996. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc. Natl. Acad. Sci. U. S. A. 93:15405-15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popham, D. L., S. Sengupta, and P. Setlow. 1995. Heat, hydrogen peroxide, and UV resistance of Bacillus subtilis spores with increased core water content and with or without major DNA-binding proteins. Appl. Environ. Microbiol. 61:3633-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poxton, I. R., J. McCoubrey, and G. Blair. 2001. The pathogenicity of Clostridium difficile. Clin. Microbiol. Infect. 7:421-427. [DOI] [PubMed] [Google Scholar]
- 31.Purdy, D., T. A. O'Keeffe, M. Elmore, M. Herbert, A. McLeod, M. Bokori-Brown, A. Ostrowski, and N. P. Minton. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46:439-452. [DOI] [PubMed] [Google Scholar]
- 32.Riggs, M. M., A. K. Sethi, T. F. Zabarsky, E. C. Eckstein, R. L. P. Jump, and C. J. Donskey. 2007. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin. Infect. Dis. 45:992-998. [DOI] [PubMed] [Google Scholar]
- 33.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. G. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779-786. [DOI] [PubMed] [Google Scholar]
- 34.Setlow, P. 2008. Dormant spores receive an unexpected wake-up call. Cell 135:410-412. [DOI] [PubMed] [Google Scholar]
- 35.Setlow, P. 2007. I will survive: DNA protection in bacterial spores. Trends Microbiol. 15:172-180. [DOI] [PubMed] [Google Scholar]
- 36.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550-556. [DOI] [PubMed] [Google Scholar]
- 37.Shimamoto, S., R. Moriyama, K. Sugimoto, S. Miyata, and S. Makino. 2001. Partial characterization of an enzyme fraction with protease activity which converts the spore peptidoglycan hydrolase (SleC) precursor to an active enzyme during germination of Clostridium perfringens S40 spores and analysis of a gene cluster involved in the activity. J. Bacteriol. 183:3742-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorg, J. A., and A. L. Sonenshein. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorg, J. A., and A. L. Sonenshein. 2009. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J. Bacteriol. 191:1115-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voth, D. E., and J. D. Ballard. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18:247-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079-1084. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, K. H., M. J. Kennedy, and F. R. Fekety. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 15:443-446. [DOI] [PMC free article] [PubMed] [Google Scholar]