Abstract
Factor H binding protein (fHBP) is a surface-exposed lipoprotein in Neisseria meningitidis, which is a component of several investigational vaccines against serogroup B meningococcus (MenB) currently in development. fHBP enables the bacterium to evade complement-mediated killing by binding factor H, a key downregulator of the complement alternative pathway, and, in addition, fHBP is important for meningococcal survival in the presence of the antimicrobial peptide LL-37. In this study, we investigate the molecular mechanisms involved in transcription and regulation of the fHBP-encoding gene, fhbp. We show that the fHBP protein is expressed from two independent transcripts: one bicistronic transcript that includes the upstream gene and a second shorter monocistronic transcript from its own dedicated promoter, Pfhbp. Transcription from the promoter Pfhbp responds to oxygen limitation in an FNR-dependent manner, and, accordingly, the FNR protein binds to a Pfhbp probe in vitro. Furthermore, expression in meningococci of a constitutively active FNR mutant results in the overexpression of the fHBP protein. Finally, the analysis of fHBP regulation was extended to a panel of strains expressing different fHBP allelic variants at different levels, and we demonstrate that FNR is involved in the regulation of this antigen in all but one of the strains tested. Our data suggest that oxygen limitation may play an important role in inducing the expression of fHBP from a dedicated FNR-regulated promoter. This implies a role for this protein in microenvironments lacking oxygen, for instance in the submucosa or intracellularly, in addition to its demonstrated role in serum resistance in the blood.
Neisseria meningitidis is a Gram-negative bacterial pathogen that is carried asymptomatically in the upper respiratory tracts of approximately 5 to 10% of the human population. Occasionally, the bacterium spreads from the nasopharynx into the bloodstream, causing bacteremia with possible progression to sepsis, meningitis, and death (41). The outcome of infection depends on the capacity of the bacterium to avoid the host immune defense and survive within the circulation (44). Therefore, prevention through vaccination can bring about a major advance in preventing meningococcal disease, which remains a major worldwide cause of morbidity and mortality. However, no vaccine is currently available to provide universal protection against all serogroups of N. meningitidis. Capsular polysaccharide-based vaccines have been developed to protect against the main disease-associated serogroups A, C, Y, and W135; tailor-made vaccines, based on outer membrane vesicles (OMV) of serogroup B epidemic strains, have been used in Norway (17), Cuba (47), and New Zealand (21) to control epidemics.
Serogroup B capsular polysaccharide is poorly immunogenic due to its similarity with glycoproteins in human neural tissues (15). Therefore, the strategy to develop a vaccine against serogroup B of N. meningitidis (MenB) has instead focused on meningococcal noncapsular surface antigens (38). One of the proteins, which is currently being investigated as a meningococcal vaccine candidate, is factor H binding protein (fHBP), previously referred to as GNA1870 or LP2086 (16, 31). This antigen was identified by genome analysis of the MC58 strain of N. meningitidis and is currently included together with other recombinant protein antigens in an investigational MenB vaccine, which has entered phase III clinical trials. Recombinant fHBP (rLP2086) is the principal antigen of another MenB vaccine containing recombinant proteins from two different variant subfamilies (16, 56). More recent studies also support the strategy to prepare native OMV vaccines from mutant strains engineered to express different fHBP alleles that would confer broad protection against meningococcal strains expressing fHBP from each of the antigenic variant groups (22, 25).
In recent years, many studies have supported the importance of this antigen as a MenB vaccine candidate and also as an important virulence factor. fHBP is able to elicit antibodies that have high bactericidal activity (31, 53) that have also been shown to confer passive protection in the infant rat model (31). It has been demonstrated that the antigen enables N. meningitidis to evade complement-mediated killing by binding factor H, a key downregulator of the complement alternative pathway (AP), allowing meningococcal survival in human blood (29, 43). As reported for Neisseria gonorrhoeae (35), the specificity of this binding is restricted to human binding factor H (19), and this important feature could explain the reason why N. meningitidis is exclusively a human pathogen. Furthermore, anti-fHBP antibodies can block binding of fH, increasing the susceptibility to killing by the complement AP (6, 29). It has been demonstrated that the antigen also confers protection against killing by the antimicrobial peptide LL-37 (46), a compound produced by human cells that may be involved in the innate host immune response against meningococcal disease.
Through many studies using monoclonal antibodies (4, 5, 53), the protective epitopes (18) and the residues involved in the binding to factor H have been identified, and the structure of the full-length fHBP has been recently solved (10, 30, 45). fHBP is a surface-exposed lipoprotein expressed in a global panel of representative meningococcal strains, albeit at different levels in different strains which can be classified as high, intermediate, and low expressers (31). In a more recent study, 98% of a panel of 1,263 invasive clinical isolates showed surface expression of fHBP at various levels, and accessibility of the protein to bactericidal antibodies was independent of capsule expression (33). Unlike the well-known role of the protein, the mechanisms that regulate and influence its expression have remained largely unexplored.
In the present study, we have focused our analysis on the mechanism of regulation of the gene encoding the fHBP protein by mimicking some environmental conditions encountered by the bacterium during infection. Transcription of the fhbp gene was studied under oxygen-restricted conditions and in response to the FNR regulatory protein. Furthermore, we broaden our analysis to a representative panel of strains expressing different variant alleles of fHBP from different clonal complexes and found that regulation of transcription is widely conserved.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The meningococcal strains (clinical isolates) used in this study were as follows: MC58, H44/76, M2197, NM008, F6124, M6190, MO445, LNP17592, M01-240185, M01-240345, 67/00, NZ98/254, M6094, NM117, ES14902, M04-240196, 2996, 961-5945, LNP17094, B3937, M01-240013, M3153, M0579, M2671, M986, 5/99, M5258, BZ3279, M5149, M2552, 1000, OX99.30304, M1239, M01-0240988, M01-0240354, and M13399. N. meningitidis strains were routinely cultured in GC-based agar medium (Difco) supplemented with Kellogg's supplement I (24) at 37°C in a 5% CO2-95% air atmosphere (microaerobic conditions) at 95% humidity. Strains were stocked in GC medium plus 15% glycerol and stored at −80°C. Each bacterial manipulation was started from an overnight culture of frozen stock. For liquid cultures, N. meningitidis strains were grown overnight on solid medium, resuspended in GC broth to an optical density at 600 nm (OD600) of 0.5, and inoculated with a 1:10 dilution into GC broth supplemented with Kellogg's supplement I. When required, erythromycin or chloramphenicol was added to a final concentration of 5 μg/ml. Escherichia coli strains DH5-α (20) and BL21(DE3) (50) were cultured in Luria-Bertani (LB) medium or on LB agar plates at 37°C, and, when required, ampicillin was added to a final concentration of 100 μg/ml.
Construction of plasmids and recombinant strains.
DNA manipulations were carried out routinely as described previously for standard laboratory methods (42). The isogenic MC58ΔfHBP knockout mutant (previously called MC58Δgna1870), in which the nmb1870 gene was truncated and replaced with an erythromycin antibiotic cassette, has been described previously (31). To knock out the nmb1869 gene in the MC58 background, the plasmid pGemTUD1869Ery was generated. Upstream and downstream flanking regions of the nmb1869 gene were amplified by PCR with the primers 1869-1 and 1869-2 and 1869-3 and 1869-4 (Table 1), respectively. Then in a second round of PCR analysis, the respective upstream and downstream fragments, which contain regions of overlap due to the design of the primers, were used in a self-priming PCR amplification for five cycles, and the corresponding united fragment was amplified using the external primers 1869-1 and 1869-4. This product was cloned into the pGEM-T (Promega) vector, and an erythromycin cassette was inserted into the SmaI site between the flanking regions, generating pGemTUD1869Ery. The plasmid was then linearized and used for allelic exchange in the MC58 strain, generating the nmb1869 knockout mutant, Δnmb1869. The fnr null mutant was produced by replacing the entire coding sequence with an erythromycin resistance cassette, as has been previously described (3). For complementation of the fnr null mutant, wild-type fnr or fnr(D148A) mutant genes under the control of the Ptac promoter were reinserted into the chromosome of Δfnr between the converging open reading frames (ORFs) NMB1428 and NMB1429, through the transformation of the Δfnr strain with pCompInd-fnr or pCompInd-fnr(D148A), respectively. pCompInd-fnr is a derivative plasmid of pCompInd (23), in which the wild-type fnr gene was amplified from the MC58 genome with primer pair FNR-for1/FNR-rev2 and cloned as a 732-bp NdeI/NsiI fragment downstream of the Ptac promoter. The pCompInd-fnr(D148A) plasmid is a derivative of pCompInd-fnr containing a site-directed mutant allele of the fnr gene, in which the GAC aspartate-148 was substituted by the GCC alanine codon. The mutation was introduced in pCompInd-fnr using the QuikChange kit (Stratagene) and the D148A-F/D148A-R primer pair. The Δfnr strain was transformed with the pCompInd-fnr or pCompInd-fnr(D148A) plasmid, and the resultant complemented mutants were called ΔfnrC and ΔfnrC(D148A), respectively. To generate recombinant strains expressing an FNR(D148A) protein from an integrated copy of the mutant gene, the pCompInd-fnr(D148A) plasmid was transformed into meningococcal isolates H44/76, 4243, F6124, M6190, LNP17592, M01-240345, NM117, LNP17094, B3937, M01-240013, M3153, 5/99, BZ232, 1000, and OX99.30304, generating the FNR(D148A)-expressing derivative version of each strain, respectively.
TABLE 1.
Oligonucleotides used in this study
| Primer | Sequencea | Site |
|---|---|---|
| FNR-for1 | attagcatatgGCTTCGCATAATACTACAC | NdeI |
| FNR-rev2 | attagatgcatCAAATGGCGTGCGAGC | NsiI |
| D148A-F | GTGAAATCGTGCGCGCCCAAGGTGTTATGCTG | |
| D148A-R | CAGCATAACACCTTGGGCGCGCACGATTTCAC | |
| 1869-1 | attaggaattcGTACATCAGTTCGAACAGGGCGTGG | EcoRI |
| 1869-2 | TGACGATTTGcccgggCTTGGGCAATAGGTAAATAAGGCGG | SmaI |
| 1869-3 | CTATTGCCCAAGcccgggCAAATCGTCAAATAACAGGTTGC | SmaI |
| 1869-4 | attcagaagcttCTGAAAGGCGGTTAAGGCGGAATGG | HindIII |
| 741FOR | CAAATCGAAGTGGACGGGCA | |
| 741REV | TGTTCGATTTTGCCGTTTCCCTG | |
| 1869rtF | CAACTGGGCTTCTCCTCTGT | |
| 1869rtR | ATTTCGCCTTCAACGGATAC | |
| 741Pf | CGTCAAATAACAGGTTG | |
| 741PE1 | GAATCAGGGCAGTGGTCAGAG | |
| 1869FW | CCAACTGCTTGATCATGCTGC | |
| 1869rev | GTGTGGAAATTCTTCGACAGCC | |
| 741F2 | TTGGCTGCGAAGGTCAGGCAG | |
| 741REV2 | GTATGTTCGCCCGCTATGTCG | |
| IG1871F | CATCAGCGGTTCCGTCCTTTAC | |
| IG1871R | GCGGATTTCCGGCAGAATCAG | |
| FNR-rev1 | attagggatccTCAAATGGCGTGCGAGC | BamHI |
| 1869PE1 | GCAGCATGATCAAGCAGTTG | |
| 1869 PF | TCATGGAGGCTGCAGACCAAG | |
| 1869 PR-2 | GCGAGCCGTCCATCATTACAG |
Capital letters indicate N. meningitidis-derived sequences, lowercase letters indicate sequences added for cloning purposes, and underlined letters indicate restriction enzyme recognition sites.
Transformation.
For transformation of naturally competent N. meningitidis, four or five single colonies of a freshly grown overnight culture were resuspended in 20 μl phosphate-buffered saline (PBS), spotted onto GC agar plates to which 5 to 10 μg of linearized plasmid DNA was added, allowed to dry, and incubated for 6 to 8 h at 37°C. Transformants were selected on plates containing the appropriate antibiotic, and single colonies were restreaked on selective medium for further analysis. Single colonies were resuspended in 50 μl PBS, placed in a boiling water bath for 5 min, and centrifuged in a benchtop centrifuge for 5 min at maximum speed. One microliter of the sample was used as a template for PCR analysis for correct double crossover transformants.
RNA preparation.
Total RNA was prepared from liquid cultures of meningococcal strains grown to an OD600 of 0.5, as described above. For altered oxygen conditions, meningococcal strains were grown in liquid broth in 16-ml tubes to mid-log phase, diluted to an OD600 of 0.1, and split into 7-ml (aerobic growth) or 15-ml (oxygen-limiting conditions) samples in 16-ml polystyrene Falcon tubes and reincubated for 30 min at 37°C. The cultures were chilled on ice for 15 min, added to an equal volume of frozen medium to keep the temperature at 4°C, and then centrifuged at 3,000 rpm in a benchtop centrifuge at 4°C. RNA was extracted from the pelleted cells as previously described (11) or was extracted for use in Northern blot analysis using the RNeasy mini kit (Qiagen).
RT-PCR analysis and primer extension.
For reverse transcription-PCR (RT-PCR) analysis, total RNA (2 μg) was reverse transcribed with random primers and SuperScript II reverse transcriptase (Invitrogen). The resulting cDNAs were amplified using primer pairs (Table 1) specific for nmb1869 (1869FW and 1869rev), for fhbp (741F2 and 741PE1), and for the intergenic regions (741FOR and 741REV2; IG1871F and IG1871R). For each primer pair, RNA-containing PCR analysis in which the reverse transcriptase step was omitted was used as a negative control for DNA contamination. Genomic DNA was amplified as positive control with the same primer sets. Primer extension was performed as previously reported (23). To ensure correct mapping of the promoter, the sequencing reaction was carried out with a T7 sequencing kit (USB Corporation) using the same primer as in the primer extension reactions and the plasmid consisting of the relevant cloned promoter. The promoter region of the fhbp gene was amplified from the MC58 genome with primers 741Pf and 741PE1 and cloned as a 215-bp fragment into pGEMT, generating pGEMTpfHBP, and the nmb1869 promoter region was cloned as a 221-bp PCR product amplified with primers 1869PF and 1869PR-2, generating pGEMTp1869.
Northern blot analysis.
Northern blot analysis was carried out using the NorthernMax kit (Ambion, Inc.) according to the manufacturer's instructions. Five micrograms of total RNA from N. meningitidis samples and 1 μg of molecular weight high-range RNA ladder (Fermentas, Inc.) were fractionated on a 0.8% agarose-formaldehyde gel and transferred onto a nylon membrane (Hybond Plus, Inc). Two PCR products consisting of 187 bp of the fhbp gene and 142 bp of the nmb1869 gene were amplified from the MC58 genome using primers 741FOR and 741REV and 1869rtF and 1869rtR (Table 1), respectively. Five picomoles of each DNA fragment was radioactively end labeled using T4 polynucleotide kinase (New England Biolabs, Inc.) and [γ-32P]ATP (5,000 Ci/mmol; Amersham) and used as fhbp and nmb1869 probes. All hybridization and wash steps were performed at 42°C.
Expression and purification of the FNR and FNR(D148A) proteins.
The fnr wild-type or fnr(D148A) mutant genes were amplified from the MC58 genome and from the plasmid preparation pCompInd-fnr(D148A), respectively, with the FNR-for1 and FNR-rev1 primer pair (Table 1). The resulting PCR products were cloned as 732-bp NdeI-BamHI fragments into the pET15b+ expression plasmid (Invitrogen), generating pET15fnr and pET15fnrD148A, which were subsequently transformed into the E. coli strain BL21(DE3) for protein expression. From an overnight culture of the strain BL21(DE3)(pET15fnr) or BL21(DE3)(pET15fnrD148A), 200 ml of LB medium was inoculated with each culture and grown to an OD600 of 0.5, and expression of the recombinant FNR and FNR(D148A) proteins containing an N-terminal histidine tag were induced by the addition of 1 mM isopropyl-d-thiogalactopyranoside (IPTG) and further incubated for 3 h. The proteins were purified from the harvested cells by Ni-nitrilotriacetic acid (Qiagen)-affinity chromatography under nondenaturing conditions according to the manufacturer's instructions. The purified protein preparations were then dialyzed overnight against 50 mM Tris-HCl (pH 8), 50 mM KCl, 10 mM MgCl, 0.01% NP-40, 1 mM dithiothreitol (DTT) at 4°C. The concentrations of the proteins were determined using the Bradford colorimetric assay (Bio-Rad), and the proteins were stored at 4°C. To generate anti-FNR antibodies, 6-week-old female CD1 mice (Charles River Laboratories) were immunized with 20 μg of FNR protein given intraperitoneally, together with complete Freund's adjuvant in three doses (days 1, 21, and 35). Bleed-out samples were taken on day 49 and used for Western blot analysis.
Western blot analysis.
Colonies from freshly grown overnight plate cultures were resuspended in PBS to an OD600 of 0.5. One milliliter was harvested by centrifugation at 8,000 × g, resuspended in 100 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (50 mM Tris-HCl [pH 6.8], 2.5% SDS, 0.1% bromophenol blue, 10% glycerol, 5% β-mercaptoethanol, 50 mM DTT). For Western blot analysis, 10 μg of each total protein sample was separated by SDS-PAGE and transferred onto a nitrocellulose filter by standard methods. Filters were blocked overnight at 4°C by agitation in blocking solution (10% skim milk and 0.05% Tween 20 in PBS) and incubated separately for 1 h at 37°C with a 1:1,000 dilution of anti-fHBP, anti-NMB1869, or anti-FNR protein serum in 3% skim milk solution. After washing, the filters were incubated with a 1:2,000 dilution of peroxidase-conjugated anti-mouse immunoglobulin (Dako) in 3% skim milk solution for 1 h, and the resulting signal was detected with the SuperSignal West Pico chemiluminescent substrate (Pierce).
DNase I footprinting.
The aniA promoter region was cloned into pGemT, generating pGemT-Nor as previously described (13). The pGemT-Nor plasmid and the pGEMTpfHBP plasmid, containing the promoter region of the fhbp gene, were digested with NcoI, dephosphorylated with calf intestine phosphatase (NEB), and then 5′-end labeled with [γ-32P]ATP (6,000 Ci/mmol; NEN). The aniA and fhbp promoter DNA was separated from the vectors by 6% polyacrylamide gel electrophoresis (Invitrogen) after digestion with BamHI and SpeI, respectively, producing probes labeled at one end only. The probes, extracted from the polyacrylamide gel, were eluted overnight in 3 ml elution buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 300 mM Na acetate [pH 5.2], 0.2% SDS) at 37°C with shaking, phenol-chloroform extracted, ethanol precipitated, and resuspended in 100 μl of water. DNase I footprinting was carried out as previously described (12) with the following variations: binding reactions were performed in 50 μl binding buffer (50 mM Tris-HCl [pH 8], 50 mM KCl, 10 mM MgCl, 0.01% NP-40, 1 mM DTT, 10% glycerol) containing 100 ng of sonicated salmon sperm DNA as nonspecific competitor DNA; DNase I digestion was carried out by addition of 2 μl DNase I (0.04 U/μl) in binding buffer containing 5 mM CaCl2 for precisely 1 min 10 s at room temperature. As a molecular weight marker, a G+A sequence reaction (32) was performed for each DNA probe and run in parallel to the corresponding footprinting reactions.
RESULTS
Analysis of the fhbp gene locus in the MC58 strain of Neisseria meningitidis.
A schematic representation of the fhbp locus is shown in Fig. 1A. The fhbp gene was originally annotated as NMB1870 in the genome sequence of strain MC58 published by TIGR (51) that assigned the start of the gene to an ATG codon located 138 bp upstream from the GTG start codon proposed in a subsequent study by Masignani and colleagues (31), which predicted a coding region of 825 bp. Downstream and in the same orientation as the fhbp gene is the ORF NMB1871, encoding a conserved hypothetical protein separated by an intergenic region of 146 bp. Transcriptional terminator analysis revealed a typical stem-loop of a rho-independent terminator (ΔG = −14.37 kcal/mol) 11 nucleotides (nt) downstream of the fhbp gene. Upstream and in the same orientation as the fhbp gene there is an ORF with amino acid homology to a fructose-bisphosphate aldolase, NMB1869 (1,064 bp), and an intergenic region of 157 bp is present between nmb1869 and the GTG starting codon of the fhbp gene. In this intergenic region, 20 nt downstream of the nmb1869 gene there is another putative rho-independent transcriptional terminator (ΔG0 = −10.45 kcal/mol).
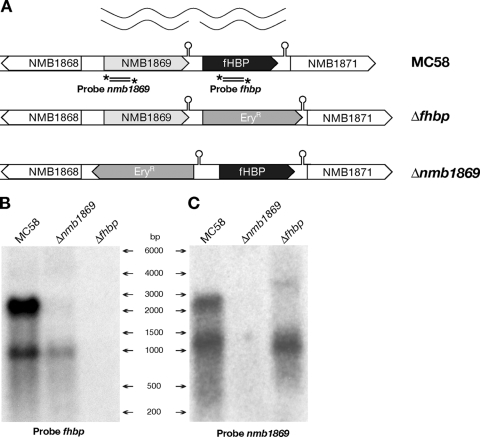
FIG. 1.
(A) Schematic representation of the fhbp locus within the genome of wild-type N. meningitidis strain MC58 and mutant strains Δnmb1869 and Δfhbp. In the null mutants, an erythromycin cassette used to replace the respective genes by allelic exchange is orientated in the same direction (Δfhbp) or in the opposite direction (Δnmb1869). Hairpin structures indicate the position of predicted rho-independent terminators. The stem-loop downstream of the fhbp gene is comprised of a 12-nt perfect palindromic sequence including the GCCGTCTGAA DNA uptake sequence (DUS) separated by 13 nt of loop and followed by a stretch of four Ts. DUS are often found in the base-paired stem of transcription terminators (48). The putative rho-independent terminator in the nmb1869-fhbp intergenic region is comprised of a similar 12-nt imperfect palindromic sequence in which the second DUS sequence has two mismatches, separated by 4 nt and followed by a stretch of three Ts. The relative positions of the radioactively labeled probes used in the Northern blot analysis are indicated under each gene. The figure provides a diagrammatic representation of the long bicistronic transcriptional unit linking the nmb1869 and fhbp genes and of the two monocistronic transcripts independently transcribed from their own dedicated promoters. (B and C) Northern blot analysis. Total RNA prepared from MC58, Δfhbp, and Δnmb1869 cultures grown to mid-log phase was run on an 0.8% denaturing agarose gel, transferred to a nylon membrane, and probed with radioactively labeled PCR products equivalent to 187 bp of the fhbp gene (B) and 142 bp of the nmb1869 gene (C). The relative positions of the molecular weight RNA ladder are shown.
To start defining the transcriptional map of the fhbp gene, we performed RT-PCR analysis on total RNA from the MC58 strain across the upstream and downstream intergenic regions. This analysis resulted in an amplification product across the upstream intergenic region but not the downstream intergenic region (data not shown), suggesting that fhbp may be cotranscribed with the upstream nmb1869 gene.
Analysis of the RNA transcripts.
To analyze the mRNAs transcribed from the locus, we performed Northern blot analysis on total RNA from the MC58 wild-type strain, the NMB1869 null mutant strain (Δnmb1869), and the fHBP null mutant strain (Δfhbp). Both mutants were generated by substituting the respective genes with an erythromycin cassette as indicated in Fig. 1A; the radioactively labeled probes used for each Northern assay are also represented. A long transcript, >2,000 nt, was detected by both the fhbp and nmb1869 probes in the wild-type strain and was absent in both mutant strains (Fig. 1B and C). The estimated size of this transcript correlates well with the predicted size of a bicistronic message and confirms the cotranscription of nmb1869 and fhbp. Interestingly, a shorter fhbp-specific RNA messenger of just less than 1,000 nt was detected in the wild-type strain and in the Δnmb1869 mutant strain (Fig. 1B), suggesting the presence of an fhbp monocistronic transcript. The presence of this shorter transcript in the Δnmb1869 mutant, where transcription of the bicistronic message has been eliminated, indicates that the fhbp gene is transcribed from its own dedicated promoter. In addition, the nmb1869 probe detected a smaller nmb1869-specific transcript of ca. 1,100 nt in the wild-type strain and in the fHBP null mutant (Fig. 1C), indicating that a monocistronic transcript of the nmb1869 upstream gene was also produced.
Taken together, these results suggest that two different promoters drive the synthesis of three separate mRNA transcripts of the nmb1869 and fhbp locus. The nmb1869 and fhbp genes are transcribed on monocistronic transcripts from their dedicated promoters and also are cotranscribed on a bicistronic transcript driven by a promoter upstream of nmb1869. The longer bicistronic transcript probably results from inefficient termination that leads to read-through of the transcriptional terminator downstream of nmb1869.
Mapping of the promoters.
To define the start point of the identified mRNAs, we carried out primer extension of total RNA extracted from N. meningitidis cultures grown to mid-log phase. An nmb1869-specific primer was hybridized to total RNA from N. meningitidis strain MC58 and elongated with reverse transcriptase. The major elongated product maps the 5′ end of the nmb1869 transcripts to a position 29 nt upstream of the ATG start codon of the nmb1869 gene, as shown in Fig. 2A. Primer extension with an fhbp-specific primer was performed with total RNA from strain MC58 (Fig. 2B) and also from the Δnmb1869 mutant (data not shown) and mapped the start of the fhbp monocistronic transcript to a position 45 nt upstream of the start codon of fhbp (Fig. 2B). Analysis of the nucleotide sequences in each case upstream of the elongated primers showed the presence of elements similar to the −10 and −35 consensus hexamers of sigma 70-dependent promoters from Escherichia coli. These sequences are likely to define the N. meningitidis Pnmb1869 and Pfhbp promoters. It is worth noting that Pnmb1869 shows a well-conserved −10 element (TATAAT) and a poorly conserved −35 region (TGGTTT), while Pfhbp shows mutation in both elements, −10 (TACCAT) and −35 (TTGATG).
FIG. 2.
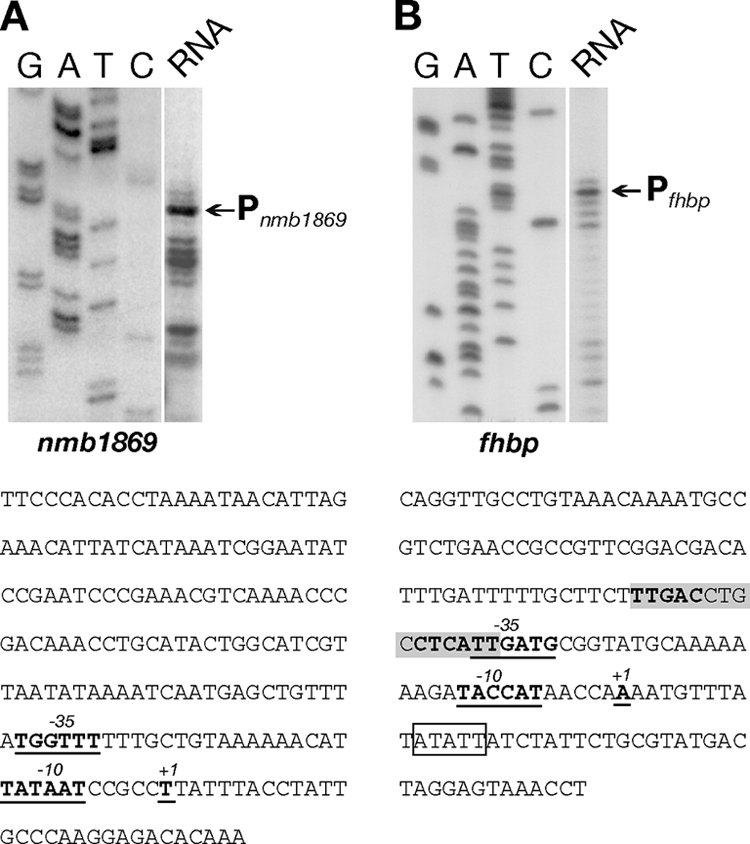
Mapping of the 5′ end of the nmb1869-specific transcript (A) and the fhbp-specific transcript (B) by primer extension. Total RNA (20 μg) prepared from cultures of the wild-type strain (MC58) grown to mid-logarithmic phase was hybridized with gene-specific primers (741PE1 and 1869PE1) (Table 1) and elongated with reverse transcriptase. Sequence reactions (G, A, T, and C) were performed with the same primers on the respective cloned promoter regions and run in parallel. The elongated primer bands mapping the major 5′ end of the corresponding gene transcripts are indicated. The corresponding +1 nt of transcriptional initiation and the upstream −10 and −35 promoter elements are underlined and in bold face in the nucleotide sequences of the respective intergenic regions shown underneath. An FNR box located at −40.5 bp is highlighted in gray. The pentanucleotide ATATT (in the box) is the target of insertion for a 181-bp AT-rich nucleotide sequence (ATR) in the genome of MenA strain Z2491 (37).
Regulation of transcription and protein levels.
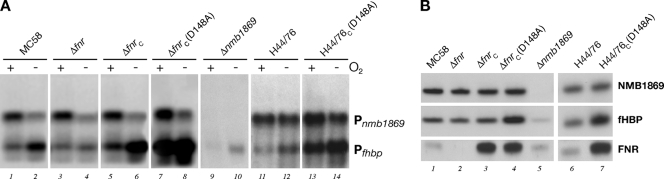
A previous study reporting microarray data of an FNR knockout in N. meningitidis (3) identified the fhbp gene as a possible member of the FNR regulon. The FNR transcription factor globally regulates gene expression in response to oxygen deprivation in many bacteria. Considering that the FNR protein is present in an active form during oxygen limitation, we analyzed transcription of the fhbp gene by FNR under different oxygen conditions. Total RNA was extracted from the wild-type strain, and the fnr null mutant was grown to mid-log phase and then exposed to aerobic conditions or oxygen limitation conditions for 30 min, respectively. Northern blot analysis was carried out to analyze the levels of the two fhbp transcripts, and the results are shown in Fig. 3A. The monocistronic transcript appeared to be upregulated during oxygen limitation in the wild type (Fig. 3A, lane 2 versus lane 1) but not in the fnr null mutant (Fig. 3A, lanes 3 and 4), indicating that FNR mediates the induction under oxygen limitation. In order to confirm the FNR-dependent regulation, we generated a complemented mutant strain expressing a single copy of the fnr gene in a heterologous location on the chromosome of strain Δfnr and assessed fhbp regulation. In the strain ΔfnrC (Fig. 3A, lanes 5 and 6), the upregulation of the fhbp monocistronic mRNA was restored under oxygen limitation conditions. In addition, our results showed that the longer bicistronic transcript was expressed at a lower level during oxygen-limiting conditions, but it did not seem to be a result of FNR-dependent regulation.
FIG. 3.
Regulation of fHBP by FNR. (A) Northern blot analysis showing the regulation of mRNA transcripts. The generation of the null mutants and Δfnr complemented strain ΔfnrC (in the MC58 and H44/76 background) and their growth under aerobic (+) and oxygen-limiting (−) conditions are described in Materials and Methods. Aliquots (5 μg) of total RNA were subjected to electrophoresis, transferred onto nylon filters, and hybridized with the fhbp-specific probe. In each lane, the amount of RNA loaded was normalized to the amount of rRNA. (B) Western blot analysis showing the FNR-regulated protein expression. Equal amounts of total protein from overnight plates cultured under microaerobic conditions were fractionated by SDS-PAGE, blotted onto nitrocellulose filters, and stained with mouse polyclonal antiserum raised against the NMB1869 protein, the fHBP protein, or the FNR protein. Where IPTG was necessary to induce the expression of FNR, it was added to plates at a final concentration of 1 mM.
We also generated an fnr-complemented mutant strain, ΔfnrC(D148A), expressing a mutant form of the FNR protein. This strain contains a site-directed mutant allele of the fnr gene, in which the GAC codon encoding aspartic acid at position 148 is substituted by the GCC alanine codon. In E. coli, the same single-amino-acid substitution at corresponding position 154 in the putative dimerization domain of FNR results in a constitutively active protein that functions as a transcriptional activator also in the presence of oxygen (26). Northern blot analysis using total RNA from this complementing strain grown during aerobic and oxygen-limiting conditions showed that the mutant FNR protein was able to promote transcription of the fhbp monocistronic mRNA also in the presence of oxygen (Fig. 3A, lanes 7 and 8), suggesting that the mutant FNR protein is constitutively active also in N. meningitidis. Furthermore, we found that knocking out the nmb1869 upstream gene, abolishing the synthesis of the bicistronic RNA messenger, reduced the level of the fhbp transcript and did not affect the FNR oxygen-dependent regulation of the monocistronic transcript (Fig. 3A, lanes 9 and 10). The reason for the low level of fhbp transcript observed in this mutant was not investigated. However, it clearly depends on the upstream transcript, which in turn could contribute to the downstream level of transcript via a putative processing event of the longer transcript. Taken together, these data indicate that transcription of fhbp is induced under oxygen limitation, likely by the dedicated FNR-regulated promoter Pfhbp.
In order to understand whether the FNR-dependent regulation of fHBP was restricted to the MC58 strain of N. meningitidis, we analyzed the transcription and the regulation of the gene in another meningococcal strain, H44/76. We found that two fhbp transcripts are synthesized also in the H44/76 strain and that the transcription of the fhbp monocistronic mRNA was upregulated in response to oxygen limitation in the wild type and also by the expression of the constitutively active FNR mutant protein (Fig. 3A, lanes 11 to 14), likely through activation of a dedicated FNR-regulated promoter. To correlate the transcriptional regulation by FNR to overall protein levels in all the strains generated, we performed Western blot analysis. Total protein extracts were prepared from overnight plate cultures grown under microaerobic conditions and immunoblotted with specific antibodies raised against the NMB1869, fHBP, and FNR proteins. As shown in Fig. 3B, fHBP expression was significantly increased in the ΔfnrC(D148A) and H44/76_CD148A strains expressing the constitutively active form of the FNR protein (Fig. 3B, lanes 4 and 7), in correlation with the Northern blotting results under aerobic conditions. Furthermore, in recombinant strains there is an overexpression of the respective FNR protein alleles expressed from the heterologous Ptac promoter compared to the FNR expression in the wild-type strain. However, only the FNR(D148A) form can induce overexpression of fHBP. This evidence strongly supports the importance of FNR activity, rather than its high level of expression, in promoting fHBP expression. NMB1869 expression remains unaltered in all strains. Moreover, the nmb1869 null mutant strain exhibited a lower level of fHBP expression, and we speculate that the abolished transcription of the bicistronic RNA messenger in this mutant was also responsible for the decrease in protein synthesis.
FNR is directly involved in promoting fHBP expression.
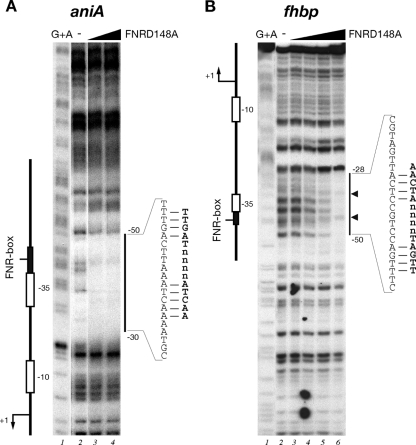
In order to test in vitro FNR binding to Pfhbp, we expressed and purified both wild-type and D148A mutant recombinant FNR proteins under aerobic conditions. To this aim, the fnr and fnr(D148A) genes were cloned into the pET15b expression vector in E. coli, and the proteins were expressed and purified by Ni2+-affinity chromatography by virtue of an N-terminally located histidine tag. We first tested the in vitro binding activity of both FNR and FNR(D148A) recombinant proteins to the aniA promoter, which has been well characterized through DNA microarray and DNA binding studies in N. meningitidis and N. gonorrhoeae to be under the direct control of FNR during oxygen limitation (3, 27, 36). A specific radioactively labeled probe containing the aniA promoter of strain MC58 was incubated with increasing concentrations of the recombinant proteins and submitted to DNase I digestion. We found that the addition of 13 nM FNR(D148A) protein resulted in complete protection of the DNA region spanning −30 to −50, with respect to the transcriptional start site, and containing the aniA-predicted FNR box consensus (Fig. 4A). Under these conditions, the wild-type protein did not result in protection of PaniA. These results support the fact that FNR purification under aerobic conditions results in an inactive form of the protein. In contrast, the aerobic purification of FNR(D148A) does not result in an oxygen-sensitive binding activity of the protein. We conclude that the FNR mutant is constitutively active for DNA binding under aerobic conditions and binds to the predicted FNR box in vitro; therefore, this mutant protein can be used to map specific meningococcal FNR binding sites in vitro.
FIG. 4.
Footprinting analysis of purified FNR(D148A) on the aniA (A) and fhbp (B) intergenic probes. The probes were labeled at one end and prepared as described in Materials and Methods. Lane 1 is a G+A sequence reaction obtained with each probe and used as a molecular weight marker. Footprinting reactions containing purified FNR(D148A) protein at final concentrations of 0 nM, 13 nM, and 40 nM in panel A (lanes 2 to 4, respectively) and at 0 nM, 8 nM, 40 nM, 200 nM, and 1 μM in panel B (lanes 2 to 6, respectively). The vertical bar on the right side of each panel indicates the area of DNase I protection in the promoter region; small arrows refer to DNase I hypersensitive sites. The numbers indicate the positions of the relevant nucleotides with respect to the +1 transcriptional start site. The nucleotides corresponding to the FNR binding site are indicated, with mismatches to the FNR box of E. coli displayed on the right.
Subsequently, we performed a footprinting experiment with increasing concentrations of the FNR(D148A) protein on a specific radioactively labeled fhbp promoter probe. The results, shown in Fig. 4B, indicate that the addition of 200 nM and 1 μM FNR(D148A) resulted in protection of nucleotides spanning from positions −28 to −50 with respect to the transcriptional start site of Pfhbp, therefore overlapping the −35 promoter element. Analysis of the promoter sequence revealed the presence of a putative FNR box consensus, TTGAC-N4-CTCAT, just overlapping the −35 hexamer, that differs by three nucleotides from the E. coli FNR box (TTGAT-N4- ATCAA) (49). These data suggest that FNR binds the fhbp promoter region to promote transcription and expression of fHBP protein.
FNR-dependent regulation of fHBP is common among meningococcal strains.
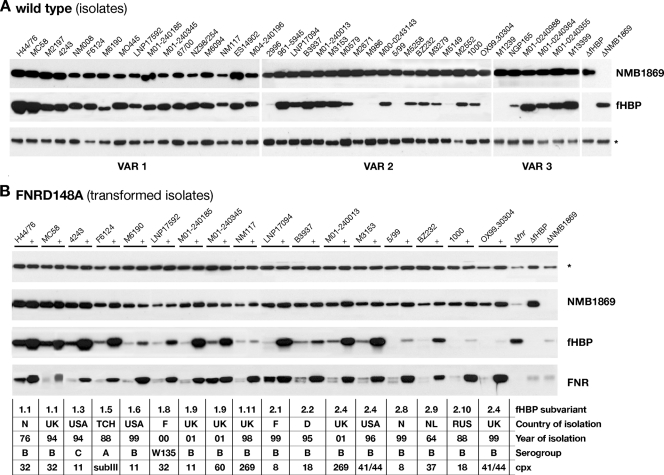
In order to investigate whether the FNR-dependent regulation of fHBP protein expression was also exhibited by other strains of meningococci, we extended our studies to a larger panel of meningococcal strains. A preliminary Western blot analysis was performed for strains expressing different fHBP variant forms (Fig. 5A). Our data showed that the fHBP antigen was expressed by all N. meningitidis strains tested, but the level of expression varied between strains (high, intermediate, and low expressers) as previously described (31), whereas the expression of the NMB1869 protein was less variable among this panel of strains.
FIG. 5.
(A) Western blot analysis of NMB1869 and fHBP protein expression in a broad panel of 40 N. meningitidis strains. Whole bacterial lysates were prepared from overnight plate cultures and subjected to immunoblot analysis with mouse polyclonal antibody against each protein. The asterisk marks a nonspecific cross-reactive band that is shown as a loading control. The fHBP protein is expressed at different levels in different strains. In this representative Western blot, it is not possible to visualize fHBP expression in some of the isolates (M2671, 5/99, OX99.30304, M1239) due to their low level of expression that was evident only with long exposure of the film (data not shown). (B) Western blot analysis of protein expression on a subset of wild-type strains and their respective FNR mutant derivative expressing the FNR(D148A) protein (+). The site-directed mutant allele of the fnr gene, fnr(D148A), was inserted under the control of the Ptac-inducible promoter in the genomes of these strains, and FNR expression is induced by growth on 1 mM IPTG. Immunoblot analysis was performed using antisera against NMB1869, fHBP, and the FNR protein. N, Norway; UK, United Kingdom; USA, United States; TCH, Chad; F, Finland; D, Denmark; NL, the Netherlands; RUS, Russia.
To understand if the fhbp genes in these N. meningitidis strains were all under FNR regulatory control, we generated isogenic strains expressing the constitutively active FNR(D148A) form by inserting the site-directed mutant allele of the fnr(D148A) gene into their genomes. The same construct used to create the MC58ΔfnrC(D148A) strain was used to transform a representative subgroup of strains. Western blot analysis was carried out for the transformed strains and their respective wild-type strains, and the results are shown in Fig. 5B. We found that the transformed strains, grown under aerobic conditions, expressed the FNR(D148A) protein at a higher level than the respective wild types, confirming the success of transformation. Importantly, the recombinant strains also exhibited an overexpression of fHBP protein. This indicates that fhbp is commonly under the control of the FNR activator protein in a large panel of strains isolated from geographically diverse origins that represent the main clonal complexes associated with disease, many of which carry fHBP variants expressed at different levels and from different variant subgroups. The only exception was represented by the NM117 strain that overexpressed the FNR protein but did not significantly overexpress the fHBP protein. We amplified and sequenced the Pfhbp promoter region from the genomes of all these strains. Analysis of the fhbp promoter sequence revealed that the FNR box sequence was perfectly conserved in all strains; however, the −10 promoter element in strain NM117 had two mutations with respect to the MC58 sequence exhibiting a TACCGC sequence, which is unlikely to act as an efficient −10 element. We speculate that the noncanonical Pfhbp promoter sequence in this strain results in lower or negligible levels of transcription in this strain even when FNR is active and conclude that the different level of fHBP expression observed among strains is not due to mutations in the FNR box.
However, taken together, these results suggest that the FNR-dependent regulation is not restricted to the fhbp gene of MC58 strain and demonstrate that the regulation of this important vaccine antigen is well conserved among diverse meningococcal isolates.
DISCUSSION
In the present study, we have analyzed the structural organization and regulation of transcription of the fhbp gene locus in N. meningitidis. We have shown that the fhbp gene and its upstream gene nmb1869 are independently transcribed from the Pfhbp and Pnmb1869 promoters and that a bicistronic mRNA can originate in vivo from the Pnmb1869 promoter. The transcriptional linking of these genes may suggest that their products could be involved in similar processes that require coregulation and coexpression. In contrast, we have shown that the FNR global regulator directly binds the Pfhbp promoter, activating fhbp transcription under oxygen limitation. The binding occurs over a putative FNR box (TTGAC-N4-CTCAT), overlapping the −35 hexamer of this promoter (Fig. 4). This is in accordance with the location of the FNR binding sequence, required to trigger promoter activation by FNR in E. coli (7, 55). In addition, we show that as with the E. coli FNR global regulator (26), a site-directed mutation in the dimerization domain of the meningococcal protein gives rise to an oxygen-insensitive FNR(D148A) mutant protein. In the background of a meningococcal strain expressing this constitutively active form, the fhbp gene was also induced in the presence of oxygen, resulting in overexpression of the fHBP protein (Fig. 3).
In N. meningitidis, FNR is essential for transcriptional activation of the aniA promoter (40) which is induced under oxygen limitation. Performing binding studies, we found that FNR binds directly to the aniA promoter, with ca. 50-fold-lower FNR concentrations than the concentration resulting in protection of the fhbp promoter. This suggests a significantly higher affinity of the FNR protein for the aniA promoter, which is agreement with it being one of the most highly FNR-regulated Neisseria genes (3, 54) and with it exhibiting a highly conserved FNR box differing by only one mismatch to the E. coli FNR consensus sequence (49). The regulation of fhbp by FNR appears to be at a more subtle level and may have more to do with a coordinated integrated response to other physiological changes required for adaptation to anaerobic growth rather than the immediate metabolic need of adapting to anaerobic conditions that is dependent on upregulation of AniA. Accordingly, Western blot analysis of the fHBP protein in MC58 total cell extracts, prepared from equivalent conditions to those used for Northern blot RNA analysis (i.e., aerobic or oxygen limitation conditions for 30 min), showed no significant differences in fHBP levels between the two oxygen conditions (data not shown) despite induction of the Pfhbp promoter. The bicistronic transcript driven from the Pnmb1869 promoter appears to be downregulated under these conditions, although in an FNR-independent manner (Fig. 3). Therefore, the cumulative effect of expression of fHBP from both differentially regulated transcripts may not give rise to a prompt differential protein expression, while the Pfhbp promoter ensures that high levels of fHBP expression are maintained.
To persist and cause disease in the human host, N. meningitidis must encounter and survive numerous complex extracellular and intracellular environments. N. meningitidis is able to live in anaerobic niches as part of its normal transmission-colonization and infectious cycles within the host. It has been shown that although N. meningitidis fails to grow under strictly anaerobic conditions, under oxygen limitation the bacterium expresses a denitrification pathway that supplements growth (1, 39). The nasopharynx can generally be thought of as an environment of fluctuating oxygen concentrations, as both strict aerobes and strict anaerobes are routinely isolated from this niche (9). N. meningitidis would be exposed to highly divergent partial pressures of oxygen as the bacteria move from the surfaces of the upper respiratory tract (21 kPa) to mucus membranes (0.4 kPa buccal fold pressure), blood (4 kPa central venous pressure), and cerebrospinal fluid (5 kPa) (2). Pathogenic Neisseria species, in a similar manner to that of E. coli, use the FNR global transcription factor to control these responses, in particular to induce the denitrification and sugar fermentation pathways (3) as an alternative to aerobic respiration. To date, only a limited number of genes have been identified as being responsive to the FNR regulator, a total of 9 transcriptional units in meningococci (3) and 14 genes in the closely related pathogenic gonococci (54). However, the finding that the FNR mutant is attenuated in the mouse and infant rat animal models indicates the importance of these responses to the virulence and pathogenesis of the organism (3).
This study (Fig. 5) and other studies (31, 33) have shown that the level of expression of the fHBP protein is very variable in diverse strains and cannot be correlated with the allele expressed or the genetic (clonal complex) or geographic association of the strain. Therefore, the question still remains as to what influences the ability of a strain to express fHBP. We hypothesize that this could be multifactorial. Obviously, fHBP variants with different amino acid sequences may have differential posttranslational effects, such as protein stability and the efficiency of secretion and lipidation on the bacterial outer membrane. From this study, we underpin the molecular factors which influence fHBP expression at the level of transcription in MC58 and can expand this to different strains. Our data show that the expression of the nmb1869-encoded protein (which is highly conserved at the amino acid level) is largely unchanged between strains, while fHBP exhibits diverse expression levels. Mutations in the nmb1869-fhbp intergenic region are likely to account for at least some of these differences. For instance, mutations in the sequences regulating initiation, regulation, or indeed termination of transcription from this locus will affect fHBP expression levels. From our analyses of strains MC58 and H44/76, the imperfect palindromic stem-loop of this terminator carries two mismatches and allows read-through of a bicistronic transcript. In the available sequences from diverse strains, we have identified terminators with only one mismatch (FAM18) (8) or up to three mismatches (NM117) in the stem-loops of this nucleotide feature. A similar element, with a perfect stem-loop downstream of the fhbp gene, functions very efficiently as a terminator in vivo, as no read-through can be detected downstream of this element by either RT-PCR or Northern blot analysis. We can predict that mutations in the rho-independent terminator downstream of the nmb1869 gene will influence the amount of the bicistronic fhbp transcript due to the Pnmb1869 upstream promoter. Furthermore, mutations in the regulatory sequences of the Pfhbp promoter, such as the mutation we observed in the −10 element of the Pfhbp promoter in strain NM117, may influence the strength of this promoter and hence the levels of expression of the monocistronic fhbp transcription. Alternatively, in the Z2491 genome there is an insertion of a 181-bp AT-rich nucleotide sequence (ATR) (37) that has similarities to an insertion element fragment at position +16 with respect to the Pfhbp promoter. This sequence may affect fHBP expression in this strain either transcriptionally (by containing promoter or terminator regulatory sequences) or posttranscriptionally, as it will be transcribed in both fhbp transcripts and may affect mRNA stability. Interestingly, in the sequences available to date, the FNR box element is conserved 100% between different strains, and our data indicate that fHBP is regulated by FNR in a representative panel of strains (Fig. 5). This implicates a role for the fHBP protein under conditions in which FNR would be active, i.e., under anaerobiosis or oxygen limitation.
The role of fHBP in binding the factor H fluid phase regulator of the complement AP, enabling the meningococcus to avoid complement-mediated killing in blood, has been well documented (29, 43, 46, 52). However, the action of complement must not be thought of as being limited to the bloodstream. Complement is important during the early stages of infection, and complement components are present on mucosal surfaces (34). Therefore, the expression of fHBP by the meningococcus in order to adsorb factor H to its surface, thereby circumventing complement, may be important also in the initial stages of colonization and invasion before infiltrating the tissues. Furthermore, the fHBP protein has an additional role in conferring resistance to the antimicrobial peptide LL-37 (46) that is constitutively expressed by leukocytes and inducibly expressed by epithelial cells (14), including the epithelial cells of the nasopharynx (28). In particular, the colonization of the human nasopharynx and the passage through the mucosa epithelium, which are the initial steps in the pathogenesis of disease due to N. meningitidis, could already expose the microorganism to different host innate immune defenses. Crossing the mucosal barrier, the pathogen may be presumed to reach an oxygen-deficient environment compared to the well-aerated upper mucosa, sense a change in oxygen concentration, and adapt rapidly to the new conditions, expressing alternative mechanisms of defense for survival. Our data suggest that in a wide panel of strains, regulatory factors are at work to ensure the expression of fHBP under these conditions where it may play an important role in resistance to disparate innate immune mechanisms. Therefore, vaccines targeting this antigen and its important virulence factor are doubly attractive, as they may also be effective in reducing carriage.
Acknowledgments
We thank Stefania Bambini, Sara Comandi, and Maurizio Comanducci for providing the meningococcal isolates used in this study. We thank the MenB Serology Group, Novartis Vaccines, for providing anti-fHBP and anti-NMB1869 antibodies. We thank Giorgio Corsi for artwork and Kate Seib for critical reading of the manuscript.
F.O. is the recipient of a Novartis fellowship from the Ph.D. program in evolutionary biology of the University of Siena, Siena, Italy.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Anjum, M. F., T. M. Stevanin, R. C. Read, and J. W. Moir. 2002. Nitric oxide metabolism in Neisseria meningitidis. J. Bacteriol. 184:2987-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archibald, F. S., and M. N. Duong. 1986. Superoxide dismutase and oxygen toxicity defenses in the genus Neisseria. Infect. Immun. 51:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartolini, E., E. Frigimelica, S. Giovinazzi, G. Galli, Y. Shaik, C. Genco, J. A. Welsch, D. M. Granoff, G. Grandi, and R. Grifantini. 2006. Role of FNR and FNR-regulated, sugar fermentation genes in Neisseria meningitidis infection. Mol. Microbiol. 60:963-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beernink, P. T., and D. M. Granoff. 2008. Bactericidal antibody responses induced by meningococcal recombinant chimeric factor H-binding protein vaccines. Infect. Immun. 76:2568-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beernink, P. T., C. LoPasso, A. Angiolillo, F. Felici, and D. Granoff. 2009. A region of the N-terminal domain of meningococcal factor H-binding protein that elicits bactericidal antibody across antigenic variant groups. Mol. Immunol. 46:1647-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beernink, P. T., J. A. Welsch, M. Bar-Lev, O. Koeberling, M. Comanducci, and D. M. Granoff. 2008. Fine antigenic specificity and cooperative bactericidal activity of monoclonal antibodies directed at the meningococcal vaccine candidate factor H-binding protein. Infect. Immun. 76:4232-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell, A. I., J. A. Cole, and S. J. Busby. 1990. Molecular genetic analysis of an FNR-dependent anaerobically inducible Escherichia coli promoter. Mol. Microbiol. 4:1753-1763. [DOI] [PubMed] [Google Scholar]
- 8.Bentley, S. D., G. S. Vernikos, L. A. Snyder, C. Churcher, C. Arrowsmith, T. Chillingworth, A. Cronin, P. H. Davis, N. E. Holroyd, K. Jagels, M. Maddison, S. Moule, E. Rabbinowitsch, S. Sharp, L. Unwin, S. Whitehead, M. A. Quail, M. Achtman, B. Barrell, N. J. Saunders, and J. Parkhill. 2007. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet. 3:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brook, I. 2003. Effects of antimicrobial therapy on the microbial flora of the adenoids. J. Antimicrob. Chemother. 51:1331-1337. [DOI] [PubMed] [Google Scholar]
- 10.Cantini, F., D. Veggi, S. Dragonetti, S. Savino, M. Scarselli, G. Romagnoli, M. Pizza, L. Banci, and R. Rappuoli. 2009. Solution structure of the factor H-binding protein, a survival factor and protective antigen of Neisseria meningitidis. J. Biol. Chem. 284:9022-9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delany, I., R. Grifantini, E. Bartolini, R. Rappuoli, and V. Scarlato. 2006. Effect of Neisseria meningitidis fur mutations on global control of gene transcription. J. Bacteriol. 188:2483-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delany, I., A. B. Pacheco, G. Spohn, R. Rappuoli, and V. Scarlato. 2001. Iron-dependent transcription of the frpB gene of Helicobacter pylori is controlled by the Fur repressor protein. J. Bacteriol. 183:4932-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delany, I., R. Rappuoli, and V. Scarlato. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52:1081-1090. [DOI] [PubMed] [Google Scholar]
- 14.Dürr, U. H., U. S. Sudheendra, and A. Ramamoorthy. 2006. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim. Biophys. Acta 1758:1408-1425. [DOI] [PubMed] [Google Scholar]
- 15.Finne, J., M. Leinonen, and P. H. Makela. 1983. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet ii:355-357. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher, L. D., L. Bernfield, V. Barniak, J. E. Farley, A. Howell, M. Knauf, P. Ooi, R. P. Smith, P. Weise, M. Wetherell, X. Xie, R. Zagursky, Y. Zhang, and G. W. Zlotnick. 2004. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect. Immun. 72:2088-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fredriksen, J. H., E. Rosenqvist, E. Wedege, K. Bryn, G. Bjune, L. O. Froholm, A. K. Lindbak, B. Mogster, E. Namork, U. Rye, et al. 1991. Production, characterization and control of MenB vaccine “Folkehelsa”: an outer membrane vesicle vaccine against group B meningococcal disease. NIPH Ann. 14:67-79. [PubMed] [Google Scholar]
- 18.Giuliani, M. M., L. Santini, B. Brunelli, A. Biolchi, B. Arico, F. Di Marcello, E. Cartocci, M. Comanducci, V. Masignani, L. Lozzi, S. Savino, M. Scarselli, R. Rappuoli, and M. Pizza. 2005. The region comprising amino acids 100 to 255 of Neisseria meningitidis lipoprotein GNA 1870 elicits bactericidal antibodies. Infect. Immun. 73:1151-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granoff, D. M., J. A. Welsch, and S. Ram. 2009. Binding of complement factor H (fH) to Neisseria meningitidis is specific for human fH and inhibits complement activation by rat and rabbit sera. Infect. Immun. 77:764-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 21.Holst, J., B. Feiring, L. M. Naess, G. Norheim, P. Kristiansen, E. A. Hoiby, K. Bryn, P. Oster, P. Costantino, M. K. Taha, J. M. Alonso, D. A. Caugant, E. Wedege, I. S. Aaberge, R. Rappuoli, and E. Rosenqvist. 2005. The concept of “tailor-made,” protein-based, outer membrane vesicle vaccines against meningococcal disease. Vaccine 23:2202-2205. [DOI] [PubMed] [Google Scholar]
- 22.Hou, V. C., O. Koeberling, J. A. Welsch, and D. M. Granoff. 2005. Protective antibody responses elicited by a meningococcal outer membrane vesicle vaccine with overexpressed genome-derived neisserial antigen 1870. J. Infect. Dis. 192:580-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ieva, R., C. Alaimo, I. Delany, G. Spohn, R. Rappuoli, and V. Scarlato. 2005. CrgA is an inducible LysR-type regulator of Neisseria meningitidis, acting both as a repressor and as an activator of gene transcription. J. Bacteriol. 187:3421-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kellogg, D. S., Jr., W. L. Peacock, Jr., W. E. Deacon, L. Brown, and D. I. Pirkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koeberling, O., S. Giuntini, A. Seubert, and D. M. Granoff. 2009. Meningococcal outer membrane vesicle vaccines derived from mutant strains engineered to express factor H binding proteins from antigenic variant groups 1 and 2. Clin. Vaccine Immunol. 16:156-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazazzera, B. A., D. M. Bates, and P. J. Kiley. 1993. The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 7:1993-2005. [DOI] [PubMed] [Google Scholar]
- 27.Lissenden, S., S. Mohan, T. Overton, T. Regan, H. Crooke, J. A. Cardinale, T. C. Householder, P. Adams, C. D. O'Conner, V. L. Clark, H. Smith, and J. A. Cole. 2000. Identification of transcription activators that regulate gonococcal adaptation from aerobic to anaerobic or oxygen-limited growth. Mol. Microbiol. 37:839-855. [DOI] [PubMed] [Google Scholar]
- 28.Lysenko, E. S., J. Gould, R. Bals, J. M. Wilson, and J. N. Weiser. 2000. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect. Immun. 68:1664-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madico, G., J. A. Welsch, L. A. Lewis, A. McNaughton, D. H. Perlman, C. E. Costello, J. Ngampasutadol, U. Vogel, D. M. Granoff, and S. Ram. 2006. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J. Immunol. 177:501-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mascioni, A., J. Jacob, F. Moy, D. Dilts, P. Fink, K. Malakian, S. Sigethy, Y. Wen, E. Novikova, G. W. Zlotnick, and D. H. Tsao. 2009. Backbone and side-chain assignment of the lipidated and non-lipidated forms of the meningococcal outer membrane protein LP2086. Biomol. NMR Assign. 3:111-113. [DOI] [PubMed] [Google Scholar]
- 31.Masignani, V., M. Comanducci, M. M. Giuliani, S. Bambini, J. Adu-Bobie, B. Arico, B. Brunelli, A. Pieri, L. Santini, S. Savino, D. Serruto, D. Litt, S. Kroll, J. A. Welsch, D. M. Granoff, R. Rappuoli, and M. Pizza. 2003. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J. Exp. Med. 197:789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. U. S. A. 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNeil, L. K., E. Murphy, X. J. Zhao, S. Guttmann, S. L. Harris, A. A. Scott, C. Tan, M. Mack, I. DaSilva, K. Alexander, K. Mason, H. Q. Jiang, D. Zhu, T. L. Mininni, G. W. Zlotnick, S. K. Hoiseth, T. R. Jones, M. W. Pride, K. U. Jansen, and A. S. Anderson. 2009. Detection of LP2086 on the cell surface of Neisseria meningitidis and its accessibility in the presence of serogroup B capsular polysaccharide. Vaccine 27:3417-3421. [DOI] [PubMed] [Google Scholar]
- 34.Meri, S., M. Jordens, and H. Jarva. 2008. Microbial complement inhibitors as vaccines. Vaccine 26(Suppl. 8):I113-I117. [DOI] [PubMed] [Google Scholar]
- 35.Ngampasutadol, J., S. Ram, S. Gulati, S. Agarwal, C. Li, A. Visintin, B. Monks, G. Madico, and P. A. Rice. 2008. Human factor H interacts selectively with Neisseria gonorrhoeae and results in species-specific complement evasion. J. Immunol. 180:3426-3435. [DOI] [PubMed] [Google Scholar]
- 36.Overton, T. W., L. Griffiths, M. D. Patel, J. L. Hobman, C. W. Penn, J. A. Cole, and C. Constantinidou. 2006. Microarray analysis of gene regulation by oxygen, nitrate, nitrite, FNR, NarL and NarP during anaerobic growth of Escherichia coli: new insights into microbial physiology. Biochem. Soc. Trans. 34:104-107. [DOI] [PubMed] [Google Scholar]
- 37.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 38.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Arico, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 39.Rock, J. D., M. R. Mahnane, M. F. Anjum, J. G. Shaw, R. C. Read, and J. W. Moir. 2005. The pathogen Neisseria meningitidis requires oxygen, but supplements growth by denitrification. Nitrite, nitric oxide and oxygen control respiratory flux at genetic and metabolic levels. Mol. Microbiol. 58:800-809. [DOI] [PubMed] [Google Scholar]
- 40.Rock, J. D., M. J. Thomson, R. C. Read, and J. W. Moir. 2007. Regulation of denitrification genes in Neisseria meningitidis by nitric oxide and the repressor NsrR. J. Bacteriol. 189:1138-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, T. Popovic, and J. M. Hughes. 2001. Meningococcal disease. N. Engl. J. Med. 344:1378-1388. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Schneider, M. C., R. M. Exley, H. Chan, I. Feavers, Y. H. Kang, R. B. Sim, and C. M. Tang. 2006. Functional significance of factor H binding to Neisseria meningitidis. J. Immunol. 176:7566-7575. [DOI] [PubMed] [Google Scholar]
- 44.Schneider, M. C., R. M. Exley, S. Ram, R. B. Sim, and C. M. Tang. 2007. Interactions between Neisseria meningitidis and the complement system. Trends Microbiol. 15:233-240. [DOI] [PubMed] [Google Scholar]
- 45.Schneider, M. C., B. E. Prosser, J. J. Caesar, E. Kugelberg, S. Li, Q. Zhang, S. Quoraishi, J. E. Lovett, J. E. Deane, R. B. Sim, P. Roversi, S. Johnson, C. M. Tang, and S. M. Lea. 2009. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature 458:890-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seib, K. L., D. Serruto, F. Oriente, I. Delany, J. Adu-Bobie, D. Veggi, B. Arico, R. Rappuoli, and M. Pizza. 2009. Factor H-binding protein is important for meningococcal survival in human whole blood and serum and in the presence of the antimicrobial peptide LL-37. Infect. Immun. 77:292-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sierra, G. V., H. C. Campa, N. M. Varcacel, I. L. Garcia, P. L. Izquierdo, P. F. Sotolongo, G. V. Casanueva, C. O. Rico, C. R. Rodriguez, and M. H. Terry. 1991. Vaccine against group B Neisseria meningitidis: protection trial and mass vaccination results in Cuba. NIPH Ann. 14:195-207. [PubMed] [Google Scholar]
- 48.Smith, H. O., M. L. Gwinn, and S. L. Salzberg. 1999. DNA uptake signal sequences in naturally transformable bacteria. Res. Microbiol. 150:603-616. [DOI] [PubMed] [Google Scholar]
- 49.Spiro, S., and J. R. Guest. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 6:399-428. [DOI] [PubMed] [Google Scholar]
- 50.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 51.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 52.Welsch, J. A., S. Ram, O. Koeberling, and D. M. Granoff. 2008. Complement-dependent synergistic bactericidal activity of antibodies against factor H-binding protein, a sparsely distributed meningococcal vaccine antigen. J. Infect. Dis. 197:1053-1061. [DOI] [PubMed] [Google Scholar]
- 53.Welsch, J. A., R. Rossi, M. Comanducci, and D. M. Granoff. 2004. Protective activity of monoclonal antibodies to genome-derived neisserial antigen 1870, a Neisseria meningitidis candidate vaccine. J. Immunol. 172:5606-5615. [DOI] [PubMed] [Google Scholar]
- 54.Whitehead, R. N., T. W. Overton, L. A. Snyder, S. J. McGowan, H. Smith, J. A. Cole, and N. J. Saunders. 2007. The small FNR regulon of Neisseria gonorrhoeae: comparison with the larger Escherichia coli FNR regulon and interaction with the NarQ-NarP regulon. BMC Genomics 8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wing, H. J., S. M. Williams, and S. J. Busby. 1995. Spacing requirements for transcription activation by Escherichia coli FNR protein. J. Bacteriol. 177:6704-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, D., Y. Zhang, V. Barniak, L. Bernfield, A. Howell, and G. Zlotnick. 2005. Evaluation of recombinant lipidated P2086 protein as a vaccine candidate for group B Neisseria meningitidis in a murine nasal challenge model. Infect. Immun. 73:6838-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]