Abstract
The DedA family genes are found in most bacterial genomes. Two of these proteins are Escherichia coli YqjA and YghB, predicted inner membrane proteins of unknown function sharing 61% amino acid identity. The E. coli single deletion mutants are largely without phenotype, but the double mutant (BC202; ΔyqjA::Tetr ΔyghB::Kanr) is characterized by incomplete cell division, temperature sensitivity, and altered phospholipid levels (K. Thompkins et al., J. Bacteriol. 190:4489-4500, 2008). In this report, we have better characterized the cell division chaining defect of BC202. Fluorescence recovery after photobleaching indicates that 58% of the cells in chains are compartmentalized by at least a cytoplasmic membrane. Green fluorescent protein fusions to the cell division proteins FtsZ, ZipA, FtsI, FtsL, and FtsQ are correctly localized to new septation sites in BC202. Periplasmic amidases AmiC and AmiA, secreted by the twin arginine transport (Tat) pathway, are localized to the cytoplasm in BC202. Overexpression of AmiA, AmiC, or AmiB, a periplasmic amidase secreted via the general secretory pathway, restores normal cell division but does not suppress the temperature sensitivity of BC202, indicating that YghB and YqjA may play additional roles in cellular physiology. Strikingly, overexpression of the Tat export machinery (TatABC) results in normal cell division and growth at elevated temperatures. These data collectively suggest that the twin arginine pathway functions inefficiently in BC202, likely due to the altered levels of membrane phospholipids in this mutant. These results underscore the importance of membrane composition in the proper function of the Tat protein export pathway.
Roughly 25 to 30% of the genes in sequenced genomes are predicted to encode integral membrane proteins (12). The functions of many of these genes, even in a well-studied organism such as Escherichia coli, remain unknown. We have reported on the functional redundancy of two highly conserved and related E. coli inner membrane proteins, YqjA and YghB (40). These proteins belong to a large family (commonly called the DedA family) found widespread in most sequenced genomes. yghB and yqjA encode predicted inner membrane proteins with multiple membrane-spanning domains and 61% amino acid identity. In addition, E. coli contains three other genes predicted to encode proteins with significant similarity to YqjA and YghB (YabI, YohD, and DedA; amino acid BLAST E value of <1 × 10−6) and two other proteins with lower degrees of similarity (YdjX and YdjZ). Currently, there are >1,000 genes in the NCBI protein database annotated as either belonging to this family or possessing significant amino acid identity to E. coli DedA/YghB/YqjA (protein BLAST E values of <0.02). No member of this family has a known function, nor is it known whether they possess common functions across phylogenetic groups.
Individually, yghB and yqjA are nonessential genes, as each single deletion mutant grows normally (2). However, BC202, an E. coli strain with targeted deletions of both yqjA and yghB, does not grow above 42°C and displays a dramatic cell division phenotype by forming chains of cells when grown at the permissive temperature of 30°C. Phase-contrast and scanning electron microscopy analysis of BC202 suggests that mutants can begin septation but are blocked at a later step in constriction (40). The cause of this phenotype is unclear.
BC202 also has alterations in membrane phospholipid composition (40). While BC202 is capable of synthesizing all classes of phospholipids at all growth temperatures, it is depleted of phosphatidylethanolamine (PE), with elevated levels of the acidic phospholipids phosphatidylglycerol (PG) and cardiolipin (CL). In some respects, BC202 resembles phosphatidylserine synthase deletion mutants, such as AD90 (pss93::Kanr), which produces no membrane PE (14). Mutants deficient in PE are viable, but they require divalent cations for growth (14) and display cell division abnormalities (28, 33). Likewise, normal growth and cell division are restored to BC202 when LB growth medium is supplemented with millimolar concentrations of divalent cations (40). Unlike many mutants defective in cell wall synthesis, BC202 is not hypersensitive to detergents or antibiotics, indicating the presence of an intact outer membrane when grown at the permissive temperature.
BC202, therefore, displays several phenotypes: a block at an apparent late stage of cell division, temperature sensitivity, and an imbalance in membrane phospholipid composition. To better understand the functions of YghB and YqjA, we have hypothesized two roles for these genes that are not mutually exclusive to explain the phenotypes of BC202. First, YqjA/YghB may play direct roles in cell division. The phospholipid phenotype may be a consequence secondary to the primary cell division defect in this scenario. Second, YqjA/YghB may play a direct role in efficient PE synthesis or controlling membrane phospholipid composition. The cell division phenotype may be a secondary consequence of the lipid imbalance. Here, we have better characterized the cell division phenotype of BC202 by using green fluorescent protein (GFP) fusions of cell division proteins and fluorescence recovery after photobleaching (FRAP) analysis. We find that while most of the cell division proteins are correctly localized to new septal rings, the periplasmic amidase AmiC is not localized to the septal ring as was reported previously (5), and this may be responsible for the observed cell division phenotype of BC202. AmiC is found mostly in the cytoplasmic compartment in BC202, as is AmiA, both of which are exported to the periplasm by the twin arginine pathway (5). The cell division defect of BC202 can be corrected by overexpression of periplasmic amidases or the TatABC operon, collectively suggesting that the Tat pathway functions inefficiently in BC202.
MATERIALS AND METHODS
Materials.
Tryptone-yeast extract was purchased from Difco. 32Pi was purchased from Perkin Elmer. Restriction enzymes, Phusion high-fidelity DNA polymerase, Taq DNA polymerase, Antartic phosphatase, and T4 DNA ligase were purchased from New England Biolabs. All DNA oligonucleotides (primers) for PCR amplification and cloning procedures were obtained from Sigma-Aldrich. Paraformaldehyde and 50% glutaraldehyde were from Sigma-Aldrich. Glass slides and coverslips were from Corning. All other chemicals were reagent grade and purchased from either Sigma-Aldrich or VWR.
Bacterial growth medium.
LB medium with 10 g/liter tryptone, 5 g/liter yeast extract, and 10 g/liter NaCl was used throughout this study. Antibiotics were used in the following concentrations, unless stated otherwise: tetracycline (Tet) at 12.5 μg/ml, kanamycin (Kan) at 30 μg/ml, chloramphenicol (Cam) at 30 μg/ml, and ampicillin (Amp) at 25 μg/ml for chromosomal Ampr or 100 μg/ml for plasmids. For GFP localization studies, LB medium was supplemented with either 10 μM (for RS102 only) or 25 μM isopropyl-1-thio-β-d-galactopyranoside (IPTG) to induce expression of chromosomal GFP fusions under the control of the modified trc promoters or with 0.1 to 1.0 mM to induce expression from the plasmids pWSK29 (41) and its derivatives and pTrcHis2-ELGFP6.1-TOPO (a gift from Naohiro Kato, LSU Department of Biological Sciences) (Table 1).
TABLE 1.
Bacterial strains and plasmids
| E. coli strain or plasmid | Relevant genotype/genetic marker(s) | Source or reference(s) |
|---|---|---|
| Strains | ||
| W3110 | Wild type, F− λ− | E. coli Genetic Stock enter, Yale University |
| BC202 | W3110, ΔyqjA::Tetr ΔyghB781::Kanr | 40 |
| EC436 | MC4100, Δ(λattL-lom)::bla lacIq P207-gfp-ftsI | 43 |
| EC438 | MC4100, Δ(λattL-lom)::bla lacIq P207-gfp-ftsL | 9, 19 |
| EC442 | MC4100, Δ(λattL-lom)::bla lacIq P207-gfp-ftsQ | 8 |
| EC448 | MC4100, Δ(λattL-lom)::bla lacIq P208-ftsZ-gfp | 43 |
| EC450 | MC4100, Δ(λattL-lom)::bla lacIq P208-zipA-gfp | 43 |
| RS101 | BC202, Δ(λattL-lom)::bla lacIq P208-ftsZ-gfp (Ampr p1vir transductant of BC202; EC448 donor) | This work |
| RS102 | BC202, Δ(λattL-lom)::bla lacIq P207-gfp-ftsI (Ampr p1vir transductant of BC202; EC436 donor) | This work |
| RS103 | BC202, Δ(λattL-lom)::bla lacIq P207-gfp-ftsL (Ampr p1vir transductant of BC202; EC438 donor) | This work |
| RS104 | BC202, Δ(λattL-lom)::bla lacIq P207-gfp-ftsQ (Ampr p1vir transductant of BC202; EC442 donor) | This work |
| RS105 | BC202, Δ(λattL-lom)::bla lacIq P208-zipA-gfp (Ampr p1vir transductant of BC202; EC450 donor) | This work |
| AD93/pDD72 | pss93::KanrrecA srl::Tn10 nadB+pss+ Camr | 14 |
| AD93/pTB32 | pss93::KanrrecA srl::Tn10 nadB+ pTB32 | This work |
| Plasmids | ||
| pWSK29 | Expression vector; lac promoter, Ampr | 41 |
| pWTD52 | pWSK29 expressing yqjA-GFPmut2 fusion | This work |
| pWTD54 | pWSK29 expressing yghB-GFPmut2 fusion | This work |
| p-yghB | pACYC184 expressing E. coli yghB; Camr | 40 |
| pTrcHis2-ELGFP6.1-TOPO | ELGFP expression vectora; Ampr | Gift from Naohiro Kato, LSU Department of Biological Sciences |
| pTB28 | AmiC-GFP expression vector; Ampr | 5 |
| pTB32 | AmiA-GFP expression vector; Ampr | 5 |
| pWSK-AmiB | pWSK29 expressing amiB; Ampr | This work |
| pBAD-TatABC | pBAD expressing TatABC operon; Ampr | This work |
ELGFP, enhanced variant of GFP (24).
Molecular biology procedures.
Standard techniques were used for cloning and analysis of DNA, standard PCR, colony PCR, transformation, agarose gel electrophoresis, and generalized transduction with P1vir phage (29). Standard PCR using Phusion DNA polymerase was performed following the protocol from Finnzymes in the Phusion HF buffer. Colony PCR was performed using Taq DNA polymerase. Competent cells were prepared according to the method of Inoue et al. (22). DNA sequencing conducted at the LSU College of Basic Science Genomics Facility confirmed the sequences of all cloned PCR products.
Construction of the chromosomal GFP fusion strains.
Generalized P1 transduction (29) was employed to transduce the Δ(λattL-lom)::bla lacIq-associated P207-gfp-ftsI, P207-gfp-ftsL, P207-gfp-ftsQ, P208-ftsZ-gfp, and P208-zipA-gfp loci from strains EC436, EC438, EC442, EC448, and EC450, respectively, into BC202, resulting in strains listed in Table 1. The W3110 transductants were selected for Amp resistance on LB-agar plates containing low levels of Amp, while the BC202 transductants were selected for Amp resistance on LB-agar plates containing Tet, Kan, and 25 μg/ml Amp. The selected transductants were further verified to contain GFP by colony PCR using primers specific for GFP: 5′-ATGACCATGATTACGCCAAGCTTG-3′ and 5′-CTTGTACAGCTCGTCCATGCCGAG-3′.
Construction and analysis of YqjA-GFP and YghB-GFP fusions.
The following strategy was applied for construction of YqjA-GFP and YghB-GFP. YqjA was amplified from pET23-YqjA (40) using the (forward) T7 promoter primer 5′-TAATACGACTCACTATAGGG-3′ and the reverse primer 5′-CTTACCGGTGCGCTACCGCCGCTACCGCCGCTACCGCCCCCCCGATTTCCATATTTCTTTTTC-3′ (AgeI site underlined). The PCR product was treated with AgeI and XbaI and was cloned into similarly treated pGFPuv (Clontech Laboratories, Inc.) (GenBank accession no. U62636). Isolated pGFPuv was treated with XbaI/EcoRI, releasing the 1.5-kb fusion genes, and subsequently cloned into the XbaI/EcoRI-treated vector pWSK29 to result in vector pWTD50. In this construct, gfpuv was replaced by gfpmut2 by amplification of gfpmut2 with the following primers: forward primer 5′-CTTACCGGTAGAAAAAATGAGTAAAGGAGAAGAACTTTTCAC-3′ (AgeI site underlined) and reverse primer 5′-CTTGAATTCTTATTTGTAGAGCTCATCCATGC-3′ (EcoRI site underlined). gfpmut2 was digested with AgeI and EcoRI and ligated into gel-purified pWTD50 (which underwent the same digestions as fusion inserts), ultimately resulting in plasmid pWTD52. GFPmut2 (10) (a gift from Jesse Stricker) gave technically better results than GFPuv. pWTD54 was produced in a manner analogous to that for pWTD52, by amplifying yghB from plasmid pET23-yghB (40) using the (forward) T7 primer described above and reverse primer 5′-CTTACCGGTGCAGAACCACCAGAACCACCAGAACCGCCGGCGTTACAGTATTTTTTTTTAATCAC-3′ (AgeI site underlined). Expression of pWTD52 and pWTD54 resulted in the fusion proteins between YqjA/YgB and GFPmut2 with short linker peptides YqjA-(GGS)3-APVEK-GFPmut2 and YghB-(GGS)3-APVEK-GFPmut2, respectively. Both plasmids independently complemented the growth and cell division defects of BC202. BC202/pWTD52 and BC202/pWTD54 were grown overnight and diluted 1:200 in fresh LB medium containing 100 μg/ml Amp and supplemented with 0, 0.1, 0.5, or 1.0 mM IPTG. Cultures were grown in a shaking incubator (30°C, 225 rpm) to an optical density at 600 nm (OD600) of ∼0.5 and analyzed as described above.
Additional cloning procedures.
amiB was amplified from the genomic DNA of parent strain W3110 by PCR using forward primer 5′-GCGCTCTAGAATGATGTATCGC-3′ (XbaI site underlined) and reverse primer 5′-GCGCAAGCTTTTAGTTTGGCAGC-3′ (HindIII site underlined). The ∼1,338-bp PCR product was purified, treated with XbaI and HindIII, and cloned between the XbaI and HindIII cloning sites of similarly treated plasmid pWSK29. The resulting construct, pWSK-AmiB, was transformed into competent BC202 cells selecting for Ampr.
The entire TatABC operon was amplified from W3110 genomic DNA using forward primer 5′-GCGCCCATGGGTGGTATCAGTATTTGG (NcoI site underlined) and reverse primer 5′-GCGCGCGAATTCTTATTCTTCAGTTTTTTCG (EcoRI site underlined). The ∼1,568-bp PCR product was purified, treated with NcoI and EcoRI, and cloned into a similarly treated plasmid, pBAD/HisA (Invitrogen). The resulting construct was termed pBAD-TatABC.
Fluorescent protein localization experiments.
For protein localization studies, the strains containing chromosomal GFP fusions were grown overnight in LB medium with 25 μg/ml Amp and diluted 1:200 in 20 ml fresh LB-Amp and either 10 μM (for RS102 only) or 25 μM IPTG for induction of GFP fusions. The cells were grown at 30°C in an incubator with shaking at 225 rpm to an OD600 of ∼0.3. The cells were fixed directly in growth medium by use of a previously described procedure (43). A portion (500 μl) of the growing cells was transferred to a microcentrifuge tube containing the fixative mixture 20 μl of 1 M NaPO4 (pH 7.4), 100 μl of 16% paraformaldehyde, and 0.4 μl of 25% glutaraldehyde and mixed. Fixation was carried out for 15 min at room temperature, followed by 45 min on ice. The fixed cells were then washed three times with 1× phosphate-buffered saline. Finally, the washed cells were resuspended in 1× phosphate-buffered saline at an OD600 of ∼1.0, and 10 μl was applied to glass slides previously coated with a thin layer of 1% agarose for cell immobilization. Cells were visualized using differential interference contrast (DIC) and fluorescence microscopy.
Localization studies of AmiA-GFP and AmiC-GFP were done using live cells. pTB28 (expressing AmiC-GFP) and pTB32 (expressing AmiA-GFP) (5) were generously provided by Thomas Bernhardt (Harvard Medical School). Wild-type W3110 and BC202 cells harboring plasmid pTB28 or pTB32 and BC202 cells harboring plasmid pTB28 or pTB32 along with plasmid p-yghB were used. The strains were grown overnight in LB medium with 100 μg/ml Amp and were diluted 1:200 in 20 ml fresh LB medium supplemented with the appropriate antibiotics and IPTG concentrations as follows. For strains harboring plasmid pTB28, 100 μM IPTG was used, while for strains harboring pTB32, 300 μM IPTG was used (for expression of AmiC-GFP and AmiA-GFP, respectively). W3110 or BC202 cells harboring pTB28 or pTB32 were grown in the presence of Amp, and BC202 cells with covering plasmid p-yghB harboring pTB28 or pTB32 were grown in the presence of Amp and Cam. The cells were grown at 30°C in an incubator with shaking at 225 rpm to an OD600 of ∼0.4. Live cells were harvested directly from the growth medium by centrifugation and resuspended in fresh LB medium at an OD600 of ∼1.0. A portion (10 μl) of the cells was applied to agarose-coated glass slides and observed using DIC and fluorescence microscopy.
Construction and growth of strain AD93/pTB32.
Strain AD93 harboring plasmid pDD72 was transformed with plasmid pTB32. pDD72 is temperature sensitive for replication (14) and was subsequently cured by growth of the strain at 43°C on LB-agar plates supplemented with 50 μg/ml Amp and 50 mM MgCl2. The absence of plasmid pDD72 from cured strain AD93/pTB32 was confirmed by the strain's sensitivity to Cam and growth only in the presence of 50 mM MgCl2. AD93/pTB32 was always grown at 37°C in LB medium supplemented with 5 μg/ml Kan, 50 μg/ml Amp, and 50 mM MgCl2. An overnight culture of AD93/pTB32 was diluted 1:50 in fresh LB medium supplemented with 300 μM IPTG to induce the expression of AmiA-GFP. Cells were grown for 3 h and analyzed as described below for other strains expressing AmiA-GFP.
Microscopy.
All fluorescence and DIC micrographs were taken using a Leica DM-RXA2 deconvolution microscope. Fluorescence microscopy was done using the Leica L5 filter set for GFP, with an excitation range of 480/40 (460 to 500 nm), an emission range of 527/30 (512 to 542 nm), and a dichroic reflector of 505 nm. All observations were recorded using a 100×, 1.40-numerical-aperture oil immersion objective lens. Images from the deconvolution microscope were captured by a cooled Cooke SensiCamQE 12-bit, 1.3-megapixel, charge-coupled-device digital camera and recorded using a workstation computer running Slidebook software (Intelligent Imaging Innovation, Denver, CO). Typical exposure times ranged from 7 to 10 s for GFP expressed in the chromosomal GFP fusion strains, 3 to 4 s for strains expressing AmiA-GFP and AmiC-GFP, and 4 to 5 s for BC202 strains expressing YqjA-GFP and YghB-GFP. DIC images and fluorescent images (for GFP only) were recorded concurrently, processed, and overlaid using the Slidebook software. Image processing in the Slidebook software involved deconvoluting the fluorescent images by using a no-neighbor deconvolution algorithm. All subsequent image editing was done using Adobe Photoshop CS2.
FRAP and confocal microscopy.
FRAP was determined with live BC202 cells expressing cytoplasmic GFP from plasmid pTrcHis2-ELGFP6.1-TOPO, which was constructed as follows. A gene encoding ELGFP6.1 in vector pELGFP6.1-SK was amplified by PCR with primers EL768 and EL769 (24). The amplified gene was then cloned into expression vector pTrcHis2-TOPO (Invitrogen, CA). The resulting clone was sequenced and designated pTrcHis2-ELGFP6.1. An overnight culture of the BC202/pTrcHis2-ELGFP6.1-TOPO strain was diluted 1:200 in fresh LB medium containing 100 μg/ml Amp and 1.0 mM IPTG. The cells were grown at 30°C in a shaking incubator up to an OD600 of ∼0.4. The cells were harvested by centrifugation and resuspended in fresh LB medium at an OD600 of ∼1.0. A portion (10 μl) of the cells was applied to an agarose-coated glass slide and observed under a confocal microscope for FRAP studies.
Confocal microscopy was performed with a Leica confocal microscope (model TCS-SP2; Leica Microsystems, Inc., IL) fitted with an HCX PL APO CS 63.0×, 1.40-numerical-aperture oil objective and driven by Leica confocal software. FRAP images were recorded by selecting sets of cells in chains and then photographing them. In order to bleach out a single cell in a chain, the size of the bleach area was set to the entire width of the selected cell, and this selected area (referred to as the region of interest [ROI]) was bleached by exposure to the 488-nm line of the Ar/Kr green laser (with the laser power set to maximum), referred to as the bleach pulse, for 3 to 4 s. Since the levels of expression of GFP vary from cell to cell, the duration of the applied bleach pulse was adjusted to achieve complete bleaching of the extremely bright variant of GFP expressed in BC202. The scan speed was set to 400 Hz. Fluorescence photobleaching and subsequent recovery were documented by photographing the cells each second for 6 s before application of the bleach pulse (prebleach images), while the bleach pulse was in effect, and for 20 to 25 s after the bleach pulse was terminated (postbleach images).
Spheroplast formation and Western blotting.
Cells harboring plasmids expressing enhanced GFP (pTrcHis2-ELGFP6.1-TOPO), AmiC-GFP (pTB28), or AmiA (pTB32) were grown in LB medium supplemented with the appropriate antibiotic to an OD600 of ∼0.5. Cells harboring pTB28 or pTB32 were grown with 0.1 or 0.3 mM IPTG additionally, respectively. Spheroplasting was carried out essentially as described previously (5), with minor modifications. A portion (30 ml) of the cells was centrifuged and resuspended with 1 ml of 25% sucrose, 30 mM Tris-Cl, pH 8.0 (SP buffer). BC202 spheroplasts lysed very easily, so vortexing was avoided and the cells were handled gently. Half of the cells were left untreated, and half of the cells were converted to spheroplasts with the addition of lysozyme (200 μg/ml) and EDTA (10 mM) followed by a 10-min incubation at room temperature. Spheroplasts were pelleted in a microcentrifuge, and the supernatant containing the periplasmic fraction was transferred to a new tube. The spheroplast pellet was resuspended in 0.5 ml SP buffer. This was followed by the addition of equal volumes of 2× SDS-PAGE loading buffer to each fraction, i.e., total, pellet, and supernatant. In some experiments, whole-cell lysates were analyzed instead of fractions. The samples were heated to 95°C for 5 min, and 10 μl of each was loaded onto a 12% SDS-PAGE gel. Western blotting was carried out as described previously (16), with primary antibody anti-GFP JL-8 (Clontech Laboratories) or anti-maltose binding protein (MBP) (New England Biolabs) used at a dilution of 1:10,000. The secondary antibody was horseradish peroxidase-conjugated goat anti-mouse IgG (Thermo Scientific), and detection was performed with an Immun-Star kit (Bio-Rad Laboratories).
RESULTS
FRAP demonstrates compartmentalization of many BC202 cells.
The nature of the cell division defect that occurs at the permissive growth temperature of 30°C in BC202 was analyzed using FRAP. In this experiment, soluble GFP was expressed from a plasmid in BC202, and recovery of fluorescence in photobleached cells was measured as an indication of the ability of cytoplasmic GFP to move from an adjacent cell into the photobleached cell. A similar approach, measuring fluorescence loss in photobleaching, has been used to characterize the chaining phenotype of an E. coli amiABC mutant (31). If the fusion of the inner membrane were completed in these chains of cells, the cytoplasm would be separated and hence soluble GFP should not diffuse into the photobleached cell from an adjacent cell. However, if inner membrane fusion were not complete, then a bleached cell should be able to recover fluorescence from adjacent cells following photobleaching. Out of 43 BC202 cells subjected to photobleaching, 18 (42%) were able to recover fluorescence from one or more of the adjacent cells in a chain, suggesting that they share a common cytoplasm, while 25 (58%) failed to recover fluorescence, suggesting that they are fully compartmentalized from their adjacent neighbors. As illustrated in a representative experiment shown in Fig. 1A and B, two cells in a chain were subjected to a bleach pulse lasting for 3 s and no subsequent fluorescence recovery was observed in the photobleached cells, indicating that they were compartmentalized with the cytoplasmic GFP and not able to move between the adjacent cells. Another representative experiment, illustrated in Fig. 1C and D, shows an occasion where the BC202 cell subjected to photobleaching exhibited subsequent fluorescence recovery from one (but not both) of its adjacent cells, indicating that the inner membrane fusion was not complete and that the cells share a common cytoplasm. Since about 58% of the observed cells were compartmentalized, we assume that the remaining 42% of the observed cells, which were able to recover fluorescence from their adjacent cells in a chain after being subjected to photobleaching, were observed at an earlier stage of cell division. From these results, we conclude that constriction can be completed in BC202, albeit at a much lower rate than that observed for parent strain W3110.
FIG. 1.
FRAP analysis. BC202 cells expressing cytoplasmic GFP from plasmid pTrcHis2-ELGFP6.1-TOPO were grown and live cells were observed using confocal microscopy as described in Materials and Methods. The region of interest (ROI), which refers to an area set equal to the width of a single cell in a chain (indicated by arrows), was monitored for changes in GFP fluorescence intensity during the entire FRAP procedure. The ROI selected for bleaching were photobleached by exposure to a bleach pulse (488-nm line of an Ar/Kr green laser set at maximum power) lasting for 3 s and further monitored for subsequent fluorescence recovery. The adjacent ROI were simultaneously monitored for any loss of fluorescence intensity. The cells were photographed and images were recorded each second throughout the entire FRAP procedure in order to monitor the fluorescence intensity. Such a time-lapse series of images of live BC202 cells in a chain subjected to a FRAP procedure is shown in panels A and C. The first row of three images in panels A and C shows prebleach images recorded at time points 0, 2, and 5 s. The bleach pulse was active from 6 to 9 s. The second and third rows of images in panels A and C show postbleach images recorded at time points 9, 12, 15, 18, 23, and 28 s. The graphs shown in panels B and D represent the fluorescence recovery kinetics for panels A and C, respectively. Panels A and B represent an instance where the cells observed in a chain are compartmentalized. ROI 1 and 2 are bleached out, and they do not recover fluorescence, while adjacent ROI 3 and 4 exhibit no loss of fluorescence over time. The fluorescence kinetics for adjacent ROI 1 and 4 are similar to that of ROI 2 and 3 but are not shown in panel B for the sake of clarity. Panels C and D represent another instance where ROI 1 is photobleached and exhibits gradual fluorescence recovery from ROI 2, which exhibits a simultaneous loss of its fluorescence, but not from ROI 3, which retains its fluorescence, indicating that ROI 1 and 2 share a common cytoplasm while ROI 3 is compartmentalized. As shown in panel D, a 20-s postbleach time was sufficient to allow for the proper intermixing of cytoplasmic contents and fluorescence recovery from adjacent cells.
FtsZ localizes normally to new septation sites in BC202.
To continue the analysis of the cell division defect of BC202, we carried out an experiment to localize GFP-tagged FtsZ in this mutant. FtsZ is a central player in bacterial cell division and is the first protein to localize to the septal ring in E. coli (42). Strain EC448 contains a chromosomal copy of FtsZ-GFP behind a lac promoter and tightly linked to an Ampr marker (43). A P1 lysate was prepared from EC448 and used to transduce BC202 to Ampr (see Materials and Methods). This strain, named RS101, was grown in the presence of 25 μg/ml Amp plus 25 μM IPTG, and cells were visualized with a fluorescence microscope. In the parent strain W3110, the majority of FtsZ localized at the midcell septal ring (data not shown), as previously described (1, 4, 6). We found that FtsZ localizes to alternate cell division sites in RS101 (Fig. 2A, arrowheads), presumably the sites of the next round of division; however, FtsZ was absent from the sites of the previous (and aborted) round of cell division. We interpret this result to mean that FtsZ is capable of correct localization to the septal ring in BC202 but disassembles and relocalizes to new sites following unsuccessful septation. FtsZ cannot reform the septal ring at the older sites, due to the inhibitory activity of MinCD (13). This is consistent with the rapid dynamics of the FtsZ ring that has been reported in studies using a thermosensitive ftsZ mutant (1).
FIG. 2.
Localization of cell division proteins in BC202 cells. (A) Strain RS101 expressing FtsZ-GFP was grown at 30°C as described in Materials and Methods. Cells were harvested and visualized using DIC (left) and fluorescence (middle) microscopy, and an overlay of both images is shown (right). (B to E) Strains RS105, RS104, RS103, and RS102, expressing chromosomally encoded ZipA-GFP, FtsQ-GFP, GFP-FtsL, and GFP-FtsI, respectively (Table 1), were grown as described in Materials and Methods and visualized using DIC and fluorescence microscopy. Shown are overlay images for each cell type. Arrowheads indicate sites of Fts protein localization. The center diagram shows the order of recruitment to the septal ring in E. coli for known cell division proteins and is modified, with the permission of the publisher, from a diagram originally presented in reference 42 to include FtsP (34, 38). The red font indicates proteins localized in BC202 in this study.
Recruitment of other divisome proteins in BC202.
In E. coli, the recruitment of cell division proteins to the septal ring occurs in a hierarchical manner, with late proteins dependent upon correct recruitment of early proteins (11, 42). We therefore used the approach described above to localize several other proteins in BC202 known to be involved in cell division. Strains EC436, EC438, EC442, and EC450 express forms of FtsI, FtsL, FtsQ, and ZipA, respectively, GFP linked to an Ampr gene (Table 1) (9, 19, 43). P1 lysates prepared from these strains were used to transduce BC202 to Ampr, resulting in strains RS102, RS103, RS104, and RS105 (Table 1). The reported order of recruitment of these proteins to the E. coli divisome and the localization of each protein in BC202 are shown in Fig. 2B to E. Each protein appears to localize correctly to new septal rings, but as seen with GFP-FtsZ, the proteins are not present at old constriction sites, probably due to the disassembly of divisome components following aborted cell division and inhibition of ring formation by the MinCD complex (13). We observed this pattern for each cell division protein analyzed in nearly all of the cell chains that were observed. Collectively, these data suggest that the block in cell division in BC202 occurs at a very late step.
Abnormal fluorescence of AmiC-GFP in live BC202 cells.
AmiC is an E. coli periplasmic amidase required for cell division and is recruited to the septal ring (5). AmiC is exported via the twin arginine transport (Tat) pathway (5), allowing for GFP to be used to visualize its cellular localization (35, 39). A plasmid expressing the fusion protein E. coli AmiC-GFP was transformed into W3110 and BC202, and its localization was visualized using fluorescence microscopy. AmiC-GFP localizes to the septal ring in W3110 cells, in agreement with what has been reported previously (5) (Fig. 3A). When this experiment was repeated with BC202, we failed to observe septal localization in spite of adequate expression of the protein, as evidenced by fluorescence that was visible throughout the entire cell (Fig. 3B). The correct localization of AmiC is restored in BC202 harboring a compatible plasmid expressing a wild-type copy of yghB (Table 1) (Fig. 3C). While it is not clear from this experiment if AmiC-GFP accumulates in the cytoplasm or periplasm in BC202, the protein is clearly not found predominantly at the septal ring, as it is in the parent strain. Inclusion of 10 mM Mg2+ in the growth medium, which rescues the growth and cell division defects of BC202 (40), fails to alter the localization of AmiC-GFP (Fig. 3D).
FIG. 3.
Localization of AmiC-GFP and AmiA-GFP fluorescence. W3110 (A and E), BC202 (B, F, D, and H), and BC202 harboring plasmid p-yghB (C and G) transformed with plasmid pTB28 (A to D) or pTB32 (E to H) were grown in LB medium supplemented with Amp (A, B, E, and F), with Amp and Cam (C and G), or with Amp and 10 mM MgCl2 (D and H) at 30°C and visualized using DIC and fluorescence microscopy. Shown are overlay images for each cell type transformed with pTB28 or the fluorescent images for each cell type transformed with pTB32 (to better illustrate the partial polar localization of AmiA-GFP).
Abnormal fluorescence of AmiA-GFP in live BC202 cells.
AmiA is another periplasmic amidase, which is exported via the Tat pathway (5). Unlike AmiC, AmiA is not a component of the septal ring, and hence it is difficult to observe distinct localization sites for AmiA. Plasmid pTB32 expressing fusion protein AmiA-GFP was transformed into W3110 and BC202, and its localization was visualized using fluorescence microscopy. AmiA was found to be localized in W3110 in such a manner that most of the cells appear to be surrounded by a fluorescent halo, with about 10 to 15% of the cells exhibiting polar localization (Fig. 3E), as was reported previously (5). This apparent polar bias of localization of AmiA in wild-type E. coli was explained by the fact that the periplasmic volume surrounding the poles is relatively large (5). However, in BC202, AmiA-GFP fluorescence was visible throughout the entire cell, again suggestive of a cytoplasmic localization (Fig. 3F). In BC202 cells expressing a copy of wild-type yghB, the localization pattern of AmiA observed was found to be similar to that of wild-type W3110 cells (Fig. 3G). As was seen with AmiC-GFP (Fig. 3D), inclusion of 10 mM Mg2+ does not visibly alter the localization of AmiA-GFP (Fig. 3H), suggesting that the effects of divalent cations on the phenotypes of BC202 are independent of the effects of the mutations on amidase export. However, Western blot analysis of whole-cell lysates does reveal a partial correction of the inefficient export of AmiA when Mg2+ is included in the growth medium (see the next section). It is possible that a strong cytoplasmic signal obscures the presence of a minor periplasmic component in these micrographs.
Cytoplasmic localization of AmiA-GFP and AmiC-GFP in BC202.
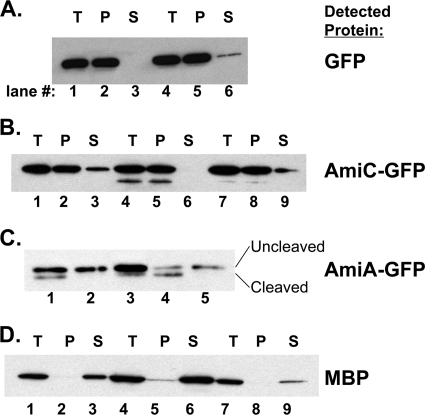
The data from Fig. 3 suggest that AmiC-GFP and AmiA-GFP are mislocalized in BC202. In order to determine the cellular compartment where the protein is found, we used spheroplasting to separate cytoplasmic from periplasmic components, followed by Western blotting with an anti-GFP antibody. When soluble GFP is expressed from a plasmid in parent strain W3110, the protein is in the pellet (or cytoplasmic) fraction, and none is found in the supernatant (or periplasmic) fraction (Fig. 4A, lanes 1 to 3). When this analysis is repeated with BC202, most of the GFP is associated with the pellet, and a small amount (less than 10%) is found in the supernatant (Fig. 4A, lanes 4 to 6). We attribute this to spontaneous lysis occurring during the spheroplasting procedure. Different spheroplasting conditions were tested, and such lysis could not be avoided but could be minimized with the high sucrose concentrations (25%) used.
FIG. 4.
Cellular fractionation and localization of AmiC-GFP and AmiA-GFP. (A) W3110 (lanes 1 to 3) and BC202 (lanes 4 to 6) transformed with plasmid pTrcHis2-ELGFP6.1-TOPO were grown in LB medium with Amp without IPTG and converted to spheroplasts as described in Materials and Methods. The total cell lysate (T), spheroplast pellet (P), and spheroplast supernatant (S) were resolved by 12% SDS-PAGE and Western blotted with anti-GFP. (B and D) W3110 (lanes 1 to 3), BC202 (lanes 4 to 6), and BC202 harboring plasmid p-yghB (lanes 7 to 9) transformed with plasmid pTB28 were grown at 30°C in LB medium containing 0.1 mM IPTG and converted to spheroplasts. The total cell lysate, spheroplast pellet, and spheroplast supernatant were resolved by 12% SDS-PAGE and Western blotted with anti-GFP (B) or anti-MBP (D). (C) W3110 (lane 1), BC202 (lane 2), BC202 grown with 10 mM MgCl2 (lane 3), BC202 harboring plasmid p-yghB (lane 4), and AD93 harboring plasmid pTB32 (lane 5) were grown at 30°C (or 37°C in the case of AD93) in LB medium containing 0.3 mM IPTG, and whole-cell lysates were analyzed by Western blotting using anti-GFP. Cleaved and uncleaved refer to signal peptidase-processed and -unprocessed forms of the protein that are expected to be found in the periplasm and cytoplasm, respectively. Longer exposures revealed no evidence of the processed protein in lanes 2 and 5 (not shown).
When W3110 cells expressing AmiC-GFP were analyzed, we found that ∼70% of the AmiC-GFP was associated with the spheroplast pellet and ∼30% was found in the supernatant (Fig. 4B, lanes 1 to 3), similarly to what was previously reported (5). In BC202, nearly all of the AmiC-GFP was found to be associated with the pellet, with none detectable in the supernatant (Fig. 4B, lanes 4 to 6), in spite of the efficient release of maltose binding protein, a periplasmic substrate of the general secretory pathway, from the same sets of cells (Fig. 4D, lanes 1 to 9). This result suggests that the mislocalization of AmiC-GFP in BC202 is caused by inefficient export from the cytoplasm. Export of AmiC-GFP could be restored in BC202 by expressing yghB from a compatible plasmid (Fig. 4B, lanes 7 to 9).
Unfortunately, attempts to release AmiA-GFP from BC202 spheroplasts resulted in visible cell lysis, due to the higher salt concentrations required (5). However, Western blotting of whole-cell lysates with anti-GFP antibody revealed that all of the AmiA-GFP expressed in BC202 is of the higher-molecular-weight form, uncleaved by periplasmic signal peptidase (Fig. 4C, lane 2). This result strongly suggests that AmiA-GFP is not exported to the periplasm in BC202, unlike with parent strain W3110 (Fig. 4C, lane 1) or BC202 harboring a wild-type copy of yghB (Fig. 4C, lane 4). Inclusion of 10 mM Mg2+ in the growth medium does partially rescue the inefficient export of AmiA (Fig. 4C, lane 3), which possibly explains the ability of divalent cations to rescue the cell division defect of BC202 (40). Overexpression of AmiA or AmiC with higher levels of IPTG (1 mM) could correct the cell division defect in BC202 (see below), so we expect the block in export to be somewhat leaky. The data from Fig. 3 and 4 collectively suggest that export of AmiA and AmiC is defective in BC202. However, the phenotypes of BC202 and Tat deletion mutants (7, 23, 37) are not identical, so we cannot conclude that the Tat pathway is nonfunctional in BC202. While Tat mutants do have cell division defects and form chains like BC202, they are also reminiscent of envA (lpxC) mutants and have severe outer membrane defects resulting in hypersensitivity to antibiotics and detergents (23, 37). In contrast, BC202 is not hypersensitive to drugs or detergents at the permissive growth temperature (40) (see Discussion).
AmiA export in PE-deficient cells.
It is possible that the effect on the Tat pathway in BC202 could be indirect. An example of an indirect role would be through effects of the reported lower levels of membrane PE (40) in BC202 on Tat function. PE is not essential for viability of E. coli, and strain AD93 (pss93::Kanr recA), which is deficient in PE, has been isolated and characterized (14, 27, 33). AD93 displays severe cell division defects, with extensive filamentation (28). AmiA-GFP was expressed in AD93 and was found to be largely cytoplasmic, based upon the absence of signal peptidase-processed protein by Western blot analysis of whole-cell lysates (Fig. 4C, lane 5). This result is consistent with the reported PE requirement for the proper Tat-dependent export of TorA (26) and suggests that the phospholipid membrane imbalance observed in BC202 may be responsible for the inefficient Tat export of amidases observed here.
Overexpression of AmiA-GFP, AmiB, and AmiC-GFP suppresses the cell division defect of BC202.
E. coli encodes three periplasmic amidases: AmiA, AmiB, and AmiC. While AmiA and AmiC are exported to the periplasm by the Tat pathway, AmiB is secreted via the general secretory pathway and does not localize to the division septum (5). Overexpression of AmiB, but not AmiA or AmiC, corrects the cell division defect of the Tat deletion mutants (5). Overexpression of AmiC-GFP from pTB28 with 1.0 mM (Fig. 5A) but not 0.1 mM (Fig. 5B) IPTG largely corrects the cell division defect of BC202, suggesting that BC202 can export at least some AmiC under these conditions. Much of the AmiC protein is still mislocalized under these conditions, emitting fluorescence throughout the cell (Fig. 5C). Overexpression of AmiA-GFP from pTB32 with 1.0 mM IPTG (Fig. 5D) was also found to largely correct the cell division defect of BC202, indicating that BC202 can also export at least some AmiA when it is overexpressed. Since overexpression of AmiB from a plasmid is capable of correcting the cell division defect in an ΔamiA ΔamiC mutant and a ΔtatC mutant (5), we overexpressed amiB from a plasmid in BC202. As shown in Fig. 5E, overexpression of AmiB also corrects the cell division defect of BC202. This suggests that YghB/YqjA play a role, either direct or indirect, in the proper localization of periplasmic amidases and that the block in export in BC202 may be partially leaky and overcome by high levels of protein expression.
FIG. 5.
Correction of cell division in BC202 overexpressing AmiC-GFP, AmiA-GFP, or AmiB. BC202 harboring plasmid pTB28 (A to C), pTB32 (D), or pWSK-AmiB (E) was grown in LB medium containing Tet, Kan, Amp, and either 1.0 mM or 0.1 mM IPTG at 30°C. DIC images are shown in panels A, B, D, and E, while panel C represents the fluorescence image corresponding to panel A. BC202 cells overexpressing amidases failed to grow at 42°C (not shown).
Overexpression of AmiA, AmiB, or AmiC, while able to correct the cell division defect in BC202, does not correct the temperature sensitivity of the strain, as these transformants still fail to grow at elevated temperatures (data not shown). It was, however, not possible to directly check amidase export at 42°C, since BC202 dies quickly due to cell lysis at this temperature (40). These results suggest that while YghB/YqjA may be required for proper cell division in E. coli, these proteins may play roles distinct from their roles in cell division that are necessary for growth at elevated temperatures.
Correction of cell division and growth phenotypes of BC202 by overexpression of TatABC.
The observation that periplasmic amidases are, at the very least, inefficiently exported to the periplasm in BC202 suggests that the Tat pathway may be affected by the mutations in yghB and yqjA. To test this, we expressed the entire tatABC operon under the control of an arabinose-inducible promoter and tested for the effect of overexpression of these genes on growth and cell division in BC202. When BC202 harboring plasmid pBAD-TatABC is grown in the presence of 0.2% arabinose (but not 0.2% glucose), there is a complete restoration of both growth at 42°C (Fig. 6A and B) and normal cell division (Fig. 6C and D). This striking result indicates that the Tat pathway functions poorly in BC202 and that this may be responsible for the observed phenotypes of the mutant. In spite of this, the phenotypes of BC202 still do not exactly match those found in Tat mutants or amidase mutants (see Discussion).
FIG. 6.
Correction of cell division and growth phenotypes of BC202 by overexpression of TatABC. (A and B) BC202 was transformed with pBAD/HisA (left side of plates) or pBAD-TatABC (right side of plates) and streaked on LB-Amp plates containing 0.2% glucose (A) or 0.2% arabinose (B). The plates were incubated at 42°C overnight. (C and D) DIC images of BC202 cells harboring plasmid pBAD-TatABC grown at 30°C in the presence of 0.2% glucose (C) or 0.1% arabinose (D).
Cellular localization of YqjA-GFP and YghB-GFP.
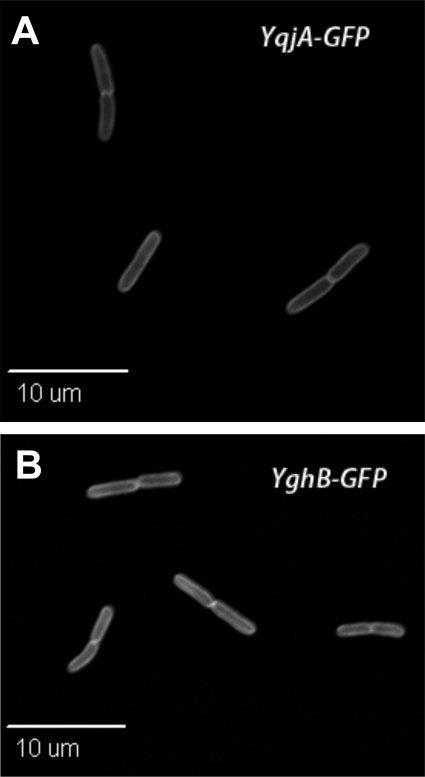
We were also interested in determining the cellular localization of YqjA and YghB. If a restricted or septal localization were observed, this would provide important insights into the function of these proteins. However, YqjA-GFP (Fig. 7A) and YghB-GFP (Fig. 7B) were evenly distributed throughout the envelope of BC202, providing little insight into protein function but not ruling out a direct involvement in cell division. The fusion proteins that we expressed were functional in that they eliminated the cell-chaining phenotype of BC202 (Fig. 7A and B) and were capable of restoring growth to BC202 at 42°C (data not shown). These results collectively suggest that YqjA and YghB are likely required for the proper export of certain periplasmic amidases and, possibly, other Tat substrates.
FIG. 7.
Localization of YqjA-GFP and YghB-GFP in BC202 cells. BC202 cells harboring plasmid pWTD52 (encoding YqjA-GFPmut2) (A) or pWTD54 (encoding YghB-GFPmut2) (B) were grown as described in Materials and Methods in the presence of 0.1 mM IPTG and analyzed using fluorescence microscopy. The plasmids encode functional proteins correcting the cell division phenotype of BC202. Western blot analysis using anti-GFP antibody with whole-cell lysates of BC202 harboring pWTD52 or pWTD54 showed only a single ∼53-kDa band (data not shown), indicating that these GFP fusions of YqjA and YghB are stable and do not degrade within the cell.
DISCUSSION
The observation that homologs of E. coli yghB and yqjA have been maintained (and in some cases duplicated) in present-day bacterial genomes suggests that these genes carry out critical functions in bacterial cellular physiology. To date, very little is known about the functions of this somewhat mysterious protein family. While individual DedA family genes are nonessential in E. coli due to functional redundancy, some organisms (such as Borrelia burgdorferi and Helicobacter pylori) have only one DedA homolog that may represent an essential gene in each of the respective organisms. Comparative sequence analysis of this protein family shows that these genes bear no sequence identity to known enzymes, transporters, channels, or two-component signal transducers.
Homologs of YqjA/YghB are found almost exclusively in bacteria (with the exception of several species of green algae). However, the products of two distantly related E. coli genes, ydjX and ydjZ, do have good sequence identity with the predicted open reading frames of many eukaryotic genomes. Intriguingly, a Caenorhabditis elegans mutant of one such gene, bus-19 (bacterially unswollen-19), has been isolated based upon its resistance to infection by Microbacterium nematophilum (44). Tvp38 of Saccharomyces cerevisiae, a YdjX homolog, was found associated with the t-SNARE Tlg2-containing Golgi/endosomal membrane compartments (21). The functions of these genes are not known, but this illustrates that the DedA family is both very ancient and very widespread in nature.
In this work, we report that the E. coli mutant BC202 (ΔyqjA::Tetr ΔyghB::Kanr) displays inefficient cell division. Using FRAP (Fig. 1), we demonstrate that BC202 cell chains are often compartmentalized by at least a fused inner membrane, indicating that the cell division block in this mutant occurs at a late stage or that cell division is extremely inefficient. Several of the known cell division proteins were localized in BC202 using fluorescent fusions and were properly recruited to the new sites of cell division but were absent from old sites (Fig. 2).
Importantly, a plasmid-expressed AmiC-GFP fusion protein was mislocalized in BC202. Fluorescence was found distributed throughout the cell (Fig. 3) and not at the septal ring as previously reported (5). Western blotting of fractionated cells shows that AmiC-GFP is largely confined to the cytoplasmic compartment of BC202 (Fig. 4B). AmiA-GFP appears to be largely confined to the cytoplasm in BC202 (Fig. 4C) as well. AmiA and AmiC are both exported via the twin arginine transport (Tat) pathway, and the cell division defect is consistent with an inability to export these substrates to the periplasm. These data point to the possibility that the Tat pathway operates inefficiently in BC202 but perhaps functions better for some substrates than others. This would explain why the phenotypes of BC202 and Tat deletion mutants are not identical although there are intriguing similarities between these classes of mutants. For example, in addition to having cell division defects, both ΔTat and BC202 mutant phenotypes are suppressed by the inclusion of 5 to 10 mM Ca2+ or Mg2+ in the growth medium (7, 40). We also show that Mg2+ partially restores the export of AmiA-GFP in BC202 (Fig. 4C, lane 3). The effects of divalent cations on these mutants are not understood.
However, mutants with mutations in the Tat pathway also display pleiotropic outer membrane defects, including hypersensitivity to antibiotics and detergents (23, 37) and resistance to P1 phage (37). These phenotypes are also shared with mutants lacking multiple amidases (20). BC202 does not display phenotypes that indicate a failure of the outer membrane permeability barrier, is sensitive to P1 phage (40), and synthesizes and exports normal amounts of lipopolysaccharide (W. T. Doerrler, unpublished results). In addition, BC202 is temperature sensitive for growth at 42°C, a phenotype not shared with ΔTatC mutants (Doerrler, unpublished). In spite of these differences, overexpression of TatABC in BC202 restores not only normal cell division but also growth at 42°C (Fig. 6), indicating that the Tat pathway is limiting in BC202 but that BC202 is not identical to Tat mutants and that YqjA/YghB may have additional cellular functions.
BC202 has alterations in membrane phospholipid levels, with decreased levels of PE and increased levels of PG and CL (40). It has been reported that the absence of membrane PE in the Δpss93 mutant causes a decreased efficiency of TorA translocation across the inner membrane by the Tat pathway (26), and we have shown that AmiA-GFP is also inefficiently exported in the PE-deficient mutant (Fig. 4C, lane 5). Therefore, we cannot rule out, and indeed it is likely, that the inefficiency of the Tat pathway in BC202 is a secondary effect of the altered membrane composition of BC202. It has been demonstrated that Tat signal peptides interact strongly with acidic phospholipids (36). Since BC202 contains up to twice as much membrane PG and CL as that found in the parent strain W3110, the Tat substrates may simply be trapped at the inner surface of the inner membrane due to these electrostatic interactions. This supports the idea that precise control of membrane composition is critically important for diverse cellular processes (45). The roles that YghB/YqjA play in controlling membrane composition are not known. A systematic analysis of export of other Tat substrates in BC202 as well as an analysis of YghB and YqjA protein-protein and/or protein-lipid interactions will shed more light on this process and the roles of these proteins with regard to the functions of the Tat pathway.
YghB and YqjA appear not to be bona fide cell division proteins, since they show diffuse inner membrane localization and are not specifically located at the septal ring (Fig. 7), like all other studied divisome proteins (42). It should be kept in mind when interpreting these results that the E. coli genome contains up to five additional genes encoding proteins with similarity to YqjA and YghB. yabI, yohD (40), and dedA (Doerrler, unpublished) can complement the growth phenotype of BC202 when overexpressed from plasmids. Therefore, an E. coli strain harboring all possible deletions of the dedA family genes will undoubtedly display a more severe phenotype and may be inviable or display increased outer membrane permeability.
Often, gene functions can be inferred by examining the mode of regulation. Since it is likely that yghB and yqjA are paralogous genes (17, 25), we hypothesize that these genes are evolving separate functions but still maintain preduplication structure and function, such that one protein is able to compensate for the loss of the other. In addition, the amino acid sequences of YghB and YqjA have three independent regions (25 residues) with greater than 80% identity (L. A. Boughner and W. T. Doerrler, unpublished). There is now accumulating evidence that demonstrates separate mechanisms of regulation for yghB and yqjA in E. coli. In a DNA microarray study, YghB is strongly induced in E. coli by the quorum-sensing molecule autoinducer 2 (AI-2), suggesting that this protein may play a role in the social behavior, establishment, and/or maintenance of bacterial biofilm communities (15). Quorum sensing is known to regulate a large subset of E. coli genes, including those involved in virulence and cell division (3). It is therefore possible that YghB is evolving to play a specialized function during cell division.
It was recently reported that expression of YqjA is induced by the CpxAR pathway and repressed by the σE pathway (30). Both of these pathways function to combat extracytoplasmic stress and are responsible for regulating distinct but overlapping sets of genes (32). The Cpx-regulated genes include those involved in envelope protein biogenesis, biofilm formation, and resistance to copper. Interestingly, this study also reported that the E. coli yqjA deletion mutant is alkaline sensitive, unable to grow at pH 9.2 (30). Therefore, YqjA may be evolving to provide functions in envelope maintenance and resistance to environmental stress. YqjA is in an operon with MzrA, which was recently identified in a screen for suppressors of the stress phenotype of a ΔbamB ΔdegP mutant of E. coli (18). The protein was found to modulate the activity of the EnvZ/OmpR two-component system. These effects, however, were independent of YqjA.
In summary, YqjA/YghB are members of a highly conserved family of inner membrane proteins. Our discovery that simultaneous deletions of yqjA and yghB result in a mutant with temperature sensitivity, cell division defects, and membrane alterations provides crucial insight into the function of these previously unstudied proteins. Here, we have characterized the cell division phenotype and show that there is a block in cell division at a late stage and that most cell division proteins appear capable of localizing to the divisome, with the noted exception of AmiC, which remains cytoplasmic. Overexpression of the components of the Tat export pathway TatABC restores normal cell division, suggesting that this pathway operates inefficiently in BC202. Future studies will be directed at identifying specific targets and roles of this widely distributed membrane protein family.
Acknowledgments
We thank David Weiss for the GFP-Fts strains listed in Table 1, Thomas Bernhardt for plasmids pTB28 and pTB32, Naohiro Kato for plasmid pTrcHis2-ELGFP6.1-TOPO, and William Dowhan for PE-deficient strain AD93. Thanks also go out to Matt Brown of the LSU Socolofsky Microscopy Facility for assistance and training and to Lisa Boughner and Kandi Thompkins for critical reading of the manuscript.
Financial support has been provided by the National Science Foundation (MCB-0841853, to W.T.D.), the LA Board of Regents [LEQSF (2005-08)-RD-A-04, to W.T.D.], and the LSU Faculty Research Grant Program (to W.T.D.).
Footnotes
Published ahead of print on 30 October 2009.
REFERENCES
- 1.Addinall, S. G., E. Bi, and J. Lutkenhaus. 1996. FtsZ ring formation in fts mutants. J. Bacteriol. 178:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassler, B. L., and R. Losick. 2006. Bacterially speaking. Cell 125:237-246. [DOI] [PubMed] [Google Scholar]
- 4.Begg, K. J., and W. D. Donachie. 1985. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J. Bacteriol. 163:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhardt, T. G., and P. A. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi, E. F., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 7.Caldelari, I., T. Palmer, and F. Sargent. 2008. Escherichia coli tat mutant strains are able to transport maltose in the absence of an active malE gene. Arch. Microbiol. 189:597-604. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395-413. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J. C., D. S. Weiss, J. M. Ghigo, and J. Beckwith. 1999. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J. Bacteriol. 181:521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 11.Dajkovic, A., and J. Lutkenhaus. 2006. Z ring as executor of bacterial cell division. J. Mol. Microbiol. Biotechnol. 11:140-151. [DOI] [PubMed] [Google Scholar]
- 12.Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew, and G. von Heijne. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321-1323. [DOI] [PubMed] [Google Scholar]
- 13.de Boer, P. A., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 14.DeChavigny, A., P. N. Heacock, and W. Dowhan. 1991. Sequence and inactivation of the pss gene of Escherichia coli. Phosphatidylethanolamine may not be essential for cell viability. J. Biol. Chem. 266:5323-5332. [PubMed] [Google Scholar]
- 15.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doerrler, W. T., and C. R. H. Raetz. 2005. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J. Biol. Chem. 280:27679-27687. [DOI] [PubMed] [Google Scholar]
- 17.Fitch, W. M. 2000. Homology a personal view on some of the problems. Trends Genet. 16:227-231. [DOI] [PubMed] [Google Scholar]
- 18.Gerken, H., E. S. Charlson, E. M. Cicirelli, L. J. Kenney, and R. Misra. 2009. MzrA: a novel modulator of the EnvZ/OmpR two-component regulon. Mol. Microbiol. 72:1408-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghigo, J. M., D. S. Weiss, J. C. Chen, J. C. Yarrow, and J. Beckwith. 1999. Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol. 31:725-737. [DOI] [PubMed] [Google Scholar]
- 20.Heidrich, C., A. Ursinus, J. Berger, H. Schwarz, and J. V. Holtje. 2002. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 184:6093-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inadome, H., Y. Noda, Y. Kamimura, H. Adachi, and K. Yoda. 2007. Tvp38, Tvp23, Tvp18 and Tvp15: novel membrane proteins in the Tlg2-containing Golgi/endosome compartments of Saccharomyces cerevisiae. Exp. Cell Res. 313:688-697. [DOI] [PubMed] [Google Scholar]
- 22.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 23.Ize, B., N. R. Stanley, G. Buchanan, and T. Palmer. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 48:1183-1193. [DOI] [PubMed] [Google Scholar]
- 24.Kato, N., D. Pontier, and E. Lam. 2002. Spectral profiling for the simultaneous observation of four distinct fluorescent proteins and detection of protein-protein interaction via fluorescence resonance energy transfer in tobacco leaf nuclei. Plant Physiol. 129:931-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonin, E. V., Y. I. Wolf, and G. P. Karev. 2002. The structure of the protein universe and genome evolution. Nature 420:218-223. [DOI] [PubMed] [Google Scholar]
- 26.Mikhaleva, N. I., C. L. Santini, G. Giordano, M. A. Nesmeyanova, and L. F. Wu. 1999. Requirement for phospholipids of the translocation of the trimethylamine N-oxide reductase through the Tat pathway in Escherichia coli. FEBS Lett. 463:331-335. [DOI] [PubMed] [Google Scholar]
- 27.Mileykovskaya, E., and W. Dowhan. 2005. Role of membrane lipids in bacterial division-site selection. Curr. Opin. Microbiol. 8:135-142. [DOI] [PubMed] [Google Scholar]
- 28.Mileykovskaya, E., Q. Sun, W. Margolin, and W. Dowhan. 1998. Localization and function of early cell division proteins in filamentous Escherichia coli cells lacking phosphatidylethanolamine. J. Bacteriol. 180:4252-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 30.Price, N. L., and T. L. Raivio. 2009. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 191:1798-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Priyadarshini, R., M. A. de Pedro, and K. D. Young. 2007. Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J. Bacteriol. 189:5334-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raivio, T. L., D. L. Popkin, and T. J. Silhavy. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol. 181:5263-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rietveld, A. G., A. J. Verkleij, and B. de Kruijff. 1997. A freeze-fracture study of the membrane morphology of phosphatidylethanolamine-deficient Escherichia coli cells. Biochim. Biophys. Acta 1324:263-272. [DOI] [PubMed] [Google Scholar]
- 34.Samaluru, H., L. SaiSree, and M. Reddy. 2007. Role of SufI (FtsP) in cell division of Escherichia coli: evidence for its involvement in stabilizing the assembly of the divisome. J. Bacteriol. 189:8044-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santini, C. L., A. Bernadac, M. Zhang, A. Chanal, B. Ize, C. Blanco, and L. F. Wu. 2001. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 276:8159-8164. [DOI] [PubMed] [Google Scholar]
- 36.Shanmugham, A., H. W. Wong Fong Sang, Y. J. Bollen, and H. Lill. 2006. Membrane binding of twin arginine preproteins as an early step in translocation. Biochemistry 45:2243-2249. [DOI] [PubMed] [Google Scholar]
- 37.Stanley, N. R., K. Findlay, B. C. Berks, and T. Palmer. 2001. Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotropic defects in the cell envelope. J. Bacteriol. 183:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarry, M., S. J. Arends, P. Roversi, E. Piette, F. Sargent, B. C. Berks, D. S. Weiss, and S. M. Lea. 2009. The Escherichia coli cell division protein and model Tat substrate SufI (FtsP) localizes to the septal ring and has a multicopper oxidase-like structure. J. Mol. Biol. 386:504-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 39:47-53. [DOI] [PubMed] [Google Scholar]
- 40.Thompkins, K., B. Chattopadhyay, Y. Xiao, M. C. Henk, and W. T. Doerrler. 2008. Temperature sensitivity and cell division defects in an Escherichia coli yghB yqjA double mutant, encoding related and conserved inner membrane proteins. J. Bacteriol. 190:4489-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 42.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 43.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yook, K., and J. Hodgkin. 2007. MosI mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 175:681-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, Y. M., and C. O. Rock. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222-233. [DOI] [PubMed] [Google Scholar]