Abstract
The λ S gene encodes a holin, S105, and an antiholin, S107, which differs by its Met-Lys N-terminal extension. The model for the lysis-defective character of S107 stipulates that the additional N-terminal basic residue keeps S107 from assuming the topology of S105, which is N-out, C-in, with three transmembrane domains (TMDs). Here we show that the N terminus of S105 retains its fMet residue but that the N terminus of S107 is fully deformylated. This supports the model that in S105, TMD1 inserts into the membrane very rapidly but that in S107, it is retained in the cytoplasm. Further, it reveals that, compared to S105, S107 has two extra positively charged moieties, Lys2 and the free N-terminal amino group, to hinder its penetration into an energized membrane. Moreover, an allele, S105ΔTMD1, with TMD1 deleted, was found to be defective in lysis, insensitive to membrane depolarization, and dominant to the wild-type allele, indicating that the lysis-defective, antiholin character of S107 is due to the absence of TMD1 from the bilayer rather than to its ectopic localization at the inner face of the cytoplasmic membrane. Finally, the antiholin function of the deletion protein was compromised by the substitution of early-lysis missense mutations in either the deletion protein or parental S105 but restored when both S105ΔTMD1 and holin carried the substitution.
In general, holins control the length of the infection cycle of double-stranded DNA phages (37). During late gene expression, the holin protein accumulates harmlessly in the bilayer until suddenly and spontaneously triggering the formation of holes in the membrane at an allele-specific time (13, 15). Holin genes are extremely diverse, but most can be grouped into two main classes based on the number of predicted transmembrane domains (TMDs): class I, with three TMDs and a predicted N-out, C-in topology, and class II, with two TMDs and a predicted N-in, C-in topology (38). Holin genes and function are subject to several levels of regulation, among which a particularly striking feature is the common occurrence of two potential translational starts, or dual-start motifs (5, 37), separated by only a few codons. Dual-start motifs are found in many holins of both of the two major classes; in nearly every case, the two starts are separated by at least one basic residue. The first dual-start motif to be characterized was that of λ S, the prototype class I holin gene (Fig. 1A and B). Translation initiation events occur at codons 1 and 3, giving rise to two products, S107 and S105, each named because of the length of its amino acid sequence; in the wild-type (wt) allele, two RNA structures define the ratio of initiations at the two start codons, resulting in an S105/S107 ratio of ∼2:1.
FIG. 1.
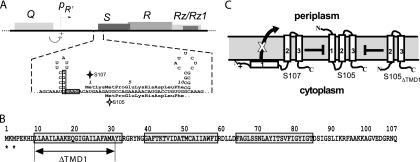
Gene, topology, and sequence of λ S. (A, top) The λ lysis cassette, including genes S, R, Rz, and Rz1, is shown, along with its promoter pR′, and Q, encoding the late gene activator. The 5′ end of the class I holin gene S has two start codons, Met1, the start for S107, and Met3, the start for S105, and two RNA structures that regulate initiations at these codons. The S105 and S107 alleles have Leu (CUG) codons in place of the Met3 and Met1 codons, respectively. (B) Primary structure of S proteins. Missense changes relevant to the text are shown. Starts for S107 and S105 are indicated by asterisks. The three TMDs are boxed (13), and the extent of the ΔTMD1 deletion is indicated. (C) Model for the membrane topology of S105, S107, and S105ΔTMD1. Topology and boundary residues for TMD1, -2, and -3 are based on Graschopf and Blasi (11) and Gründling et al. (13), respectively.
Although they differ only by the Met-Lys N-terminal extension of S107, the two proteins have opposing functions; S105 is the holin and S107 the antiholin. The antiholin function is reflected by four principal features: first, when the Met3 start is inactivated, the mutant allele, designated S107 (Fig. 1A), is lysis defective (26); second, the S107 protein binds and inhibits S105 specifically (3, 16); third, when S107 is produced in stoichiometric excess over S105, lysis is blocked for several times the length of the normal infection cycle (3, 4, 7, 16); and fourth, S107 antiholin function, i.e., inhibition of S105 hole formation, can be instantly subverted by collapsing the proton motive force, most easily done by addition of energy poisons to the medium (3). The predicted N-out, C-in topology and the requirement for the energized membrane led to a model in which S107 is initially inserted in the membrane with only two TMDs, with TMD1 being blocked from insertion by the presence of the positively charged residue, Lys2, whereas S105 has three TMDs (Fig. 1C) (39). From this perspective, S105-S107 complexes, which are approximately twice as numerous as the S105 homodimers, are defective in triggering hole formation. An appealing feature of this model is that when an S105-mediated hole formation event does occur in a cell, the resultant collapse of the membrane potential allows insertion of TMD1 of S107 into the membrane, instantly tripling the amount of active holin by making the previously inactive pool of S105-S107 complexes functional (38).
Some genetic and physiological evidence for the topology of the λ S proteins has been obtained using gene fusions. First, a fusion of the S gene at codon 105 with lacZ generates a functional, membrane-inserted β-galactosidase chimera, indicating, as expected, the cytoplasmic disposition of the highly charged C terminus of the S protein (40). Second, Graschopf and Bläsi (12) demonstrated that S-mediated hole formation could be obtained with constructs where a secretory signal sequence was fused to the N termini of both S105 and S107. Lysis required the cleavage of the signal sequence by leader peptidase, and export of the signal-S107 form was slower than for the signal-S105 form. However, evidence for the topology of native forms of S has not been available. Moreover, no basis for the inhibitory character of S107 has been established. In the simplest view, the antiholin function could be due to the absence of TMD1 from the bilayer or the ectopic localization of TMD1 in the cytoplasm, or both. Here, we report studies directed at dissecting the precise role of topology in S107 function and correlating antiholin activity with its ability to heterodimerize with S105. The results are discussed in terms of a general model for the formation of the holin lesion and the role of dynamic membrane topology in its temporal regulation.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, plasmids, culture growth, and phage titration.
The strains and plasmids used in this work are described in Table 1. Media, growth conditions, and inductions of the λ lysis genes from a prophage and/or plasmids have been described previously (7, 13, 15, 30). Bacterial cultures were grown in standard LB medium supplemented with ampicillin (100 μg/ml) (LB-Ap), kanamycin (40 μg/ml) (LB-Km), and chloramphenicol (10 μg/ml) (LB-Cm), or some combination, when appropriate. The ability of S alleles to be triggered by membrane depolarization was assessed by adding dinitrophenol (DNP) to a final concentration of 5 mM. CHCl3 was used at a 1% final concentration. N-Ethylmaleimide (NEM) and 1-10 phenanthroline were purchased from Sigma-Aldrich (St. Louis, MO). RY17456, which complements all lysis gene defects because it carries the lysis genes of phage 21 under the control of the λ late promoter, pR′, on plasmid pTP2 (Table 1), was used for determination of the efficiency of plating (EOP).
TABLE 1.
Bacteriophages, strains, and plasmids used in this study
| Phage, strain, or plasmid designation | Genotype and/or relevant features | Source or referencea |
|---|---|---|
| Phages | ||
| λΔ(SR) | Δ(stf-tfa)::cat cI857 Δ(SR) | 30 |
| λ81 | λcI Ram54 Ram60; the Ram54 and Ram60 mutations, previously unsequenced, were found to be RQ26am and RW73am, respectively | Stock |
| λ84 | λimm434cI RQ26am RW73am | Stock |
| λS105 | Δ(stf-tfa)::cat cI857 SM1L | 29 |
| λS105A52V | Same as λS105, but with an Ala52Val change in S105 | |
| λS107 | Δ(stf-tfa)::cat cI857 SM3L | |
| λS105ΔTMD1 | Isogenic to λS105 except codons for TMD1 (nt 27-90, codons 9-30 of S105 are deleted) (Fig. 1C) | |
| λSam | λSW56am (also known as Sam7) | |
| Strains | ||
| MC4100 | E. coli K-12 F−araD139 Δ(argF-lac)U169 rpsL15 relA1 flbB3501 deo pstF25 rbsR | Stock |
| BL21(DE3) slyD::Tn10 | E. coli B hyper-expression strain (Novagen), converted to slyD mutant by P1 transduction | 2 |
| RY17303 | MC4100ΔtonA | Stock |
| RY17366 | MC4100ΔtonA [λΔ(SR)]] | Stock |
| MG1655 | F−ilvG rfb-50 rph-1 | Stock |
| RY16504 | MG1655ΔtonA lacIq ΔlacY pQ | |
| MDS12 | MG1655 with 12 deletions, totaling 376,180 nt, including all cryptic prophages | 20 |
| RY17341 | MDS12ΔtonA | |
| RY17456 | MDS12ΔtonA pTP2 | |
| Plasmids | ||
| pQ | λ Q gene cloned into pZS*24; inducible with arabinose and IPTG (isopropyl-β-d-thiogalactopyranoside); Kmr | 14 |
| pS105 | S105 R Rz Rz1 genes of λ cloned into pBR322, Apr | 15 |
| pSwt | pS105 S+ | 16 |
| pKB1 | pS105 Sam7 | 15 |
| pS107 | pS105 SM3L | 16 |
| pS105ΔTMD1 | pS105 S105ΔTMD1 | |
| pS105ΔTMD1,A52G | pS105 S105ΔTMD1,A52G | |
| pλ81 | pS105 S+ RQ26am RW73am | |
| pλ81-2 | pS105 S+ RQ26am RW73am RzQ100am RzW38am | |
| pS105ΔTMD1,A52V | pS105 S105ΔTMD1,A52V | |
| pS105RamRzamRz1am | pS105 RQ26am RW73am RzQ100am RzW38am, plus a unique KpnI site constructed in R | |
| pS105RzamRz1am | pS105 RQ26am RW73am RzQ100am RzW38am | |
| pS107RamRzamRz1am | pS107 RQ26am RW73am RzQ100am RzW38am | |
| pS105ΔTMD1RamRzamRz1am | pS105ΔTMD1RQ26am RW73am RzQ100am RzW38am | |
| pS105A52VRamRzamRz1am | pS105 S105A52V RQ26am RW73am RzQ100am RzW38am | |
| pSamRamRzamRz1am | pS105SW56am RQ26am RW73am RzQ100am RzW38am | |
| pTP2 | Isogenic to pS105, but with the lysis genes S21, R21, Rz21, and Rz121 of lambdoid phage 21; the presence of this plasmid in an indicator strain complements defects in all λ lysis genes for plaque formation. | See supporting information, Table 1 of Park et al. (24) |
| pET-S105τ94 | pETlla S105τ94; product has GGH6GG oligohistidine tag inserted between residues 94 and 95 | 30 |
| pET-S107τ94 | Isogenic to pET-S105τ94 except in S107τ94 | |
| pET-S105ΔTMD1τ94 | Isogenic to pET-S105τ94 except in S105ΔTMD1 | |
| pET-22B-def-CHT | pET22B (Novagen), encoding oligohistidine-tagged E. coli deformylase | 27 |
Unless otherwise indicated, strains, phages, and plasmids were constructed in this study. Stock, laboratory stock.
DNA manipulations, mutagenesis, and sequencing.
Isolation of plasmid DNA, DNA amplification by PCR, DNA transformation, site-directed mutagenesis, and DNA sequencing were performed as previously described (13). Splicing by overlapping extension (SOE) PCR was performed as described previously (17). Oligonucleotides for PCR were obtained from Integrated DNA Technologies, Coralville, IA, and were used without further purification; sequences of the oligonucleotides used are available on request. Enzymes were purchased from New England Biolabs, except for Pfu polymerase, which was from Stratagene. Automated fluorescence sequencing was performed at the Laboratory for Plant Genome Technology at Texas AgriLife.
Plasmid construction and generation of recombinant bacteriophage.
The parental plasmid used was pS105 (Table 1), which is a medium-copy-number vector carrying the entire λ lysis cassette downstream of its native pR′ promoter (Fig. 1A). pS105 carries the S105 allele, which produces the S105 holin but not the S107 antiholin. The plasmid pS105ΔTMD1 was constructed from pS105 by site-directed mutagenesis, with nucleotides (nt) 27 to 90 of the S105 gene deleted. The plasmid pλ81 is identical to pS105 but has the lysis cassette from λ81, carrying the Ram54 Ram60 endolysin allele, cloned into the corresponding EcoRI and ClaI sites (λ nt 44972 to 46448), with a silent mutation (C to T at λ nt 44594) to ablate the second AatII site in the R gene. The EcoRV-ClaI fragment from pS105RzamRz1am containing a portion of the R gene and nonsense alleles of the Rz Rz1 gene pair was cloned into pλ81, creating pλ81-2. The S105 gene from pS105 was then cloned into pλ81-2 via SOE PCR to create pS105RamRzamRz1am, carrying the nonsense alleles of all the lysis genes except S. Ths plasmid was used to create pS107 RamRzamRz1am, pS105A52V RamRzamRz1am, pS105ΔTMD1RamRzamRz1am, and pSamRamRzamRz1am by site-directed mutagenesis.
Lysis gene alleles were transferred to the phage context by recombination. For nonlytic alleles of S105 (such as S105ΔTMD1), the cognate plasmids were transformed into RY17366 and individual transformants were grown up and induced as previously described (14). CHCl3 was added to the cultures 2 h after induction. Lysates were cleared by centrifugation (3,000 rpm in a clinical centrifuge for 10 min) and used to reinfect early-log cultures of RY17341 at an input multiplicity of infection (MOI) of 5. Two hours after infection, CHCl3 was added and the lysate again cleared. This enrichment step was repeated twice more, and the final lysate was used to lysogenize RY17303. For plaque-forming alleles, the method was the same except that the cleared lysates were directly plated on RY17341. Phage from purified plaques was used to lysogenize RY17303. Lysogens were selected by plating infected cells on LB-Cm plates at 30°C, screened by PCR for the presence of a single prophage as described by Powell et al. (25), and checked for the presence of R+ by cross-streaking with the hetero-immune Ram phage λ84.
Purification of S proteins.
His-tagged versions of S105, S107, and S105ΔTMD1 were purified as described previously (9), using the overexpression plasmids pET-S105τ94, pET-S107τ94, and pET-S105ΔTMD1, τ94. The last plasmid, encoding the His-tagged deletion protein, was constructed by site-directed mutagenesis of pET-S105τ94. N-terminal sequencing of these proteins was performed at the Protein Chemistry Laboratory at the Texas Agricultural Experiment Station.
Purification of Escherichia coli deformylase.
The plasmid pET-22B-def-CHT, encoding oligohistidine-tagged deformylase and kindly provided by Dehua Pei, was used to produce the deformylase in BL21(DE3) slyD::Tn10, as described previously (27). The enzyme was purified by immobilized metal affinity chromatography to a final concentration of 5 mg/ml. The activity of the purified enzyme was determined using the tri-peptide formyl-Met-Ala-Ser (Sigma) as the substrate. The appearance of N-terminal amines was monitored using 2,4,6-trinitrobenzenesulfonic acid (10).
Deformylation of S105.
To remove the formyl group on S105, 50 μl of purified S105 (50 μg) was added to 110 μl of 0.1 M MOPS (morpholinepropanesulfonic acid), 0.25 M KCl (pH 7.2), 5 μl catalase (10 mg/ml in water; Sigma), and 5 μl of purified deformylase at 5 mg/ml. The reaction mixture was incubated at 30°C for 20 m, and the reaction was stopped by trichloroacetic acid (TCA) precipitation. The protein pellet was resuspended in 40 μl of 1% Empigen BB (Calbiochem, La Jolla, CA), 20 mM BES [N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid], 0.1 M NaCl, pH 7. After addition of 20 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer, the samples were boiled for 5 min and subjected to 16% SDS-PAGE. The S105 band was excised and used for N-terminal sequencing by the Protein Chemistry Laboratory at Texas A&M University.
Protein analysis.
For TCA precipitations, 1-ml culture aliquots were added to 111 μl of cold, 6.1 N trichloroacetic acid and then placed on ice for 30 min. The precipitate was collected by centrifugation (18,000 × g in a tabletop microcentrifuge for 30 min) and washed once with acetone, with complete resuspension of the pellet. Pellets were air dried, resuspended in sample loading buffer, and then analyzed by SDS-PAGE as previously described (14), except that the gels used for Fig. 7 were not precast. Western blotting with anti-S antibodies (13) and oxidative disulfide bridge formation by Cu-phenanthroline treatment of membranes (16) were carried out as previously described. Relative band intensities were determined by analysis using ImageJ (http://rsb.info.nih.gov/ij/docs/intro.html).
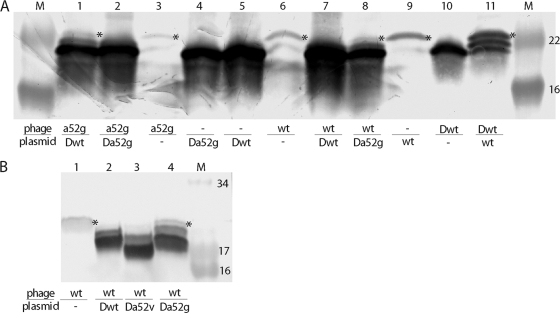
FIG. 7.
Allele-specific inhibition by S105ΔTMD1 correlates with S105 homodimer formation. Membranes prepared from inductions with the indicated prophage and/or transactivation plasmid were treated with Cu-phenanthroline, resolved by SDS-PAGE, and analyzed by Western blotting. An asterisk to the right of a band indicates the position of the disulfide-linked S105 homodimer. Below the panels, the S alleles on the phages and plasmids are indicated by the following abbreviations: wt, S105; a52g, S105A52G; Δ, ΔTMD1; Δwt, S105ΔTMD1; Δa52g, S105ΔTMD1,A52G. M, molecular mass markers, with approximate masses (in thousands) indicated to right of the blot in each panel. (A) Results of 12 to 20% Tris-glycine SDS-PAGE. Lanes: 1, λS105A52G, plasmid S105ΔTMD1; 2, λS105A52G, plasmid S105ΔTMD1,A52G; 3, λS105A52G; 4, plasmid S105ΔTMD1,A52G; 5, plasmid S105ΔTMD1; 6, λS105; 7, λS105, plasmid S105ΔTMD; 8, λS105, plasmid S105ΔTMD,A52G; 9, plasmid S105; 10, λS105ΔTMD1; 11, λS105ΔTMD1, plasmid S105. (B) Results of 16% Tris-Tricine SDS-PAGE. Lanes: 1, λS105; 2, λS105, plasmid S105ΔTMD1; 3, λS105, plasmid S105ΔTMD1,A52V; 4, λS105, plasmid S105ΔTMD1,A52G.
Phage accumulation after the induction of λ lysogens.
At various times after induction, 1-ml aliquots were treated with 1% CHCl3 to release progeny phage and the titer was determined by plating on lawns of RY17456. Plaques from triplicate platings were counted after 12 to 16 h of incubation at 37°C, and the averages from two experiments were used.
Viability assays.
Two-milliliter samples of induced cultures of RY16504 carrying plasmids with the S105 allele indicated in Fig. 2 and nonsense alleles of the other three lysis genes were placed on ice. Half of each sample was serially diluted on ice and plated in triplicate on LB-Km-Ap, and the remainder was precipitated with TCA, as previously described (33), and analyzed by immunoblotting. Colonies from three separate experiments were counted after 12 to 16 h of incubation at 37°C.
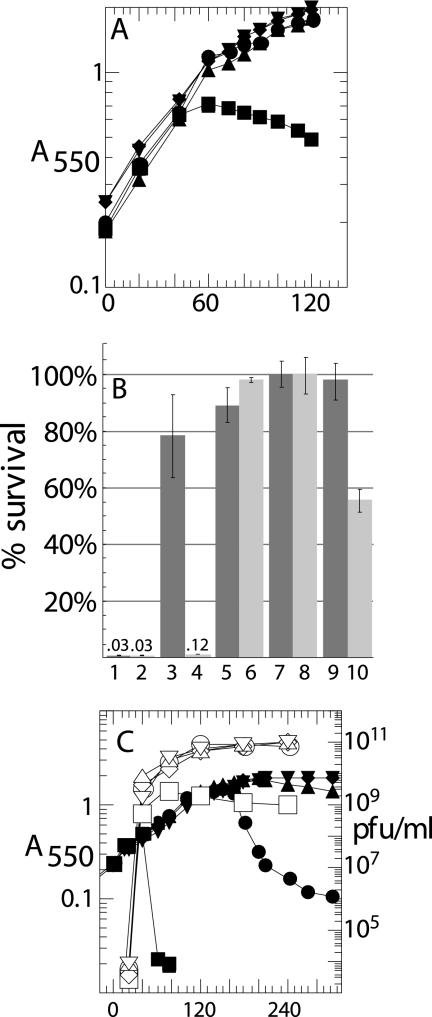
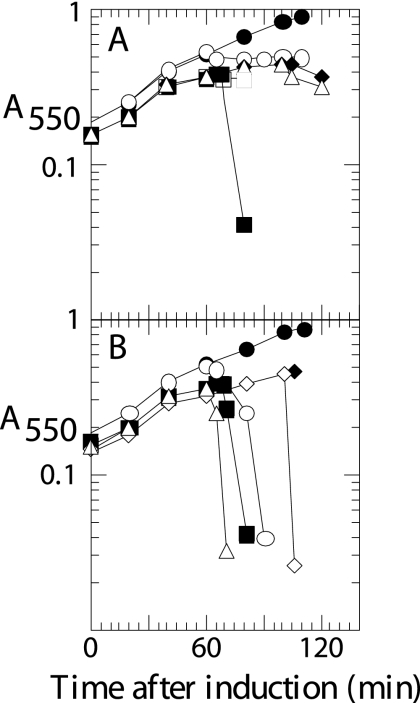
FIG. 2.
S105ΔTMD1 is an absolute defective holin. (A and C) Cultures of the nonsuppressing hosts RY16504 (lacIq) (A) and RY17303 (Δlac) (C) carrying the indicated prophages and/or plasmids were induced at time zero and monitored for A550 (filled symbols) as described in Materials and Methods. The abscissa is time after induction (min) for all panels except B. (A) Plasmids pS105RamRzamRz1am (▪), pS107RamRzamRz1am (•), pS105A52VRamRzamRz1am (▴), pS105ΔTMD1RamRzamRz1am (⧫), and pS105amRamRzamRz1am (▾). (B) Titers of surviving phage in cultures from panel A, plated at 60 min (odd-numbered bars) or 120 min (even-numbered bars). S alleles S105 (bars 1 and 2), S107 (bars 3 and 4), SA52V (bars 5 and 6), S105W56am (bars 7 and 8), and S105ΔTMD1 (bars 9 and 10). Numbers above bars 1, 2, and 4 indicate percent survival. (C) Prophages λS105 (▪), λS107 (•), λS105A52V (▴), λS105ΔTMD1Δ (⧫), and λS105W56am (▾). Accumulations of phage (PFU) are also shown with corresponding open symbols.
Chemical mutagenesis of phage.
Hydroxylamine mutagenesis of λS105ΔTMD1 phage was performed as described previously (8). Following mutagenesis, the titers of the samples were determined by plating on lawns of the complementing indicator, RY17456. PFU (108) were plated at an MOI of ∼0.1 to 0.2 on RY17341 on each of 10 to 15 plates. No plaques were observed in three experiments, with a total of ∼5 × 109 virions plated. Ethylmethane sulfonate (EMS) mutagenesis of λS105ΔTMD1 was performed as described previously (22), with the following adaptations. A logarithmic culture of RY17434 was induced at an A550 of ∼0.2, and EMS was added after 15 min. The culture was grown for 4 h at 37°C, harvested by centrifugation, washed in λ dilution buffer, resuspended in 2 ml of λ dilution buffer, and disrupted in a French pressure cell (Spectronic Instruments, Rochester, NY) at 16,000 lb/in2 (1 lb/in2 = 6.89 kPa). The titer of the lysate was determined on lawns of RY17456. A control culture of RY17341 was treated identically, except that instead of being passed through a French pressure cell, the culture was serially diluted and plated on LB plates and LB plates containing 100 μg/ml rifampin to measure survival and mutation frequency. An aliquot containing 108 PFU was plated at an MOI of ∼0.1 to 0.2 on RY17341 on each of 12 plates. The experiment was repeated twice, and no plaques were observed.
RESULTS
The N terminus of S105, but not S107, retains its fMet residue.
Membrane proteins that achieve N-out topology without a cleavable signal sequence are relatively uncommon, but one example, Lep, or signal peptidase II, has been studied in detail. The N terminus of Lep was found to retain its N-terminal formyl group, as a consequence of its rapid externalization to the periplasm, thereby avoiding the normal processing by cytoplasmic polypeptide deformylase, Def (35). If the model indicating that S105 and S107 have different topologies is correct, the N terminus of S105, but not S107, might also escape deformylation. To test this idea, the S105 and S107 proteins were purified and subjected to N-terminal sequencing. The N-terminal sequence for S107 was found to correspond exactly to that predicted from the dual-start motif and was obtained at a high repetitive yield (Table 2). In contrast, no sequence information was obtained for S105, suggesting that its α-amino group was blocked. When S105 was pretreated with purified E. coli Def, a sequence consistent with translational initiation at the Met3 codon was obtained (Table 2), demonstrating that S105 indeed retains the N-terminal fMet residue. This directly supports the model for S105 and S107 topology and suggests that the N terminus of S105 exits the cytoplasm rapidly enough to escape cytoplasmic deformylation. Moreover, this suggests that the inability of S107 to assume an S105-like topology in the energized inner membrane is due to two extra positive charges in the protonated free amino groups at Lys2 and the N terminus, compared to none in S105.
TABLE 2.
Sequencing of purified S proteins
| Samplea | Repetitive yield (%) | Sequence(s) |
|---|---|---|
| S105 | 0 | |
| S107* | 82 | MKLPEK |
| Def-treated S105 | >95 | XXHDLLA, XXXHDLLAb |
| S105ΔTMD1 | 91 | PEKHD |
Purified His-tagged proteins. S107* is the product of the S107τ94 allele, in which codon 3, the start codon of the S105 reading frame, is converted to a CTG (Leu) (30).
TMD1 of S is essential for holin activity.
In view of the topological difference between S105 and S107, it was of interest to know whether TMD1 is required for the holin function of S105, the antiholin function of S107, or both. To address these questions, we deleted codons 9 to 30, which encode TMD1, from S105 to make the allele S105ΔTMD1. We also constructed the S105ΔTMD1 equivalents of S105A52G and S105A52V, which are, respectively, early-lysis and lysis-defective alleles of S105 (14, 18). S105ΔTMD1 proved to be nonlytic in the context of the phage and also nonlethal when induced from a plasmid (Fig. 2A and B). After 1 h of induction, the plasmid-borne deletion allele had no effect on host viability and was indistinguishable from a null SW56am allele, the lysis-defective S105A52V allele, and the S107 allele, which produces only the S107 antiholin. Moreover, after prolonged incubation, the deletion allele, like the null allele SW56am, retained its nonlethal character, whereas a significant proportion of cells expressing S107 were killed (Fig. 2B, last four bars), presumably reflecting the eventual penetration of the membrane by the N-terminal domain of S107. When the deletion allele was recombined onto the phage, not only was there an absolute lysis defect, but progeny accumulated for hours after induction, indicating a complete absence of membrane toxicity (Fig. 2C). These phenotypic defects were not due to the mislocalization or instability of the S105ΔTMD1 protein, which accumulated exclusively in the membrane (data not shown) and to higher levels than either S105 or S107 (Fig. 3A). The higher expression is consistent with the loss, in the deletion of codons 9 to 30 in SΔTMD1, of a regulatory RNA secondary structure spanning codons 11 through 16 (Fig. 1A), previously shown to be required for regulation of the S105 reading frame (4, 23, 26). Moreover, N-terminal sequencing of purified S105ΔTMD1 protein revealed that its N terminus was unblocked, indicating that it was retained in the cytoplasm (Table 2). Thus, deletion of TMD1 did not affect the topology of the remainder of the protein.
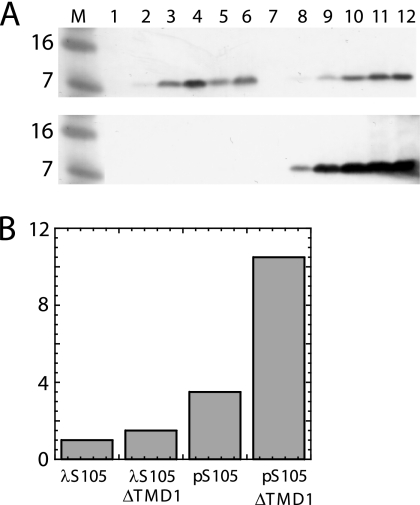
FIG. 3.
Accumulation of S gene products after induction. (A) RY17303 cultures carrying λS105 (lanes 1 to 6, upper blot), λS107 (lanes 7 to 12, upper blot), λSam (lanes 1 to 6, lower blot), or λS105ΔTMD1 (lanes 7 to 12, lower blot) were induced and samples taken by TCA precipitation at 0, 15, 30, 45, 60, and 90 min after induction (respectively for each grouping of lanes 1 to 6 and 7 to 12), resolved by 16% Tris-Tricine SDS-PAGE, and analyzed by Western blot analysis using anti-S antibodies. The λS105 sampling was curtailed after culture lysis. Lane M, molecular weight markers (in thousands). (B) Quantification of S105 and S105ΔTMD1. Experiments were performed as described for panel A, except RY17303 carried the indicated prophage or plasmid, samples were taken at 15 min after induction, and band intensities (y axis) were quantified, relative to the amount in the λS105 sample.
When plated on wt E. coli K-12 lawns, λS105ΔTMD1 generated plaques but at a much lower frequency (efficiency of plating [EOP], ∼1.5 × 10−8) than λSW56am, which plates with an EOP of ∼2 × 10−5 under these conditions. However, the plaques derived from λS105ΔTMD1 proved to be pseudorevertants derived from recombination with cryptic prophage elements, resulting in replacement of the S105ΔTMD1 gene with that of a heterologous functional holin (data not shown; also see Kaiser [19]). On strain MDS12, from which all the vestigial prophage elements of the E. coli chromosome have been deleted (20), no plaque-forming phage could be isolated, even with extensive chemical mutagenesis, from more than 5 × 109 virions (not shown; see Materials and Methods). The much higher reversion rate of λSW56am under the same conditions suggests that the deletion allele cannot be converted to lytic function even with several mutations, despite its intrinsically higher expression level. We conclude that the N-terminal TMD of S105 is essential for its holin function, although not for its localization or synthesis.
The antiholin character of S107 is due to the absence of TMD1 from the bilayer.
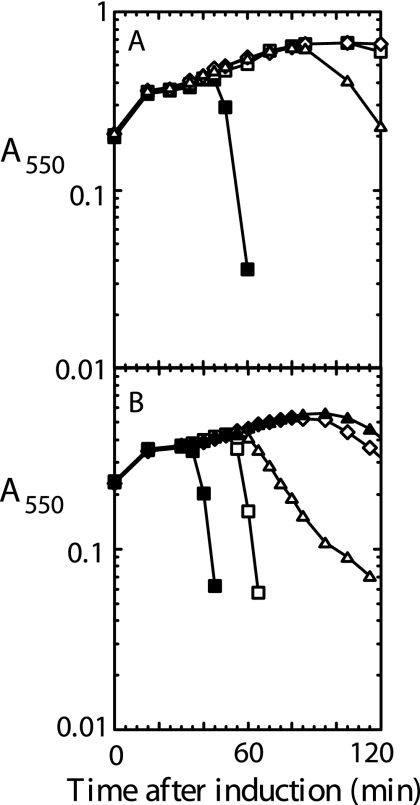
The antiholin character of S107 could be due to the lack of TMD1 in the bilayer or the presence of the ectopically localized TMD1 in the cytoplasm, or both. To address this issue, we next decided to test the S105ΔTMD1 protein for antiholin function. Accordingly, either the S105ΔTMD1 or the S107 allele in its normal transcriptional context was introduced on a medium-copy-number plasmid into a lysogen carrying the λS105 prophage. Lysogenic induction of these strains results in transactivation of the plasmid genes, concomitant with the onset of late gene expression in the prophage. The presence of pS107 or pS105ΔTMD1 in trans resulted in identical levels of inhibition of lysis (Fig. 4). Thus, the S105ΔTMD1 allele is dominant negative and its product can function as an antiholin. Unlike S107 and consistent with the observation that TMD1 is essential for holin function, S105ΔTMD1 could not be triggered by the addition of energy poisons (Fig. 4A, open circles). Moreover, unlike with S107, which is sensitive to energy poisons (Fig. 4B), the ability of S105ΔTMD1 to inhibit the function of S105 could not be subverted by addition of DNP (Fig. 4A, open squares and triangles). These results suggest that S107 antiholin function is determined by the lack of TMD1 in the membrane, and its leakiness and its ability to be triggered by energy poisons reflect the imperfect barrier of the bilayer for transit of TMD1.
FIG. 4.
S105ΔTMD1 is an untriggerable antiholin. Cultures carrying the indicated prophage, with or without the indicated transactivation plasmid, were induced and monitored as described for Fig. 2. (A) Inductions with or without pS105ΔTMD1RamRzamRz1am in trans were carried out with λS105 (▪), λS105ΔTMD1 (•), λS105ΔTMD1 with DNP added at 40 min (○), λS105 plus pS105ΔTMD1 (⧫), λS105 plus pS105ΔTMD1RamRzamRz1am with DNP added at 40 min (□), and λS105 plus pS105ΔTMD1RamRzamRz1am with DNP added at 80 min (Δ). (B) Inductions with or without pS107RamRzamRz1am in trans were carried out with λS105 (▪), λS107 (•), λS107 with DNP added at 40 min (○), λS105 plus pS107RamRzamRz1am (⧫), λS105 plus pS107RamRzamRz1am with DNP added at 40 min (Δ), and λS105 plus pS107RamRzamRz1am with DNP added at 80 min (⋄).
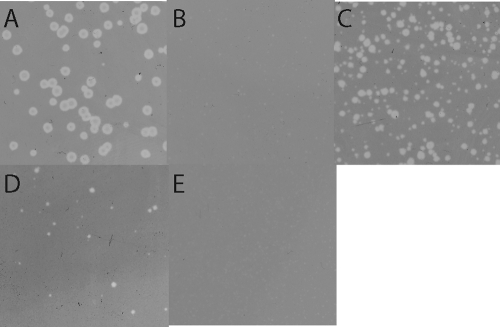
The S105ΔTMD1-mediated block was also reflected in the inability of λS105 to plate (EOP, ∼10−2) on lawns carrying the plasmid pS105ΔTMD1, although it plated near normal efficiency (EOP, ∼0.6) on lawns carrying pS107 (Fig. 5). Presumably, this reflects the eventual collapse of the proton motive force in the infected cells, which, with S107 but not S105ΔTMD1, allows penetration of TMD1 into the bilayer and thus conversion to the functional holin form. Even if this occurred very late in each infection cycle, the ability to form plaques would be buffered by the greatly increased burst size (26). Part of the stricter barrier to plaque formation in cells carrying the plasmid-borne S105ΔTMD1 allele may be due to increased production of the deletion product, relative to the level of S105 produced by the infecting phage. Indeed, quantitative Western blotting revealed that the amount of S105ΔTMD1 produced from the transactivation plasmid construct was about 7-fold higher than the amount of S105 produced from the phage context (Fig. 3B). When the deletion allele was transferred to the context of the phage and S105 to the plasmid, spontaneous lysis was observed (Fig. 6A), but the lengthy delay of the triggering time (from 30 min with pS105 alone to ∼65 min for pS105 in trans to the S105ΔTMD1 prophage) indicates the antiholin character of S105ΔTMD1. These results indicate that S107 antiholin function derives from the absence of TMD1 in the membrane rather than from its ectopic localization in the cytoplasm.
FIG. 5.
Allele-specific inhibition of plaque formation in lawns carrying S105ΔTMD1. Dilutions of λS105 were plated on lawns of RY17341 carrying transactivating plasmids with the indicated S allele. (A) SW56am; (B) S105ΔTMD1; (C) S107; (D) S105ΔTMD1,A52G; (E) S105ΔTMD1,A52V.
FIG. 6.
Allele-specific inhibition of λS105 lysis by S105ΔTMD1. RY17303 cultures bearing the indicated prophage and carrying the transactivation plasmid with the indicated allele were induced and monitored as described for Fig. 2. (A) All cultures had λS105 with the plasmid allele SW56am (▪), S105ΔTMD1 (□), S105ΔTMD1,A52V (⋄), or S105ΔTMD1,A52G (Δ). (B) ▪, λS105A52G with SW56am; □, λS105 with SW56am; Δ, λS105A52G with S105ΔTMD1; ▴, λS105 with S105ΔTMD1; ⋄, λS105A52G with S105ΔTMD1,A52G.
The dependence of holin and antiholin function on TMD2-TMD2 interactions.
Previous studies have indicated that the S107 antiholin inhibits S105 by a homotypic TMD2-TMD2 interaction, detected by the formation of disulfides between Cys51 residues as a result of treating membrane fractions with Cu2+-phenanthroline (Cu-Phe). This is also the case for the deletion allele (Fig. 7). For the inductions with the S105 allele on the prophage and the deletion allele on the plasmid that do not ultimately result in lysis, S105-S105ΔTMD1 heterodimers and S105ΔTMD1 homodimers could be detected, but no S105 homodimers could (Fig. 7A, lane 7, and B, lanes 2 and 3). In contrast, S105 homodimers could be detected in inductions which do result in lysis: with S105 alone, in either the plasmid or phage context, and with S105 on the plasmid and the deletion allele on the prophage (Fig. 7A and B). Interestingly, two mutations in TMD2 that cause a lack of plaque-forming ability, A52V and A52G, have opposite effects on the antiholin character of the deletion protein. A52G, which causes catastrophically early lysis at or near the end of the eclipse period in the context of S105 (18), decreases the ability of S105ΔTMD1 to inhibit S105, as reflected in lysis profiles (Fig. 6A) and in plating ability (Fig. 5D), suggesting a weakened TMD2-TMD2 interaction because of the Ala-Gly mismatch at position 52 between the altered deletion protein and wt S105. In contrast, A52V, which blocks plaque formation in the context of S105 because it is an absolute lysis-defective mutation, has no effect on the antiholin character of S105ΔTMD1 (Fig. 5E and 6A). In this case, the TMD2-TMD2 interaction is apparently unimpaired. Further, for A52G, this defect in inhibition could be suppressed by the same change in S105 (Fig. 6B), suggesting that resolving the mismatch restored the TMD2-TMD2 interaction. Moreover, restoration of inhibition was correlated with the absence of holin homodimers after Cu-Phe treatment (Fig. 7A, lane 2).
DISCUSSION
The dual-start motif is one of the most notable features of holin genes and serves to highlight the importance of the regulation of lysis timing, since the two translational start codons give rise to two polypeptides that, despite near identity in sequence, have opposing functions as the holin and antiholin. Here, we provide biochemical evidence for the difference in topology, present data that define the role of that difference in regulating hole formation, and describe experiments that relate antiholin function to homotypic TMD2-TMD2 interactions.
The N terminus of TMD1 is periplasmic.
S105 has three TMDs with N-out and C-in (Fig. 1B and C). Here we show that the native S105 protein retains the formyl moiety on its N terminus but that S107 is quantitatively deformylated. Leader peptidase, another N-out integral membrane protein with multiple TMDs, has also been shown to retain its N-formyl group. This finding supports the difference in topology between S105 and S107 and indicates that TMD1 of S105 enters the bilayer so rapidly after synthesis that it avoids processing by deformylase. It should be noted that the single positive charge of the Lys2 residue need only retard the entry of the N terminus of S107 into the membrane long enough for Def-mediated deformylation, which leaves S107 with two positive charges at its N terminus and thus presumably even more refractory to export against the polarity of the energized membrane. S alleles modified to have three or more basic residues after codon 1 are defective and not able to be triggered by energy poisons (31), suggesting that there is a limit to the ability to impose potential-dependent dynamic character on TMD1. A finely tuned balance between the retention and export of the N-terminal TMD is also reflected in the properties of the S2168-S2171 holin-antiholin pair encoded by the pinholin gene, S21 (24). Although the topological change is different, in that TMD1 escapes from the membrane rather than entering it, it is still required for hole formation. However, the single charge confers on the longer form a somewhat more subtle delay of about 20 min in the topological change. To obtain an antiholin form that cannot be triggered, it was necessary to append an oligopeptide sequence containing two positive charges to the N terminus of the shorter form. The more drastic effect of the second charge in this case may reflect the more difficult energetics of removing a TMD from the bilayer rather than inserting it.
The antiholin character of S107 is due to the absence of TMD1 from the bilayer.
The relationship of the functional and topological differences between S105 and S107 has previously not been addressed. That is, the antiholin character of S107 could be due to the presence of the ectopically located TMD1 or to the absence of TMD1 from the bilayer, or both. Here we show that S105ΔTMD1 has the same antiholin character as S107, strongly indicating that it is the absence of TMD1 from the membrane that underlies antiholin function. Moreover, the antiholin effects of the deletion protein are more stable in vivo and cannot be subverted by collapsing the membrane potential, further supporting the notion that it is the membrane penetration of TMD1 that defines the ability of uncouplers and energy poisons to collapse antiholin inhibition. The absolute-lysis-defective character of the deletion protein also suggests that TMD1 is required for hole formation.
Similar antiholin properties have been reported for a truncation of the class I holin, Hol, of the Listeria phage A118 (36). Clones of hol with a deletion of the 13 N-terminal codons of the 96-codon reading frame were shown to be nonfunctional when they were substituted for the S gene in a λ chimera. However, no evidence for the topological disposition of TMD1 in the parental or deletion allele was obtained, which is problematic because the deletion still leaves a potential 19-amino-acid N-terminal TMD. Moreover, expression of this A118 hol deletion allele from the same plasmid-borne λ pR′ context used here in the experiments with S and SΔTMD1 was lethal to the host. Finally, the limitations of Listeria phage genetics did not allow rigorous determination of whether or not the shorter protein was produced from either the parental or deletion allele in Listeria.
The results presented here indicate that the antiholin character of S105ΔTMD1 derives from the loss of TMD1, which eliminates hole-forming activity while retaining the ability to bind specifically to S105. The requirement for TMD1 is not surprising, since missense changes in TMD1 abrogate hole formation without affecting accumulation (26). However, it should be noted that it is not necessary for an antiholin to have TMD-TMD interactions to block hole formation; in phage T4, the antiholin, gprI, is a soluble periplasmic protein when it binds to the periplasmic domain of the holin, gpt (34).
Protein-protein interactions in the timing of lysis.
Antiholin inhibition is not the central modality by which holin function is temporally regulated. With the phage λ, 21, and T4 systems, representing class I, II, and III holins, it has been shown that elimination of the antiholin leaves the intrinsic timing function intact. Antiholin function provides fine-tuning of the timing, in the cases of λ (3, 26) and phage 21 (1, 6), or a mechanism for environmental signals for overriding the intrinsic timing, in the case of T4 (28). The key feature of holin function is saltatory triggering of hole formation at a time built in to the primary structure. Moreover, single missense changes can confer drastic differences on the triggering time, best illustrated by opposing phenotypes of the early-lysis S105A52G and nonlytic S105A52V alleles. Here we have presented results suggesting that an Ala-Gly mismatch at position 52 impairs the homotypic TMD2-TMD2 interaction involved in dimerization and that resolution of the mismatch, i.e., to the mutant Gly-Gly or to the wt Ala-Ala, restores this interaction. Similar phenotypic “mismatch” sensitivity was observed when the original library of lysis-defective S mutants was tested for their dominant/recessive character (26). A number of these lysis-defective mutants exhibited an unexpected form of dominance, in that they accelerated lysis timing in the presence of the wt allele. Since S encodes both the holin and antiholin, the existence of the “antidominant” alleles was most easily explained by invoking differential effects on holin and antiholin forms. The results presented here suggest instead that intimate helical packing of TMD2 defines the dimerization step that is required for both the hole formation pathway and antiholin function.
Acknowledgments
We thank the members of the Young laboratory, past and present, for their helpful criticisms and suggestions, especially Angelika Gründling and Ing-Nang Wang, who did the original constructions with and characterizations of the S deletion.
This work was supported by PHS grant GM27099 to R.Y., the Robert A. Welch Foundation, the Program for Membrane Structure and Function, and a Program of Excellence grant from the Office of the Vice President for Research at Texas A&M University.
Footnotes
Published ahead of print on 6 November 2009.
REFERENCES
- 1.Barenboim, M., C. Y. Chang, F. dib Hajj, and R. Young. 1999. Characterization of the dual start motif of a class II holin gene. Mol. Microbiol. 32:715-727. [DOI] [PubMed] [Google Scholar]
- 2.Bernhardt, T. G., I. N. Wang, D. K. Struck, and R. Young. 2001. A protein antibiotic in the phage Qβ virion: diversity in lysis targets. Science 292:2326-2329. [DOI] [PubMed] [Google Scholar]
- 3.Bläsi, U., C. Y. Chang, M. T. Zagotta, K. Nam, and R. Young. 1990. The lethal λ S gene encodes its own inhibitor. EMBO J. 9:981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bläsi, U., K. Nam, D. Hartz, L. Gold, and R. Young. 1989. Dual translational initiation sites control function of the λ S gene. EMBO J. 8:3501-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bläsi, U., and R. Young. 1996. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol. Microbiol. 21:675-682. [DOI] [PubMed] [Google Scholar]
- 6.Bonovich, M. T., and R. Young. 1991. Dual start motif in two lambdoid S genes unrelated to lambda S. J. Bacteriol. 173:2897-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, C. Y., K. Nam, and R. Young. 1995. S gene expression and the timing of lysis by bacteriophage λ. J. Bacteriol. 177:3283-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 9.Deaton, J., C. G. Savva, J. Sun, A. Holzenburg, J. Berry, and R. Young. 2004. Solubilization and delivery by GroEL of megadalton complexes of the lambda holin. Protein Sci. 13:1778-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fields, R. 1972. The rapid determination of amino groups with TNBS. Methods Enzymol. 25:464-468. [DOI] [PubMed] [Google Scholar]
- 11.Graschopf, A., and U. Bläsi. 1999. Functional assembly of the lambda S holin requires periplasmic localization of its N-terminus. Arch. Microbiol. 172:31-39. [DOI] [PubMed] [Google Scholar]
- 12.Graschopf, A., and U. Bläsi. 1999. Molecular function of the dual-start motif in the λ S holin. Mol. Microbiol. 33:569-582. [DOI] [PubMed] [Google Scholar]
- 13.Gründling, A., U. Bläsi, and R. Young. 2000. Biochemical and genetic evidence for three transmembrane domains in the class I holin, λ S. J. Biol. Chem. 275:769-776. [DOI] [PubMed] [Google Scholar]
- 14.Gründling, A., U. Bläsi, and R. Young. 2000. Genetic and biochemical analysis of dimer and oligomer interactions of the λ S holin. J. Bacteriol. 182:6082-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gründling, A., M. D. Manson, and R. Young. 2001. Holins kill without warning. Proc. Natl. Acad. Sci. U. S. A. 98:9348-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gründling, A., D. L. Smith, U. Bläsi, and R. Young. 2000. Dimerization between the holin and holin inhibitor of phage lambda. J. Bacteriol. 182:6075-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 18.Johnson-Boaz, R., C. Y. Chang, and R. Young. 1994. A dominant mutation in the bacteriophage lambda S gene causes premature lysis and an absolute defective plating phenotype. Mol. Microbiol. 13:495-504. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser, K. 1980. The origin of Q-independent derivatives of phage lambda. Mol. Gen. Genet. 179:547-554. [DOI] [PubMed] [Google Scholar]
- 20.Kolisnychenko, V., G. Plunkett III, C. D. Herring, T. Feher, J. Posfai, F. R. Blattner, and G. Posfai. 2002. Engineering a reduced Escherichia coli genome. Genome Res. 12:640-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livingston, D. M., and P. Leder. 1969. Deformylation and protein biosynthesis. Biochemistry 8:435-443. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Nam, K., U. Bläsi, M. T. Zagotta, and R. Young. 1990. Conservation of a dual-start motif in P22 lysis gene regulation. J. Bacteriol. 172:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, T., D. K. Struck, J. F. Deaton, and R. Young. 2006. Topological dynamics of holins in programmed bacterial lysis. Proc. Natl. Acad. Sci. U. S. A. 103:19713-19718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powell, B. S., M. P. Rivas, D. L. Court, Y. Nakamura, and C. L. Turnbough, Jr. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raab, R., G. Neal, C. Sohaskey, J. Smith, and R. Young. 1988. Dominance in lambda S mutations and evidence for translational control. J. Mol. Biol. 199:95-105. [DOI] [PubMed] [Google Scholar]
- 27.Rajagopalan, P. T., S. Grimme, and D. Pei. 2000. Characterization of cobalt (II)-substituted peptide deformylase: function of the metal ion and the catalytic residue Glu-133. Biochemistry 39:779-790. [DOI] [PubMed] [Google Scholar]
- 28.Ramanculov, E. R., and R. Young. 2001. An ancient player unmasked: T4 rI encodes a t-specific antiholin. Mol. Microbiol. 41:575-583. [DOI] [PubMed] [Google Scholar]
- 29.Smith, D. L., C. Y. Chang, and R. Young. 1998. The λ holin accumulates beyond the lethal triggering concentration under hyper-expression conditions. Gene Expr. 7:39-52. [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, D. L., D. K. Struck, J. M. Scholtz, and R. Young. 1998. Purification and biochemical characterization of the lambda holin. J. Bacteriol. 180:2531-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steiner, M., and U. Bläsi. 1993. Charged amino-terminal amino acids affect the lethal capacity of lambda lysis proteins S107 and S105. Mol. Microbiol. 8:525-533. [DOI] [PubMed] [Google Scholar]
- 32.Takeda, M., and R. E. Webster. 1968. Protein chain initiation and deformylation in B. subtilis homogenates. Proc. Natl. Acad. Sci. U. S. A. 60:1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran, T. A., D. K. Struck, and R. Young. 2007. The T4 RI antiholin has an N-terminal signal anchor release domain that targets it for degradation by DegP. J. Bacteriol. 189:7618-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran, T. A. T., D. K. Struck, and R. Young. 2005. Periplasmic domains define holin-antiholin interactions in T4 lysis inhibition. J. Bacteriol. 187:6631-6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Heijne, G. 1989. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341:456-458. [DOI] [PubMed] [Google Scholar]
- 36.Vukov, N., I. Moll, U. Bläsi, S. Scherer, and M. J. Loessner. 2003. Functional regulation of the Listeria monocytogenes bacteriophage A118 holin by an intragenic inhibitor lacking the first transmembrane domain. Mol. Microbiol. 48:173-186. [DOI] [PubMed] [Google Scholar]
- 37.Wang, I. N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799-825. [DOI] [PubMed] [Google Scholar]
- 38.Young, R. 2002. Bacteriophage holins: deadly diversity. J. Mol. Microbiol. Biotechnol. 4:21-36. [PubMed] [Google Scholar]
- 39.Young, R., I. N. Wang, and W. D. Roof. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8:120-128. [DOI] [PubMed] [Google Scholar]
- 40.Zagotta, M. T., and D. B. Wilson. 1990. Oligomerization of the bacteriophage lambda S protein in the inner membrane of Escherichia coli. J. Bacteriol. 172:912-921. [DOI] [PMC free article] [PubMed] [Google Scholar]