Abstract
Reduced ferredoxin is an intermediate in the methylotrophic and aceticlastic pathway of methanogenesis and donates electrons to membrane-integral proteins, which transfer electrons to the heterodisulfide reductase. A ferredoxin interaction has been observed previously for the Ech hydrogenase. Here we present a detailed analysis of a Methanosarcina mazei Δech mutant which shows decreased ferredoxin-dependent membrane-bound electron transport activity, a lower growth rate, and faster substrate consumption. Evidence is presented that a second protein whose identity is unknown oxidizes reduced ferredoxin, indicating an involvement in methanogenesis from methylated C1 compounds.
The aceticlastic pathway of methanogenesis creates approximately 70% (10) of the biologically produced methane and is of great ecological importance, as methane is a potent greenhouse gas. Organisms using this pathway to convert acetate to methane belong exclusively to the genera Methanosarcina and Methanosaeta. The two carbon atoms of acetate have different fates in the pathway. The methyl moiety is converted to methane, whereas the carbonyl moiety is further oxidized to CO2 and the electrons derived from this oxidation step are used to reduce ferredoxin (Fd) (6). During methanogenesis from methylated C1 compounds (methanol and methylamines), one-quarter of the methyl groups are oxidized to obtain electrons for the reduction of heterodisulfide (27). A key enzyme in the oxidative part of methylotrophic methanogenesis is the formylmethanofuran dehydrogenase, which oxidizes the intermediate formylmethanofuran to CO2 (7). The electrons are transferred to Fd. It has been suggested that reduced ferredoxin (Fdred) donates electrons to the respiratory chain with the heterodisulfide (coenzyme M [CoM]-S-S-CoB) as the terminal electron acceptor and that the reaction is catalyzed by the Fdred:CoM-S-S-CoB oxidoreductase system (7, 24). The direct membrane-bound electron acceptor for Fdred is still a matter of debate; for the Ech hydrogenase, a reduced ferredoxin-accepting, H2-evolving activity has been observed for Methanosarcina barkeri (20), which implies that the H2:CoM-S-S-CoB oxidoreductase system is involved in electron transport (13). Direct electron flow from the Ech hydrogenase to the heterodisulfide reductase has not been shown to date (20, 21). In contrast to M. barkeri, Methanosarcina acetivorans lacks the Ech hydrogenase (11). It can nevertheless grow on acetate, which is why another complex present in this organism, the Rnf complex, is thought to be involved in the aceticlastic pathway of methanogenesis as an acceptor for Fdred (8, 10, 17). The Methanosarcina mazei genome, however, contains genes coding for the Ech hydrogenase, but this species lacks the Rnf complex (5).
To investigate whether the Ech hydrogenase is the only means by which M. mazei channels electrons from Fdred into the respiratory chain, a mutant lacking the Ech hydrogenase (M. mazei Δech mutant) was constructed. Electron transport experiments using Fdred as the electron donor and CoM-S-S-CoB as the electron acceptor were conducted with wild-type and mutant membranes to gain deeper insight into the actual membrane-bound protein complexes that accept electrons from Fdred. Furthermore, an in-depth characterization of the growth and trimethylamine (TMA) consumption of the Δech mutant was performed, which provided insight into the in vivo role of Ech hydrogenase.
MATERIALS AND METHODS
Purification of proteins.
Ferredoxin was purified from Clostridium pasteurianum DSM 525T essentially as described by Mortenson (22). The last steps (dialysis and crystallization) were replaced by ultrafiltration. The ferredoxin content was determined using the bicinchoninic acid method (26). CO dehydrogenase/acetyl-CoA synthase (CODH/ACS) was purified from Moorella thermoacetica ATCC 39073 as described by Ragsdale et al. (25), except for the following modifications. The extract was not heat treated or fractionated with ammonium sulfate. The first DEAE-cellulose column was eluted with a step gradient consisting of 0.1 to 0.5 M NaCl. CODH/ACS was then purified further using Q-Sepharose anion-exchange and phenyl-Sepharose hydrophobic interaction columns. Active fractions were concentrated and buffer exchanged into 50 mM Tris-HCl (pH 7.6) using Amicon ultracentrifuge concentrators in an anaerobic chamber (Vacuum Atmospheres). Immediately after purification, the enzyme had a CO oxidation-methyl viologen reduction specific activity of 273 U/mg at 37°C.
Membrane preparations.
M. mazei DSM 7222 and derivatives of this strain were grown anaerobically at 37°C in Methanosarcina medium (DSM medium 120) with TMA as the substrate (12). Subsequent steps were performed anaerobically in an anaerobic chamber (Coy Laboratory Products, United States) under a 97% N2-3% H2 atmosphere. In the late exponential phase the cells were harvested and lysed by resuspending them in phosphate buffer (40 mM KH2PO4/K2HPO4 [pH 7.0], 5 mM dithioerythritol [DTE], 1 μg ml−1 resazurin) and incubated with DNase I for 30 to 60 min at 4°C. The cell lysate was ultracentrifuged (1 h, 45,000 rpm), the supernatant was discarded, and the membrane pellet was homogenized in 20 mM phosphate buffer (KH2PO4/K2HPO4, pH 7.0) containing 20 mM MgSO4, 500 mM sucrose, 5 mM DTE, and 1 μg ml−1 resazurin. The ultracentrifugation step was repeated, the supernatant was discarded, and the membrane pellet was homogenized in the buffer described above. The protein content was determined using the Bradford method (3).
Enzyme assays.
The photometric tests were performed anaerobically in 1.5-ml glass cuvettes with rubber stoppers using a V-550 UV/visible spectrophotometer (Jasco, Germany). Benzylviologen-dependent heterodisulfide reductase activity was determined by the decrease in absorption at 575 nm using 600 μl phosphate buffer [40 mM KH2PO4/K2HPO4 [pH 7.0] reduced with Ti(III) citrate] containing 625 nmol benzylviologen (ɛ575 = 8.9 mM−1 cm−1), 300 nmol Na2S2O4, 50 μg M. mazei membrane, and 50 nmol CoM-S-S-CoB.
F420H2 dehydrogenase activity was determined by determining the increase in absorption at 420 nm using 600 μl phosphate buffer (40 mM KH2PO4/K2HPO4 [pH 7.0], 5 mM DTE) containing 15 nmol F420H2 (ɛ420 = 40 mM−1 cm−1), 50 μg M. mazei membrane, 300 nmol metronidazole, and 180 nmol methylviologen (18). F420 was isolated and reduced as described by Abken et al. (1). One unit of activity was defined as 1 μmol electrons transported per min. CoM-S-S-CoB (synthesized as described previously [9, 23]) was quantified in anaerobic glass vials with rubber stoppers containing 250 μl phosphate buffer [40 mM phosphate and 1 μg ml−1 resazurin, reduced with Ti(III) citrate] flushed with N2-CO (5% CO [purity, 1.8] and 95% N2 [purity, 5.0]) for 1 min. For analysis of CoM-S-S-CoB reduction, 100 nmol CoM-S-S-CoB, 8.9 μg C. pasteurianum ferredoxin, 150 μg M. mazei membrane, and 75 μg M. thermoacetica CO dehydrogenase/acetyl-CoA synthase were added. For CoM-SH/CoB-SH quantification (28) 20-μl samples were taken every 10 min for 1 h and directly used in a modified Ellman's assay; 950 μl Tris buffer (150 mM, pH 8.1) was mixed with the sample and 100 μl Ellman's reagent [5 mM 5,5′-dithiobis(2-nitrobenzoic acid (ɛ412 = 13.6 mM−1 cm−1] in 50 mM sodium acetate buffer, pH 5.0], and the absorption at 412 nm was immediately measured.
Alternatively, H2:heterodisulfide oxidoreductase activity was determined by replacing CO, CO dehydrogenase/acetyl-CoA synthase, and ferredoxin with H2. In this analysis, only 50 μg membrane preparation was used, and 1 U of activity was defined as the reduction of 1 μmol CoM-S-S-CoB per min.
Generation of the M. mazei Δech mutant.
The M. mazei Δech mutant was generated by homologous recombination using the techniques described by Metcalf et al. (19). Up- and downstream regions of the ech gene region were cloned into the two multiple cloning sites of pJK3 (19), linearized with ApaI, and transformed into M. mazei. Instead of the gene, a puromycin resistance (pac) cassette was inserted. Puromycin-resistant colonies were picked and screened for gene knockout. Gene knockout was confirmed by sequencing and Southern blotting. Homologous recombination led to deletion of echA to echE (mm2320 to mm2324) and part of echF (mm2325).
For construction of the M. mazei Δech mutant, primers 5′-CCTACTCGAGGAGTGAATCAGCGAATAGAG-3′ and 5′-GACCGAATTCACAACGTATCCTCCGACCTA-3′ (upstream region of mm2320) with XhoI and EcoRI restriction sites and primers 5′-CCAGGAGCTCGGTATGCTAAACCTTGTATT-3′ and 5′-CAAGGGATCCCACTACAAATGTTGTCTCC-3′ (mm2325 and downstream region) with SacI and BamHI restriction sites were used.
Growth analysis and substrate quantification.
The M. mazei wild type and the Δech mutant were grown as described above in 50-ml cultures with TMA as the growth substrate. Growth of the Δech mutant was also observed with methanol, TMA plus H2, or TMA plus H2 and CO2, but not with acetate as the substrate. The optical density at 600 nm (OD600) was measured with a Helios Epsilon Vis photometer (Thermo Scientific) in the presence of sodium dithionite. At different time points 700-μl samples were taken from TMA-grown cultures. Cells and medium were separated by silicone oil centrifugation (100 μl silicone oil, 5 min, 14,000 rpm) to thoroughly separate cells from the medium.
TMA, dimethylamine (DMA), and monomethylamine (MMA) were quantified as described by Krätzer et al. (15). The concentrations of these compounds were measured in triplicate, and the results were compared to standard curves.
RESULTS AND DISCUSSION
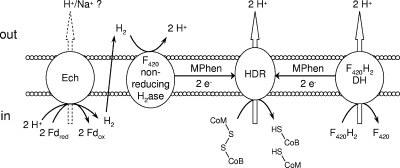
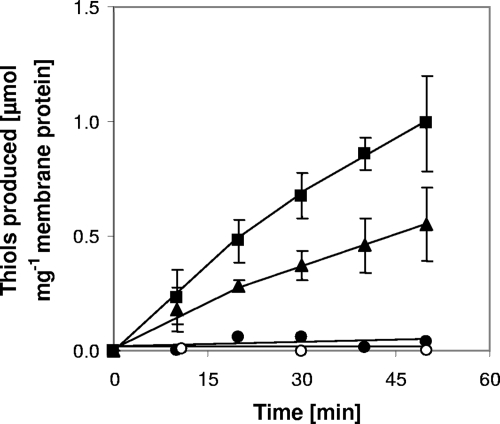
To investigate ferredoxin-mediated membrane-bound electron transport in M. mazei, an in vitro assay with washed membrane preparations of the M. mazei wild-type and Δech mutant strains grown on trimethylamine was performed. In this assay, electrons were transferred from the initial substrate CO to M. thermoacetica CO dehydrogenase/acetyl-CoA synthase (CODH/ACS), which reduces C. pasteurianum ferredoxin (Fd). Fd donates electrons to different membrane-bound complexes (described in detail below), which are predicted to reduce methanophenazine (1). Methanophenazine is the electron donor for the cytochrome b subunit of the heterodisulfide reductase (Fig. 1), which reduces the terminal electron acceptor CoM-S-S-CoB (4). The activity of the Fdred:heterodisulfide oxidoreductase system, described by the following equation, was about 24 mU/mg membrane protein and was confirmed with three independent membrane preparations obtained from different M. mazei cultures (Fig. 2): 2Fdred + CoM-S-S-CoM + 2H+ → 2Fdox + HS-CoM + HS-CoB. This value was similar to the values obtained by Peer (24), who obtained an activity of 23 mU/mg for the Methanosarcina thermophila CO:CoM-S-S-CoB oxidoreductase system. The apparent requirement for Fd in the M. mazei system makes it a suitable tool to investigate Fd-dependent reduction of heterodisulfide. CoM-S-S-CoB reduction was clearly dependent on the presence of CO, CODH/ACS, Fd, and membranes. If one component was omitted, heterodisulfide-reducing activity could not be measured (Fig. 2). Chemical degradation of heterodisulfide could not be observed in the reaction mixtures.
FIG. 1.
Proposed model of membrane-bound electron transfer in M. mazei. Proton translocation has been shown for the heterodisulfide reductase (14) and F420H2 dehydrogenase (2). Proton translocation for the Ech hydrogenase is assumed. H2ase, hydrogenase; DH, dehydrogenase; HDR, heterodisulfide reductase; MPhen, methanophenazine.
FIG. 2.
Fd-dependent heterodisulfide reduction with wild-type and Δech mutant cytoplasmic membranes. The assay mixtures (total volume, 250 μl) contained 100 nmol CoM-S-S-CoB, 8.9 μg Fd, 150 μg membrane, and 75 μg CODH under an atmosphere consisting of N2 (95%) and CO (5%). ▪, wild-type membranes; ▴, membranes from Δech mutant; •, control without Fd; ○, control without CO (100% N2). Other control experiments (omission of CO dehydrogenase, membranes, or heterodisulfide) were performed, and it was found that heterodisulfide reduction was abolished.
According to the current model (Fig. 1), Fdred produced in the aceticlastic pathway, as well as in the methylotrophic pathway, is oxidized by the Ech hydrogenase, which releases molecular hydrogen (20). The Ech hydrogenase is membrane bound and consists of six subunits (EchA to EchF). Hydrogen produced in the first reaction is reoxidized by the F420 nonreducing hydrogenase (Fig. 1), and electrons are subsequently shuttled to the heterodisulfide reductase, which reduces CoM-S-S-CoB (14). In M. barkeri, Fdred-oxidizing and H2-evolving activities have been observed for the purified Ech hydrogenase, which supports the hypothesis described above (20). The coupling of Ech hydrogenase activity and CoM-S-S-CoB reduction, however, has not been shown to date. To analyze this process in more detail, we constructed an M. mazei Δech mutant and used the membrane fraction in the CoM-S-S-CoB reduction assays. The activity of the M. mazei Δech mutant membrane fraction was about 50% of the activity of the wild-type membrane fraction (Fig. 2). The decrease in activity confirms that in M. mazei the Ech hydrogenase also accepts electrons from Fdred. However, the Fdred-oxidizing activity was not completely absent in the M. mazei Δech mutant and was one-half of the wild-type activity. Hence, it is tempting to speculate that there is at least one membrane-bound protein that is different from the Ech hydrogenase and accepts electrons from Fdred, allowing the cell to obtain energy by electron transport phosphorylation during aceticlastic growth as well as methylotrophic growth. In M. acetivorans, genes for the Ech hydrogenase are not present, but the organism can nevertheless grow on acetate, in which Fdred oxidation by a membrane-bound protein complex is crucial. Proteome analysis suggested that another membrane-bound complex, referred to as Rnf, is involved in acetate metabolism and hence in the respiratory chain and the oxidation of Fdred (16, 17). In contrast, M. mazei possesses an Ech hydrogenase, but the genomic data indicate that the Rnf complex is not present.
To ensure correct functioning of the Fd-independent parts of the respiratory chain (Fig. 1), the H2:heterodisulfide reductase activity assay, benzylviologen-dependent heterodisulfide reductase assay, and F420H2 dehydrogenase activity assay were performed with the wild-type strain and deletion mutant membrane preparations. The H2:heterodisulfide reductase exhibited an activity of approximately 150 mU/mg protein in the wild type and in the ech mutant, indicating that there was a fivefold increase compared to the CO system. The higher activity of the H2:CoM-S-S-CoB oxidoreductase system probably resulted from limited interaction of the nonnative reaction components with the Ech hydrogenase in our test system. Also, the benzylviologen-dependent heterodisulfide reductase and F420H2 dehydrogenase activities were similar for all M. mazei membranes tested. Hence, deletion of the Ech hydrogenase had no influence on the H2- and F420H2-dependent electron transport systems in M. mazei (Fig. 1).
The M. mazei Δech mutant did not grow on acetate, as observed previously by Meuer et al. (21) for an M. barkeri Δech mutant. When trimethylamine (TMA) was used as a growth substrate, distinct differences in growth behavior were observed. TMA is demethylated stepwise to dimethylamine (DMA) and monomethylamine (MMA), portions of which are excreted into the growth medium. DMA occurs only as an intermediate product at very low concentrations in the culture supernatant, whereas MMA accumulates and is used only when TMA and DMA are completely consumed (15). The respective methyl groups are transferred to CoM via substrate-specific methyl transferases, yielding methyl-CoM (15). Three-quarters of the methyl moieties are reduced to CH4, leading to the formation of heterodisulfide (CoM-S-S-CoB) (Fig. 1). One-quarter of the methyl-CoM is oxidized to CO2, yielding reducing equivalents or electrons that are transferred to F420 and ferredoxin, respectively (27) (Fig. 1). F420H2 is oxidized by the membrane-bound F420H2 dehydrogenase (Fig. 1), whereas Fdred is recycled by the Fdred:heterodisulfide oxidoreductase system, which is Ech independent in the Δech mutant.
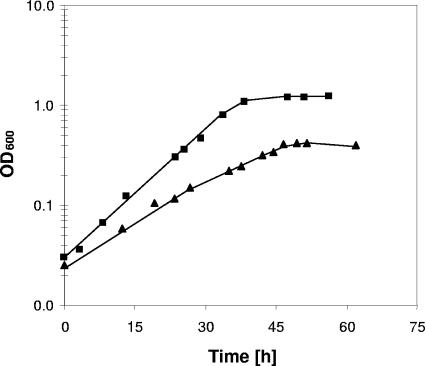
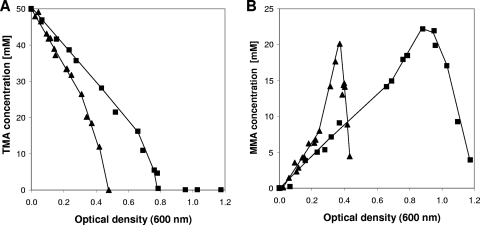
When growth of the M. mazei wild-type strain with TMA as the substrate was compared with growth of the Δech mutant with the same substrate, apparent doubling times of 7.7 h for the wild type and 9.1 h for the Δech mutant were observed. Furthermore, the wild type grew to an OD600 of 1.2, whereas the Δech mutant stopped growing at an OD600 of around 0.5 (Fig. 3). The TMA consumption rate of the Δech mutant was twofold higher than that of the wild type; TMA was completely consumed at an OD600 of 0.48 for the Δech mutant and at an OD600 of 0.80 for the wild type (Fig. 4A). As mentioned above, MMA accumulates in the culture supernatant as a product from TMA and DMA breakdown and is used only when TMA and DMA are completely metabolized. Thus, the MMA concentration in the culture supernatant first increases to a certain level and then decreases. The MMA formation rate was 2.7-fold higher in the Δech mutant than in the wild type. The MMA degradation rate showed the same tendency; it was 2.5-fold higher in the Δech mutant than in the wild type (Fig. 4B).
FIG. 3.
Growth curves for the wild type (▪) and Δech mutant (▴) during growth on TMA.
FIG. 4.
Substrate utilization by the wild type (▪) and the Δech mutant (▴) during growth with 50 mM TMA as the substrate. (A) TMA consumption. (B) MMA production and consumption.
In summary, the electron transport deficiency of the Δech mutant is reflected in its growth behavior, as it grows slower but uses more substrate to generate less biomass. Taken together, the data indicate that there is a deficient energy coupling site in the Δech mutant; thus, an H+- or Na+-translocating activity of the Ech hydrogenase, as proposed (but not demonstrated) by Meuer et al. (20), is very likely. This hypothesis also explains why the Δech mutant strain does not grow on acetate. The free energy change associated with methanogenesis from 1 mol acetate allows synthesis of only a little more than 1 mol ATP (ΔGo′ = −36 kJ/mol CH4 [6]), but 1 mol ATP is used in acetate activation to form acetyl-CoA. Therefore, this methanogen lives close to the thermodynamic limit, and the loss of the Ech hydrogenase as an ion-translocating enzyme would lead to severe deficiencies in energy conservation. The second yet unidentified ferredoxinred-oxidizing protein obviously cannot compensate for this defect.
Overall, the results described here indicate that the ferredoxin-mediated membrane-bound electron transport chain is more complex than previously thought and that the Ech hydrogenase, as well as another unknown membrane-bound protein, is involved in aceticlastic and methylotrophic methanogenesis in M. mazei.
Acknowledgments
We thank Elisabeth Schwab for technical assistance and Paul Schweiger for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (grant De488/9-1).
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Abken, H. J., M. Tietze, J. Brodersen, S. Bäumer, U. Beifuss, and U. Deppenmeier. 1998. Isolation and characterization of methanophenazine and the function of phenazines in membrane-bound electron transport of Methanosarcina mazei Gö1. J. Bacteriol. 180:2027-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäumer, S., T. Ide, C. Jacobi, A. Johann, G. Gottschalk, and U. Deppenmeier. 2000. The F420H2 dehydrogenase from Methanosarcina mazei is a redox-driven proton pump closely related to NADH dehydrogenases. J. Biol. Chem. 275:17968-17973. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brodersen, J., G. Gottschalk, and U. Deppenmeier. 1999. Membrane-bound F420H2-dependent heterodisulfide reduction in Methanococcus voltae. Arch. Microbiol. 171:115-121. [DOI] [PubMed] [Google Scholar]
- 5.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Bäumer, C. Jacobi, H. Brüggemann, T. Lienard, A. Christmann, M. Bömeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 6.Deppenmeier, U., and V. Müller. 2008. Life close to the thermodynamic limit: how methanogenic archaea conserve energy. Results Probl. Cell Differ. 45:123-152. [DOI] [PubMed] [Google Scholar]
- 7.Deppenmeier, U. 2002. The unique biochemistry of methanogenesis. Prog. Nucleic Acid Res. Mol. Biol. 71:223-283. [DOI] [PubMed] [Google Scholar]
- 8.Ding, Y. H., and J. G. Ferry. 2004. Flavin mononucleotide-binding flavoprotein family in the domain Archaea. J. Bacteriol. 186:90-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellermann, J., A. Kobelt, A. Pfaltz, and R. K. Thauer. 1987. On the role of N-7-mercaptoheptanoyl-O-phospho-l-threonine (component B) in the enzymatic reduction of methyl-coenzyme M to methane. FEBS Lett. 220:358-362. [DOI] [PubMed] [Google Scholar]
- 10.Ferry, J. G., and D. J. Lessner. 2008. Methanogenesis in marine sediments. Ann. N. Y. Acad. Sci. 1125:147-157. [DOI] [PubMed] [Google Scholar]
- 11.Galagan, J. E., et al. 2002. The genome of Methanosarcina acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hippe, H., D. Caspari, K. Fiebig, and G. Gottschalk. 1979. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc. Natl. Acad. Sci. U. S. A. 76:494-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hovey, R., S. Lentes, A. Ehrenreich, K. Salmon, G. Gottschalk, R. P. Gunsalus, and U. Deppenmeier. 2005. DNA microarray analysis of Methanosarcina mazei during growth on methanol and acetate. Mol. Genet. Genomics 273:225-239. [DOI] [PubMed] [Google Scholar]
- 14.Ide, T., S. Bäumer, and U. Deppenmeier. 1999. Energy conservation by the H2:heterodisulfide oxidoreductase from Methanosarcina mazei Gö1: identification of two proton-translocating segments. J. Bacteriol. 181:4076-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krätzer, C., P. Carini, R. Hovey, and U. Deppenmeier. 2009. Transcriptional profiling of methyltransferase genes during growth of Methanosarcina mazei on trimethylamine. J. Bacteriol. 191:5108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lessner, D. J., and J. G. Ferry. 2007. The archaeon Methanosarcina acetivorans contains a protein disulfide reductase with an iron-sulfur cluster. J. Bacteriol. 189:7475-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, L., Q. Li, L. Rohlin, U. Kim, K. Salmon, T. Rejtar, R. P. Gunsalus, B. L. Karger, and J. G. Ferry. 2007. Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J. Proteome Res. 6:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahlmann, A., U. Deppenmeier, and G. Gottschalk. 1989. Methanofuran-b is required for CO2 formation from formaldehyde by Methanosarcina barkeri. FEMS letters. 61:115-120. [Google Scholar]
- 19.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. R. Sowers, and R. S. Wolfe. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. U. S. A. 94:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meuer, J., S. Bartoschek, J. Koch, A. Künkel, and R. Hedderich. 1999. Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur. J. Biochem. 265:325-335. [DOI] [PubMed] [Google Scholar]
- 21.Meuer, J., H. C. Kuettner, J. K. Zhang, R. Hedderich, and W. W. Metcalf. 2002. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. U. S. A. 99:5632-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortenson, L. E. 1964. Purification and analysis of ferredoxin from Clostridium pasteurianum. Biochim. Biophys. Acta 81:71-77. [DOI] [PubMed] [Google Scholar]
- 23.Noll, K. M., K. L. Rinehart, Jr., R. S. Tanner, and R. S. Wolfe. 1986. Structure of component B (7-mercaptoheptanoylthreonine phosphate) of the methylcoenzyme M methylreductase system of Methanobacterium thermoautotrophicum. Proc. Natl. Acad. Sci. U. S. A. 83:4238-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peer, C. W., M. H. Painter, M. E. Rasche, and J. G. Ferry. 1994. Characterization of a CO:heterodisulfide oxidoreductase system from acetate-grown Methanosarcina thermophila. J. Bacteriol. 176:6974-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ragsdale, S. W., L. G. Ljungdahl, and D. V. Der Vartanian. 1983. Isolation of carbon monoxide dehydrogenase from Acetobacterium woodii and comparison of its properties with those of the Clostridium thermoaceticum enzyme. J. Bacteriol. 155:1224-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 27.Thauer, R. K., A. K. Kaster, H. Seedorf, W. Buckel, and R. Hedderich. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6:579-591. [DOI] [PubMed] [Google Scholar]
- 28.Zahler, W. L., and W. W. Cleland. 1968. A specific and sensitive assay for disulfides. J. Biol. Chem. 243:716-719. [PubMed] [Google Scholar]