Abstract
Salmonella plasmid pFPTB1 includes a Tn3-like transposon and a Xer recombination site, fpr, which mediates site-specific recombination at efficiencies lower than those required for stabilizing a plasmid by dimer resolution. Mutagenesis and comparative studies with mwr, a site closely related to fpr, indicate that there is an interdependence of the sequences in the XerC binding region and the central region in Xer site-specific recombination sites.
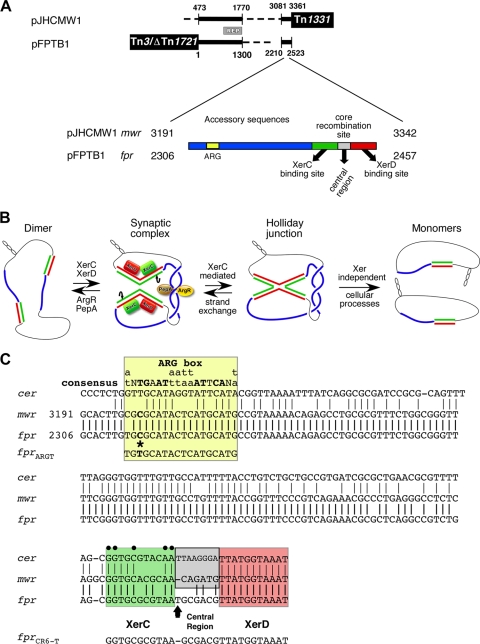
Xer site-specific recombination stabilizes many plasmids by resolving dimers created through recombination events (17, 18). Most plasmids' Xer recombination sites consist of a core recombination site (CRS) that includes two 11-nucleotide binding sites for XerC and XerD, separated by a 6- to 8-nucleotide central region, and a stretch of about 180 bp known as accessory sequences (AS) that bind the architectural proteins PepA and ArgR (Fig. 1A). These elements form a synaptic complex (11, 14) where XerC is activated by interaction with XerD and catalyzes the exchange of the first pair of strands, which results in the formation of a Holliday junction (3, 6) that, in the case of cer (ColE1) or mwr (pJHCMW1), is resolved by Xer-independent processes (Fig. 1B) (2).
FIG. 1.
(A) Comparative diagrams of pFPBT1 and pJHCMW1. The black lines represent regions of homology. The Tn3-like transposons in both plasmids are shown at the correct locations but are not to scale. The gray bar between the plasmid maps identifies the replication (REP) regions, which share 97% homology. The numbers indicate the coordinates in the GenBank database (pJHCMW1, accession number AF479774; pFPBT1, accession number AJ634602). The location and a diagram of the Xer site-specific recombination site are shown below the plasmid diagrams. The different regions of the Xer site-specific recombination site, shown with different colors, are not drawn to scale. ARG, ARG box, ArgR binding site. (B) Schematic representation of dimer resolution mediated by the Xer site-specific recombination reaction. For clarity, the proteins are shown only in the synaptic complex (red rectangle, XerD; green rectangle, XerC; brown oval, PepA; yellow oval, ArgR). Blue lines represent AS, and the CRS is the only region shown with a double line (green and red) representing the two DNA strands. The green line represents the DNA strand exchanged by XerC. Only the Xer-independent pathway of resolution of the Holliday junction (demonstrated for cer and mwr) is shown. (C) Comparison of the nucleotide sequences of mwr, fpr, and cer. The ARG box and different regions of the CRS are individually boxed. The ARG box consensus sequence is shown at the top. Nucleotides mutated in different derivatives are indicated by an arrow (central region) or a star (ARG box). Black dots identify the most conserved nucleotides among several sites (8).
The pJHCMW1 plasmid, originally isolated from Klebsiella pneumoniae, includes mwr and the Tn3-like transposon Tn1331, which transposes through a replicative pathway (15, 20, 22). The efficiency of Xer-mediated dimer resolution at mwr when Escherichia coli is cultured in L broth is below the levels needed to stabilize the plasmid; instead, stabilization is mediated by the Tn1331 resolvase acting at the res site (13, 21). However, the efficiency of Xer-mediated dimer resolution at mwr is substantially increased when the cells are cultured in low-osmolarity broth (4, 13). The low levels of dimer resolution observed when cells are cultured in L broth seem to be due to a weak interaction of the substandard mwr ARG box with ArgR, hindering proper formation of the synaptic complex. When the cells are cultured in low-osmolarity growth medium, an increase in the density of negative supercoiling results in an increased stability of the synaptic complex and/or efficiency of catalysis by XerC, leading to a significantly higher efficiency of dimer resolution (23). These characteristics make pJHCMW1 a very unusual plasmid that includes a Xer recombination site that, under certain conditions, is unable to confer stability by resolution of dimers; instead, that function is performed by the cointegrate resolution system of Tn1331. In this study, we report that another plasmid, Salmonella Typhimurium plasmid pFPTB1 (12), includes a Xer recombination site with high homology to mwr (Fig. 1C), from here on called fpr (pFPTB1 Xer recombination site), and a copy of the replicative transposon Tn3-ΔTn1721 (Fig. 1A and C). Plasmid pFPTB1 is the second case reported in which stabilization by dimer resolution is provided by the insertion of a replicative transposon rather than a resident Xer recombination site, suggesting that rather than being exceptional, pJHCMW1 and pFPTB1 may be part of a group of plasmids with these characteristics.
The E. coli strains and plasmids used in this study are described in Table 1. Plasmid pFPRTT1 was generated by inserting a synthetic DNA fragment with the fpr site sequence (coordinates 2221 to 2520, accession number AJ634602) from pFPTB1 (12) into the EcoRV site of pUC57 (accession number Y14837). Plasmids pTTT1 through pTTT6 were generated by site-directed mutagenesis using the QuikChange II XL kit (Stratagene). Lennox L broth (containing 2% [wt/vol] agar in the case of solid medium) is called high-osmolarity medium (containing 0.5% NaCl; osmolality, 209 mmol/kg); for low-osmolarity growth medium, NaCl was omitted (osmolality, 87 mmol/kg) (13, 23). In vivo resolution assays were carried out as described by Pham et al. (13). Although pFPTB1 has been isolated from S. Typhimurium, we decided it was appropriate to carry out the in vivo experiments with E. coli because it has been shown before that the Xer recombination proteins of S. Typhimurium can substitute for and are highly homologous to the corresponding proteins of E. coli (7), K. pneumoniae Xer recombination proteins share high homology with those of E. coli and can complement mutants, we have observed no difference in levels of resolution with some sites such as dif and cer and minimal differences with mwr (4), and pJHCMW1-like replicons such as pGY1 (Salmonella) (9), pVI678 (E. coli; accession number NC_008597), and pTKH11 (Klebsiella) (25) are being found in nature across the members of the family Enterobacteriaceae rather than in one specific genus.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic or genotype | Relevant characteristics of Xer recombination sites | Reference or origin |
|---|---|---|---|
| E. coli | |||
| DS941 | AB1157 recF143lacIqlacZΔM15; used as a host in dimer resolution assays | 19 | |
| DS981 | DS941 xerC2::aph; used to purify dimers | 5 | |
| JC8679 | DS945 recBCsbcA (hyperrecombinogenic); used to generate dimers | 17 | |
| Plasmids | |||
| pES | Recombinant plasmid containing the pJHCMW1 mwr site | ASmwr, CRSXerCmwr,crmwr | 13 |
| pKS492 | Recombinant plasmid containing the ColE1 cer fragment | AScer, CRScer | 16 |
| pFPRTT1 | Recombinant plasmid containing the pFPBT1 fpr site (nucleotides 2221-2520, accession no. AJ634602) | ASfpr, CRSXerCfpr,crfpr | This work |
| pTTT1 | Mutant derivative of pFPRTT1 containing fprARGT | ASfprARGT, CRSXerCfpr,crfpr | This work |
| pTTT2 | Mutant derivative of pFPRTT1 containing fprCR6-T | ASfpr CRSXerCfpr,crCR6-T | This work |
| pTTT3 | Mutant derivative of pFPRTT1 containing fprARGT/CR6-T | ASfprARGT, CRSXerCfpr,crCR6-T | This work |
| pTTT4 | Mutant derivative of pFPRTT1 containing fprARGT/CRmwr | ASfprARGT, CRSXerCfpr,crmwr | This work |
| pTTT5 | Mutant derivative of pFPRTT1 containing fprARGT/CR6-T/XerCmwr | ASfprARGT, CRSXerCmwr,crCR6-T | This work |
| pTTT6 | Mutant derivative of pFPRTT1 containing fprARGT/XerCmwr/CRmwr | ASfprARGT, CRSXerCmwr,crmwr | This work |
| pUC18 | Plasmid vector; lacks a Xer recombination site; used as a control in stability experiments | Accession no. L09136 | |
| pUC57 | Plasmid vector that differs from pUC18 in the multiple cloning site; used as a vector to clone fpr | 24; accession no. Y14837 |
The backbones of pJHCMW1 and pFPTB1 share homology in regions essential for their stable inheritance and include loci with the characteristics of Xer recombination sites (mwr and fpr, respectively) (Fig. 1A). Both mwr and fpr include an ARG box that is substandard due to the presence of a C instead of a T nucleotide in one of the highly conserved positions (Fig. 1C). We have shown before that replacing the C in mwr with a T increased the efficiency of Xer recombination, probably due to better binding of ArgR to the ARG box, facilitating formation of the synaptic complex (Fig. 1B) (13, 23). The AS and the XerC binding site differ by only seven and two nucleotides, respectively (Fig. 1C). The central regions not only have different nucleotide sequences but also differ in length. The mwr central region consists of six nucleotides, but the fpr site central region possesses seven nucleotides, a rare feature (2, 8).
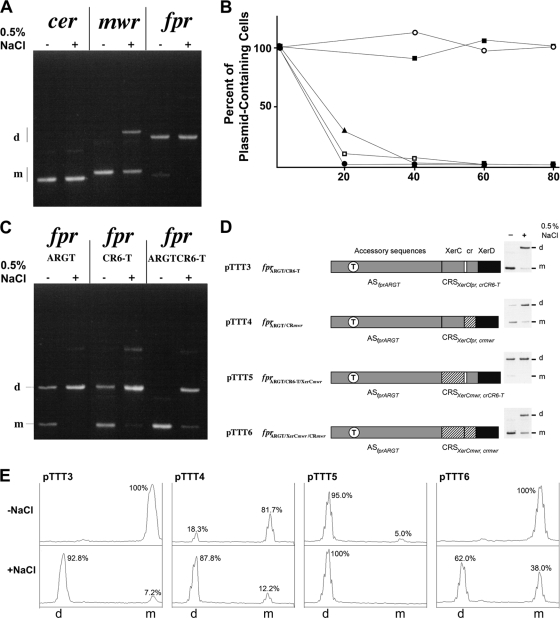
Figure 2A shows the plasmid content of E. coli cells transformed with dimers of plasmid pKS492, pES, or pFPRTT1, which includes cer, mwr, or fpr, respectively, after culturing in low- or high-osmolarity growth medium. While the resolution of pFPRTT1 dimers was marginal, ca. 50% and 100% of the pES dimers were resolved at high and low osmolarities, respectively, and 100% of the pKS492 dimers were resolved under both conditions. Stabilization assays (Fig. 2B) showed that pES and pFPRTT1 were lost at approximately the same rate as plasmid pUC18, which lacks a Xer recombination site, indicating that the levels of recombination at mwr or fpr were not high enough to stabilize the plasmids. Conversely, pKS492 was stably maintained. As expected, the natural pJHCMW1 plasmid was also stably maintained through the action of the Tn1331 resolvase.
FIG. 2.
Dimer resolution and plasmid stability. (A) Dimers of plasmids pKS492 (cer), pES (mwr), and pFPRTT1 (fpr) were introduced by transformation into E. coli DS941. The cells were cultured in low-osmolarity medium (− 0.5% NaCl) or high-osmolarity medium (+ 0.5% NaCl) in the presence of 100 μg of ampicillin per ml for 20 generations. Plasmid DNA was isolated and subjected to agarose gel electrophoresis. The locations of dimers (d) and monomers (m) are indicated at the left. (B) Stability of plasmids harboring different Xer recombination sites. Plasmids were introduced by transformation into hyperrecombinogenic E. coli JC8679, which was cultured under nonselective conditions for the indicated number of generations, and the plasmid content was analyzed. The plasmids tested were pJHCMW1 (empty circles), pKS492 (filled squares), pFPRTT1 (filled triangles), pUC18 (empty squares), and pES (filled circles). (C) Dimers of plasmids pTTT1 (fprARGT), pTTT2 (fprCR6-T), and pTTT3 (fprARGT/CR6-T) were introduced by transformation into E. coli DS941. The cells were cultured in low- or high-osmolarity medium in the presence of 100 μg of ampicillin per ml for 20 generations. Plasmid DNA was isolated and subjected to agarose gel electrophoresis. The locations of dimers and monomers are indicated at the left. (D) Resolution of dimers containing hybrid sites. Hybrid sites contain regions from mwr (cross-hatched) or fpr (gray). The XerD binding sites are identical in both sites (black). The AS are those from fpr but with the T substitution that enhances the ARG box. The gray central region with a white gap represents the fpr modified central region. The results of dimer resolution assays are shown to the right. (E) Quantification of the gels shown in panel C. The bands were quantified using the ImageJ software (http://rsb.info.nih.gov/ij) (1).
To identify nucleotides responsible for the difference in recombination efficiency observed between fpr and mwr, three fpr mutant derivatives were generated. (i) fprARGT (plasmid pTTT1) contains a replacement of the C nucleotide in the fpr ARG box with a T (asterisk in Fig. 1C), resulting in a derivative with AS including an ARG box with the appropriate consensus sequence (ASfprARGT) and the wild-type CRS (CRSXerCfpr,crfpr). (ii) fprCR6-T (plasmid pTTT2) contains a deletion of a T (identified by an upward arrowhead in Fig. 1C) in the CRS (CRSXerCfpr, crCR6-T), generating a six-nucleotide central region from here on referred to as the fpr modified central region. (iii) fprARGT/CR6-T (ASfprARGTCRSXerCfpr, crCR6-T, plasmid pTTT3) is a derivative including mutations i and ii described above. Although derivatives fprARGT and fprCR6-T showed a small increase in efficiency of recombination with respect to fpr, the cumulative effect of both substitutions was necessary to reach levels comparable to those observed for mwr (note that dimers harboring mwr or fprARGT/CR6-T were 100% resolved at low osmolarity [Fig. 2A and C]). These results indicate that a more efficient ARG box leads to an increase in the efficiency of Xer site-specific recombination in fpr, as was known to occur with mwr. However, this increase was smaller than that found for the same modification in mwr, in which case the increase was to the levels observed for cer (100% resolution at high and low osmolarities) (13). Although the change in efficiency after modification of a nucleotide in the ARG box could also be due to the fact that the ARG box in the actual host is different from that in E. coli, we think that this is not the case because the ARG box has been found to be highly conserved between genomes (10). The results also show that the fpr seven-nucleotide central region is detrimental for the recombination reaction because reducing the number of nucleotides to six resulted in an increase in the efficiency of dimer resolution (compare Fig. 2A fpr to Fig. 2C fprCR6-T). However, this enhancing effect on the recombination levels, although measurable, is not as pronounced as in the case of mwr (compare Fig. 2A and C) and is not enough to stabilize the plasmid. These experiments also showed that the recombination efficiency of the fpr derivatives is dependent on the osmolarity of the milieu (Fig. 2C).
A series of hybrid sites combining XerC binding sites and central regions was generated to further characterize fpr and mwr. These hybrid sites consisted of ASfprARGT and combinations of the fpr or mwr XerC binding site (XerCfpr or XerCmwr) with the mwr or fpr modified central region (crmwr or crCR6-T). A diagram of the different derivatives and the levels of dimer resolution are shown in Fig. 2D and E, respectively. Replacement of the modified fpr central region with that of mwr generated fprARGT/CRmwr (ASfprARGTCRSXerCfpr, crmwr, plasmid pTTT4). Resolution levels for pTTT4 dimers were similar to those for pTTT3 dimers at 0.5% NaCl but were significantly lower when the cells were cultured in low-osmolarity medium. This result could be interpreted to mean that the fpr modified central region confers slightly higher recombination efficiency on otherwise identical Xer recombination sites. However, comparison of the efficiencies of resolution of dimers of pTTT5 (fprARGT/CR6-T/XerCmwr, ASfprARGT, CRSXerCmwr, crCR6-T) and pTTT6 (fprARGT/CR6-T/XerCmwr/CRmwr, ASfprARGT, CRSXerCmwr, crmwr) showed that in the presence of the mwr XerC binding region, the site including the mwr central region is substantially more efficient as a recombination target than that including the fpr modified central region (Fig. 2D and E). These results indicate that there does not seem to be a best central region; instead, there are more efficient combinations of XerC binding regions and central region sequences.
Conclusions.
Plasmid pFPTB1 includes fpr, a Xer recombination site that shows a recombination efficiency lower than that required for stabilizing the plasmid by dimer resolution. Instead, stabilization must occur through the activity of the resolvase encoded by a resident replicative transposon. To the best of our knowledge, these characteristics were described only in one natural plasmid, pJHCMW1. The findings described here may indicate that pJHCMW1 is not exceptional but that there may be a group of plasmids with these properties. Furthermore, the dimer resolution levels mediated by fpr derivatives at high and low osmolarities also suggest that osmoregulation of mwr is not a unique phenomenon. The results obtained with the hybrid sites indicate that there is an interdependence of the sequences in the XerC binding region and the central region in Xer site-specific recombination sites; i.e., there is not a best central region, but instead, there are more efficient combinations of the XerC binding regions and central region sequences.
Acknowledgments
This work was supported by Public Health Service grant 2R15AI047115 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (to M.E.T.), and a grant from the Wellcome Trust (to D.J.S.). T.T. was supported by grant MHIRT 2T37MD001368 from the National Center on Minority Health and Health Disparities, National Institutes of Health.
We thank A. Soler Bistué for comments and suggestions.
Footnotes
Published ahead of print on 4 December 2009.
REFERENCES
- 1.Abramoff, M., P. Magelhaes, and S. Ram. 2004. Image processing with Image. J. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Arciszewska, L. K., R. A. Baker, B. Hallet, and D. J. Sherratt. 2000. Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J. Mol. Biol. 299:391-403. [DOI] [PubMed] [Google Scholar]
- 3.Blakely, G., G. May, R. McCulloch, L. Arciszewska, M. Burke, S. Lovett, and D. J. Sherratt. 1993. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell 75:351-361. [DOI] [PubMed] [Google Scholar]
- 4.Bui, D., J. Ramiscal, S. Trigueros, J. S. Newmark, A. Do, D. J. Sherratt, and M. E. Tolmasky. 2006. Differences in resolution of mwr-containing plasmid dimers mediated by the Klebsiella pneumoniae and Escherichia coli XerC recombinases: potential implications in dissemination of antibiotic resistance genes. J. Bacteriol. 188:2812-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colloms, S. D., P. Sykora, G. Szatmari, and D. J. Sherratt. 1990. Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J. Bacteriol. 172:6973-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallet, B., L. Arciszewska, and D. J. Sherratt. 1999. Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: an enzymatic switch in site-specific recombination. Mol. Cell 4:949-959. [DOI] [PubMed] [Google Scholar]
- 7.Hayes, F., S. A. Lubetzki, and D. J. Sherratt. 1997. Salmonella typhimurium specifies a circular chromosome dimer resolution system which is homologous to the Xer site-specific recombination system of Escherichia coli. Gene 198:105-110. [DOI] [PubMed] [Google Scholar]
- 8.Hayes, F., and D. J. Sherratt. 1997. Recombinase binding specificity at the chromosome dimer resolution site dif of Escherichia coli. J. Mol. Biol. 266:525-537. [DOI] [PubMed] [Google Scholar]
- 9.Huang, H., J. Li, X. L. Yang, Y. G. Wang, Y. P. Wang, J. S. Tao, Y. Z. Huang, and X. L. Zhang. 2009. Sequence analysis of the plasmid pGY1 harbored in Salmonella enterica serovar Paratyphi A. Biochem. Genet. 47:191-197. [DOI] [PubMed] [Google Scholar]
- 10.Makarova, K. S., A. A. Mironov, and M. S. Gelfand. 2001. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2:RESEARCH0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minh, P. N., N. Devroede, J. Massant, D. Maes, and D. Charlier. 2009. Insights into the architecture and stoichiometry of Escherichia coli PepA*DNA complexes involved in transcriptional control and site-specific DNA recombination by atomic force microscopy. Nucleic Acids Res. 37:1463-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasquali, F., C. Kehrenberg, G. Manfreda, and S. Schwarz. 2005. Physical linkage of Tn3 and part of Tn1721 in a tetracycline and ampicillin resistance plasmid from Salmonella Typhimurium. J. Antimicrob. Chemother. 55:562-565. [DOI] [PubMed] [Google Scholar]
- 13.Pham, H., K. J. Dery, D. J. Sherratt, and M. E. Tolmasky. 2002. Osmoregulation of dimer resolution at the plasmid pJHCMW1 mwr locus by Escherichia coli XerCD recombination. J. Bacteriol. 184:1607-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reijns, M., Y. Lu, S. Leach, and S. D. Colloms. 2005. Mutagenesis of PepA suggests a new model for the Xer/cer synaptic complex. Mol. Microbiol. 57:927-941. [DOI] [PubMed] [Google Scholar]
- 15.Sarno, R., G. McGillivary, D. J. Sherratt, L. A. Actis, and M. E. Tolmasky. 2002. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 46:3422-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stirling, C. J., G. Szatmari, G. Stewart, M. C. Smith, and D. J. Sherratt. 1988. The arginine repressor is essential for plasmid-stabilizing site-specific recombination at the ColE1 cer locus. EMBO J. 7:4389-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Summers, D., and D. J. Sherratt. 1984. Multimerization of high copy number plasmids causes instability: ColE1 encodes a determinant essential for plasmid monomerization and stability. Cell 36:1097-1103. [DOI] [PubMed] [Google Scholar]
- 18.Summers, D. K., C. W. Beton, and H. L. Withers. 1993. Multicopy plasmid instability: the dimer catastrophe hypothesis. Mol. Microbiol. 8:1031-1038. [DOI] [PubMed] [Google Scholar]
- 19.Summers, D. K., and D. J. Sherratt. 1988. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 7:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tolmasky, M. E. 2007. Aminoglycoside-modifying enzymes: characteristics, localization, and dissemination, p. 35-52. In R. Bonomo and M. E. Tolmasky (ed.), Enzyme-mediated resistance to antibiotics: mechanisms, dissemination, and prospects for inhibition. ASM Press, Washington, DC.
- 21.Tolmasky, M. E., S. Colloms, G. Blakely, and D. J. Sherratt. 2000. Stability by multimer resolution of pJHCMW1 is due to the Tn1331 resolvase and not to the Escherichia coli Xer system. Microbiology 146:581-589. [DOI] [PubMed] [Google Scholar]
- 22.Tolmasky, M. E., and J. H. Crosa. 1987. Tn1331, a novel multiresistance transposon encoding resistance to amikacin and ampicillin in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 31:1955-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trigueros, S., T. Tran, N. Sorto, J. Newmark, S. D. Colloms, D. J. Sherratt, and M. E. Tolmasky. 2009. mwr Xer site-specific recombination is hypersensitive to DNA supercoiling. Nucleic Acids Res. 37:3580-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 25.Wu, S. W., K. Dornbusch, G. Kronvall, and M. Norgren. 1999. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type beta-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC beta-lactamase. Antimicrob. Agents Chemother. 43:1350-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]