Abstract
Mycoplasma mobile binds to solid surfaces and glides smoothly and continuously by a unique mechanism. A huge protein, Gli521 (521 kDa), is involved in the gliding machinery, and it is localized in the cell neck, the base of the membrane protrusion. This protein is thought to have the role of force transmission. In this study, the Gli521 protein was purified from M. mobile cells, and its molecular shape was studied. Gel filtration analysis showed that the isolated Gli521 protein forms mainly a monomer in Tween 80-containing buffer and oligomers in Triton X-100-containing buffer. Rotary shadowing electron microscopy showed that the Gli521 monomer consisted of three parts: an oval, a rod, and a hook. The oval was 15 nm long by 11 nm wide, and the filamentous part composed of the rod and the hook was 106 nm long and 3 nm in diameter. The Gli521 molecules form a trimer, producing a “triskelion” reminiscent of eukaryotic clathrin, through association at the hook end. Image averaging of the central part of the triskelion suggested that there are stable and rigid structures. The binding site of a previously isolated monoclonal antibody on Gli521 images showed that the hook end and oval correspond to the C- and N-terminal regions, respectively. Partial digestion of Gli521 showed that the molecule could be divided into three domains, which we assigned to the oval, rod, and hook of the molecular image. The Gli521 molecule's role in the gliding mechanism is discussed.
Mycoplasmas are commensal and occasionally parasitic bacteria with small genomes that lack a peptidoglycan layer (31). Several mycoplasma species form membrane protrusions, such as the headlike structure in Mycoplasma mobile and the attachment organelle in Mycoplasma pneumoniae (15, 19, 21, 22, 25, 33, 34, 36). On solid surfaces, these species exhibit gliding motility in the direction of the protrusion; this motility is believed to be involved in the pathogenicity of mycoplasmas (12, 13, 16, 20, 21). Interestingly, mycoplasmas have no surface flagella or pili, and their genomes contain no genes related to other known bacterial motility systems. In addition, no homologs of motor proteins that are common in eukaryotic motility have been found (11).
M. mobile, which was isolated from the gills of a freshwater fish in the early 1980s, is a fast gliding mycoplasma (14). It glides smoothly and continuously on glass at an average speed of 2.0 to 4.5 μm/s, or three to seven times the length of the cell per second, exerting a force of up to 27 pN (8, 9, 24, 25, 32). Previously, we identified huge proteins involved in this gliding mechanism that are localized at the so-called cell neck, the base of the membrane protrusion (17, 26, 30, 35, 37, 39); we also visualized the putative machinery and the binding protein (1, 18, 23) and identified both the direct energy source used and the direct binding target (10, 27, 38). The force generated by the gliding machinery may be supported from inside the cell by a cytoskeletal “jellyfish” structure (28, 29). On the basis of these results, we proposed a working model, called the centipede or power stroke model, where cells are propelled by “legs” composed of Gli349 that repeatedly catch and release sialic acids fixed on the glass surface (5, 19, 21). These legs are driven by the force exerted by P42 through Gli521 molecules, which is supported by the jellyfish structure, based on energy from ATP hydrolysis.
The Gli521 protein, which has an unusually high molecular mass (521 kDa), is suggested to have the role of force transmission, because a monoclonal antibody against this protein stops gliding, keeping the cells on a solid surface (35). About 450 molecules are estimated to be clustered in the gliding machinery with other component proteins, although their alignment has not been clarified (35, 37, 39). In this study, we isolated the Gli521 protein and studied its molecular shape using electron microscopy (EM) and biochemical analyses in order to understand the gliding mechanism.
MATERIALS AND METHODS
Isolation of Gli521 protein.
M. mobile strain 163K (= ATCC 43663) was grown in Aluotto medium at 25°C to an optical density at 600 nm of around 0.1 (2, 26). Most procedures were done at 4°C unless indicated otherwise. Cells from 1 liter of culture were centrifuged at 14,000 × g for 10 min and washed twice with phosphate-buffered saline (PBS) consisting of 75 mM sodium phosphate (pH 7.3) and 68 mM NaCl. The cells were suspended to an optical density at 600 nm of 20 in 10 mM Tris-HCl (pH 8.0), 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and then were mixed with Triton X-100 (1%, vol/vol). After gentle shaking for 1 h, the suspension was ultracentrifuged at 450,000 × g for 30 min. The supernatant was fractionated by salting out with ammonium sulfate (20% saturation) at room temperature, and the insoluble fraction was recovered by centrifugation at 22,000 × g for 15 min. The pellet was dissolved and then dialyzed overnight using 10 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 7.0) containing 0.1% Triton X-100. The insoluble fraction was removed by centrifugation at 22,000 × g for 15 min. The soluble fraction was loaded onto a HiLoad 16/60 Superdex 200 pg set (GE Healthcare, Milwaukee, WI) in an AKTA purifier (GE Healthcare), eluted with a buffer consisting of 0.1% Triton X-100, 0.2 M NaCl, and 10 mM Tris-HCl (pH 8.0) at a flow rate of 1 ml/min at 4°C, and fractionated into 1-ml aliquots. The homogeneity of the Gli521 protein fraction was estimated by densitometry of SDS-PAGE gels stained with Coomassie brilliant blue using a scanner (GT-9800F; Epson, Nagano, Japan) and the analysis software Image-J, version 1.42 (National Institutes of Health). The Gli521 protein fraction was dialyzed against 10 mM ammonium acetate (pH 6.5) overnight and concentrated using Biomax-50 (Millipore, Bedford, MA) to obtain a concentration of 0.3 mg/ml. This protein preparation was kept at −20°C in 33% (vol/vol) glycerol and 0.3 M ammonium acetate (pH 6.5), if necessary, and used within 1 month.
Analytical gel filtration.
The Gli521 protein fraction (0.05 mg protein in 0.1 ml) was applied to a TSK G5000PWXL gel set (Tosoh, Tokyo, Japan) in a series 1200 high-performance liquid chromatograph (Agilent Technologies, Palo Alto, CA) and was eluted with a buffer consisting of 0.2 M NaCl and 10 mM Tris-HCl (pH 7.0) at a flow rate of 1 ml/min at 20°C, and the absorbance at 280 nm was monitored. Thyroglobulin, ferritin, glucose oxidase, and chymotrypsinogen were used as size standards.
Rotary shadowing EM.
Protein samples under different conditions were diluted to obtain a concentration of 20 μg/ml in 33% (vol/vol) glycerol and 0.3 M ammonium acetate and sprayed onto a freshly cleaved mica surface as described previously (1, 3). Skeletal muscle myosin and clathrin were gifts from T. Arata at Osaka University and were commercially available (Sigma). No detergents were added to myosin and clathrin samples. Whole micrographs were digitized as 8-bit images using a GT-9800F scanner. Each particle image was picked and analyzed by using EMAN, version 1.6 (http://ncmi.bcm.tmc.edu/∼stevel/EMAN/doc/).
Partial digestion of the Gli521 protein.
Purified Gli521 protein in PBS was digested at 25°C for 15 min with various concentrations of proteinase K and analyzed by SDS-PAGE after the reaction was stopped by addition of 10 mM PMSF. The resulting fragments were excised and subjected to in-gel digestion and peptide mass fingerprinting (7). Mass spectra were obtained in the reflectron mode with a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (AXIMA CFR Plus; Shimadzu/Kratos, Kyoto, Japan) operated in delayed extraction mode. The resultant peptides were identified by using an algorithm for peptide identification, Peptide Map (http://prowl.rockefeller.edu). The partial digests were also subjected to Edman degradation after the protein bands were transferred to a nylon membrane.
RESULTS
Isolation of the Gli521 protein.
The Gli521 protein was isolated by a three-step biochemical procedure: (i) solubilization, (ii) salting out, and (iii) gel filtration (Fig. 1). Briefly, a protein fraction was extracted from cultured mycoplasma cells with 1% Triton X-100 and used in the salting-out process. The Gli521 protein was enriched in the precipitate from 20% ammonium sulfate saturation. The precipitate was dissolved, dialyzed with a buffer, and used for gel filtration for fractionation by molecular mass. The fractions from gel filtration containing mostly the Gli521 protein were concentrated and used in the following experiments. First, 0.1% Triton X-100 was added to buffers for all the preparation procedures, because the Gli521 protein easily forms aggregates in the absence of detergent. The homogeneity of the Gli521 specimen was estimated to be 99.5%, based on the band intensity in SDS-PAGE gels.
FIG. 1.

Protein profiles of fractions from Gli521 purification procedures. Lane 1, whole-cell lysate; lane 2, Triton-insoluble fraction; lane 3, Triton-soluble fraction; lane 4, precipitate with ammonium sulfate (20% saturation); lane 5, Gli521 fraction eluted by gel filtration; lane 6, concentrated Gli521 protein fraction. The protein fractions were applied to an SDS-6% PAGE gel and stained with Coomassie brilliant blue. Molecular masses are indicated on the left. The Gli521 protein band is indicated by a triangle on the right.
Self-assembly of proteins examined by analytical gel filtration.
To examine the possibility of self-assembly, the isolated Gli521 protein was used in an analytical gel filtration assay (Fig. 2). Gli521 prepared in the presence of 0.1% Triton X-100 was subjected to gel filtration directly or after replacement of Triton X-100 with 0.1% Tween 80 by dialysis. In both experiments, the elution pattern showed two peaks at calculated molecular masses of 3,630 kDa and 720 kDa, based on the retention volume. The peaks at 3,630 kDa and 720 kDa were major peaks in the presence of Triton X-100 and Tween 80, respectively. The molecular mass of mature Gli521 is estimated to be 516 kDa, based on genetic information and Edman analyses (35). Therefore, the isolated Gli521 protein should be mainly in oligomeric and monomeric forms in Triton X-100 and Tween 80, respectively. The differences in the molecular mass values, especially for the monomers, may be due to the characteristic molecular shape of the protein, as shown by EM imaging (see below).
FIG. 2.
Gel filtration assay of isolated Gli521 protein. Gli521 prepared in the presence of 0.1% Triton X-100 was subjected to gel filtration directly (dotted line) or after the 0.1% Triton X-100 was replaced by 0.1% Tween 80 by dialysis (solid line). Samples (50 μl) were applied and analyzed with a flow rate of 1 ml/min at room temperature.
EM images of the Gli521 molecule.
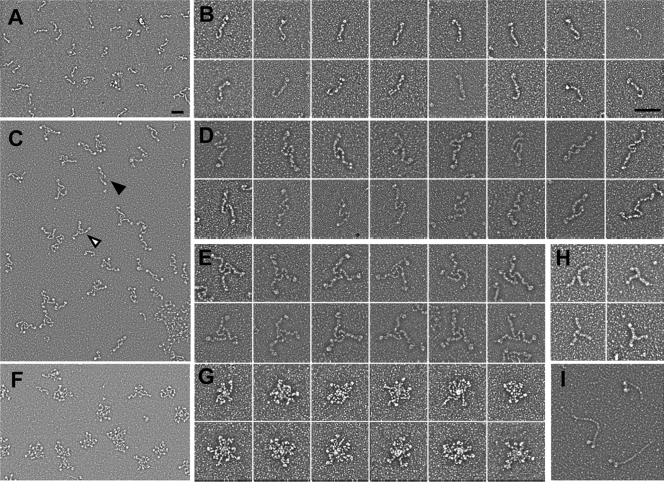
Gli521 protein samples in a buffer containing 0.1% Triton X-100 or 0.1% Tween 80 were used for rotary shadowing EM (Fig. 3). In EM images obtained under monomer-rich conditions with Tween 80, flexible rods were found, and their density changed depending on the protein concentration used, indicating that the flexible rod is the Gli521 monomer (Fig. 3A). The monomer molecule had a hook at one end and an oval at the other end (Fig. 3B), and the total length was about 121 ± 21 nm.
FIG. 3.
EM images of Gli521 molecule. Gli521 protein samples in a buffer containing Tween 80 (monomer-rich conditions) or Triton X-100 (oligomer-rich conditions) were subjected to rotary shadowing and observed. (A) Field image under monomer-rich conditions. (B) Images of a monomer under monomer-rich conditions. (C) Field image under oligomer-rich conditions. The filled and open triangles indicate a representative dimer and trimer, respectively. (D and E) Images of a dimer (D) and trimer (E) under oligomer-rich conditions. (F) Field image under oligomer-rich conditions, showing rosette forms. (G) Images of a rosette form under oligomer-rich conditions. (H and I) EM images of clathrin-triskelion (H) and myosin (I) obtained by a procedure similar to that used for Gli521. Different magnifications were used to obtain panels A, C, and F and to obtain panels B, D, E, G, H, and I. Bars, 100 nm.
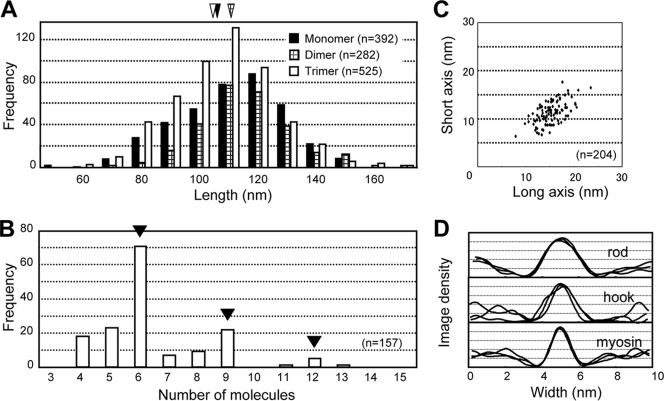
The assembled forms were found frequently in the images obtained under oligomer-rich conditions with Triton X-100, and the ratio of the apparent monomeric, dimeric, trimeric, and rosette forms was 2.3:1.6:1:1.4 (Fig. 3C to E). Detailed observations suggested that the molecules associate apparently at the hook end. No specific associations could be found for other parts of the molecule. The lengths of each arm excluding the oval for the dimer (111 ± 15 nm) and trimer (103 ± 17 nm) were similar to the length of the monomer (106 ± 18 nm), showing that the protein molecules associate at a rather small area near the terminus (Fig. 4A). The trimer shape was similar to the shape of the “triskelion” of eukaryotic “clathrin,” as shown in Fig. 3H, although the amino acid sequences of Gli521 and clathrin do not share any similarity (6, 35).
FIG. 4.
Dimensions of Gli521 molecules. (A) Length of filamentous part (rod plus hook). Filled, cross-hatched, and open triangles indicate the average lengths of the monomer (106 ± 18 nm), dimer (111 ± 15 nm), and trimer (103 ± 17 nm), respectively. (B) Numbers of ovals in individual rosettes. (C) Dimensions of the oval. A total of 204 ovals obtained under oligomeric conditions were analyzed. The average lengths were 15.2 ± 2.6 and 11.2 ± 2.3 nm for the long and short axes, respectively. (D) Sectional image density profile of the rod and hook of Gli521 and the rod of myosin. Three separate molecules were traced.
The rosette forms apparently include larger numbers of protein subunits, but the individual subunits cannot be traced (Fig. 3F and G). However, determination of the numbers of the oval part suggested that the rosettes preferentially contain six or nine monomers (Fig. 4B). This in turn suggested that the rosettes resulted from stacking of trimers.
On average, the long and short axes of the oval were 15.2 ± 2.6 and 11.2 ± 2.3 nm long, respectively (Fig. 4C). The diameters of the rods and hooks of 20 molecules were 5.1 ± 0.3 nm and 4.9 ± 1.3 nm, respectively. Generally, in metal-shadowed EM images, filamentous structures look thicker than the original structures, because the EM shadowing method coats the protein molecule with metal particles to enhance contrast. Therefore, we obtained molecular images of skeletal muscle myosin using the same method and compared them with the images of Gli521 (Fig. 3I and 4D). The diameter of the myosin rod was 3.8 ± 0.4 nm, which was about 1.8 nm greater than the actual diameter (2.0 nm) determined in previous structural studies (41), suggesting that the filament diameters obtained with the rotary shadowing used here can be estimated to be 1.8 nm greater. These results suggest that the diameters of the rods and hooks of Gli521 are approximately 3.3 and 3.1 nm, respectively.
The central part of the Gli521 triskelion appeared to be rigid compared to the other parts (Fig. 5). To confirm this, image averaging was applied to the central part of the trimer. We picked 108 images manually and subjected them to image classification. An average image was obtained from 64 picked images, as shown in Fig. 5C. This image was suggested to have three times rotational symmetry, called C3 symmetry, because overlaying images rotated 120° and 240° on the original image did not change its appearance significantly, as shown in Fig. 5D. These observations suggest that the central part of the trimer has a stable and rigid structure.
FIG. 5.
Image averaging for the central part of trimeric Gli521 molecules. (A) Typical whole image of trimeric Gli521 protein. The central part encircled by a dashed line was picked manually (B) and used for image averaging (C and D). (C) A total of 108 images of the central part were used for image averaging, and 64 images were integrated by using EMAN, an image-averaging software program. (D) The average image was rotated 120° and 240° and overlaid onto the original image. Bars, 100 nm.
Assignment of an amino acid sequence to the molecular image of Gli521 based on the binding position of MAbR19.
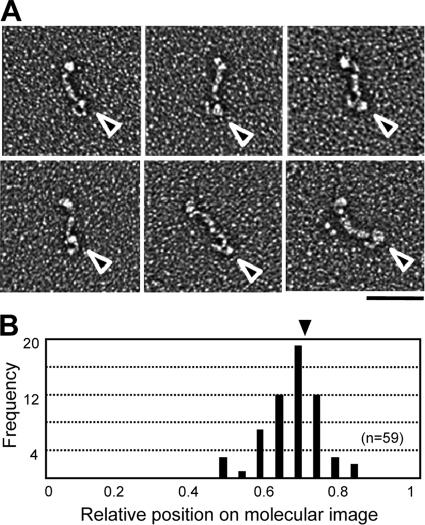
To determine the relationship between the molecular shape of Gli521 and its amino acid sequence, the binding position of a monoclonal antibody, MAbR19, was examined using the EM images under monomeric conditions. MAbR19 binds to an epitope that localizes in a region comprising amino acids 3736 to 3863 of the 4,727 amino acids in Gli521 (35, 40). In the present study, the Gli521 protein was mixed with MAbR19 at a 1:1 molar ratio, rotary shadowed, and observed by EM (Fig. 6A). Gli521 molecules were found to be bound by the antibody at a frequency of approximately 1/30. All of the images of the bound protein showed that MAbR19 bound to the hook. Statistical analysis of the binding position showed that it is distributed around 0.29 of total length, including the oval, from the hook end (Fig. 6B). Approximately 1 of 30 Gli521 molecules had extra electron density near the hook when antibody had been added, whereas less than 1 of 400 Gli521 molecules exhibited this feature in the absence of antibody.
FIG. 6.
Binding positions of monoclonal antibody on molecular images of Gli521. (A) EM images of the monomeric form of the Gli521 molecule associated with the monoclonal antibody, MAbR19. The triangles indicate the antibody bound to Gli521. Bar, 100 nm. (B) Distribution of binding positions of the antibody on Gli521 images. The central positions of the antibody are shown as ratios based on the total length for 59 molecules. The shorter distance to an end was measured. The average position is indicated by a triangle.
Considering that the epitope is located at position 0.20 from the C terminus in the total amino acid sequence of the mature molecule (35), the hook end and oval correspond to the C- and N-terminal regions, respectively.
Domain structure of the Gli521 protein suggested by partial digestion.
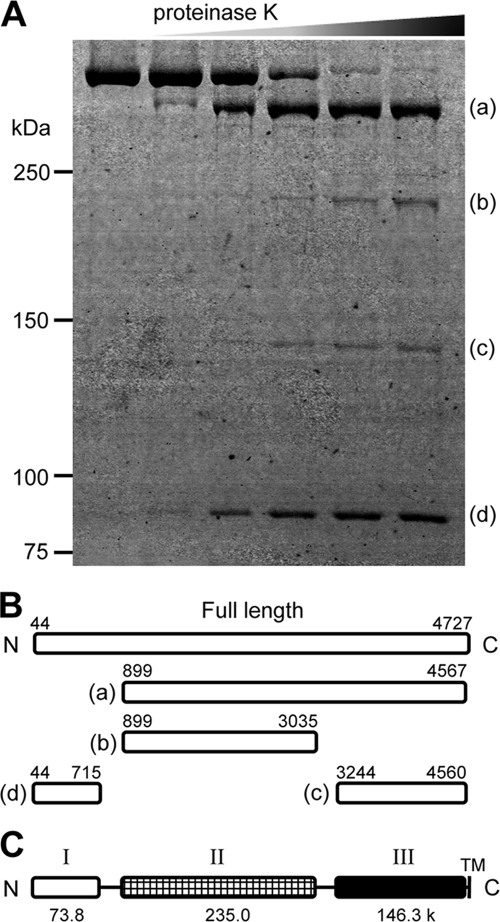
Gli521 was partially digested by proteinase K and subjected to SDS-PAGE, and four major products were detected in the gel at 401, 226, 143, and 85 kDa, as shown in Fig. 7A (fragments a, b, c, and d, respectively). Peptide mass fingerprinting by MALDI-TOF mass spectrometry showed that peptide fragments a, b, c, and d correspond to at least amino acids 926 to 4567, 926 to 3035, 3297 to 4560, and 76 to 715, respectively, in the amino acid sequence encoded by the Gli521 open reading frame (ORF). The peptides detected by peptide mass fingerprinting accounted for 9.2%, 3.9%, 3.1%, and 5.2% of the lengths of regions a, b, c, and d, respectively. The results of Edman analysis showed that the N termini of the fragments started at amino acids 899, 899, 3244, and 44, respectively (Fig. 7B). These results showed that the Gli521 molecule can be divided into three domains containing amino acids 44 to 715, 899 to 3035, and 3244 to 4560, which we call domains I, II, and III, respectively (Fig. 7C). The molecular masses of these three domains, which are connected by flexible parts, were calculated to be 73.8, 235.0, and 146.3 kDa, respectively, based on the amino acid sequences. The digestion pattern suggests that the linking region between domains I and II (I-II linker) is more sensitive to protease digestion than the linking region between domains II and III (II-III linker), suggesting that the I-II linker is more flexible than the II-III linker.
FIG. 7.
Partial digestion of Gli521. (A) Protein profiles of Gli521 digested with various concentrations of proteinase K. Gli521 (0.3 μg) was treated with 0.015, 0.06, 0.15, 0.3, and 0.6 μg proteinase K and subjected to SDS-PAGE. The resultant peptide fragments, fragments a to d, were identified by peptide mass fingerprinting. (B) Mapping of digests on the amino acid sequence. The N-terminal 43 amino acids predicted from the genetic information are processed in the mature form. The N-terminal amino acid in each fragment was determined by Edman analysis. The C-terminal positions were based on the C-terminal end of the peptide detected by peptide mass fingerprinting. (C) Schematic diagram of Gli521 domains. Gli521 consists of three domains, domains I, II, and III. The molecular mass (in kDa) is indicated below each domain. TM, transmembrane segment.
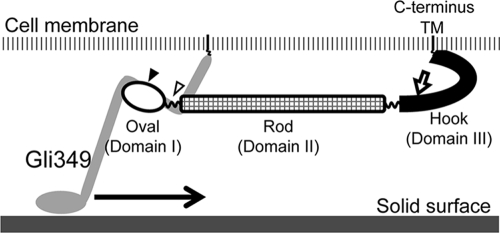
Considering the length of each part in EM images, the molecular mass of each domain, and the assignment of the N terminus on the EM images, domains I, II, and III are suggested to correspond to the oval, rod, and hook structures, respectively, in the molecular images (Fig. 8).
FIG. 8.
Schematic diagram of the Gli521 molecule. The transmembrane segment (TM) is anchored in the cell membrane. The filled and open triangles indicate the positions of mutations P476R and S859R, which rescue the inhibition of cell binding to solid surfaces caused by a monoclonal antibody targeted to Gli349, the “leg” protein. The region including these mutations may interact with the Gli349 molecule. The putative leg movement is indicated by an arrow. MAbR19 binds to the position indicated by an open arrow and stops gliding motility (40).
Mycoplasma pulmonis is closely related to M. mobile and can also glide (4). This species has an ortholog of Gli521, which is encoded by three ORFs in the genome database of M. pulmonis (MYPU_2120, MYPU_2130, and MYPU_2140). However, we previously sequenced the corresponding regions of another strain of M. pulmonis and found that the ortholog is encoded as a single peptide (35). We tried to determine the domain structures by comparing the amino acid sequences of Gli521 and the M. pulmonis ortholog, but the structures were not suggested by sequence similarities.
DISCUSSION
Assembly of Gli521 molecules into triskelion.
The monomeric molecular image of Gli521 consists of three parts: an oval, a rod, and a hook. These parts can be seen in all assembled forms, the monomer, the dimer, the trimer, and the rosette. However, the oval appears to be slightly less obvious in the images obtained in the presence of Tween 80 (Fig. 3B). The analytical gel filtration assay showed that Tween 80 has the ability to dissociate the oligomers of Gli521 to monomers. This detergent may also be able to reduce the intramolecular binding and loosen the oval structure.
Although Gli521 dimer, trimer, and rosette forms were observed by EM, none of the corresponding peaks were detected in the analytical gel filtration assay even under the oligomeric conditions (Fig. 2). Instead, a broad peak was detected at a position corresponding to a molecular weight higher than that of the monomer. This broad peak appears to include the dimer, trimer, and rosette forms. The whole Gli521 molecule is a flexible rod that should behave differently than globular particles. This feature may be enhanced in dimeric and trimeric forms and affect the separation of peaks in the gel filtration assay.
EM images of the dimer and trimer suggested that the molecules bind to one another at the hook end corresponding to the C-terminal region (Fig. 3 and 5). The analyses of the amino acid sequence of Gli521 did not suggest that there are any coiled-coil structures, which are sometimes involved in protein-protein interactions. A transmembrane segment was predicted at the C terminus near the hook domain. The Gli521 molecules may bind to each other at the transmembrane segment, as observed in the clathrin triskelion (6).
Assembly into gliding machinery on the cell surface.
Most of the Gli521 protein molecule is predicted to be exposed, because the gliding motility of M. mobile is stopped by addition of a monoclonal antibody and only one transmembrane segment is predicted to be near the C terminus (35, 40). As the detailed structure of the whole gliding machinery has not been elucidated, we cannot know if Gli521 actually forms a trimer on the surface. However, the protein molecules should be aligned regularly because about 450 molecules each of Gli123, Gli349, and Gli521 are clustered on the surface of the cell neck, which is 350 nm square (37, 39). The self-associating activity of isolated Gli521 proteins may be important for assembly of the gliding machinery. The molecules may be aligned in a directed manner, because the cells glide unidirectionally along the cell axis. Therefore, the rod and oval may be aligned along the cell axis, although the center of the triskelion forms a rigid part on the cell, because these parts are linked with flexible hinges (Fig. 8).
Genetic studies have suggested that Gli521 is necessary for the localization of Gli349 (39). The P476R and S859R substitutions in the amino acid sequence of Gli521 rescue the inhibition of binding to solid surfaces caused by adding the monoclonal antibody targeted to Gli349 (Fig. 8) (40). These observations suggest that Gli349 binds to the N-terminal region, which corresponds to the oval part of Gli521.
Role of the Gli521 molecule in the gliding mechanism.
We have proposed a working model, called the centipede or power stroke model, focusing on the movement of the “leg,” Gli349 (5, 19, 21). In this model, a novel ATPase, P42, transmits movements through the gear protein, Gli521, to the legs, which are composed of Gli349. The legs pull the cell forward, repeating the catch, pull, and release of sialic acids on the solid surface. MAbR19 binds to the hook and stops the gliding motility, suggesting that the hook has actual movement. P42 may interact with, and cause movement of, the hook end of Gli521. This movement could be transmitted through the rod, which functions as a crank, to the oval, which interacts with the legs composed of Gli349. In this model, repeated pulling of the Gli349 legs results in cell movement.
Acknowledgments
This work was supported by a grant-in-aid for scientific research (A) and by a grant-in-aid for scientific research on the priority areas Applied Genomics, Structures of Biological Macromolecular Assemblies, and System Cell Engineering by Multi-Scale Manipulation from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M.M.) and by a grant from the Institution for Fermentation, Osaka, Japan (to M.M.).
Footnotes
Published ahead of print on 13 November 2009.
REFERENCES
- 1.Adan-Kubo, J., A. Uenoyama, T. Arata, and M. Miyata. 2006. Morphology of isolated Gli349, a leg protein responsible for glass binding of Mycoplasma mobile gliding via glass binding, revealed by rotary shadowing electron microscopy. J. Bacteriol. 188:2821-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58. [Google Scholar]
- 3.Arata, T. 1998. Electron microscopic observation of monomeric actin attached to a myosin head. J. Struct. Biol. 123:8-16. [DOI] [PubMed] [Google Scholar]
- 4.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., J. Neu, M. Miyata, and G. Oster. 2009. Motor-substrate interactions in Mycoplasma motility explains non-Arrhenius temperature dependence. Biophys. J. 97:2930-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fotin, A., Y. Cheng, P. Sliz, N. Grigorieff, S. C. Harrison, T. Kirchhausen, and T. Walz. 2004. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature 432:573-579. [DOI] [PubMed] [Google Scholar]
- 7.Hellman, U., C. Wernstedt, J. Gonez, and C. H. Heldin. 1995. Improvement of an “in-gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224:451-455. [DOI] [PubMed] [Google Scholar]
- 8.Hiratsuka, Y., M. Miyata, T. Tada, and T. Q. P. Uyeda. 2006. A microrotary motor powered by bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:13618-13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiratsuka, Y., M. Miyata, and T. Q. P. Uyeda. 2005. Living microtransporter by uni-directional gliding of Mycoplasma along microtracks. Biochem. Biophys. Res. Commun. 331:318-324. [DOI] [PubMed] [Google Scholar]
- 10.Jaffe, J. D., M. Miyata, and H. C. Berg. 2004. Energetics of gliding motility in Mycoplasma mobile. J. Bacteriol. 186:4254-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaffe, J. D., N. Stange-Thomann, C. Smith, D. DeCaprio, S. Fisher, J. Butler, S. Calvo, T. Elkins, M. G. FitzGerald, N. Hafez, C. D. Kodira, J. Major, S. Wang, J. Wilkinson, R. Nicol, C. Nusbaum, B. Birren, H. C. Berg, and G. M. Church. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jordan, J. L., H. Y. Chang, M. F. Balish, L. S. Holt, S. R. Bose, B. M. Hasselbring, R. H. Waldo III T. M. Krunkosky, and D. C. Krause. 2007. Protein P200 is dispensable for Mycoplasma pneumoniae hemadsorption but not gliding motility or colonization of differentiated bronchial epithelium. Infect. Immun. 75:518-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirchhoff, H. 1992. Motility, p. 289-306. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, DC.
- 14.Kirchhoff, H., and R. Rosengarten. 1984. Isolation of a motile mycoplasma from fish. J. Gen. Microbiol. 130:2439-2445. [DOI] [PubMed] [Google Scholar]
- 15.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 16.Krunkosky, T. M., J. L. Jordan, E. Chambers, and D. C. Krause. 2007. Mycoplasma pneumoniae host-pathogen studies in an air-liquid culture of differentiated human airway epithelial cells. Microb. Pathog. 42:98-103. [DOI] [PubMed] [Google Scholar]
- 17.Kusumoto, A., S. Seto, J. D. Jaffe, and M. Miyata. 2004. Cell surface differentiation of Mycoplasma mobile visualized by surface protein localization. Microbiology 150:4001-4008. [DOI] [PubMed] [Google Scholar]
- 18.Metsugi, S., A. Uenoyama, J. Adan-Kubo, M. Miyata, K. Yura, H. Kono, and N. Go. 2005. Sequence analysis of the gliding protein Gli349 in Mycoplasma mobile. Biophysics 1:33-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyata, M. 2008. Centipede and inchworm models to explain Mycoplasma gliding. Trends Microbiol. 16:6-12. [DOI] [PubMed] [Google Scholar]
- 20.Miyata, M. 2005. Gliding motility of mycoplasmas—the mechanism cannot be explained by current biology, p. 137-163. In A. Blanchard and G. Browning (ed.), Mycoplasmas: pathogenesis, molecular biology, and emerging strategies for control. Horizon Bioscience, Norfolk, United Kingdom.
- 21.Miyata, M. 2007. Molecular mechanism of mycoplasma gliding—a novel cell motility system, p. 137-175. In P. Lenz (ed.), Cell motility. Springer, New York, NY.
- 22.Miyata, M., and H. Ogaki. 2006. Cytoskeleton of Mollicutes. J. Mol. Microbiol. Biotechnol. 11:256-264. [DOI] [PubMed] [Google Scholar]
- 23.Miyata, M., and J. Petersen. 2004. Spike structure at the interface between gliding Mycoplasma mobile cells and glass surfaces visualized by rapid-freeze-and-fracture electron microscopy. J. Bacteriol. 186:4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyata, M., and A. Uenoyama. 2002. Movement on the cell surface of the gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215:285-289. [DOI] [PubMed] [Google Scholar]
- 26.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311-1320. [DOI] [PubMed] [Google Scholar]
- 27.Nagai, R., and M. Miyata. 2006. Gliding motility of Mycoplasma mobile can occur by repeated binding to N-acetylneuraminyllactose (sialyllactose) fixed on solid surfaces. J. Bacteriol. 188:6469-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakane, D., and M. Miyata. 2007. Cytoskeletal “jellyfish” structure of Mycoplasma mobile. Proc. Natl. Acad. Sci. U. S. A. 104:19518-19523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakane, D., and M. Miyata. 2009. Cytoskeletal asymmetrical dumbbell structure of a gliding mycoplasma, Mycoplasma gallisepticum, revealed by negative-staining electron microscopy. J. Bacteriol. 191:3256-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohtani, N., and M. Miyata. 2007. Identification of a novel nucleoside triphosphatase from Mycoplasma mobile: a prime candidate for motor of gliding motility. Biochem. J. 403:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosengarten, R., and H. Kirchhoff. 1987. Gliding motility of Mycoplasma sp. nov. strain 163K. J. Bacteriol. 169:1891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seto, S., and M. Miyata. 2003. The attachment organelle formation represented by localization of cytadherence protein and formation of the electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seto, S., A. Uenoyama, and M. Miyata. 2005. Identification of 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 187:3502-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu, T., and M. Miyata. 2002. Electron microscopic studies of three gliding mycoplasmas, Mycoplasma mobile, M. pneumoniae, and M. gallisepticum, by using the freeze-substitution technique. Curr. Microbiol. 44:431-434. [DOI] [PubMed] [Google Scholar]
- 37.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uenoyama, A., and M. Miyata. 2005. Gliding ghosts of Mycoplasma mobile. Proc. Natl. Acad. Sci. U. S. A. 102:12754-12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uenoyama, A., and M. Miyata. 2005. Identification of a 123-kilodalton protein (Gli123) involved in machinery for gliding motility of Mycoplasma mobile. J. Bacteriol. 187:5578-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uenoyama, A., S. Seto, D. Nakane, and M. Miyata. 2009. Regions on Gli349 and Gli521 protein molecules directly involved in movements of Mycoplasma mobile gliding machinery, suggested by use of inhibitory antibodies and mutants. J. Bacteriol. 191:1982-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, W. C., Y. J. Peng, W. Q. He, N. Lv, C. Chen, G. Zhi, H. Q. Chen, and M. S. Zhu. 2008. Identification and functional characterization of an aggregation domain in long myosin light chain kinase. FEBS J. 275:2489-2500. [DOI] [PubMed] [Google Scholar]