Abstract
Conjugative transfer from Clostridium symbiosum to enterococci of Tn1549, which confers VanB-type vancomycin resistance, has been reported. This indicates the presence of a transfer origin (oriT) in the element. Transcription analysis of Tn1549 indicated that orf29, orf28, orfz, and orf27 were cotranscribed. A pACYC184 derivative containing 250 bp intergenic to orf29-orf30 of Tn1549 was mobilized in Escherichia coli recA::RP4::Δnic provided that orf28 and orf29 were delivered simultaneously. These open reading frame (ORF) genes were able to promote mobilization in trans, but a cis-acting preference was observed. On the basis of a mobilization assay, a minimal 28-bp oriT was delimited, although the frequency of transfer was significantly reduced compared to that of a 130-bp oriT fragment. The minimal oriT contained an inverted repeat and a core, which was homologous to the cleavage sequence found in certain Gram-positive rolling-circle replicating (RCR) plasmids. While Orf29 was a mobilization accessory component similar to MobC proteins, Orf28 was identified as a relaxase belonging to a new phyletic cluster of the MOBp superfamily. The nick site was identified within oriT by an oligonucleotide cleavage assay. Closely related oriTs linked to mobilization genes were detected in data banks; they were found in various integrative and conjugative elements (ICEs) originating mainly from anaerobes. These results support the notion that Tn1549 is a member of a MOBp clade. Interestingly, the Tn1549-derived constructs were mobilized by RP4 in E. coli, suggesting that a relaxosome resulting from DNA cleavage by Orf28 interacted with the coupling protein TraG. This demonstrates the capacity of Tn1549 to be mobilized by a heterologous transfer system.
Tn1549 is a 34-kb transposon which confers vancomycin resistance in Enterococcus spp. (22). This entirely sequenced element, which harbors the vanB operon, is transferred passively among enterococci by conjugative plasmids or by the exchange of large chromosomal fragments (9, 17, 22). Tn1549 has also been reported in Gram-positive anaerobes (18, 46), and transfer from Clostridium symbiosum to Enterococcus has been demonstrated (28). This element is likely to be a conjugative transposon consisting of three functional modules involved in transposition, transfer, and antibiotic resistance. The transposition module encodes an excisionase and an integrase which cooperatively direct excision of the element. Excision is followed by circularization, resulting in a heteroduplex at the joined ends according to the tyrosine recombinase model of the Tn916 prototype. Analysis of Enterococcus transconjugants indicated that acquisition of VanB-type resistance was due to transposition of Tn1549 (28). However, it remains to be established if Tn1549 is a conjugative or a mobilizable transposon, since a single retransfer event has been obtained but in the presence of Tn916 (28). In both instances, whether active or passive transfer is being considered, presence of a transfer origin (oriT) is required. Identification of oriT is thus a prerequisite to the elucidation of the transfer process.
oriT is a short specific sequence necessary for conjugative transfer of any mobile genetic element to a recipient cell provided that a conjugation machinery is present. This machinery involves (i) mating-pair formation (Mpf) between the donor and the recipient strains by an exocellular pilus system in Gram-negative bacteria, whereas, in the majority of Gram-positive bacteria, the means to achieve intimate donor-recipient cell contact remains to be identified, and (ii) formation of a channel structure for secretion (14, 30, 35, 36, 41). DNA relaxases are key enzymes in the initiation of the conjugative transfer by catalyzing single-strand cleavage of a phosphodiester bond at the nick site (nic) in a specific and reversible manner (2, 3, 5, 34). Many relaxases need an accessory protein, such as MobC, which stimulates strand opening and strand separation, to enhance their activity (52). Conjugative plasmids possess the genes required to promote their self-transfer. Mobilizable elements possess an oriT and a cognate relaxase to initiate the relaxosome but need a helper conjugative system which supplies Mpf consisting of the DNA transport machinery for intercellular transfer. Direct interaction between the relaxosome and the transfer machinery occurs by TraG coupling proteins. This family of essential proteins includes TraG of RP4, TrwB of R388, TraD of F, and TraG and VirD4 of Ti in Gram-negative bacteria and TrsK of pGO1 in Gram-positive bacteria (14, 23, 25, 32). Determination of the nucleotide sequence of tra regions of conjugative plasmids and of entire genomes from Gram-positive bacteria has revealed homology to genes encoding TraG/TrwB/VirD4 and to the conjugative transfer ATPase VirB4 involved in the transfer DNA translocation process. Homology has also been detected with integrative and conjugative elements (ICEs) that include conjugative transposons (33). Conjugative transfer of ICEs occurs by a mechanism similar to that of conjugative plasmids (25). In contrast to plasmid oriTs, little is known about the initiation of conjugation in ICEs. Recently, the oriTs from three ICEs, Tn916, ICEBs1 of Bacillus subtilis, and an element of the SXT/R391 family, have been characterized (12, 29, 39). Similarly to conjugative plasmids, ICEs encode their own relaxase to cleave the nic site. Examination of Tn1549 has shown that Orf28 displays similarity with the MobA relaxase of pC221 (31% identity) and that Orf29 is similar to MobC of pC221 (21% identity). In this study we show that the product of orf28 acts as a relaxase and we have identified its cognate oriT.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The origins and properties of bacterial strains and plasmids are listed in Table 1. E. coli recA::RP4::Δnic was used as a donor in the mating experiments and was grown on Luria-Bertani (LB) agar supplemented with 0.02% arabinose and 1 mM diaminopimelic acid (DAP). E. coli BL21λ DE3 was used as a host for protein expression and purification.
TABLE 1.
Bacterial strains and plasmidsb
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| E. coli | ||
| SM10 | F−tonA21 lacY1 supE44 Mu λ− [RP4-2 (Tc::Mu)]Km Tra+ | 43 |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcr BC) φ80lacZΔM15 ΔlacX74 deoR rec araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| recA::RP4::Δnic | chrRP4::Δnic Km DAP− | 1 |
| C1a | rep+ilv+ Nal | 40 |
| BL21λ DE3 | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Invitrogen |
| E. faecalis | ||
| JH2-2::Tn1549 | Fus Rif Vm | 28 |
| Plasmids | ||
| pACYCGm | Derivative of pACYC184 in which the cat-tet fragment was replaced by aac(3)-IV; Gm Apra | Laboratory |
| pCM105 | pACYCGm Ω250-bp EcoRI-HindIII PCR intergenic fragment orf29-orf30 from Tn1549 | This study |
| pCM105-1 | pCM105 (C141→T)a | This study |
| pCM105-2 | pCM105 (G152G154→ T152C154) | This study |
| pCM105-3 | pCM105 (G159C160→ T159G160) | This study |
| pCM105-4 | pCM105 (G194C195→ T194G195) | This study |
| pCM105-5 | pCM105 (C216C218→ G216G218) | This study |
| pCM106 | pACYCGm Ω28-bp oriTTn1549 | This study |
| pCM107 | pACYCGm Ω130-bp EcoRI-HindIII PCR part of the PCR intergenic fragment orf29-orf30 from Tn1549 | This study |
| pBAD/HisA | Expression vector, araBAD promoter; Amp | Invitrogen |
| pCM108 | pBAD/HisA Ω1,328-bp NcoI-XhoI PCR fragment (rlxTn1549) | This study |
| pUC18Gm | Derivative of pUC18 in which blaT is replaced by aac(3)-IV; Gm Apra | This study |
| pCM109 | pUC18Gm Ω1,879-bp EcoRI-XbaI PCR fragment (intergenic orf30-orf29 region, orf29, and orf28) of Tn1549 | This study |
| pCM110 | pUC18Gm Ω2,664-bp EcoRI-XbaI PCR fragment (intergenic region orf30-orf29, orf29, orf28, orfz, and orf27) of Tn1549 | This study |
| pGB2 | Spc Str | 15 |
| pCM111 | pGB2 Ω372-bp EcoRI-HindIII PCR fragment (mobC) of Tn1549 | This study |
| pET28a(+) | His tag expression vector; Km | Invitrogen |
| pCM112 | pET28a+ Ω354-bp EcoRI-NdeI PCR fragment (mobC) of Tn1549 | This study |
| pCM113 | pET28a+ Ω1,325-bp NcoI-XhoI PCR fragment (rlx) of Tn1549 | This study |
| pCM114 | pET28a+ Ω741-bp NcoI-XhoI PCR fragment (N247rlx) of Tn1549 | This study |
| pCR2.1 | PCR cloning vector; Amp Km | Invitrogen |
| pCM115 | pCR2.1 Ω2,374 bp PCR fragment (intergenic region orf30-orf29 mobC rlx orfz) of Tn1549 | This study |
Sequence was from GenBank accession number AF192329.1, but numbering refers to the orf30 termination codon.
Amp, ampicillin; Apra, apramycin; Fus, fusidic acid; Gm, gentamicin; Km, kanamycin; Nal, nalidixic acid; Rif, rifampin; Str, streptomycin; Vm, vancomycin.
Plasmid methodology, enzymes, and oligonucleotides.
Plasmid DNA purification (Qiagen), digestion with restriction endonucleases (New England Biolabs, Invitrogen), amplification of DNA by PCR with Pfu (Stratagene) or Taq (Invitrogen) DNA polymerases, ligation of DNA fragments with T4 DNA ligase (Invitrogen), and cloning with a TOPO-TA cloning kit (Invitrogen) were performed according to the manufacturers' instructions. Enterococcus faecalis JH2-2::Tn1549 total DNA was prepared with an Illustra kit (GE Healthcare). Recombinant plasmid DNA was introduced into E. coli Top10 or E. coli recA::RP4::Δnic by electrotransformation. The oligonucleotides used (purchased from Invitrogen) are listed in Table 2.
TABLE 2.
Oligonucleotides
| Oligonucleotidea | Sequence (5′-3′) | Position (start, end)b |
|---|---|---|
| RlxNco (−) | AGC CAT GGCAGTTACAAAGATACATCCc | −21857, −21835 |
| RlxXho (+) | TACTCGAGTAGCTCTTGCGTCCATTCCTTTCCC | 20532, 20556 |
| RlxNXho (+) | TACTCGAGCGCTCTGGTGATAACGCGGCC | 21116, 21137 |
| RlxpBADXho (+) | TACTCGAGTTCATAGCTCTTGCGTCCATTCCTTTCCC | 20529, 20556 |
| C1 (+) | AAGGCAGGGACGGGGCAGGCCGTTc | 22212, 22233 |
| C2 (−) | GCCGCGTCCGTTTTGCTTGTTGG | −22209, −22187 |
| FmutNic (+) | TAAGCATAGGGAGTGTAGCC | 22263, 22245 |
| RmutNic (−) | CTACGTTACTGTTTCTCCAC | −22265, −22284 |
| MutAF (+) | CACTCC CGGAAAAACGGCCTG | 22245, 22225 |
| MutA1R (−) | GCTACAGTACCTATGCTTGCTACGTTACTG | −22246, −22275 |
| MutA2R (−) | CATACACTCCCTATGCTTGCTACGTTACTG | −22248, −22275 |
| MutBF (+) | ATCGCGAAACCCCTGAATGGAAAGG | 22158, 22176 |
| MutBR (−) | TTCCCCAACAAGCAAAACGGAC | −22204, −22183 |
| MobCNdeI (−) | GACATATGGCAAACAGAAAGC G | −22147, −22131 |
| MobCFHindIII (−) | ATAAGCTTGGAAAGGACGGTGAAGCA | −22165, −22147 |
| MobCREcoRI (+) | ATGAATTCTCGATGTAGTCAA GGG C | 21793, 21812 |
| 30R (−) | ATAAGCTTACTTTTAGCGGGAGCTGC | −22389, −22370 |
| 29F (+) | ATGAATTCACCGTCCTTTCCATTCAG | 22150, 22170 |
| O30DF (−) | CTAAGCTTGAAACAGTAACGTAGCAAGC | −22279, −22260 |
| 27FEcoRI (+) | ATGAATTCATGC CGGAAAAGCCTTGAA | 20129, 20148 |
| 28RHindIII (−) | ATAAGCTTATGAGTATTCCACGAATGA | −20533, −20514 |
| 184OriF/EcoRI (+) | CTGAATTCAGCAAGCATAGGGAGTGTAGCCACTCCCTGCAGAAGCTTTGACACAGTGCCG | 22266, 22239 |
| 184R/EcoRI (−) | TAGAATTCCGGATGAGCATTCATCAGG | |
| HTH-EcoRI (−) | TGAATTCTGGCATCCTCTGTTTTACT | −22404, −22385 |
| RlxXbaI (+) | AATCTAGATACTCATAGCTCTTGCGTCC | 20525, 20545 |
| YXbaI (+) | ATCTAGACTATCTCTGTTCCAGCGGGAAG | 19742, 19763 |
| Ori34F (+)d | TAACGTAGCAAGCATAGGGAGTGTAGCCACTCCC | 22272, 22239 |
| Ori34R (−)d | GGGAGTGGCTACACTCCCTATGCTTGCTACGTTA | −22239, −22272 |
| 28RpACY (−) | ACTCCACCGCTGATGACATCAG | |
| 28FpACY (+) | CTTTATTCACATTCTTGCCCG | |
| 27f (+)e | GCTCTAAGTGCGTAGGTTTC | 20166, 20185 |
| 28f2 (+)e | AAACTTTCCCGGCTTGATCT | 21231, 21250 |
| 28f1 (+)e | CCGCAGGAAATCATCAAAAT | 21273, 21292 |
| 28r1 (−)e | GTGCAAGGAAAACGGATTGT | −21435, −21416 |
| 29f2 (+)e | CAGGGACGCGCAGCACGATT | 22108, 22127 |
| 29r2 (−)e | GAGATACAGAAGATCGGCGT | −21967, −21948 |
| 30f (+)e | CGATGATCGGCAGGTATTCT | 22470, 22489 |
| 30r (−)e | ATTGACTGGCGCTATCTTGC | −22668, −22649 |
| Race 1(+)e | GATGATCGGCAGGTATTCTT | 22471, 22490 |
| Race 2(+)e | ATAAGCTGTATGACAACCGG | 22595, 22614 |
+, sense primer; −, antisense primer.
Nucleotide numbering according to GenBank accession number AF192329.1.
Restriction sites used in cloning are underlined. 5′-end phosphorylated nucleotides are in bold.
Oligonucleotides used in nicking assay.
Oligonucleotides used in RT-PCR and 5′ RACE assays.
Reverse transcription.
JH2-2::Tn1549 total RNA was extracted using the RiboPure bacteria kit (Ambion) according to the manufacturer's instructions. To rule out contamination by genomic DNA, total RNA was treated with RNase-free DNase (DNase I) (Invitrogen). The quality and quantity of RNA were measured using the microchip analysis system of an Agilent 2100 bioanalyzer apparatus (Agilent Technologies). Reverse transcription was performed using 2.5 μg total heat-denatured DNA-free RNA, 0.8 μl random primer (3 μg/μl), 1 μl 10 mM deoxynucleoside triphosphates, 4 μl 5× first-strand buffer, 1 μl 0.1 M dithiothrietol, 1 μl RNaseOUT (40 U/μl), 1 μl of SuperScript III reverse transcriptase (200 U/μl) (Invitrogen), and H2O to a total volume of 20 μl. An aliquot of the cDNA (1 μl of the reverse transcription [RT] reaction mixture) was amplified by PCR with 20 pmol each of primer pairs 30r-29f2, 29r2-28f2, and 28r1-27f in order to determine a putative cotranscription of orf30, orf29, orf28, orfz, and orf27. Negative controls were performed with 1 μl DNA-free RNA.
Identification of the transcriptional start site for orf30 and for the relaxase operon.
Reverse transcription and rapid amplification of cDNA ends (RACE) were performed using the 5′RACE system kit (Invitrogen) on RNA extracted from E. faecalis JH2-2::Tn1549. Briefly, for the determination of the transcriptional start site of orf30, after a reverse transcription step using an orf30-specific primer, Race1, dC-tailed DNA was amplified by PCR with another orf30-specific primer, Race2, and a dG-anchor oligonucleotide primer. PCR products were then cloned into pCR2.1 (Invitrogen) and further sequenced. The same protocol was applied to identify the transcriptional start site of the orf29-orf28-orfz-orf27 operon by the use of (i) orf28-specific primer 28f2 for the reverse transcription step, (ii) orf28-specific primer 28f1 plus dG-anchor oligonucleotide primer in a first PCR, and (iii) orf29-specific primer 29f2 and dG-anchor oligonucleotide primer in the nested PCR step. The PCR products were cloned in pCR2.1 and sequenced.
Plasmid constructions.
To localize oriT, a 250-bp EcoRI-HindIII PCR fragment containing the intergenic orf29-orf30 region was cloned in pACYCGm, resulting in pCM105. pCM106 was obtained by PCR amplification with primers 184OriF/EcoRI and 184R/EcoRI and pCM105 DNA as a template. An EcoRI-HindIII PCR fragment obtained with primers 29F and O30DF from E. faecalis JH2-2::Tn1549 was cloned in pACYCGm, resulting in plasmid pCM107.
The mobC-like and relaxase genes amplified with primers MobcFHindIII plus MobcREcoRI and RlxNco plus RlxpBADXho were cloned in pGB2 and pBAD/His vectors to generate pCM111 and pCM108, respectively.
A 1,879-pb EcoRI-XbaI-PCR fragment containing the intergenic region orf29-orf30, the mobCTn1549 gene, and the rlx gene was obtained with primers HTH-EcoRI and RlxXbaI and E. faecalis JH2-2::Tn1549 DNA and cloned in pUC18Gm to generate plasmid PCM109. A 2,662-bp EcoRI-XbaI PCR fragment, containing the intergenic region orf29-orf30, the mobCTn1549 gene, the rlx gene, orfz, and orf27 was obtained with primers HTH-EcoRI and YXbaI and E. faecalis JH2-2::Tn1549 DNA as a template and was cloned in pUC18Gm to generate plasmid pCM110. To purify MobCTn1549, the relaxase (442 amino acids) and its active N-terminal domain (247 amino acids) proteins were expressed as 6×His-tagged fusions. The EcoRI-NdeI PCR fragment obtained with primers MobcREcoRI and MobCNdeI, the Xho-NcoI PCR fragment obtained with primers RlxNco and RlxXho, and the Xho-NcoI PCR fragment obtained with primers RlxNco and RlxNXho were cloned into pET28a(+). The sequences of the recombinant plasmids were verified to rule out error during DNA polymerization.
Construction of orf29-orf30 intergenic region (ID) mutants.
Site-directed mutagenesis with pCM105 DNA as a template was performed within the intergenic orf29-orf30 portion to identify nucleotides essential in the mobilization process. Plasmid mutants called pCM105-1 to -5 were obtained with the primer pairs MutAF/MutA1R, MutAF/MutA2R, MutBF/MutBR, C1/C2, and FmutNic/RmutNic, respectively. Each PCR fragment was ligated by T4 ligase and transformed into E. coli Top10 (Invitrogen). The presence of the expected mutations was confirmed by DNA sequencing. The various plasmid mutants were then introduced into E. coli recA::RP4::Δnic(pCM108, pCM111) by electrotransformation.
Conjugation experiments.
E. coli recA::RP4::Δnic was successively transformed with plasmids pCM108 and pCM111. The resulting strain was then transformed independently with the following plasmids: pCM105 and its mutant derivatives, pCM106, and pCM107. Each transformant was used as a donor in conjugation experiments with E. coli C1a as a recipient. Mating experiments were carried out on LB agar plates supplemented with 0.02% arabinose and 1 mM DAP incubated at 37°C for 24 h. Transconjugants were selected on LB agar plates containing gentamicin (8 μg/ml) and nalidixic acid (40 μg/ml). Quantification of donors was obtained by selection on LB agar plates with 1 mM DAP supplemented with ampicillin (100 μg/ml), streptomycin (20 μg/ml), and gentamicin (8 μg/ml). To evaluate the efficiency of cis-acting mobilization proteins, pCM109 and pCM110 were introduced separately into E. coli recA::RP4::Δnic. Both transformants were used as donors in conjugation experiments to E. coli C1a. Mating experiments were carried out on LB agar plates supplemented with DAP (1 mM). Transconjugants were selected on gentamicin (8 μg/ml) and nalidixic acid (40 μg/ml), and donors were selected on gentamicin (8 μg/ml) and 1 mM DAP. Mating frequencies were calculated by dividing the number of transconjugants by the number of donor cells and expressed as the means of results of three independent experiments.
Purification of relaxase, N-terminal relaxase, and MobC proteins.
Plasmids pCM112, pCM113, and pCM114 were introduced into E. coli BL21αDE3. The transformants were grown in LB broth containing kanamycin (50 μg/ml) at 37°C. Protein expression was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM), and each recombinant protein was then purified by immobilized metal affinity chromatography (TALON) resin (Clontech Laboratories, Inc) and was dialyzed after elution, as recommended by the manufacturer.
Electrophoretic mobility shift assays (EMSAs).
A 250-bp fragment encompassing the putative oriT sequence was amplified by PCR with primers 29F and 30R from JH2-2::Tn1549 total DNA used as a template. A 96-bp PCR fragment comprising the 28-bp oriT sequence (minimal oriT) was obtained with primers 28RpACY and 28FpACY and pCM106 DNA as a template; the remaining 68 bp corresponded to the plasmid flanking sequences. Purified fragments were end labeled with [γ-32P]ATP (3,000 Ci mmol−1) and T4 polynucleotide kinase (New England Biolabs). Binding of relaxase or mobilization protein to DNA was carried out in a 20-μl buffer (20 mM Tris-HCl [pH 7.4], 5 mM MgCl2, 50 mM KCl, 25% glycerol, and 0.1 mg/ml sonicated salmon sperm DNA) and 5 nM labeled DNA. The reaction mixtures were incubated at 25°C for 30 min. Samples were loaded onto an 8% polyacrylamide gel (29:1 acrylamide-bisacrylamide, 0.5× TBE) and electrophoresed for 2 h at 120 V. The gels were then dried and visualized by autoradiography.
Oligonucleotide cleavage.
Ori34F and Ori34R oligonucleotides were labeled at the 5′ end using [γ-32P]ATP (3,000 Ci mmol−1) and polynucleotide kinase (New England Biolabs). Unbound [γ-32P]ATP was eliminated from the mixture by purification with the mini Quick Spin DNA column (Roche). Nick site cleavage was carried out as previously described (38).
RESULTS
Organization of the mobilization region of Tn1549.
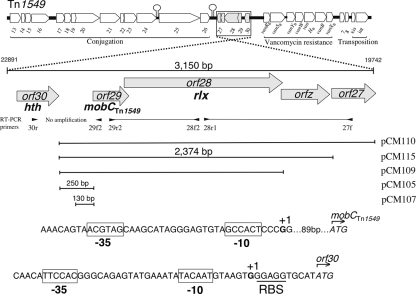
The sequence of Tn1549 is available (GenBank accession number AF192329), and a putative relaxase gene (rlx) was attributed to orf28, which is part of a four-gene cluster transcribed in the same orientation and located upstream from the vanB operon (Fig. 1). orf28 overlaps orf29 and orfz by 39 and 19 bp, respectively. The orf30 product displays homology with helix-turn-helix XRE-family-like proteins, which include regulatory proteins. Orf29 is a member of the MobC superfamily of bacterial mobilization proteins and shares 20% and 30% identity with MobC of pC221 and MbeC of ColE1, respectively (45). Orf28 belongs to the ColE1 superfamily of relaxases and displays 31% identity with MobA of pC221, which belongs to the MOBP7 clade (11). The motif [LF]xxx[GS]xNxNQxAxxxN, described as conserved in mobilization-associated proteins from Gram-positive and Gram-negative bacteria (50), is present in MobCTn1549 except for the [L/F] residue replaced by an isoleucin residue, which is likely an isofunctional amino acid. In contrast, the orfz and orf27 products do not match to any recognized protein family.
FIG. 1.
The schematic representation of Tn1549 and of its functional modules. Coordinates of the 3,150-bp fragment including the mobilization module are indicated according to GenBank accession no. AF192329.1. Open arrows represent ORFs and indicate the sense of transcription. Large stem-loops are indicated. The relaxase gene cluster is enlarged and inverted to facilitate analysis. The primers used for amplification of transgene fragments from the cDNA are indicated by arrowheads. The resulting PCR products are shown by a thin dark line, which corresponds to a cotranscription. The inserts of plasmids indicated on the right are represented by horizontal bars. Initiation codons of orf29 and orf30 are italicized. The beginning of transcription of orf30 and of the mobilization operon detected by 5′ RACE are indicated in bold, and the −35 and −10 motifs are boxed.
Transcriptional analysis of the relaxase-associated genes.
The cDNA corresponding to total RNA from E. faecalis JH2-2::Tn1549 was used for PCR to determine if the orfs of the mobilization region were cotranscribed (Fig. 1). Amplification of transgene fragments was obtained from cDNA with primer pairs 29r2-28f2 and 28r1-27f, leading to ca. 730-bp and 1,270-bp fragments, respectively. In addition, no amplification was detected with primers 29f2 and 30r located, respectively, in orf29 and orf30. These results summarized in Fig. 1 indicated that orf29, orf28, orfz, and orf27 constitute an operon and that there is no cotranscription of orf30.
Mapping of the transcriptional start site.
Amplification and sequencing of the 5′ RACE products indicated that the end of the mRNA transcribed from orf30 was found 12 bp upstream from the translational initiation site, suggesting −35 (TTCCAC) and −10 (TACAAT) as promoter sequences separated by 17 bp (Fig. 1). The mRNA for orf29 was found to begin 91 bp upstream from the start codon, consistent with a promoter composed of −35 (ACGTAG) and −10 (GCCACT) motifs separated by a 17-bp sequence. The −10 (GCCACT) motif was also proposed by computer analysis, since the underlined bases are in agreement with binding sequences of the σ70 factor of the E. coli RNA polymerase; in contrast, the −35 motif was uncommon. The −10 promoter sequence was within the hairpin A, which in addition to being part of the oriT could play a role in the regulation of the mobilization operon. However, an artifact due to RT stopping at the secondary structure should not be completely excluded.
Mobilization experiments with E. coli.
In a preliminary assay, a 2,374-bp PCR fragment of Tn1549 beginning from the intergenic region orf29-30 and ending at the end of orfz was cloned in pCR2.1. The recombinant plasmid, pCM115, was mobilizable in E. coli SM10 by the transfer apparatus of RP4 (data not shown). This result demonstrated that the cloned fragment contained an oriT, and the fact that no homology was detected with the oriT region of RP4 suggests that this fragment also encoded mobilization functions. In order to localize the oriT and to establish if the MobCTn1549 accessory protein was necessary for transfer, a mobilization assay was carried out with E. coli recA::RP4::Δnic(pCM111, pCM108). It consisted of the mobilization of pACYCGm derivatives in which several portions of the 2,374-bp PCR fragment of Tn1549 were cloned, while the relaxase was delivered in trans. The experiments were also carried out in the absence or in the presence of orf29 (mobCTn1549) under the control of a constitutive promoter provided by pGB2. Mobilization was obtained with a pACYCGm derivative carrying a 250-bp orf30-orf29 intergenic fragment, provided that both rlxTn1549 and mobCTn1549 genes were present (Fig. 2B). Induction by arabinose increased transfer frequency up to 100-fold (data not shown). In contrast, no transfer was observed in the absence of MobCTn1549.
FIG. 2.
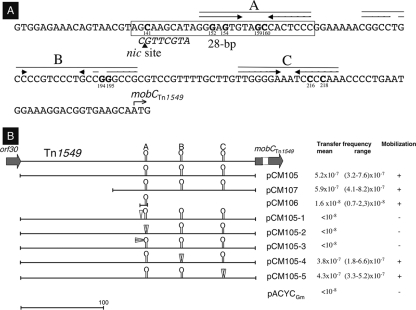
(A) Portion of the orf29-orf30 intergenic region upstream from the mobC gene. The inverted repeats of hairpins A, B, and C are indicated by arrows. The 28-bp oriT is boxed, and the nic core site on the antisense strand is italicized. The nic site is indicated by an arrowhead. The initiation codon of the mobC gene is shown by a bent arrow. Nucleotides which have been substituted in pCM105 derivatives are in bold, and their positions are indicated. Numbering is from GenBank accession no. AF192329.1, but for the sake of simplicity in reading refers to the orf30 termination codon. (B) Mobilization of pACYCGm derivatives in E. coli recA:RP4::Δnic. Expression of the rlx gene was induced by 0.02% arabinose. The intergenic 250-bp orf29-orf30 fragment is shown with hairpins A, B, and C. Mutations are shown by arrowheads; when they were predicted to decrease the stability of the hairpins, the latter were deleted. Frequencies of transfer were the means of results of three independent mating experiments.
Directed mutagenesis in the oriT region.
Detailed analysis of the 250-bp oriT fragment showed the presence of three hairpin structures upstream from the mobCTn1549 gene which were designated A, B, and C. Computer-generated models of the secondary structure formed by the inverted repeats A, B, and C indicated the free enthalpy of the thermodynamically most stable forms to be −6.25, −4.98, and −5.42 kcal mol−1, respectively. Interestingly, inverted repeat A was preceded on the complementary strand by a 8-base-pair stretch with a RYGCTTG′C cleavage motif conserved in rolling-circle replicating (RCR) plasmids such as pNZ4000 of Lactococcus lactis (50) and pC221 and pC223 of Staphylococcus aureus (10, 11). The impact of the hairpin structures on the activity of oriT was evaluated by directed mutagenesis of pCM105 to generate in every hairpin two base substitutions (G152G154→T152C154, G194C195→T194G195, andC216C218→G216G218) that disrupt the secondary structure (Fig. 2A). In addition, the substitution G159C160→T159G160 was achieved to alter the head of hairpin A. The consequence on transfer was tested by mobilization in E. coli as described above. Disruption of inverted repeat A or alteration at the head of the stem-loop abolished transfer. In contrast, disruption of the B and C hairpins did not significantly affect mobilization (Fig. 2B). When the predicted nic cleavage site was altered by a substitution (C141→T), transfer was abolished.
Identification of a minimal oriT.
A 130-bp fragment just upstream from mobCTn1549 was cloned in the pACYCGm vector, and the resulting plasmid, pCM107, was found to be as efficient as pCM105 in the mobilization assay (Fig. 2B). Taken together, these results suggest that hairpin A and the adjacent conserved cleavage site were part of the oriT. The corresponding 28-bp sequence was introduced following PCR in pACYCGm to delimit the minimal oriT. This fragment was sufficient for mobilization of pACYCGm in E. coli recA::RP4::Δnic[pCM108(rlx), pCM111(mobCTn1549)], although at a diminished frequency (ca. 30-fold) compared to the 250-bp orf29-30 intergenic fragment (Fig. 2B); pACYCGm used as a control could not be transferred.
Mobilization of pUC18Gm by the cis-acting mobC and rlx genes.
The ability for certain proteins to act in trans can be strongly reduced. Although conjugation proteins are not known to be cis acting, TraA cis-acting preference has been recently demonstrated for the relaxase of Rhizobium etli (38). In order to examine the possibility that rlx exhibits a higher efficiency in cis, a 1,879-bp fragment which includes the entire orf30-29 intergenic region, mobCTn1549, and rlx was cloned in pUC18Gm. The recombinant plasmid, pCM109, introduced by electrotransformation in E. coli recA::RP4::Δnic, transferred to E. coli C1a at a frequency of 7.2 × 10−4. Such an efficiency was never obtained when the mobCTn1549 and rlx genes were provided in trans despite the induction of rlx by arabinose. The frequency of transfer was found to be almost identical when the experiments were performed with pCM110, which contains a 2,664-bp fragment delimited by the terminal codons of orf29 and orf27. These data indicate that the presence of orfz and orf27 did not influence mobilization.
Electrophoresis mobility shift assay.
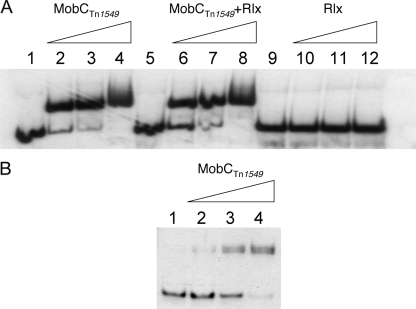
The mobC and rlx genes of Tn1549 were amplified by PCR and cloned in pET28a(+) expression vectors in order to express 6×His-tagged fusion proteins. In addition, the N-terminal domain (247 amino acids) of the relaxase (Rlx) known to support the transesterase activity was also expressed as a 6×His-tagged fusion (27). The proteins were purified by cobalt ion chelating chromatography, and RlxH6 and RlxN247H6 soluble forms were used to test activity. The relaxosome accessory protein MobCTn1549 (Mw, 12,486) is basic (calculated pI = 10.30) like most DNA-binding proteins and was found to bind to the 250-bp orf29-orf30 intergenic fragment and to the 28-bp minimal oriT (Fig. 3). In contrast, RlxH6 alone was unable to bind to these fragments (Fig. 3). It has been proposed that accessory proteins, such as MobC of R1162, denature the oriT strands, providing a single-stranded substrate for the relaxase (52) whereas certain relaxases are unable to nick covalently closed circular (CCC) DNA (5).
FIG. 3.
Electrophoresis mobility shift assay. (A) The labeled 250-bp orf29-orf30 intergenic fragment was incubated with increasing micromolar amounts of purified proteins MobCTn1549, Rlx, and a mix of MobC Tn1549 and Rlx. (B) The 28-bp minimal oriT fragment was cloned in pACYCGm, and the PCR product was amplified as part of a 96-bp fragment including flanking sequences of the cloning site. The PCR product was labeled and incubated with increasing micromolar amounts of purified MobCTn1549 protein.
Site-specific cleavage of oligodeoxyribonucleotides by Rlx.
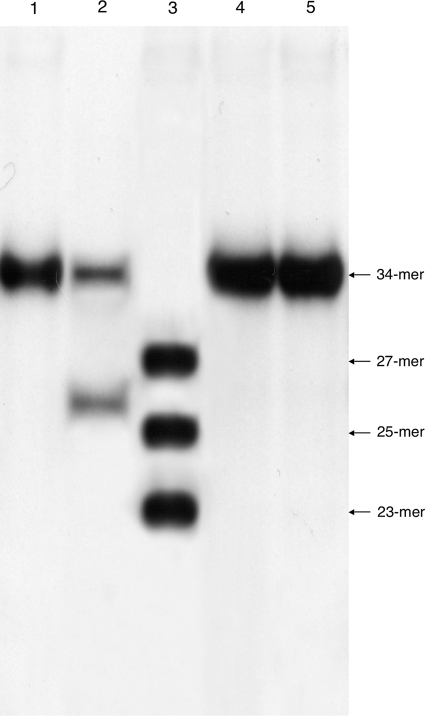
To identify precisely the nic site, cleavage assays were performed with single-stranded oligodeoxyribonucleotides spanning the transfer origin's nic region incubated with RlxN247. The reaction mixtures were separated by electrophoresis in a polyacrylamide gel (Fig. 4). RlxN247 cleaved the Ori34R-34-mer oligonucleotide, yielding the 26-mer 5′-32P-GGGAGTGGCTACACTCCCTATGCTTG plus CTACGTTA-3′, but not the complementary oligonucleotide Ori34F-34-mer, indicating that single-strand DNA nicking occurred on the antisense strand upstream from mobCTn1549 and rlx genes and that, as expected, nic was located at the end of the core site. To study the role of MobCTn1549, the cleavage assay of the Ori34R oligonucleotide was carried out using several quantities of relaxase (0.5 to 12.5 μM) in the absence or in the presence of MobCTn1549 protein (10 μM). Cleavage was more efficient when the concentration of relaxase was increased; in contrast, the reaction was not influenced by the presence of MobCTn1549 (data not shown).
FIG. 4.
In vitro cleavage of oriT by RlxTn1549. Reaction mixtures contained 5′-labeled oligonucleotides (10 nM) incubated with or without RlxN247 (12.5 μM). Reactions were carried out at 37°C for 3 h as described in Materials and Methods. Lane 1, Ori34R-34-mer (5′-32P-GGGAGTGGCTACACTCCCTATGCTTGCTACGTTA); lane 2, Ori34R-34-mer plus RlxN247; lane 3, mix of oligonucleotides 27-mer, 25-mer, and 23-mer used as size standards; lane 4, Ori34F-34-mer (5′-32P-TAACGTAGCAAGCATAGGGAGTGTAGCCACTCCC); lane 5, Ori34F-34-mer plus RlxN247. Oligonucleotide control sizes are indicated by arrows.
Identification of oriT closely related to that of Tn1549.
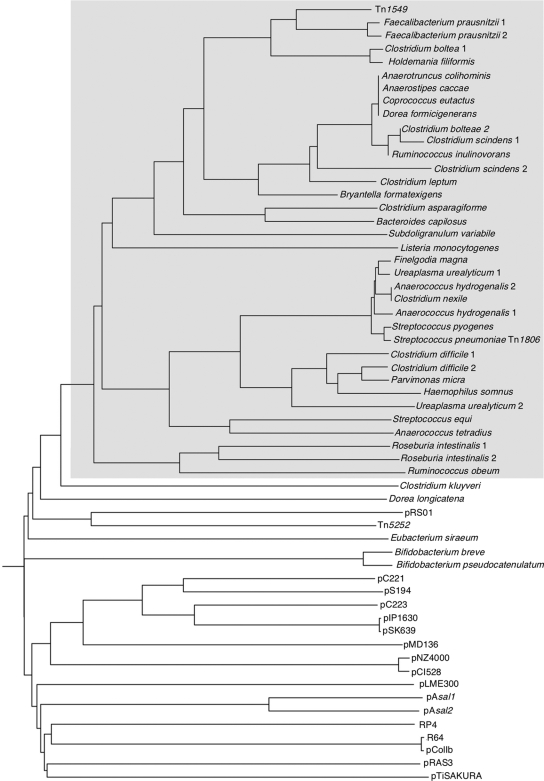
oriTTn1549 did not display homology with other oriTs from plasmids and transposons (16, 20, 29) except as mentioned at the 8-bp core site found to be conserved with the cleavage motif of RCR staphylococcal plasmids. To test the possibility that oriTTn1549 could be a member of a large family, putative oriTs were screened by BLAST in GenBank on the basis of DNA sequence homology with oriTTn1549 in a mobilization gene environment and an identical organization with the mobilization module of Tn1549. A total of 39 potential oriTs consisting of a consensus motif, C(a)T(a)RTGCTTG′CT, adjacent to an inverted repeat were found to be associated with both putative mobC and rlx genes as part of ICEs. The genetic elements were distributed in various bacterial species belonging to the Firmicutes, and they were also present in Ureaplasma species and Gram-negative bacteria such as Bacteroides and Eubacterium species and Haemophilus somnus. In all instances, these putative oriTs included a consensus core with the nicking motif composed of 11 conserved nucleotides linked to a variable inverted repeat and were between 28 and 32 bp in size, except in Eubacterium siraeum (36 bp). Conserved nucleotides were also found in the inverted repeats but to a lesser extent than in the core site (Fig. 5). Several putative oriTs were common to different genera; e.g., the oriTTn1549 was detected in Faecalibacterium prausnitzii M21/2 (Fig. 5). Anaerococcus hydrogenalis DSM 7454, Clostridium bolteae ATCC BAA-613, Clostridium difficile 630, Clostridium scindens ATCC35704, F. prausnitzii M21/2, Finegoldia magna ATCC29328, and Roseburia intestinalis L1-82 contained two distinct oriT mobilization modules. In H. somnus 129PT, a putative oriT was detected just upstream from a mobC-like gene in the absence of a relaxase gene. In contrast, H. somnus 2336 harbors a relaxase gene which most probably originated from clostridia (Fig. 6). Since relaxases are key enzymes in the transfer process, a phylogenetic tree was drawn to establish the relationship between the relaxases associated with the putative oriTs. The relaxases of Anaerostipes caccae DSM1462, Anaerotruncus colihominis DSM 17241, Coprococcus eutactus ATTC 27759, and Dorea formicigenerans ATTC 27755, which are part of a conserved ICE, were found to be identical (Fig. 6). These bacteria also shared a putative minimal oriT (Fig. 5). C. bolteae ATCC BAA-613 harbors two relaxases, one of which displays 98% identity with this relaxase common to these bacteria. An almost identical relaxase was also found as the product of one of the two intron relaxase genes of C. scindens ATCC 35704, whereas the other product was distinct (79% identity). Analysis of the environment of the relaxase genes revealed a sequence highly conserved on ca. 29 kb which was part of an ICE. The presence of this element in these various bacteria supports the notion of recent horizontal transfer. The relaxases of F. magna ATCC 29328, Ureaplasma urealyticum ATCC 33175, A. hydrogenalis DSM 7454, Clostridium nexile DSM 1787, Tn1806 from Streptococcus pneumoniae AP200, and S. pyogenes MGAS10750 were very similar (≥96% identity) and thus indicate origination from a common ancestor.
FIG. 5.
Alignment of oriT regions of the MOBp superfamily. (a) Putative oriT related to oriTTn1549 detected by BLAST searches. Only motifs found in the vicinity of the mobilization genes were examined. Inverted repeats are indicated by arrows: dark, complementary bases; gray, no complementary bases. Conserved nucleotides in the inverted repeats are in gray boxes. Bacterial sources are derived from GenBank accession numbers and are cited in phylogenetic order according to the similarity of their cognate relaxases with RlxTn1549 (Fig. 6): ABXA01000002.1 and ABXA01000003.1 (A. hydrogenalis DSM 7454), ACGC01000033.1 (A. tetradius ATCC 35098), ABAX03000014.1. (A. caccae DSM 14662), ABGD02000024.1 (A. colihominis DSM 17241), AAXG02000015.1 (B. capillosus ATCC 29799), ACCG01000027.1 (B. breve DSM 20213), ABXX02000003.1 (B. pseudocatenulatum DSM 20438), NZ_ACCL01000033.1 (B. formatexigens DSM 14469), NZ_ACCJ01000017.1 (C. asparagiforme DSM 15981), ABCC02000025.1 (C. bolteae ATCC BAA-613), NC_009089.1 (C. difficile 630), NC_011837.1 (C. kluyveri NBRC 12016), ABCB02000021.1 (C. leptum DSM 753), ABWO01000059.1 (C. nexile DSM 1787), ABFY02000002.1 (C. scindens ATCC 35704), ABEY02000028.1 (C. eutactus ATCC 27759), AAXA02000014.1 (D. formicigenerans ATCC 27755), AAXB02000001.1 (D. longicatena DSM 13814), ABCA03000042.1 (E. siraeum DSM 15702), ABED02000029.1 (F. prausnitzii M212-2), NC_010376.1 (F. magna ATCC 29328), NC_008309.1 (H. somnus 129PT), AADR01000004.1 (L. monocytogenes 4b H7858), ABEE02000016.1 (P. micra ATCC 33270), ABYJ01000019.1 (R. intestinalis L1-82), AAVO02000008.1 (R. obeum ATCC 29174), NZ_ACFY01000051.1 (R. inulinivorans DSM 16841), AM909652 (S. equi ICESe2), EF469826 (S. pneumoniae AP200 Tn1806), NC_008024.1 and FM162351.1 (S. pyogenes MAGS10750 and C1), NZ_ACBY01000108.1 (S. variabile), AAYQ2000002.1 (U. urealyticum serovar 9 ATCC 33175), ABEV01000002.1 (U. urealyticum serovar 13 ATCC 33698). The putative oriT of B. pseudocatenulatum is identical to that of B. breve. Core cleavage sequences are indicated in bold. Consensus nick sequences are indicated; Uppercase letters, conserved bases; lowercase letters, bases present in the majority of sequences. (b) Core cleavage sequences of RCR Staphylococcus plasmids and consensus sequence. (c) Core cleavage of ColE1 and pRA2 and consensus sequence.
FIG. 6.
Phylogenetic relationship among proteins belonging to the 3H family of MOBp-superfamily relaxases. The tree was constructed from sequences available in data banks by use of the neighbor-joining method. The origin of relaxase-containing plasmids, transposons, or the bacterial host is indicated at each branch. The phyletic group including RlxTn1549 is gray shadowed. Accession numbers of relaxases: Tn1549, AAF72355; A. hydrogenalis 1, ZP_03303665.1; A. hydrogenalis 2, ZP_03303763.1; A. tetradius, NC_ACGC01000033.1; A. caccae, ZP_02419708.1; A. colihominis, NZ_ABGD02000024.1; B. capillosus, ZP_02036891; B. breve, ZP_03619377; B. pseudocatenulatum, NZ_ABXX02000003; B. formatexigens, ZP_03686097; C. asparagiforme DSM 15981, NZ_ACCJ01000017.1; C. bolteae, ZP_02085866.1 and ZP_02085146.1; C. difficile, YP_001088377.1 and YP_001086893.1; C. kluyveri, YP_0013933676.1; C. leptum, ZP_02081957.1; C. nexile, ZP_03288837.1; C. scindens, ZP_02429884.1 and ZP_02432986.1; C. eutactus, ZP_02207623.1; D. formicigenerans, ZP_02235231.1; D. longicatena, ZP_01994204.1; E. siraeum, ZP_02422210.1; F. prausnitzii, ZP_02093051.1 and ZP_02090960.1; F. magna, YP_001692251.1; H. somnus, YP_001784516.1; H. filiformis, ZP_03636025.1; YP_0011784516.1; L. monocytogenes, ZP_00229823.1; P. micra, ZP_02093865.1; R. intestinalis, ZP_03483057.1 and ZP_03485669.1; R. inulinovorans, ZP_03752747.1; R. obeum, ZP-01964421.1 and ZP_01962871.1; S. equi, CAP20357; S. pneumoniae AP200 (Tn1806), EF469826; S. pyogenes, CAQ56294.1; S. variabile, ZP_03776736.1; U. urealyticum serovar 9, ZP_03079552.1; U. urealyticum serovar 13, ZP_02696018.1; ColIbP-9, NP_052501.1; pAH82, NP_862611.1; pAsal1, CAD48422.1; pAsal2, NP_710170.1; pC221, CAA26106; pC223, NP_943089; pCI1528, AAB28188.1; pFN1, NP_862674.1; pIP1629, AAD02378.1; pIP1630, AAD02405.1; pLME300, CAD59910; pMD136, NP_037562.1; pNZ4000, NP_053037; pRAS3, NP_387457.1; pRS01, AAB06502.1; pS194, NP_976272; pSK639, NP_976278; RP4, Q00191; R64, B38529; Tn5252, NP_358551.
DISCUSSION
Emergence of VanA-type resistance in S. aureus has required plasmid acquisition, combined with transposition of a Tn1546-like element, or acquisition of broad-host-range conjugative plasmids such as Inc18 plasmids bearing the transposon (53). In contrast, VanB-type Tn1549 is able to disseminate vancomycin resistance among various bacterial genera. Although the transfer origins of conjugative plasmids have been extensively analyzed, only a few oriT genes of ICEs have been characterized, i.e., Tn916 (39), ICEBs1 of B. subtilis (29), and ICESt1 and ICESt3 of Streptococcus thermophilus (12). In these elements, the four oriT regions contain a highly conserved sequence comprised of a GC-rich inverted repeat separated by ca. 10 bp. In Tn1549, a 2,374-bp fragment which includes the orf29-30 intergenic domain, orf29 (mobCTn1549), orf28 (rlx), and orfz was shown to contain both an oriT and the genes for mobilization proteins. In certain plasmids a single protein is required for mobilization, such as MobM of pMV158 in Streptococcus agalactiae (19, 24), but in most instances the relaxase needs for DNA cleavage an accessory protein, MobC, which cooperates for strand separation by binding at the oriT, generating local distortion of the helix which extends to the nic site (52). In the mobilization assay of pACYCGm derivatives performed with E. coli recA::RP4::Δnic by providing rlxTn1549 in trans, transfer did not occur in the absence of mobCTn1549. Furthermore, mobilization was enhanced when mobilization genes were delivered in cis. Such a cis preference has been reported with TraA, the relaxase of symbiotic plasmid pRetCF2d from Rhizobium etli (38).
A functional oriT was identified on a 130-bp fragment of Tn1549. As often reported, minimal changes in base composition in the oriT have dramatic consequences for mobilization (3). In Tn1549, the alteration of the putative secondary structure A by directed mutagenesis affected mobilization. Interestingly, a minimal oriT limited to a 28-bp sequence was found to be sufficient for mobilization of pACYCGm. This minimal oriT was a particularly short sequence, consistent with the capacity of MobCTn1549 to bind to the minimal oriT as demonstrated by EMSA together with the ability of RlxTn1549 to cleave short oligodeoxynucleotides. The fact that the transfer frequency was significantly diminished compared to that of the orf29-30 intergenic region suggests the presence of specific features in the neighboring environment of oriT that would enhance the binding of MobCTn1549 and Rlx. Likewise, the 130-bp fragment just upstream from mobCTn1549 contained the binding motifs, since this fragment was as efficient as the entire intergenic orf29-30 fragment for mobilization of pACYCGm. RlxTn1549 did not bind to oriT alone, but in contrast the minimal oriT was found to be sufficient for binding of MobCTn1549. Most of the oriTs consist of a short cleavage sequence (nic core) associated to an immediately adjacent variable hairpin (37). The nic site of Tn1549 was identified at the 3′ end of a 10-bp arm (core) adjacent to a 7-bp perfect inverted repeat sequence (hairpin A). This sequence shared eight conserved bp with the consensus motif reported in RCR plasmid, whereas nic was found in the antisense strand upstream from the relaxase.
Relaxases cleave specifically a single-strand DNA at oriT and play a central role in initiation of the relaxosome and DNA transfer of conjugative and mobilizable plasmids. It has been suggested that most mobilizable plasmid relaxases are evolutionarily related (37). The relaxase from Tn1549 is a member of the MOBP7 clade of MOBp superfamily relaxases (21). MOBp represents the largest relaxase family and has been divided into seven clades (21). MOBP7 is a large clade with profound branches. In this study, we propose to distinguish from the phylogenetic tree a new phyletic cluster in MOBP7 with relaxase of Tn1549 as a prototype (Fig. 6). In this cluster, the putative relaxases from ICEs originate from various anaerobes, Listeria monocytogenes, Tn1806 from S. pneumoniae, S. pyogenes MGAS10750, ICESe2 from Streptococcus equi, and Ureaplasma species, and share more than 40% identity. A second cluster includes RCR plasmids from Staphylococcus, Streptococcus, and Lactococcus species, Tn5252 from S. pneumoniae, and ICEs from Bifidobacterium pseudocatenulatum and Ruminococcus obeum. The third cluster corresponds to plasmids from Gram-negative bacteria. Interestingly, these relaxases were associated with a MobC protein and an oriT closely related to that of Tn1549. oriTTn1549 is likely a member of a family mainly spread in ICEs from anaerobes which shared a consensus 11-nucleotide sequence nic core consisting of a MWRTGCTTG′CT motif. A large portion of this motif is common to the consensus oriT RYGCTTG′C characterized in certain RCR plasmids of Gram-positive bacteria. The detection of sequences identical to oriTTn1549 just upstream from mobC-like and rlx genes allows us to propose a putative transfer origin for various transposons, such as the mobile elements carrying the erythromycin resistance determinant erm(TR) in S. pneumoniae (Tn1806) (8) and S. pyogenes (ICE 10750-RD.2) (4) or involved in iron acquisition in S. equi (ICESe2) (26). Interestingly, these elements are known to be mobilizable, a feature that could be shared by other elements which encode relaxases closely related to RlxTn1549 and carry this common transfer origin. The relationship between plasmid relaxases has been extensively described, and relaxases have been recently proposed as suitable markers for plasmid evolution (21). In contrast, relaxases from ICEs have not been classified and, in addition, the data banks have been recently enriched with numerous genomic sequences from anaerobic bacteria. The phyletic tree drawn in Fig. 6 shows that, in a similar manner, relaxases from ICEs represent a useful tool to understand lateral gene transfer mediated by these elements, which contributes to the diversification and adaptation of microorganisms. The relaxases related to rlxTn1549 were mostly confined to the Firmicutes except those found in Bacteroidetes (Bacteroides capillosus), proteobacteria (H. somnus) and Tenericutes (U. urealyticum), which likely resulted from genetic exchange between phyla.
Trans-Gram genetic exchanges have occurred under natural conditions, as exemplified by the acquisition by Gram-negative bacteria of genes from Gram-positive bacteria (49, 51). Evidence that transfer can be mediated by conjugation was demonstrated by the use of shuttle vectors (48). In these models the tra functions were provided with their cognate oriT. However, mobilization by heterologous systems can be the result of interplay between compatible transfer mechanisms. Mobilization of Bacteroides plasmids by IncP plasmids in E. coli was reported in the late 1980s (42); nevertheless, mobilization of a transposon from Gram-positive bacteria such as Tn1549 by RP4, as obtained from constructs in E. coli, remains an intriguing feature. The interaction between the relaxosome of mobilizable plasmids and the transfer apparatus of bacteria is thought to be mediated by type IV family coupling proteins (6). These proteins, by their capacity to direct this interaction, are crucial for the specificity of mobilization. It has been shown with the IncW plasmid R388 that membrane protein TrwB is responsible for coupling the relaxosome with the DNA transport apparatus (6, 47). In general, the coupling proteins are specific (31). However, several interactions in heterologous mobilization systems have been reported; for example, the RP4 traG gene can replace the R388 trwB gene for mobilization of plasmid ColE1 (7). Similarly, the Tn4555 mobATn gene and the adjacent oriTTn from Bacteroides are mobilized by IncPα plasmids such as RK2 (44). The oriT of this element is related to a plasmid family from Gram-positive bacteria. This plasmid family also includes pMV158 of S. agalactiae, pT181 of S. aureus, pIP823 of L. monocytogenes, and the mobilizable transposon Tn4451 of C. difficile (13, 16). Interestingly, mobilization of pMV158 and pIP823 by functions provided by RP4 has been reported (19). Similarly, the oriT-relaxase complex of Tn1549 is likely able to interact in E. coli with the TraG coupling protein for mobilization. This is an indirect but consistent argument, suggesting that conjugative transfer of Tn1549 from C. symbiosum to Enterococcus spp. occurred by mobilization by a helper element. If this hypothesis is true, conjugative transfer of Tn1549 obtained from an E. faecium transconjugant harboring Tn916 is due to the latter transposon. In the same manner, transfer of Tn1549 from anaerobes could be associated with the presence in the donors of resident ICEs or conjugative plasmids. It remains to be established if the mobilization systems related to the oriTTn1549 family are also mobilizable by the RP4 transfer apparatus or by other elements.
Acknowledgments
This work was supported by a grant from ANR contract MIME 2007 026-02.
We thank Didier Mazel for the gift of strain E. coli recA::RP4::Δnic. We also thank Patrice Courvalin and Fernando de la Cruz for helpful discussions.
Footnotes
Published ahead of print on 4 December 2009.
REFERENCES
- 1.Babic, A., A.-M. Guérout, and D. Mazel. 2008. Construction of an improved RP4 (RK2)-based conjugative system. Res. Microbiol. 159:545-549. [DOI] [PubMed] [Google Scholar]
- 2.Becker, E. C., and R. J. Meyer. 2000. Recognition of oriT for DNA processing at termination of a round of conjugal transfer. J. Mol. Biol. 300:1067-1077. [DOI] [PubMed] [Google Scholar]
- 3.Becker, E. C., and R. J. Meyer. 2003. Relaxed specificity of the R1162 nickase: a model for evolution of a system for conjugative mobilization of plasmids. J. Bacteriol. 185:3538-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beres, S., and J. M. Musser. 2007. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One 2(8):e800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrd, D. R., and S. W. Matson. 1997. Nicking by transesterification: the reaction catalysed by a relaxase. Mol. Microbiol. 25:1011-1022. [DOI] [PubMed] [Google Scholar]
- 6.Cabezón, E., and F. de la Cruz. 2006. TrwB: an F(1)-ATPase-like molecular motor involved in DNA transport during bacterial conjugation. Res. Microbiol. 157:299-305. [DOI] [PubMed] [Google Scholar]
- 7.Cabezón, E., E. Lanka, and F. de la Cruz. 1994. Requirements for mobilization of plasmids RSF10 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J. Bacteriol. 176:4455-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camilli, R., M. Del Grosso, F. Iannelli, and A. Pantosti. 2008. New genetic element carrying the erythromycin resistance determinant erm(TR) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 52:619-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carias, L. L., S. D. Rudin, C. J. Donskey, and L. B. Rice. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caryl, J. A., M. C. A. Smith, and C. D. Thomas. 2004. Reconstitution of a staphylococcal plasmid-protein relaxation complex in vitro. J. Bacteriol. 186:3374-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caryl, J. A., and C. D. Thomas. 2006. Investigating the basis of substrate recognition in the pC221 relaxosome. Mol. Microbiol. 60:1302-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceccarelli, D., A. Daccord, M. René, and V. Burrus. 2008. Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J. Bacteriol. 190:5328-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charpentier, E., G. Gerbaud, and P. Courvalin. 1999. Conjugative mobilization of the rolling-circle plasmid pIP823 from Listeria monocytogenes BM4293 among Gram-positive and Gram-negative bacteria. J. Bacteriol. 181:3368-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christie, P. J. 2004. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim. Biophys. Acta 1694:219-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. [DOI] [PubMed] [Google Scholar]
- 16.Crellin, P. K., and J. I. Rood. 1998. Tn4451 from Clostridium perfringens is a mobilizable transposon that encodes the functional Mob protein, TnpZ. Mol. Microbiol. 27:631-642. [DOI] [PubMed] [Google Scholar]
- 17.Dahl, K. H., T. P. Røkenes, E. W. Lundblad, and A. Sundsfjord. 2003. Nonconjugative transposition of the vanB-containing Tn5382-like element in Enterococcus faecium. Antimicrob. Agents Chemother. 47:786-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domingo, M. C., A. Huletsky, A. Bernal, R. Giroux, D. K. Boudreau, F. J. Picard, and M. G. Bergeron. 2005. Characterization of a Tn5382-like transposon containing the vanB2 gene cluster in a Clostridium strain isolated from human faeces. J. Antimicrob. Chemother. 55:466-474. [DOI] [PubMed] [Google Scholar]
- 19.Farias, M. E., and M. Espinosa. 2000. Conjugal transfer of plasmid pMV158: uncoupling of the pMV158 origin of transfer from the mobilization gene mobM, and modulation of pMV158 transfer in Escherichia coli mediated by IncP plasmids. Microbiology 146:2259-2265. [DOI] [PubMed] [Google Scholar]
- 20.Francia, M. V., A. Varsaki. M. P. Garcillán-Barcia, A. Latorre. C. Drainas, and F. de la Cruz. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28:79-100. [DOI] [PubMed] [Google Scholar]
- 21.Garcillán-Barcia, M. P., M. V. Francia, and F. de la Cruz. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33:657-687. [DOI] [PubMed] [Google Scholar]
- 22.Garnier, F., S. Taourit, P. Glaser, P. Courvalin, and M. Galimand. 2000. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology 146:1481-1489. [DOI] [PubMed] [Google Scholar]
- 23.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guzmán, L. M., and M. Espinosa. 1997. The mobilization protein, MobM, of the streptococcal plasmid pMV158 specifically cleaves supercoiled DNA at the plasmid oriT. J. Mol. Biol. 266:688-702. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton, C. M., L. H. Lee, P.-L. Li, D. M. Cook, K. R. Piper, S. Beck von Bodman, E. Lanka, W. Ream, and S. K. Farrand. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 182:1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heather, Z., M. T. G. Holden, K. F. Steward, J. Parkhill, L. Song, G. L. Challis, C. Robinson, N. Davis-Poynter, and A. S. Waller. 2008. A novel streptococcal integrative conjugative element involved in iron acquisition. Mol. Microbiol. 70:1274-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopec, J., A. Bergmann, G. Fritz, E. Grohmann, and W. Keller. 2005. TraA and its N-terminal relaxase domain of the Gram-positive plasmid pIP501 show specific oriT binding and behave as dimers in solution. Biochem. J. 387:401-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Launay, A., S. A. Ballard, P. D. R. Johnson, M. L. Grayson, and T. Lambert. 2006. Transfer of vancomycin resistance transposon Tn1549 from Clostridium symbiosum to Enterococcus spp. in the gut of gnotobiotic mice. Antimicrob. Agents Chemother. 50:1054-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, C. A., and A. D. Grossman. 2007. Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J. Bacteriol. 189:7254-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Llosa, M., F. X. Gomis-Rüth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 31.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. U. S. A. 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moncalián, G., E. Cabezón, I. Alkorta, M. Valle, F. Moro, and J. M. Valpuesta. 1999. Characterization of ATP and DNA binding activities of Trw, the coupling protein essential in plasmid R388 conjugation. J. Biol. Chem. 274:36117-36124. [DOI] [PubMed] [Google Scholar]
- 33.Osborn, A. M., and D. Böltner. 2002. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid 48:202-212. [DOI] [PubMed] [Google Scholar]
- 34.Pansegrau, W., and E. Lanka. 1996. Mechanisms of initiation and termination reactions in conjugative DNA processing. J. Biol. Chem. 271:13068-13076. [DOI] [PubMed] [Google Scholar]
- 35.Pansegrau, W., W. Schröder, and E. Lanka. 1993. Relaxase (TraI) of IncPα plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc. Natl. Acad. Sci. U. S. A. 90:2925-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pansegrau, W., W. Schröder, and E. Lanka. 1994. Concerted action of three distinct domains in the DNA cleavage-rejoining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J. Biol. Chem. 269:2782-2789. [PubMed] [Google Scholar]
- 37.Parker, C., E. Becker, X. Zhang, S. Jandle, and R. Meyer. 2005. Elements in the co-evolution of relaxases and their origins of transfer. Plasmid 53:113-118. [DOI] [PubMed] [Google Scholar]
- 38.Pérez-Mendoza, D., M. Lucas, S. Muñoz, J. A. Herrera-Cervera, J. Olivares, F. de la Cruz, and J. Sanjuán. 2006. The relaxase of the Rhizobium etli symbiotic plasmid shows nic site cis-acting preference. J. Bacteriol. 188:7488-7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rocco, J. M., and G. Churchward. 2006. The integrase of the conjugative transposon Tn916 directs strand- and sequence-specific cleavage of the origin of conjugal transfer, oriT, by the endonuclease Orf20. J. Bacteriol. 188:2207-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasaki, I., and G. Bertani. 1965. Growth anomalities in Hfr derivatives of Escherichia coli strain C. J. Gen. Microbiol. 40:365-376. [DOI] [PubMed] [Google Scholar]
- 41.Schröder, G., and E. Lanka. 2005. The mating pair formation system of conjugative plasmids—a versatile secretion machinery for transfer of proteins and DNA. Plasmid 54:1-25. [DOI] [PubMed] [Google Scholar]
- 42.Shoemaker, N. B., C. Getty, E. Guthrie, and A. Salyers. 1986. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 166:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engeneering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784-794. [Google Scholar]
- 44.Smith, C. J., and A. C. Parker. 1998. The transfer origin for Bacteroides mobilizable transposon Tn4555 is related to a plasmid family from gram-positive bacteria. J. Bacteriol. 180:435-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, M. C. A., and C. D. Thomas. 2004. An accessory protein is required for relaxosome formation by small staphylococcal plasmids. J. Bacteriol. 186:3363-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stinear, T. P., D. C. Olden, P. D. R. Johnson, J. K. Davies, and M. L. Grayson. 2001. Enterococcal vanB resistance locus in anaerobic bacteria in human faeces. Lancet 357:855-856. [DOI] [PubMed] [Google Scholar]
- 47.Tato, I., I. Matilla, I. Arechaga, S. Zunzunegui, F. de la Cruz, and E. Cabezon. 2007. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA. J. Biol. Chem. 282:25569-25576. [DOI] [PubMed] [Google Scholar]
- 48.Trieu-Cuot, P., C. Carlier, and P. Courvalin. 1988. Conjugative plasmid transfer from Enterococcus faecalis to Escherichia coli. J. Bacteriol. 170:4388-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trieu-Cuot, P., G. Gerbaud, T. Lambert, and P. Courvalin. 1985. In vivo transfer of genetic information between Gram-positive and Gram-negative bacteria. EMBO J. 4:3583-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Kranenburg, R., M. Kleerebezem, and W. M. de Vos. 2000. Nucleotide sequence analysis of the lactococcal EPS plasmid pNZ4000. Plasmid 43:130-136. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Y., G.-R. Wang, A. Shelby, N. B. Shoemaker, and A. A. Salyers. 2003. A newly discovered Bacteroides conjugative transposon, CTnGERM1, contains genes also found in gram-positive bacteria. Appl. Environ. Microbiol. 69:4595-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, S., and R. Meyer. 1997. The relaxosome protein MobC promotes conjugal plasmid mobilization by extending DNA strand separation to the nick site at the origin of transfer. Mol. Microbiol. 25:509-516. [DOI] [PubMed] [Google Scholar]
- 53.Zhu, W., N. C. Clark, L. K. McDougal, J. Hageman, L. C. McDonald, and J. B. Patel. 2008. Vancomycin-resitant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 52:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]