Abstract
ArcBA is a two-component regulatory system of Escherichia coli involved in sensing oxygen availability and the concomitant transcriptional regulation of oxidative and fermentative catabolism. Based on in vitro data, it has been postulated that the redox state of the ubiquinone pool is the determinant for ArcB kinase activity. Here we report on the in vivo regulation of ArcB activation, as determined using a lacZ reporter specifically responsive to phosphorylated ArcA. Our results indicate that upon deletion of a ubiquinone biosynthetic enzyme, regulation of ArcB in the anaerobic-aerobic transition is not affected. In contrast, interference with menaquinone biosynthesis leads to inactivation of ArcB during anaerobic growth; this phenotype is fully rescued by addition of a menaquinone precursor. This clearly demonstrates that the menaquinones play a major role in ArcB activation. ArcB shows a complex pattern of regulation when E. coli is titrated through the entire aerobiosis range; ArcB is activated under anaerobic and subaerobic conditions and is much less active under fully aerobic and microaerobic conditions. Furthermore, there is no correlation between ArcB activation and the redox state of the ubiquinone pool, but there is a restricted correlation between the total cellular ubiquinone content and ArcB activity due to the considerable increase in the size of the ubiquinone pool with increasing degrees of aerobiosis. These results lead to the working hypothesis that the in vivo activity of ArcB in E. coli is modulated by the redox state of the menaquinone pool and that the ubiquinone/ubiquinol ratio in vivo surely is not the only determinant of ArcB activity.
Two-component systems are employed by prokaryotes to respond to changing environmental and intracellular conditions. The ArcBA (anoxic redox control) system is a two-component system in Escherichia coli that functions as the aerobiosis-sensing device that tunes the activity of catabolic pathways to variations in oxygen availability. Its first component, ArcB, functions as the sensor that relays a signal via signal-dependent kinase activity to the second component, the response regulator ArcA (17, 20). Upon signal perception, ArcB is phosphorylated at the conserved His-292 residue. The phosphoryl group is subsequently transferred intramolecularly to the conserved Asp-576 residue, after which it is transferred once more intramolecularly to His-717, which is located in the phosphoryl transfer domain. Then the latter residue transfers the phosphoryl group to ArcA (12, 23). The extent of phosphorylation of ArcA determines the expression of operons involved in a wide variety of mostly catabolic pathways that are operative under different redox growth conditions (3, 14, 24). Thus, the ArcBA system is important for the organism's ability to distribute energy generation for fermentation and respiration (3).
ArcB can form intermolecular disulfide bonds via Cys-180 and Cys241, which are located in the PAS domain of the protein. The kinase activity of ArcB is highly dependent on this covalent linkage. A disulfide bond formed between two Cys-180 residues results in a 85% reduction in kinase activity, and a bond between two Cys-241 residues results in a 15% reduction (26). It has been shown in vitro that the cysteine residues can be oxidized by ubiquinone (26), and hence the latter redox carrier was postulated to regulate the kinase activity of ArcB in vivo. Consistent with this hypothesis, maximal kinase activity of ArcB with ArcA has been shown to occur under anaerobic conditions (25); nevertheless, it has also been suggested that significant levels of ArcA∼P are present in aerobic cells (18, 19).
Previous studies on ArcB have used lacZ reporters that could be or are known to be subject to regulation by additional factors (e.g., FNR for the cydA-lacZ reporter used by Georgellis et al. [11, 26, 27]). In this study, a PcydA(−176+1)-lacZ reporter that is not responsive to FNR but is dependent on ArcA∼P (1) was used to characterize the factors that are involved in the in vivo regulation of ArcB activation in relation to (decreasing) oxygen availability (1). Our findings indicate that the menaquinone pool plays an important role in ArcB activation.
The ArcBA system exhibits maximal activity under high-microaerobisis conditions (equivalent to 80% aerobiosis according to the quantitative definition formulated by Alexeeva et al. [1]) and fully anaerobic conditions and much lower levels of activity under low-microaerobisis conditions (20% aerobiosis) and fully aerobic conditions. We show that additional regulation of ArcB kinase activity by the redox state of the menaquinone pool is prevalent under microaerobic to anaerobic conditions. Neither the catabolites known so far to affect ArcB activity in vitro (d-lactate, acetate, and pyruvate [12, 17, 19]) nor the ArcB-specific phosphatase SixA regulated ArcB activity in microaerobic conditions.
Based on these observations we concluded that regulation of the ArcBA system in vivo in our microaerobic conditions is controlled by both the menaquinone and the ubiquinone pools.
MATERIALS AND METHODS
Strains and plasmids used in this study.
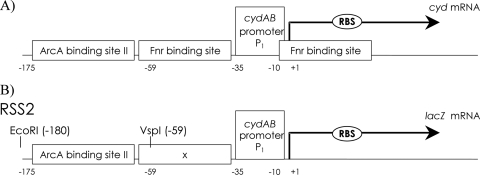
To create pSal1 (Table 1), a 176-bp DNA fragment from the upstream region (Fig. 1A) of the cydAB operon (cydA−176+1) (corresponding to bases 512 to 687 in the GenBank accession number J03939 sequence; bases 1 to −176 relative to the start site of cydAB P1 transcription) was amplified by PCR with primers Cyd(E)-174 (5′ T ATG AAT TCT TTT TAT CTT TAA TTG CCA ACC G) and Cyd(Bam)+1 (5′ ATA GGA TCC CGA GAA CAA TTT ATC TCT TTT TGA TGC C) using E. coli chromosomal DNA from strain MC4100 as the template. The PCR product was digested with BamHI and EcoRI and cloned in the corresponding sites of plasmid pQE30.
TABLE 1.
E. coli strains, phages, and plasmids used in this study
| Strain, phage, or plasmid | Genotypea | Reference |
|---|---|---|
| Strains | ||
| BW25113 | K-12 wild-type | 4 |
| ASA12 | MC4100 recA, λRSS2[φ(′cydA-lacZ)] | 1 |
| ASA32 | MC4100 recA ΔarcA, λRSS2[φ(′cydA-lacZ)] | 1 |
| JA001 | BW25113, λRSS2[φ(′cydA-lacZ)] | This study |
| JA022 | JW2257 ΔmenB, λRSS2[φ(′cydA-lacZ)] | This study |
| JA023 | JW5713 ΔubiC, λRSS2[φ(′cydA-lacZ)] | This study |
| JA029 | JW2337 ΔsixA, λRSS2[φ(′cydA-lacZ)] | This study |
| JA032 | JW5536 ΔarcB, λRSS2[φ(′cydA-lacZ)] | This study |
| BL21(DE3) | F-−ompT [lon] hsdSb (rb− mb−; E. coli B strain) with DE3, a prophage carrying the T7 RNA polymerase gene | Novagen, United States |
| MC4100 | F-−araD139 (argF-lac)U169 rpsL150 relA1 deoC1 flb-5301 pstF1 | 7a |
| RM101 | MC4100 Δfnr | 35a |
| RM3133 | MC4100 ΔarcA::tet | 2 |
| ASA22 | RM101 Δfnr, λRSS2φ(P1cydA−176+1lacZ) | This study |
| Phages | ||
| λRS45 | lacZ′ lacY+imm21ind+ | 38 |
| λRSS2 | λRS45 Kmr P1cydA−176+1lacZ+lacY+lacA+ | This study |
| Plasmids | ||
| pETarcA-1 | pET28a KmrarcA+ | This study |
| pQE30 | Apr | Qiagen |
| pRS551 | Kmr AprlacZ+lacY+lacA+ | 38 |
| pSal1 | pQE30 P1cydA−176+1 | This study |
| pSal2 | pQE30 P1cydA−176+1 | This study |
| pRSS2 | pRS551 P1cydA−176+1lacZ+lacY+lacA+ | This study |
P1cydA−176+1 is the cydA promoter P1 from base −176 to base 1 relative to the transcription start containing five base pair substitutions in the Fnr box (Fig. 1B).
FIG. 1.
(A) Schematic representation of part of the transcriptional regulatory elements upstream of the cydAB operon (not to scale). (B) Schematic representation of the constructed ArcA∼P-dependent promoter, based on P1 of cydAB (not to scale).
To create pSal2, PCR-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene) was performed using primers cydmut1 (5′ CAT AAT TTG TAG GAA ATT AAT TTT AAC AAT GTA TAA GTC TTG G) and cydmut2 (5′ CCA AGA CTT ATA CAT TGT TAA AAT TAA TTT CCT ACA AAT TAT G) and plasmid pSal1 as the template according to the manufacturer's instructions. This resulted in the introduction of point mutations (underlined) in the essential base pairs of the consensus sequence of the FNR-binding site (GGAATTGATATTTATCAATGTA to GGAAATTAATTTTAACAATGTA). Through these mutations a VspI restriction site was introduced. Successful mutagenesis was confirmed by restriction analysis and sequencing using the semiautomated DNA sequencing technique.
To construct the operon fusions, the 176-bp EcoRI-BamHI fragment of plasmid pSal2 was ligated into EcoRI-BamHI-digested lacZ operon fusion vector pRS551 (38), resulting in pRSS2. The fusion (Fig. 1B) was then transferred to the λ transducing phage λRS45 (the MC1061 strain bearing pRSS2 was infected with λRS45) as described previously (35), yielding λRSS2. Lysate containing λRSS2 was used to lysogenize strains MC4100, yielding ASA12. The reporter from ASA12 was subsequently P1 phage transduced into BW25113, JW5713, JW2237, JW5536, RM101, and RM3133, yielding JA001, JA023, JA029, JA032, ASA22, and ASA32, respectively.
Measurement of enzyme activity.
β-Galactosidase activity in permeabilized cells was measured by a method originally described by Miller (30) and modified by Giacomini et al. (13).
Overproduction and purification of His6-ArcA.
For overproduction of His6-ArcA, 500-ml cultures of E. coli strain BL21 transformed with pETArcA-1 were grown in 2-liter conical flasks with vigorous shaking (200 rpm) on a rotary shaker at 37°C in PB medium (20 g·liter−1 tryptone, 10 g·liter−1 yeast extract, 5 g·liter−1glucose, g·liter−1NaCl, 8.7 g·liter−1 K2HPO4; pH 7). Kanamycin was routinely included at a final concentration of 50 μg·ml−1 for plasmid maintenance. When the culture attained an optical density at 600 nm (OD600) of approximately 0.4, induction of arcA expression was initiated by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. After growth for 2.5 h, the cells were harvested by centrifugation, and the cell pellet was stored at −20°C until use. All subsequent steps were performed at 4°C. Next, the cell pellet was resuspended in 4 ml of buffer A (0.5 M NaCl, 20 mM Tris-HCl; pH 7.9), containing 1.3 mg·ml−1 lysozyme, 30 μg·ml−1 DNase and RNase. After 30 min of incubation at room temperature, the cells were disrupted by sonication. The resulting cell lysate was clarified by centrifugation at 15,000 rpm for 30 min. Phenylmethylsulfonyl fluoride (PMSF) was added to the lysate to a final concentration of 0.1 mM in order to prevent protein degradation. The cell lysate was then applied to a 1.5-ml Ni-nitrilotriacetic acid-agarose column (Qiagen) equilibrated with buffer A. After the column was washed with 10 ml of 10 mM imidazole (in buffer A [see above]), the protein was eluted with 50 mM imidazole (in buffer A) and collected in five 2-ml fractions. These fractions were immediately dialyzed against 50 mM Tris-HCl (pH 7.5), 0.1 mM dithiothreitol (DTT), 0.1 mM EDTA. After dialysis glycerol was added to a final concentration of 10% (vol/vol), and the fractions were analyzed by SDS-PAGE. Addition of 0.1 mM DTT plus 0.1 mM EDTA and 10% (wt/vol) glycerol to the protein solution after dialysis was essential to prevent precipitation of ArcA during thawing after storage at −20°C. Protein concentrations were measured by the microbiuret method (28), after precipitation with trichloroacetic acid (6). Bovine serum albumin was used as a standard.
In vitro phosphorylation of His6-ArcA.
His6-ArcA protein was phosphorylated by incubating the protein in TEGD buffer (50 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 10% glycerol) to which 5 mM MgCl2 and 50 mM (final concentration) carbamoyl phosphate were added as described by Drapal and Sawers (9). The mixtures were incubated for 90 min at 25°C, and the phosphorylated ArcA was used immediately for DNA-binding assays. For calculations it was assumed that this procedure yielded 100% phosphorylation of the protein. In parallel, similar incubations with mixtures lacking carbamoyl phosphate were conducted to prepare unphosphorylated protein used for DNA-binding reactions.
Preparation of radioactively labeled DNA fragments.
Plasmid pSal2 (see above) contains a 121-bp EcoRI-VspI fragment with ArcA-binding site II of the cydAB promoter region (8, 25) from position −59 to position −175 relative to the start of cydAB P1 (Fig. 1A), which also corresponds to bases 512 to 628 in the GenBank accession number J03939 sequence. EcoRI-VspI-digested pSal2 was separated on a 12.5% (wt/vol) polyacrylamide gel. The 121-bp DNA fragment was eluted from the gel by incubation overnight at 65°C in elution buffer (1 mM sodium acetate, 10 mM magnesium acetate, 0.5 mM EDTA [pH 8.0], 0.1% [wt/vol] SDS) and recovered by ethanol precipitation in the presence of 10 mM MgCl2 and 0.3 M sodium acetate. DNA concentrations were determined spectrophotometrically. End labeling of the fragment was performed with the Klenow fragment of E. coli DNA polymerase and [α-32P]dATP (3,000 Ci/mmol; Amersham) in OPA buffer (10 mM Tris-acetate [pH 7.5], 10 mM MgCl2, 50 mM potassium acetate). The labeled DNA fragments were not separated from unincorporated nucleotides but were used directly in mobility shift experiments.
Gel retardation assays.
The labeled 121-bp DNA fragment was used in all retardation assays at a concentration of 0.32 nM. Protein samples were combined with 32P-end-labeled DNA substrates in 10-μl reaction mixtures containing 12 mM HEPES-NaOH (pH 7.9), 4 mM Tris-HCl (pH 7.9), 60 mM KCl, 1 mM EDTA, 1 mM DTT, 12% (wt/vol) glycerol, 300 μg/ml bovine serum albumin (BSA), and 1 μg poly(dI-dC)-poly(dI-dC) (Sigma). After incubation for 15 min at 30°C, 10× loading buffer (50% [wt/vol] glycerol, 0.2% [wt/vol] bromphenol blue, 0.2% [wt/vol] xylene cyanol) was added to 10% (wt/vol) of the final volume, and the mixture was directly applied to a gel using PhastSystem sample applicators (Pharmacia).
Gel retardation assays were performed essentially as described by Ramanujam et al. (32) with 8 to 25% (wt/vol) gradient polyacrylamide gels (Pharmacia), using a PhastSystem at 4°C and the following separation method: sample application down 1.2 using 0 V·h; sample application up 1.2 using 2 V·h; separation 1.1 using 400 V, 10 mA, 2.5 W, 4°C, and 10 V·h; separation 1.2 using 400 V, 1 mA, 2.5 W, 4°C, and 2 V·h; and separation 1.3 using 400 V, 1 mA, 2.5 W, 4°C, and 268 V·h.
After completion of the run, the lower part of the gel, containing unincorporated [α-32P]dATP from the DNA labeling reactions, was removed to prevent the signal from the nucleotides from interfering with the signal from the labeled DNA fragments. The results were visualized by exposing the gel to film with an intensifying screen overnight at −70°C. Quantification of bound and unbound DNA fragments was performed by densitometric analysis using ImageMaster 1D Prime, version 2.0 (Pharmacia Biotech).
Western blotting.
A rabbit anti-ArcA polyclonal antiserum was produced for this study by immunization of a rabbit with highly purified His6-ArcA protein. The antiserum was checked for cross-reactions, and its titer was determined by Western blot analysis using cell extracts of the MC4100 and RM3133 (ΔarcA) E. coli strains. Equal amounts of cell extract (20 μg total protein per lane) resolved on a 12.5% (wt/vol) SDS-PAGE gel were blotted onto nitrocellulose in a Trans-Blot semidry cell (Bio-Rad) and subsequently immunolabeled as described by Towbin et al. (39). The rabbit anti-ArcA antibody was used at a dilution of 1:10,000. The secondary antibody, horseradish peroxidase-conjugated goat anti-rabbit IgG (Bio-Rad), was used at a dilution of 1:3,000 for subsequent visualization by a color reaction. Amounts of ArcA (with the purified ArcA protein used as a reference) were quantified using densitometric analysis (Image Master 1D prime, version 2.0; Pharmacia Biotech).
Continuous cultures.
Cells were grown in Applikon-type fermentors (1-3, 10) at a dilution rate of 0.15 ± 0.01 h−1 under glucose-limited conditions. A simple salts medium described by Evans et al. (10) was used, but instead of citrate, nitriloacetic acid (2 mM) was used as the chelator. Selenite (30 μg/liter) and thiamine (15 mg/liter) were added to the medium. Glucose was used as the single carbon and energy source at a final concentration of 45 mM in the feed. The pH was maintained at 7.0 ± 0.1 by titration with sterile 4 M NaOH, and the temperature was set to 35°C. The oxygen supply was varied as described previously (3). In addition to β-galactosidase activities, in all cultures the steady-state specific rates of fermentation product formation and glucose and O2 consumption were measured as described by Alexeeva et al. (3) to determine the percentage of aerobiosis. In previous experiments the chemostat cultures were calibrated to quantify oxygen availability (2). Essentially, 0% aerobiosis reflects fully anaerobic conditions and 100% aerobiosis is the minimal oxygen input rate required for completely aerobic catabolism (Alexeeva et al. [2] provide a quantitative definition of microaerobiosis).
Batch culture.
In batch cultures, the composition of the medium was similar to the composition of the medium described above, except that sodium phosphate (pH 7) was used at a concentration of 100 mM instead of 10 mM to increase the buffering capacity of the medium. Glucose (final concentration, 1% [wt/vol]) was sterilized separately. High levels of aeration of cultures during aerobic growth were obtained by shaking 10-ml cultures in 100-ml Erlenmeyer flasks at 180 rpm. For anaerobic growth 15-ml cultures in sealed Greiner tubes (15 ml) were used. Inoculated cells were obtained from cultures pregrown under aerobic or anaerobic conditions (after dilution to an OD600 of ∼5 × 10−3) and allowed to double about seven times to mid-log exponential phase (final OD600 for anaerobic conditions, 0.4 to 0.6; final OD600 for aerobic conditions, 0.8 to 1.2) prior to measurement of β-galactosidase enzyme activity. Batch cultures (100 ml) used for comparison of the redox state of the ubiquinone pool with the cydA−176+1-lacZ expression level (Fig. 2) were grown in 1-liter Erlenmeyer flasks stirred at a rate of 250 rpm. Other conditions were similar to those described above. The strains were maintained in vials in LB medium with 30% (wt/vol) glycerol at −70°C.
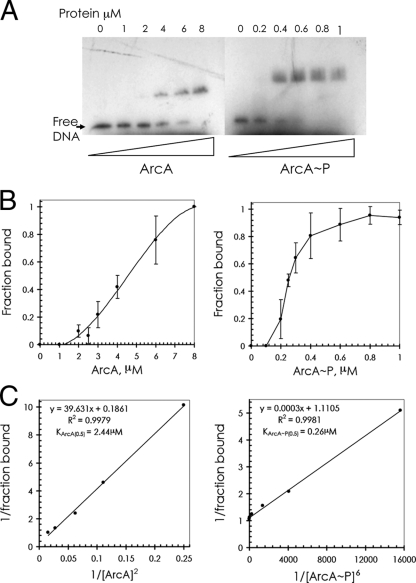
FIG. 2.
Concentration dependence of binding of ArcA and phosphorylated ArcA (ArcA∼P) to the DNA fragment containing ArcA-binding site II (from position −59 to position −175 relative to the start of cydAB P1). (A) Radiolabeled DNA fragment (0.32 nM) incubated with different amounts of either phosphorylated or unphosphorylated ArcA. Protein concentrations are indicated above the lanes. (B) Quantitative evaluation of the results of the gel retardation assays shown in panel A. Each point indicates the mean of four to eight independent experiments. (C) Double-reciprocal plot for binding of ArcA and ArcA∼P to the DNA fragment with n = 2 for ArcA and n = 6 for ArcA∼P.
Analysis of carbon fluxes.
Steady-state bacterial dry weight was measured as described previously (2). Glucose, pyruvate, lactate, formate, acetate, succinate, and ethanol contents were determined by high-performance liquid chromatography (HPLC) (LKB) with a REZEX organic acid analysis column (Phenomenex) at a temperature of 45°C with 7.2 mM H2SO4 as the eluent, using an RI 1530 refractive index detector (Jasco) and AZUR chromatography software for data integration. The carbon balance for all data is >92% (mean, 96%), as calculated from the glucose consumption and product formation rates.
Quinone extraction.
Two-milliliter samples from a culture were quenched with 6 ml of ice-cold 0.2 M HClO4 in methanol or with only methanol. Next, 6 ml of petroleum ether (boiling point, 40 to 60°C) was added rapidly to the mixture, which was vortexed for 1 min. After the mixture was centrifuged (900 × g, 2 min), the upper petroleum ether phase was removed, transferred to a test tube, and evaporated to dryness under a flow of nitrogen. Another 3 ml of petroleum ether was added to the lower phase, and the vortexing and centrifugation steps were repeated. The upper phases were combined. After evaporation to dryness, extracts could be stored for at least 7 days under nitrogen at −20°C. Immediately before use, the extracted quinone-quinol mixture was resuspended with a glass rod in 80 μl ethanol and fractionated by HPLC (Pharmacia LKB 2249 gradient pump system with an LKB 2151 variable-wavelength monitor) using a reversed-phase Lichrosorb (Chrompack, Bergen op Zoom, The Netherlands) 10 RP 18 column (size, 250 mm; interna1 diameter, 4.6 mm). The column was equilibrated with ethanol-methanol (1:1, vol/vol) or pure methanol (HPLC grade), and the ethanol-methanol mixture was used as the mobile phase. The flow rate was 2 ml/min at 50°C. Detection of the quinones was performed at 290 nm for ubiquinones and at 248 nm for menaquinones. The amount of each quinone species was calculated from the relevant peak area, using ubiquinone-10 (UQ10) and menaquinone-4 as standards and the method described by Shestopalov et al. (37). The methanol, ethanol, and petroleum ether used were analytical grade.
Peaks were identified by UV/visible and mass spectral analysis. A UV/visible spectrum of demethylmenaquinone-8 (DMK8) was kindly provided by A. V. Bogachev (Moscow University, Moscow, Russia). For mass spectral analysis fractions collected from the HPLC were evaporated under nitrogen and redissolved in 90% (wt/vol) acetonitrile, 1% (vol/vol) formic acid (LC grade; Merck, Frankfurt, Germany). Then fractions were analyzed by off-line electrospray mass spectrometry using coated Picotips (Econo12; New Objective, Woburn, MA) with an electrospray ionization quantitative time of flight mass spectrometer (Micromass; Waters, Manchester, United Kingdom). Ions selected for tandem mass spectrometry collided with argon in the hexapole collision cell.
RESULTS
Specific interaction of His6-ArcA∼P with the regulatory region of the cydAB operon.
In order to construct an ArcA∼P-dependent reporter system that responds exclusively to this regulator, we amplified a 176-bp DNA fragment from the cydAB regulatory region of E. coli (cydA−176+1). This DNA fragment contains an ArcA-binding site (site II), an FNR-binding site, and the cydAB promoter P1 (8). The FNR-binding site in this regulatory region was selectively inactivated by site-directed mutagenesis (Fig. 1; also see Materials and Methods). This fragment was fused with the lacZ gene and introduced into E. coli strain MC4100 to create strain ASA12 (see Materials and Methods).
To verify that the reporter construct indeed responds solely to the phosphorylation state of ArcA in vivo and that FNR does not interfere with this response, strain ASA12 (wild type with cydA−176+1-lacZ) and strain ASA22 (Δfnr cydA−176+1-lacZ) (see Materials and Methods) were grown in anaerobic and aerobic batch conditions, respectively. The expression of the reporter was found to be twofold higher in anaerobic conditions than in aerobic conditions for both strains (data not shown). In contrast, the change in expression of the reporter in strain ASA11, which expresses FNR and contains the reporter with the FNR-binding site, was less than 1.5-fold (data not shown).
The interaction of the ArcA protein with the mutated cydAB regulatory region was quantified in vitro by performing gel retardation assays. Relative binding affinities of ArcA and ArcA∼P were determined using a 32P-end-labeled fragment derived from pSal2 (Fig. 1 and 2A). ArcA was phosphorylated by incubation with carbamoyl phosphate. A single retarded complex was observed for each binding reaction. Fractions of DNA retarded by ArcA or ArcA∼P were quantified by densitometric analysis and plotted (Fig. 2B) as a function of protein concentration. For ArcA∼P, an apparent Kd(a) [the ArcA∼P concentration at which one-half of the amount of the DNA fragment is bound, designated KArcA∼P(0.5)] was estimated to be approximately 0.25 μM, whereas the corresponding value for ArcA [KArcA(0.5)] was estimated to be approximately 4.5 μM. The sigmoid shapes of the binding curves suggest that binding of neither ArcA nor ArcA∼P to the regulatory DNA fragment obeys simple binding kinetics (i.e., a hyperbolic [bound fraction]/[substrate] function). Analysis of the data essentially as described by Keleti (22) and Segel (36) by fitting a double-reciprocal plot of 1/Y versus 1/[S], where Y is the fraction of bound DNA and [S] is the ArcA(∼P) concentration, to the equation Y = Ymax[S]n/(Km + [S]n) provided the best fits (R2), with a value of 2 for ArcA and a value of 6 for ArcA∼P (Fig. 2C), strongly suggesting that ArcA and ArcA∼P bind to the DNA fragment with different stoichiometries. The Km values for binding derived from the double-reciprocal plot with the coordinates {1/Y; 1/[S]n} when n is 2 for ArcA and 6 for ArcA∼P were 2.44 μM and 0.26 μM, respectively. Comparable stoichiometries (when n is 3 for ArcA and 6 for ArcA∼P) and Km values (4.3 μM for ArcA and 0.27 μM for ArcA∼P) were obtained by analysis of the data using a Hill plot (not shown). Together, these results suggest that ArcA∼P binds to the DNA fragment as a hexamer, which is in accordance with the observations of Jeon et al. (21).
Only when ArcA was phosphorylated was a sequence-specific interaction with the DNA fragment containing ArcA-binding site II of the cydAB regulatory region observed. This was concluded from the lack of an effect of addition of a large excess of competing fragments [poly(dI-dC)-poly(dI-dC) or pQE30] with a random sequence to the binding assays with ArcA∼P. In contrast, addition of a 50-fold molar excess of either competitor DNA in the binding assay with nonphosphorylated ArcA did prevent sequence-specific complex formation. These competing fragments did not have an effect when phosphorylated ArcA was used (data not shown). Similar observations with respect to the specificity of ArcA and ArcA∼P binding to the same binding site have been made previously by Lynch and Lin (25).
ArcB activation by various quinone species.
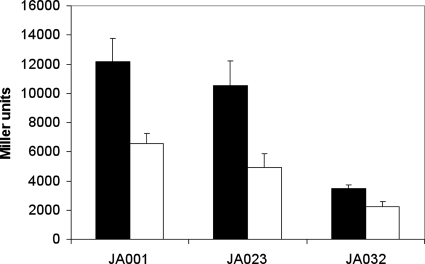
The specific ArcA∼P-dependent lacZ reporter construct (1) (see above) was transduced with phage P1 into strains BW25113 (wild type), JW5536 (ΔarcB) (4), and JW5713 (ΔubiC), generating strains JA001, JA032, and JA023, respectively. The resulting strains were grown in batch culture in minimal medium supplemented with glucose as the sole carbon and energy source. In the wild-type derivative JA001, cydA−176+1-lacZ expression was twofold greater under anaerobic conditions compared to aerobic conditions (Fig. 3), whereas for strain JA032, which lacks the ArcB histidine kinase, the levels of expression were not significantly different under anaerobic and aerobic conditions. These findings confirm that the anaerobic regulation mediated by ArcA is dependent on the activity of the histidine kinase ArcB and therefore presumably on the phosphorylation state of ArcA.
FIG. 3.
Activity of the ArcA∼P-dependent-lacZ reporter construct (λRSS2). The construct was tested in the wild-type (JA001), ΔubiC (JA023), and ΔarcB (JA032) backgrounds after batch culture growth in mineral medium supplemented with glucose. Cultures were grown aerobically (open bars) and anaerobically (filled bars).
Georgellis et al. (11) have reported that ubiquinone and menadione have an effect on the histidine kinase activity of ArcB in vitro, suggesting that the oxidation state of the quinone pool may play a crucial role in ArcB regulation in vivo. To further discern the role that ubiquinone and menaquinone have in regulation of ArcB, we constructed strain JA023, which lacks ubiC and is therefore unable to synthesize ubiquinone. The difference between the aerobic and anaerobic levels of expression of the cydA−176+1-lacZ reporter was similar to the difference found for the wild-type strain. This result strongly suggests that aerobic inhibition of the ArcB kinase in vivo can be due to other effectors and thus is regulated not only by ubiquinone.
Previously, we showed (5) that the ubiquinone pool of E. coli is gradually reduced during growth in batch culture when the OD600 reaches values higher than 1.0. One would therefore expect that an increase in cydA−176+1-lacZ expression would parallel the increase in the ratio of ubiquinol concentration to ubiquinone concentration if this expression is governed solely by this ratio. To test this hypothesis, strain ASA12 was grown in aerobic batch culture, as described previously (5). As anticipated, a considerable increase in the ubiquinol/ubiquinone ratio was observed at the later stages of growth; however, this increase was not accompanied by a significant increase in the level of cydA−176+1-lacZ expression (see Fig. S1 in the supplemental material). Together, these observations indicate that there was no apparent regulation by the ubiquinol/ubiquinone ratio under the conditions used in these experiments.
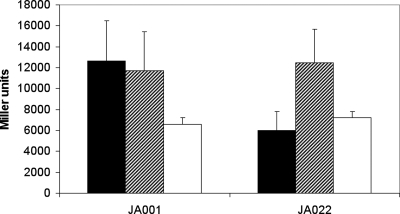
Besides addition of ubiquinone, addition of menadione has been reported to influence the in vitro autophosphorylation rate of ArcB, although higher concentrations of the latter quinone were required to obtain half-maximal inhibition (50 μM, compared to 5 μM for ubiquinone). To investigate whether demethylmenaquinone plays a role in the regulation of ArcB in vivo, strain JW2257 (ΔmenB) was lysogenised with the P1 phage lysate obtained from strain ASA12, resulting in strain JA022. The latter strain's inability to synthesize demethylmenaquinones yielded a surprising phenotype: loss of the activation response of cydA−176+1-lacZ expression to anaerobiosis (Fig. 4). Moreover, addition of 2 μM 1,4-dihydroxy-2-naphthoic acid, an intermediate in the menaquinone biosynthetic pathway that is synthesized downstream of the block in this pathway introduced by deletion of menB (encoding naphthoate synthase), to anaerobic batch cultures restored the wild-type phenotype. Independent analyses of cellular quinone levels verified that addition of naphthoic acid resulted in significant restoration of the intracellular demethylmenaquinone and menaquinone pools (see Fig. S2 in the supplemental material) but had no effect on the redox state of the ubiquinone pool (data not shown). This clearly shows that menaquinone (or the menaquinone pool) also has a role in the regulation of ArcB activation.
FIG. 4.
Activity of the ArcA∼P-dependent-lacZ reporter construct (λRSS2). The construct was tested in the wild type (JA001) and ΔmenB (JA022) backgrounds after growth in anaerobic batch cultures in medium with (bars with diagonal lines) or without (filled bars) 2 μM 1,4-dihydroxy-2-naphthoic acid or after aerobic growth without 1,4-dihydroxy-2-naphthoic acid (open bars).
ArcB activation under microaerobic conditions.
Previously, we have described a controlled aerobiosis system based on glucose-limited chemostat cultures (see reference 2 for details concerning the experimental setup) that allows growth of steady-state cultures at any ratio of aerobic catabolism to anaerobic fermentation. To monitor the degree of ArcA phosphorylation in vivo in relation to quantified and steady-state oxygen availability, strain ASA12, an MC4100 derivative which contains the reporter system described above for strain JA001 and its relatives, was grown in glucose-limited chemostat cultures at a growth rate of 0.15 h−1 with controlled oxygen input rates ranging from 0 to 110% aerobiosis (see Materials and Methods). Under these conditions a rather complex pattern of cydA−176+1-lacZ expression was observed, with maximal values at 0% and 85% aerobiosis. The maximal values were approximately 2.5-fold higher than the minimal values measured at 20% aerobiosis and under fully aerobic conditions (Fig. 5). The expression levels of the cydA−176+1-lacZ reporter under the latter two conditions were not significantly different. In contrast, in similar experiments with strain ASA32, which lacks ArcA, the level of cydA−176+1-lacZ expression increased linearly with increasing oxygen availability, and there was an approximately 1.5-fold increase when anaerobic growth conditions were compared to fully aerobic growth conditions (see Fig. S3 in the supplemental material).
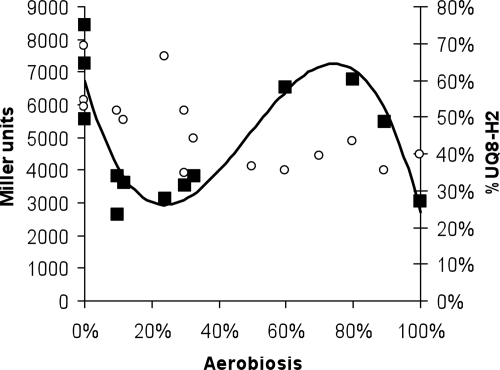
FIG. 5.
Activity of the ArcA∼P-dependent-lacZ reporter construct (ASA12) (filled squares) and redox state of the UQ8 pool (open circles) under various aerobiosis conditions. The construct was tested in the wild-type ASA12 background after glucose-limited continuous growth in mineral medium supplemented with various amounts of oxygen.
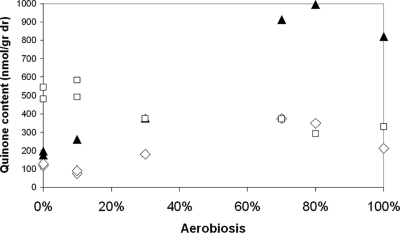
In view of the in vitro results of Georgellis et al. (11) and Malpica et al. (26), the cellular content of the ubiquinol-ubiquinone pools was measured for all steady-state chemostat cultures by HPLC-UV analysis that allowed separation of the reduced and oxidized quinone species (5). As expected, the ubiquinol/ubiquinone ratio gradually increased toward anaerobiosis (Fig. 5) and exhibited a pattern that does not resemble that of the changes observed for cydA−176+1-lacZ expression. Furthermore, in accordance with previous reports for aerobic and anaerobic batch conditions (37), the absolute cellular content of ubiquinol and ubiquinone combined increased gradually with increasing oxygen availability up to 80% aerobiosis (Fig. 6), which was followed by a decrease from 80 to 100% aerobiosis. Thus, it appears that the size of the ubiquinol pool correlates with cydA−176+1-lacZ expression throughout the range from 20 to 100% aerobiosis but not at aerobiosis levels below 20% (i.e., at very low oxygen supply rates).
FIG. 6.
Contents of total UQ8 (black triangles), DMK8 (open diamonds), and MK8 pool (open squares) under various aerobiosis conditions. Strain ASA12 was tested in glucose-limited continuous growth conditions in mineral medium supplemented with various amounts of oxygen. The values are data from single experiments.
Alternative effectors of ArcB activity under microaerobiosis conditions.
Matsubara and Mizuno (27) have identified a phosphatase (SixA) that selectively dephosphorylates ArcB∼P. One possible explanation for the decreased ArcB-dependent activation of the cydA−176+1-lacZ reporter in the range from 0 to 80% aerobiosis is SixA-mediated dephosphorylation of ArcB. Therefore, the ArcA∼P reporter system was inserted by P1 phage transduction into strain JW2337, which lacks sixA, and strain BW25113 (the corresponding wild-type strain), resulting in strains JA029 and JA001, respectively. These two strains were grown in glucose-limited chemostat cultures at a constant growth rate (dilution rate, 0.15 h−1) with variable but controlled rates of oxygen input through the entire aerobiosis range. No phenotypic differences were observed between these two strains with respect to cydA−176+1-lacZ expression (see Fig. S4 in the supplemental material), and therefore the possibility of a role for SixA in the regulation of ArcB activity under these aerobiosis conditions was excluded.
Catabolic intermediates, such as pyruvate, acetate, and lactate, have been shown in vitro to modulate ArcBA activity (20, 34). The experimental setup used to vary the degree of aerobiosis allows calculation of metabolic fluxes. The acetate flux showed a linear decrease toward aerobiosis, as shown previously by Alexeeva (1). Furthermore, fluxes toward lactate and pyruvate were virtually absent (data not shown). We therefore concluded that these metabolites do not affect ArcB activation under these aerobiosis conditions.
Due to the low maximal growth rate (<0.2 h−1) of strain JA022, no comparative study in chemostat cultures with different oxygen availabilities could be carried out. To assess the role of the demethylmenaquinones in the regulation of ArcB activation, the cellular menaquinone and demethylmenaquinone contents were determined (Fig. 6). Unfortunately, we could not isolate these menaquinone species in their reduced forms due to (auto)oxidation during sample processing (data not shown). The cellular menaquinone content increased substantially between 100% and 80% aerobiosis and then increased only slightly further toward anaerobiosis. The demethylmenaquinone pool exhibited a similar pattern in the high-aerobiosis region, but there was a gradual decrease toward anaerobiosis. Therefore, the aerobiosis ranges in which there was increasing ArcB activation (i.e., from 100 to 80% and from 20 to 0%) correlate with an increase in the size of the menaquinone pool.
DISCUSSION
The regulatory signals that activate and the mechanisms that underlie the function of the ArcBA two-component system have been the subject of numerous studies (17, 19-21, 24, 26, 27, 34). ArcBA is at the core of the catabolic network of E. coli, and hence a detailed understanding of its role should give us valuable insight into the regulation of energy conservation in bacteria. This not only is of fundamental scientific importance but also is essential to a range of biotechnological applications for obvious reasons.
The results presented here show that the ArcB regulation system is not a straightforward linear system and also show that there is a nonlinear response system with regard to oxygen availability. Indisputably, the redox state of the ubiquinone pool and/or the concentration of ubiquinone per se (11, 12, 26) plays an important role in transducing environmental signals and may be a key factor in the regulatory network that governs catabolism. However, the complexity of the regulatory network is illustrated by the fact that no correlation was found between ArcB activation and oxygen availability (Fig. 5). If a single redox-active compound regulates ArcB activation, a sigmoidal relationship with the degree of aerobiosis would be expected. This study used steady-state growth conditions with different, but controlled, changes in the rate of oxygen supply. This facilitated description of these conditions in terms of redox state, the size of the ubiquinone pool, and the total size of the menaquinone pools and allowed correlation of the information obtained with ArcB kinase activity by use of a quantifiable reporter system. Here we provide evidence that our reporter system does indeed function according to its design and monitors the degree of ArcB activation. This allowed us to conduct a more detailed in vivo analysis of this process than has been reported previously (26, 34). Unfortunately, we have not successfully resolved the relative concentrations of the oxidized and reduced forms of menaquinone. Unidentified components presumably cause a high and variable rate of (auto)oxidation of the menaquinols, even when the cell extracts are acidified. The considerably more negative midpoint potential of menaquinone than of ubiquinone (−80 mV versus 110 mV [40]) may contribute to this sensitivity to autooxidation. It is relevant to note in this respect that Grammel and Ghosh (15) recently described an analysis of the redox state of the two quinone pools of Rhodospirullum rubrum (i.e., rhodoquinone and ubiquinone). These workers were unable to detect significant amounts of rhodoquinol in vivo. Rhodoquinol has a midpoint potential that is close to that of menaquinol (i.e., −63 mV [29]). We noted, however, that the decreasing rate of succinate production (1) at the lowest levels of aerobiosis is consistent with the assumption that oxidized menaquinone is formed at the lowest rate of oxygen supply (i.e., <20% aerobiosis).
The observation made with both batch and chemostat cultures that significant variation in the ubiquinol/ubiquinone ratio is not reflected in alteration of ArcB kinase activity therefore justifies the conclusion that this protein histidine kinase must be under control of an additional factor.
Previously, we have presented evidence indicating that the ArcBA system is a subtle microaerobic sensory and regulatory system (3) rather than an aerobic-anaerobic detection device. We argued that such a system is needed, given the fact that key enzymes of the fermentative and respiratory modes of energy conservation may be inactivated by the presence of traces of oxygen and under highly reducing conditions, respectively. Consequently, our physiological analyses prompted us to suggest that the current model of ArcB regulation is an oversimplification and that a more complex regulatory system is expected to operate. Differential regulation of the SixA system (with respect to oxygen availability) would be an elegant means to fine-tune ArcB kinase activity. However, our results show that this is not the case (i.e., no difference in expression of cydA−176+1-lacZ was observed between strain JA029 [ΔsixA] and JA001 [wild type]).
Previous work (1) has shown that the steady-state cellular concentration of NADH increases substantially in both the upper (100 to 80%) and lower (20 to 0%) aerobiosis ranges, with a twofold increase at both transitions. This NADH pattern coincides with the changes in the cydA−176+1-lacZ expression in the same aerobiosis ranges. These data therefore do not exclude the interpretation that NADH functions as an additional signal input. The lack of a high level of anaerobic cydA−176+1-lacZ expression in a mutant deficient in menaquinone biosynthesis does indicate, however, that NADH does not function as an activation signal as such, since the concentration of NADH in such a mutant is expected to even increase under anaerobic conditions compared to the concentration in the wild-type strain. Rather, alternative electron acceptors, like fumarate, may lower the level of ArcB activation under these conditions.
Demethylmenaquinone was not tested here or by other workers (11) as an in vitro regulatory signal for ArcB autophosphorylation activity due to the lack of commercially available demethylmenaquinone. Georgellis et al. (11) showed that both ubiquinone (UQ0) and menadione (MK3) influence the in vitro phosphorylation rate of the ArcB kinase. However, the concentrations required for half-maximal inhibition are higher for menadione (50 μM) than for ubiquinone (5 μM). Strikingly, these authors observed that ubiquinol and menadiol did not activate ArcB autophosphorylation. Such an effect, however, may have been masked by isolation of the ArcB kinase in its activated form.
The fact that the midpoint potential of the menaquinones is significantly more negative than that of ubiquinones (see above) makes it likely that in the range of aerobiosis, moving from anaerobic conditions, the menaquinone-H2/menaquinone ratio decreases significantly before the same transition occurs for the ubiquinone couple. One possibility is that this menaquinone transition takes place between 0 and 20% aerobiosis. The resulting increase in menaquinone concentration may then explain the corresponding decrease in ArcB activation. The increase in ArcB activation between 20 and 80% aerobiosis (as well as the decrease between 80 and 100% aerobiosis) correlates with the cellular concentration of ubiquinol (while simultaneously the concentration of the menaquinone pool decreases), suggesting that in the higher aerobiosis ranges it is the ubiquinol couple that governs the activation level of ArcB. This complex mode of regulation of ArcB is supported by the relative affinities of ArcB for menaquinones and ubiquinones, which is higher for the ubiquinones in vitro (11). This working hypothesis is shown in Fig. 7, and it extends the view that the ubiquinone-ubiquinol couple interacts with the PAS domain of ArcB (26) to include an interaction of this domain with the menaquinone-menaquinol couple. Accordingly, combined regulation by the redox state of the ubiquinone pool and menaquinone pool provides a consistent hypothesis to explain the observed complex regulation of ArcB activation at variable rates of oxygen supply. Menaquinols would then be the dominant activators under anaerobic conditions, where the size of the ubiquinone pool is approximately five times less than the size of the menaquinone pool. Given the fact that menaquinones are involved in the transfer of electrons to alternative acceptors (7, 16, 31, 33) and the fact that the cell maintains a subtle balance between aerobic respiration, anaerobic respiration, and fermentation, this work shows that ArcB can be viewed as a master regulator that governs both anaerobic and aerobic respiration. In this respect, the system should be considered a redox-sensing system rather than an oxygen-sensing system.
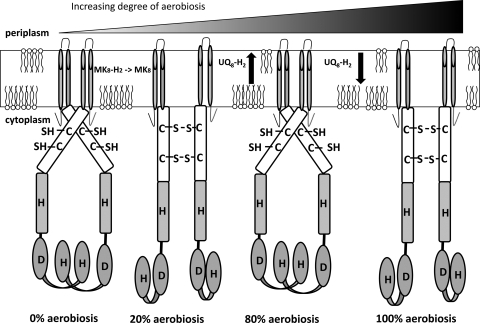
FIG. 7.
Simplified model for modulation of ArcB activity by the degree of aerobiosis. Upon a shift from anaerobic growth conditions to low-aerobiosis growth conditions, the menaquinone pool oxidizes rapidly, resulting in an inactive ArcB kinase. Too little ubiquinol is present to prevent binding of the oxidized form of ubiquinone. A further increase in aerobiosis to high-microaerobiosis conditions (80% aerobiosis) results in an increase in the total ubiquinone pool and therefore an increase in ubiquinol, allowing binding of ubiquinol, which brings back the cysteine in the reduced form. In completely aerobic conditions the content of the quinone pool decreases, which results in oxidation of the key cysteines and in inactivation of ArcB. Adapted from the work of Malpica et al. (26). H, histidine kinase domain; D, receiver domain.
Supplementary Material
Footnotes
Published ahead of print on 20 November 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexeeva, S. 2000. Molecular physiology of responses to oxygen in Escherichia coli. Ph.D. thesis. University of Amsterdam, Amsterdam, The Netherlands.
- 2.Alexeeva, S., B. de Kort, G. Sawers, K. J. Hellingwerf, and M. J. de Mattos. 2000. Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. J. Bacteriol. 182:4934-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexeeva, S., K. J. Hellingwerf, and M. J. Teixeira de Mattos. 2003. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J. Bacteriol. 185:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bekker, M., G. Kramer, A. F. Hartog, M. J. Wagner, C. G. de Koster, K. J. Hellingwerf, and M. J. de Mattos. 2007. Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 153:1974-1980. [DOI] [PubMed] [Google Scholar]
- 6.Bensadoun, A., and D. Weinstein. 1976. Assay of proteins in the presence of interfering materials. Anal. Biochem. 70:241-250. [DOI] [PubMed] [Google Scholar]
- 7.Butler, C. S., S. A. Fairhurst, S. J. Ferguson, A. J. Thomson, B. C. Berks, D. J. Richardson, and D. J. Lowe. 2002. Mo(V) co-ordination in the periplasmic nitrate reductase from Paracoccus pantotrophus probed by electron nuclear double resonance (ENDOR) spectroscopy. Biochem. J. 363:817-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. U. S. A. 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotter, P. A., S. B. Melville, J. A. Albrecht, and R. P. Gunsalus. 1997. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol. Microbiol. 25:605-615. [DOI] [PubMed] [Google Scholar]
- 9.Drapal, N., and G. Sawers. 1995. Purification of ArcA and analysis of its specific interaction with the pfl promoter-regulatory region. Mol. Microbiol. 16:597-607. [DOI] [PubMed] [Google Scholar]
- 10.Evans, C. G. T., D. Herbert, and D. W. Tempest. 1970. The continuous culture of microorganisms. Construction of a chemostat., vol. 2. Academic Press, London, United Kingdom.
- 11.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 12.Georgellis, D., A. S. Lynch, and E. C. Lin. 1997. In vitro phosphorylation study of the arc two-component signal transduction system of Escherichia coli. J. Bacteriol. 179:5429-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giacomini, A., V. Corich, F. J. Ollero, A. Squartini, and M. P. Nuti. 1992. Experimental conditions may affect reproducibility of the beta-galactosidase assay. FEMS Microbiol. Lett. 79:87-90. [DOI] [PubMed] [Google Scholar]
- 14.Govantes, F., J. A. Albrecht, and R. P. Gunsalus. 2000. Oxygen regulation of the Escherichia coli cytochrome d oxidase (cydAB) operon: roles of multiple promoters and the Fnr-1 and Fnr-2 binding sites. Mol. Microbiol. 37:1456-1469. [DOI] [PubMed] [Google Scholar]
- 15.Grammel, H., and R. Ghosh. 2008. Redox-state dynamics of ubiquinone-10 imply cooperative regulation of photosynthetic membrane expression in Rhodospirillum rubrum. J. Bacteriol. 190:4912-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingledew, W. J., and R. K. Poole. 1984. The respiratory chains of Escherichia coli. Microbiol. Rev. 48:222-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iuchi, S., D. C. Cameron, and E. C. Lin. 1989. A second global regulator gene (arcB) mediating repression of enzymes in aerobic pathways of Escherichia coli. J. Bacteriol. 171:868-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iuchi, S., and E. C. Lin. 1993. Adaptation of Escherichia coli to redox environments by gene expression. Mol. Microbiol. 9:9-15. [DOI] [PubMed] [Google Scholar]
- 19.Iuchi, S., and E. C. Lin. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. U. S. A. 85:1888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iuchi, S., Z. Matsuda, T. Fujiwara, and E. C. Lin. 1990. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol. Microbiol. 4:715-727. [DOI] [PubMed] [Google Scholar]
- 21.Jeon, Y., Y. S. Lee, J. S. Han, J. B. Kim, and D. S. Hwang. 2001. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J. Biol. Chem. 276:40873-40879. [DOI] [PubMed] [Google Scholar]
- 22.Keleti, K. 1990. Kinetics of regulatory enzymes, p. 266-284. In Basic enzyme kinetics. MIR, Moscow, Russia.
- 23.Kwon, O., D. Georgellis, and E. C. Lin. 2000. Phosphorelay as the sole physiological route of signal transmission by the arc two-component system of Escherichia coli. J. Bacteriol. 182:3858-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, X., and P. De Wulf. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279:12588-12597. [DOI] [PubMed] [Google Scholar]
- 25.Lynch, A. S., and E. C. Lin. 1996. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J. Bacteriol. 178:6238-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malpica, R., B. Franco, C. Rodriguez, O. Kwon, and D. Georgellis. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. U. S. A. 101:13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsubara, M., and T. Mizuno. 2000. The SixA phospho-histidine phosphatase modulates the ArcB phosphorelay signal transduction in Escherichia coli. FEBS Lett. 470:118-124. [DOI] [PubMed] [Google Scholar]
- 28.Matsui, H., T. Kurosaki, M. Tokuda, and O. Hatase. 1983. Modulation of optical density by sulfhydryl reagents in microbiuret method: a modified method for protein determination in the presence of sulfhydryl reagents. Acta Med. Okayama 37:125-129. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto, J., K. Sakamoto, N. Shinjyo, Y. Kido, N. Yamamoto, K. Yagi, H. Miyoshi, N. Nonaka, K. Katakura, K. Kita, and Y. Oku. 2008. Anaerobic NADH-fumarate reductase system is predominant in the respiratory chain of Echinococcus multilocularis, providing a novel target for the chemotherapy of alveolar echinococcosis. Antimicrob. Agents Chemother. 52:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Potter, L., H. Angove, D. Richardson, and J. Cole. 2001. Nitrate reduction in the periplasm of gram-negative bacteria. Adv. Microb. Physiol. 45:51-112. [DOI] [PubMed] [Google Scholar]
- 32.Ramanujam, P., S. Fogerty, W. Heiser, and J. Jolly. 1990. Fast gel electrophoresis to analyze DNA-protein interactions. Biotechniques 8:556-563. [PubMed] [Google Scholar]
- 33.Richardson, D. J., B. C. Berks, D. A. Russell, S. Spiro, and C. J. Taylor. 2001. Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell. Mol. Life Sci. 58:165-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez, C., O. Kwon, and D. Georgellis. 2004. Effect of d-lactate on the physiological activity of the ArcB sensor kinase in Escherichia coli. J. Bacteriol. 186:2085-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawers, G. 1993. Specific transcriptional requirements for positive regulation of the anaerobically inducible pfl operon by ArcA and FNR. Mol. Microbiol. 10:737-747. [DOI] [PubMed] [Google Scholar]
- 35a.Sawers, G., and B. Suppmann. 1992. Anaerobic induction of pyruvate formate-lyase gene expression is mediated by the ArcA and FNR proteins. J. Bacteriol. 174:3474-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segel, I. 1993. Multisite and allosteric enzymes. John Wiley & Sons, Inc., New York, NY.
- 37.Shestopalov, A. I., A. V. Bogachev, R. A. Murtazina, M. B. Viryasov, and V. P. Skulachev. 1997. Aeration-dependent changes in composition of the quinone pool in Escherichia coli. Evidence of post-transcriptional regulation of the quinone biosynthesis. FEBS Lett. 404:272-274. [DOI] [PubMed] [Google Scholar]
- 38.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 39.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.