Abstract
In the nitrate-responsive, homodimeric NarX sensor, two cytoplasmic membrane α-helices delimit the periplasmic ligand-binding domain. The HAMP domain, a four-helix parallel coiled-coil built from two α-helices (HD1 and HD2), immediately follows the second transmembrane helix. Previous computational studies identified a likely coiled-coil-forming α-helix, the signaling helix (S helix), in a range of signaling proteins, including eucaryal receptor guanylyl cyclases, but its function remains obscure. In NarX, the HAMP HD2 and S-helix regions overlap and apparently form a continuous coiled-coil marked by a heptad repeat stutter discontinuity at the distal boundary of HD2. Similar composite HD2-S-helix elements are present in other sensors, such as Sln1p from Saccharomyces cerevisiae. We constructed deletions and missense substitutions in the NarX S helix. Most caused constitutive signaling phenotypes. However, strongly impaired induction phenotypes were conferred by heptad deletions within the S-helix conserved core and also by deletions that remove the heptad stutter. The latter observation illuminates a key element of the dynamic bundle hypothesis for signaling across the heptad stutter adjacent to the HAMP domain in methyl-accepting chemotaxis proteins (Q. Zhou, P. Ames, and J. S. Parkinson, Mol. Microbiol. 73:801-814, 2009). Sequence comparisons identified other examples of heptad stutters between a HAMP domain and a contiguous coiled-coil-like heptad repeat sequence in conventional sensors, such as CpxA, EnvZ, PhoQ, and QseC; other S-helix-containing sensors, such as BarA and TorS; and the Neurospora crassa Nik-1 (Os-1) sensor that contains a tandem array of alternating HAMP and HAMP-like elements. Therefore, stutter elements may be broadly important for HAMP function.
Transmembrane signaling in homodimeric bacterial sensors initiates upon signal ligand binding to the extracytoplasmic domain. In methyl-accepting chemotaxis proteins (MCPs), the resulting conformational change causes a displacement of one transmembrane α-helix (TM α-helix) relative to the other. This motion is conducted by the HAMP domain to control output domain activity (reviewed in references 33 and 39).
Certain sensors of two-component regulatory systems share topological organization with MCPs. For example, the paralogous nitrate sensors NarX and NarQ contain an amino-terminal transmembrane signaling module similar to those in MCPs, in which a pair of TM α-helices delimit the periplasmic ligand-binding domain (Fig. 1) (24) (reviewed in references 32 and 62). The second TM α-helix connects to the HAMP domain. Hybrid proteins in which the NarX transmembrane signaling module regulates the kinase control modules of the MCPs Tar, DifA, and FrzCD demonstrate that NarX and MCPs share a mechanism for transmembrane signaling (73, 74, 81, 82).
FIG. 1.
NarX modular structure. Linear representation of the NarX protein sequence, from the amino (N) to carboxyl (C) termini, drawn to scale. The four modules are indicated at the top of the figure and shown in bold typeface, whereas domains within each module are labeled with standard (lightface) typeface. The nomenclature for modules follows that devised by Swain and Falke (67) for MCPs. Overlap between the HAMP domain HD2 and S-helix elements is indicated in gray. The three conserved Cys residues within the central module (62) are indicated. TM1 and TM2 denote the two transmembrane helices. Helices H1 to H4 of the periplasmic domain (24), and the transmitter domain H, N, D, G (79), and X (41) boxes, are labeled. The HPK 7 family of transmitter sequences, including NarX, have no F box and an unconventional G box (79). The scale bar at the bottom of the figure shows the number of aminoacyl residues.
The HAMP domain functions as a signal conversion module in a variety of homodimeric proteins, including histidine protein kinases, adenylyl cyclases, MCPs, and certain phosphatases (12, 20, 77). This roughly 50-residue domain consists of a pair of amphiphilic α-helices, termed HD1 and HD2 (formerly AS1 and AS2) (67), joined by a connector (Fig. 2A). Results from nuclear magnetic resonance (NMR) and electron paramagnetic resonance (EPR) spectroscopy, Cys and disulfide scanning, and mutational analysis converge on a model in which the HD1 and HD2 α-helices form a four-helix parallel coiled-coil (7, 20, 30, 42, 67, 75, 84). The mechanisms through which HAMP domains mediate signal conduction remain to be established (30, 42, 67, 84) (for commentary, see references 43, 49, and 50).
FIG. 2.
HAMP domain extensions. (A) Sequences from representative MCPs (E. coli Tsr and Salmonella enterica serovar Typhimurium Tar) and S-helix-containing sensors (E. coli NarX, NarQ, and BarA, and S. cerevisiae Sln1p). The HAMP domain, S-helix element, and the initial sequence of the MCP adaptation region are indicated. Flanking numbers denote positions of the terminal residues within the overall sequence. Sequential heptad repeats are indicated in alternating bold and standard (lightface) typeface. Numbering for heptad repeats in the methylation region and S-helix sequences has been described previously (4, 8). Numbers within the HD1 and HD2 helices indicate interactions within the HAMP domain (42). Residues at heptad positions a and d are enclosed within boxes, residues at the stutter position a/d are enclosed within a thickly outlined box, and residues in the S-helix ERT signature are in bold typeface. (B) NarX mutational alterations. Deletions are depicted as boxes, and missense substitutions are shown above the sequence. Many of these deletions were reported previously (10) and are presented here for comparison. The phenotypes conferred by the alterations are indicated as follows: impaired induction, black box; constitutive and elevated basal, light gray box; reversed response, dark gray box; wild-type, white box; null, striped box.
Coiled-coils result from packing of two or more α-helices (27). The primary sequence of coiled-coils exhibits a characteristic heptad repeat pattern, denoted as a-b-c-d-e-f-g (52, 61), in which positions a and d are usually occupied by nonpolar residues (reviewed in references 1, 47, and 80). For example, the coiled-coil nature of the HAMP domain can be seen in the heptad repeat patterns within the HD1 and HD2 sequences (Fig. 2A).
Coiled-coil elements adjacent to the HAMP domain have been identified in several sensors, including Saccharomyces cerevisiae Sln1p (69) and Escherichia coli NarX (60). Recently, this element was defined as a specific type of dimeric parallel coiled-coil, termed the signaling helix (S helix), present in a wide range of signaling proteins (8). Sequence comparisons delimit a roughly 40-residue element with a conserved heptad repeat pattern (Fig. 2A). Based on mutational analyses of Sln1p and other proteins, the S helix is suggested to function as a switch that prevents constitutive activation of adjacent output domains (8).
The term “signaling helix” previously was used to define the α4-TM2 extended helix in MCPs (23, 33). Here, we use the term S helix to denote the element described by Anantharaman et al. (8).
The NarX and NarQ sensors encompass four distinct modules (Fig. 1): the amino-terminal transmembrane signaling module, the signal conversion module (including the HAMP domain and S-helix element), the central module of unknown function, and the carboxyl-terminal transmitter module (62). The S-helix element presumably functions together with the HAMP domain in conducting ligand-responsive motions from the transmembrane signaling module to the central module, ultimately regulating transmitter module activity.
Regulatory output by two-component sensors reflects opposing transmitter activities (reviewed in reference 55). Positive function results from transmitter autokinase activity; the resulting phosphosensor serves as a substrate for response regulator autophosphorylation. Negative function results from transmitter phosphatase activity, which accelerates phosphoresponse regulator autodephosphorylation (reviewed in references 64 and 65). We envision a homogeneous two-state model for NarX (17), in which the equilibrium between these mutually exclusive conformations is modulated by ligand-responsive signaling.
Previous work from our laboratory concerned the NarX and other HAMP domains (9, 10, 26, 77) and separately identified a conserved sequence in NarX and NarQ sensors, the Y box, that roughly corresponds to the S helix (62). Therefore, we were interested to explore the NarX S helix and to test some of the predictions made for its function. Results show that the S helix is critical for signal conduction and suggest that it functions as an extension of the HAMP HD2 α-helix in a subset of sensors exemplified by Sln1p and NarX. Moreover, a stutter discontinuity in the heptad repeat pattern was found to be essential for the NarX response to signal and to be conserved in several distinct classes of HAMP-containing sensors.
MATERIALS AND METHODS
Plasmid and strains.
Phenotypes were determined for narX alleles in plasmid pVJS2474 (10), which contains the ColE1-type replication origin from plasmid pBR322 (13). The narX‡ allele on this plasmid was modified to contain several unique, silent restriction endonuclease sites. Small, readily sequenced DNA segments were subcloned after mutagenesis to reconstruct an intact narX gene, thereby reducing the likelihood of spurious mutations elsewhere in the gene.
NarX positive function (autokinase plus phosphotransfer) was monitored in host strain VJS5054 [λΦ(narG-lacZ)250 ΔnarX242 ychN2084::Ω-Cm narQ251::Tn10d(Tc) narL+ pcnB1 zad-981::Tn10d(Km)] (76). This strain carries the pcnB1 allele, which results in an average plasmid copy number of approximately one (44).
NarX negative function (phosphatase) was monitored in host strain VJS4033 [λΦ(narG-lacZ)250 ΔnarX242 ychO2084::Ω-Cm narQ251::Tn10d(Tc) narL505 pcnB+] (76). The pcnB+ allele, which permits the normal average plasmid copy number of about 20, allows for increased sensitivity in assigning phenotypes (C. E. Noriega, H.-Y. Lin, L.-L. Chen, and V. Stewart, unpublished data).
Phenotypes.
During anaerobic growth, expression of the narGHJI operon encoding respiratory nitrate reductase is induced about 100-fold by nitrate. This wide dynamic range encompasses a variety of distinct phenotypes. Induction by nitrate requires the narL+ gene and, in the narX narQ double null strains employed, plasmid-encoded NarX sensor function. Test strains carry a monocopy Φ(narG-lacZ) gene fusion at attλ (58). The induction ratio is calculated as the LacZ specific activity from the nitrate-supplemented culture divided by activity from the unsupplemented culture.
(i) Constitutive phenotype.
The constitutive phenotype denotes mutants for which Φ(narG-lacZ) expression was similar in both cultures, with an induction ratio of 1.3 or less and uninduced values 35 to 75% of the wild-type induced level. Mutants with the strong constitutive phenotype had uninduced values 80% or more of the wild-type induced level. Others use the term “locked-on” for these phenotypes (6, 25).
(ii) Elevated basal phenotype.
The elevated basal phenotype denotes mutants for which induced Φ(narG-lacZ) expression was at least 30% of the wild-type level, with an induction ratio of 1.4 or more and uninduced values of about 7.5 to 30% of the wild-type induced level. Mutants with the strong elevated basal phenotype had uninduced values 35% or more of the wild-type induced level.
(iii) Impaired induction phenotype.
The impaired induction phenotype denotes mutants for which induced Φ(narG-lacZ) expression was less than 40% of the wild-type level, with an induction ratio of 12 or more and essentially wild-type uninduced level. Mutants with the strongly impaired induction phenotype had induced values essentially equal to the wild-type uninduced level and an induction ratio of 3.3 or less. Others use the term “locked-off” for these phenotypes (6, 25).
(iv) Null phenotype.
The null phenotype was distinguished from the strong impaired induction phenotype by testing for NarX negative function in strain VJS4033. The NarL(V88A) protein expressed by this strain can be phosphorylated by a Nar-independent route. In narX null strains, Φ(narG-lacZ) expression from uninduced cultures is equal to the fully induced level, whereas in narX+ strains uninduced expression is lower due to NarX phosphatase activity (58, 76). Additionally, sensor protein was undetectable by Western blot analysis of membrane fractions from two null mutants, narX Δ(Y227-L258) and narQ Δ(Y226-L257) (36) (J. A. Appleman and V. Stewart, unpublished data).
(v) Reversed response phenotype.
The reversed response phenotype denotes mutants for which the induction ratio is 0.2 or less (10).
Culture media and conditions.
Defined, complex, and indicator media for genetic manipulations were used as described previously (48). Defined medium to grow cultures for enzyme assays was buffered with 3-(N-morpholino)propanesulfonic acid (MOPS) as described previously (63). Glucose was added at 80 mM. NaNO3 (40 mM) was added as indicated. TY broth contained tryptone (0.8%), yeast extract (0.5%), and NaCl (0.5%).
Plasmid-containing strains were cultured in medium prepared by mixing equal volumes of defined (MOPS) and complex (TY) media. This medium produces a favorable combination of rapid growth and robust response to nitrate (10). Ampicillin (Ap; 100 μg ml−1) was added to maintain selection for plasmid-bearing cells.
Culture densities were monitored with a Klett-Summerson photoelectric colorimeter (Klett Manufacturing Co., New York, NY) equipped with a number 66 (red) filter. Cultures to monitor Φ(narG-lacZ) expression were grown at 37°C to the early exponential phase (about 25 to 35 Klett units). Anaerobic cultures were grown in filled screw-cap tubes as described previously (63).
Enzyme assay.
β-Galactosidase activities were measured in CHCl3-sodium dodecyl sulfate-permeabilized cells. Specific activities are expressed in arbitrary units (Miller units) (53). Assays were performed at room temperature at approximately 21°C. Reported values are averaged from at least two independent cultures grown on different days. Each culture was assayed in duplicate, with one assay reaction mixture containing twice as much cell extract as the other.
DNA cloning and sequencing.
Standard methods were used for PCR, restriction endonuclease digestion, ligation, and transformation of DNA (48). Restriction enzymes and T4 DNA ligase were from New England Biolabs, Inc. (Beverly, MA). DNA for sequencing was isolated from purified plasmid minipreps (Qiagen, Valencia, CA). DNA sequencing was performed by Davis Sequencing (Davis, CA) or by the College of Biological Sciences Automated DNA Sequencing Facility (University of California, Davis, CA).
Site-specific mutagenesis.
Oligonucleotide-directed deletions and single-nucleotide substitutions were generated by a modification of the QuikChange PCR protocol (Stratagene, La Jolla, CA) as described previously (10). In cases where phenotypes were unexpected or confusing, we used oligonucleotide-directed mutagenesis to change the alteration back to the wild-type sequence (true reversion) in order to ensure that the wild-type phenotype was restored. This provided reassurance that the mutant allele did not contain other, undetected alterations contributing to the phenotype.
Computational analyses.
Genome database searches employed the BLAST programs (5) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Identification of potential coiled-coil regions used the COILS and PCOILS programs (37), accessed through the MPI Bioinformatics Toolkit (15). The MTDIK matrix was used, and both weighted and unweighted analyses were performed (37). Both programs return analyses with scanning window sizes of 14, 21, and 28 residues.
RESULTS
Sequence features.
The S-helix element, located between two signaling domains, is predicted to form a dimeric parallel coiled-coil (8). A defining feature is denoted as the “ERT signature” in the central heptad, with conserved residues Glu (or another polar residue) at S-helix position 19 (heptad position c), Arg or Lys at position 20 (heptad position d), and Thr at position 21 (position e) (numbering shown in Fig. 2A) (8). Other conserved residues include Asn (or another polar residue) at S-helix position 10 (heptad position a), Glu at position 14 (heptad position e), Val at position 17 (position a), and Leu at positions 13 (position d) and 24 (position a). Sequence outside this region is less conserved, except for the pattern of hydrophobic residues that predominate at heptad positions a and d. We therefore denote the region spanning positions 10 through 24 as the S-helix conserved core (Fig. 2A).
As identified by Anantharaman et al. (8), the NarX S helix encompasses residues Asn-218 through Arg-257 (Fig. 2A), between the upstream HAMP domain and downstream central module. This sequence includes characteristic S-helix features, including the ERT signature (NarX residues Glu-236, Lys-237, and Thr-238) as well as S-helix conserved core residues Leu-230, Glu-231, Val-234, and Leu-241 (Fig. 2A).
An earlier analysis of Nar sensor domain architecture defined a 32-residue element, spanning residues Tyr-227 through Leu-258, as strongly conserved in NarX and NarQ sequences (62). This element was termed the Y box because of the nearly invariant residue Tyr-227, which lies at S-helix position 10. Other noted features were the heptad repeat pattern, the charged residue Lys-237 at heptad position d (in the ERT signature), and the nearly invariant residues Glu-231 and Thr-238. Thus, the Y box corresponds to positions 10 to 40 of the S helix (Fig. 2A).
It is suggested that some S-helix modules likely merge at their termini with adjacent helical segments (8). Indeed, inspection reveals that the six amino-terminal residues of the NarX and NarQ S-helix elements overlap with the six carboxyl-terminal residues of their adjacent HAMP domain HD2 helices (Fig. 2A). The identical six-residue overlap is present in other sensors, including S. cerevisiae Sln1p and E. coli BarA (Fig. 2A). Similarly, the S-helix distal portion (Fig. 2A) likely forms part of the NarX central module, a GAF-like domain (see Discussion).
Prior computational analysis identified potential coiled-coils spanning E. coli NarX residues Leu-212 through Leu-251 and NarQ residues Asn-216 through Arg-246 (60). Similar results were obtained with the programs COILS and PCOILS, run as described in Materials and Methods. Although exact boundaries varied somewhat according to the parameters and scanning window size, the region encompassing NarX HD2 residue Leu-212 through the S-helix core sequence was consistently identified has having a high probability of forming a coiled-coil (results not shown).
Inspection reveals that the HAMP-S-helix overlap includes a discontinuity within the heptad repeat pattern, termed a stutter, equivalent to a three-residue deletion (19). We denote the stutter in Fig. 2A as position a/d, because the residue at this position (Leu-223) lies at heptad position a with respect to the upstream sequence but at heptad position d with respect to the downstream sequence.
Large deletions.
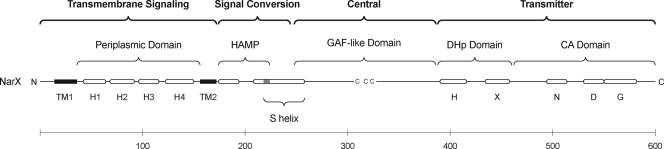
The Δ(L178-R206) deletion (29 residues) removes the HD1 and connector elements from the HAMP domain, whereas the Δ(L178-L216) deletion (39 residues) removes most of the HAMP domain. Both conferred the constitutive phenotype (Fig. 3). The Δ(P181-N217) deletion (37 residues), which also removes most of the HAMP domain, conferred the strong elevated basal level phenotype, as did the Δ(H243-R256) deletion (14 residues), which removes the distal portion of the S-helix element. Finally, the Δ(Y227-L241) deletion (15 residues), which removes the S-helix conserved core, conferred the strong constitutive phenotype. The constitutive and strong constitutive phenotypes are similar to those conferred by seven- and ten-residue deletions within the HD1 and connector elements, and the strong elevated basal level phenotypes are similar to those conferred by the Δ(M209-N218) and Δ(M209-M219) deletions (10 and 11 residues, respectively) within the HD2 element (10) (examples presented in Fig. 3).
FIG. 3.
Positive-function phenotypes conferred by HAMP and S-helix deletions. Expression of the Φ(narG-lacZ) reporter is normalized to the induced wild-type value, which corresponds to about 2,000 Miller units. Hatched bars show data from cultures grown in the absence of nitrate (uninduced). Filled bars show data from cultures grown in the presence of nitrate (induced). Data for mutants indicated with asterisks are from reference 10.
In contrast, two other large deletions conferred the null phenotype (in vivo phosphotransfer data not shown), and therefore likely cause severe structural perturbations. The Δ(W182-S220) deletion (39 residues) removes most of the HAMP domain, including the first three residues of the S helix (10), and the Δ(Y227-L258) deletion (32 residues), designed to eliminate the Y-box element, removes most of the S helix (Fig. 2B). In vivo negative-function data for the Δ(Y227-L258) deletion are shown in Table 1; those for the Δ(W182-S220) deletion are presented in reference 10. (Note that this deletion erroneously is depicted as including residue Pro-181 in Fig. 2 of reference 10.) The congruent Δ(Y226-L257) deletion of NarQ also conferred the null phenotype (data not shown).
TABLE 1.
Effects of HAMP and S-helix alterations on NarX negative function: NarL(V88A)-dependent Φ(narG-lacZ) expressiona
| Alterationb | LacZ sp act |
Induction ratioc | |
|---|---|---|---|
| − Nitrate | + Nitrate | ||
| None (vector only) | 1,270 | 1,150 | 0.9 |
| None (wild type) | 200 | 1,440 | 7.2 |
| Δ(S220-L223) | 330 | 140 | 0.4 |
| Δ(L223-S226) | 170 | 74 | 0.4 |
| Δ(Y227-L258) | 2,050 | 1,150 | 0.6 |
| Δ(V234-G240) | 7 | 150 | 21 |
| Δ(K237-H243) | 7 | 890 | 127 |
| Δ(K244-F250) | 56 | 1,390 | 25 |
| Δ(L251-R257) | 2,660 | 1,390 | 0.5 |
| R233E | 18 | 1,350 | 75 |
| L241A | 280 | 1,370 | 4.9 |
Strains were cultured anaerobically to the mid-exponential phase in enriched medium (TY broth plus a 1:1 mixture of MOPS-glucose). Nitrate was added as indicated at 40 mM.
Alteration on the indicated narX allele on plasmid pVJS2474 derivative in strain VJS4033 [Φ(narG-lacZ) ΔnarX242 narQ251::Tn10 narL505 pcnB+].
The induction ratio is calculated as the LacZ specific activity from the nitrate-supplemented culture divided by activity from the unsupplemented culture (minus nitrate).
Heptad deletion scan.
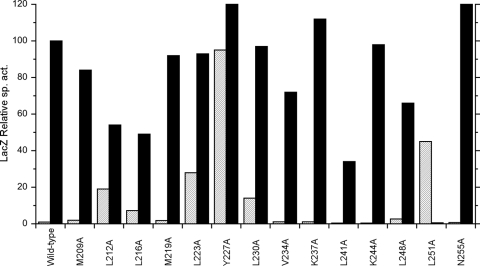
Previously, our laboratory constructed a series of overlapping seven-residue deletions to scan through the HAMP domain HD1, connector, and HD2 elements (10). In an α-helix, seven residues correspond to two full turns (56), so these heptad deletions are expected to leave the overall coiled-coil structure intact, albeit shorter. This contrasts to the larger deletions described above, most of which disrupt the heptad repeat pattern and therefore likely disrupt the coiled-coil structure.
We extended this heptad deletion scan across the S-helix element. Patterns of Φ(narG-lacZ) expression are shown in Fig. 3, which also includes results from previous analysis of HD2 deletions for comparison. The Δ(L223-V229) and Δ(Y227-R233) deletions conferred constitutive phenotypes, the Δ(L230-E236) and Δ(L248-A254) deletions conferred elevated basal level phenotypes, the Δ(L241-I247) deletion mutant was essentially wild-type, and the Δ(L251-R257) deletion conferred the null phenotype (Table 1).
Strikingly, three S-helix heptad deletions, Δ(V234-G240), Δ(K237-H243), and Δ(K244-F250), conferred strong impaired induction phenotypes. These mutants were differentiated in the phenotypic test for negative function (Table 1). The Δ(V234-G240) and Δ(K237-H243) deletions, which both remove the ERT signature motif, exhibited strong negative-function phenotypes comparable to those of missense mutants devoid of autokinase activity (C. E. Noriega, H.-Y. Lin, L.-L. Chen, and V. Stewart, unpublished data). In contrast, the negative-function phenotype for the Δ(K244-F250) mutant was similar to that of the wild type. Thus, the Δ(V234-G240) and Δ(K237-H243) mutant proteins appear to be strongly biased toward the ligand-free signaling conformation.
Stutter deletions.
We constructed a pair of four-residue deletions, each with residue Leu-223 at one endpoint (Fig. 2B). These deletions remove the stutter, thereby forming a continuous heptad repeat pattern from the HD2 element through the end of the S helix. These deletions conferred the strong impaired induction phenotype (Fig. 3), quite different from the elevated basal level and constitutive phenotypes conferred by the heptad deletions Δ(M219-E225) and Δ(L223-V229) which remove residue Leu-223 but leave the stutter pattern intact (Fig. 2B). Additionally, the four-residue stutter deletion mutants exhibited strong negative function that, unlike all other mutants tested, was indifferent to added inducer (Table 1). This suggests that these mutant proteins are essentially immobilized in the ligand-free signaling conformation. Therefore, the stutter is indispensable for NarX response to nitrate.
Heptad Ala scan.
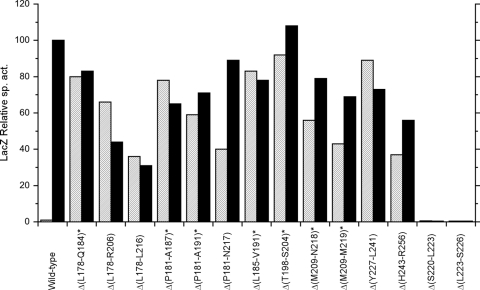
We used site-specific mutagenesis to substitute Ala at each heptad position a or d from Met-209 in the HD2 element through Asn-255 near the end of the S helix (Fig. 2B). Alanine-scanning mutagenesis was developed originally to infer functional contributions of aminoacyl side chain atoms beyond the α carbon (28). Our further goal here was to identify alterations that destabilize the coiled-coil structure. Formation of parallel coiled-coils is governed in large part by interaction of hydrophobic “knobs” at heptad positions a and d with their corresponding “sockets” on the partner helix (27). Accordingly, the large aliphatic residues (Leu, Val, and Ile) predominate at these positions, although polar and charged residues account for about 20% of the total (reviewed in references 1, 47, and 80). In particular, Leu residues prevail at the d heptad positions in parallel two-stranded coiled-coils due to perpendicular packing geometry (38). We reasoned that Ala substitutions at heptad positions a and d could act to destabilize the corresponding regions of the HD2-S-helix coiled-coil and thereby provide informative phenotypes, although cognizant that effects of individual residues are extremely dependent on context (45). Recently, similar logic was employed as part of a detailed analysis of the Tar adaptation region (68).
Ala substitution mutants displayed the range of phenotypes excluding null (see Fig. 5). All five of the elevated basal level or constitutive phenotype alterations lie within the first seven heptad positions in the region tested (Fig. 2B). Two of these (L212A and L216A) are at positions critical for HD2 function (10, 84). The remainder exhibited the wild-type phenotype, with two notable exceptions. Ala substitution at residue Leu-241 (at the end of the S-helix conserved core) conferred the impaired induction phenotype (Table 1), whereas substitution at Leu-251 (in the S-helix distal portion) conferred the reversed response phenotype. The basis for the reversed response phenotype is unknown (10).
FIG. 5.
Positive-function phenotypes conferred by HAMP and S-helix Ala scan substitutions. Expression of the Φ(narG-lacZ) reporter is normalized to the induced wild-type value, which corresponds to about 2,000 Miller units. Hatched bars show data from cultures grown in the absence of nitrate (uninduced). Filled bars show data from cultures grown in the presence of nitrate (induced).
The position corresponding to NarX residue Lys-244, at heptad position d, is occupied by Gln, Ala, Thr, or Ser in NarX and NarQ sequences from other species, so the wild-type phenotype conferred by the K244A substitution is not altogether surprising. Accordingly, we also substituted Lys-244 with Leu, the residue generally preferred at coiled-coil heptad positions d. Indeed, the K244L substitution conferred a mild impaired induction phenotype (Table 2).
TABLE 2.
Effects of S-helix missense substitutions on NarX positive function: Φ(narG-lacZ) expressiona
| Substitution(s)b | LacZ sp act |
Induction ratioc | |
|---|---|---|---|
| − Nitrate | + Nitrate | ||
| None (wild type) | 12 | 1,660 | 142 |
| E231A | 16 | 1,800 | 111 |
| E231V | 29 | 1,330 | 45 |
| E231R | 17 | 1,470 | 88 |
| R233E | 10 | 370 | 36 |
| E231R + R233E | 12 | 370 | 30 |
| E236K | 50 | 1,660 | 33 |
| K237E | 170 | 1,850 | 11 |
| T238A | 14 | 1,560 | 111 |
| T238F | 730 | 1,550 | 2.1 |
| K244L | 7 | 690 | 98 |
| W252R | 940 | 950 | 1.0 |
Strains cultured anaerobically to the mid-exponential phase in enriched medium (TY broth plus a 1:1 mixture of MOPS-glucose). Nitrate was added as indicated at 40 mM.
Substitution(s) at the indicated narX allele on plasmid pVJS2474 derivative in strain VJS5054 [Φ(narG-lacZ) ΔnarX242 narQ251::Tn10 pcnB1].
The induction ratio is calculated as the LacZ specific activity from the nitrate-supplemented culture divided by activity from the unsupplemented culture (minus nitrate).
Missense substitutions.
Finally, we employed site-specific mutagenesis in order to examine specific residues within the S helix.
(i) The ERT signature.
NarX residues Glu-236, Lys-237, and Thr-238 form the ERT signature at S-helix positions 19, 20, and 21, respectively.
The Arg residue at S-helix position 20 (Arg-838) is critical for control of human guanylyl cyclase 2D activity. Substitution with Lys has no effect, whereas substitution with Glu or Asp results in elevated activity that is regulated only weakly by signal ligand (59). The corresponding K237E substitution in NarX similarly conferred the elevated basal phenotype (Table 2). Surprisingly, the K237A change had little effect (see Fig. 5). Nevertheless, some other S-helix sequences, including that from BarA, have Ala at this position (Fig. 2B).
In S. cerevisiae Sln1p, substitution of large hydrophobic residues for the Thr residue at S-helix position 21 (Thr-550; Fig. 2A) results in hyperactive kinase activity, whereas substitution of Asp has little effect (69). Likewise, substitution of Met at position Thr-839 in human guanylyl cyclase 2D causes increased basal activity (59). The NarX T238F change caused the elevated basal phenotype, whereas the T238A substitution had little effect (Table 2).
Finally, the NarX E236K mutant, substituted at S-helix position 19, exhibited a mild elevated basal phenotype (Table 2).
(ii) Tyr-227.
Residue Tyr-227 at S-helix position 10 (heptad position a) is strongly conserved in NarX and NarQ sequences (62). This position in other S-helix sequences is usually occupied by a polar residue, such as Asn (8). As noted above, substitution of Ala for residue Tyr-227 (Y227A) resulted in the strong constitutive phenotype (see Fig. 5). Confoundingly, however, the congruent NarQ Y226A mutant behaved essentially as the wild type did (data not shown).
(iii) Glu-231 and Arg-233.
Residue Glu-231 at S-helix position 14 is strongly conserved in NarX and NarQ sequences (62). In S. cerevisiae Sln1p, substitution of Val at residue Glu-243 results in strongly hyperactive kinase activity (69). However, the congruent change in NarX (E231V) conferred the wild-type phenotype (Table 2), as did substitution of Ala (E231A).
NarX residues Glu-231 and Arg-233 represent oppositely charged residues at adjacent heptad positions e and g, which are suggested to form interhelical interactions to help stabilize the S-helix coiled-coil (8). (Note, however, that salt bridges in coiled-coils usually result from interaction between position g in one heptad and position e of the partner chain in the successive heptad [reviewed in reference 80].) We reasoned that, if residues Glu-231 and Arg-233 interact directly through their opposite charges, single substitutions to the opposite charge at these positions would each display the same phenotype, whereas the double substitution would display a suppressed phenotype (83). Thus, we made the single substitutions E231R and R233E, as well as the double substitution E231R plus R233E. The E231R change resulted in an essentially wild-type phenotype, whereas the R233E change caused the impaired induction phenotype (Tables 1 and 2). Notably, the double mutant (E231R plus R233E) displayed the same phenotype as the R233E single mutant (Table 2).
(iv) Trp-252.
Finally, in human guanylyl cyclase A (natriuretic peptide receptor), substitution of Arg at S-helix position 35 (Leu-849) causes loss of catalytic activity, presumably by interfering with dimerization (57). The congruent change in NarX (W252R) caused the constitutive phenotype (Table 2). This position is occupied by Tyr or Phe in most other NarX and NarQ sequences. Interestingly, the N276S substitution at the corresponding position in BarA was isolated as an impaired induction mutant (70).
DISCUSSION
The homodimeric HAMP domain is a four-helix parallel coiled-coil, in which the two helices of each monomer are offset by one helical turn (42, 67, 84). Therefore, the carboxyl-terminal end forms the beginning of a dimeric parallel coiled-coil extruding from the domain. We propose that the S-helix element functions as a contiguous coiled-coil extension of the HAMP domain in NarX. Analogous coiled-coil extensions may function in other sensor subclasses as described below.
The HAMP-S-helix stutter is essential.
The heptad repeat pattern at the distal boundary of the HAMP HD2 element exhibits a stutter, equivalent to a three-residue deletion within a series of tandem heptads (19) (Fig. 2A). The four-residue deletions Δ(S220-L223) and Δ(L223-S226), which introduce a continuous heptad repeat pattern, resulted in a uniquely strong version of the impaired induction phenotype (Fig. 2B and Table 1). In contrast, the seven-residue deletions Δ(L223-V229) and Δ(M219-E225), which leave a stutter in place, resulted in constitutive and strong elevated basal phenotypes, respectively. Thus, this discontinuity was critical for signal transmission.
Remarkably, the identical stutter marks the junction between the HAMP domain and helix 1 of the adjacent coiled-coil adaptation (methylation) region in MCPs (Fig. 2A). The MCP adaptation region forms part of an extended four-helix antiparallel coiled-coil (78), whereas the S helix is a dimeric parallel coiled-coil. Nevertheless, the conserved heptad stutter suggests that HAMP domain signaling may operate in analogous fashion through a contiguous coiled-coil extension in both MCPs and NarX.
Recently, Zhou et al. suggested a dynamic bundle model to account for signal propagation across the HAMP domain and adaptation region in Tsr and other MCPs (84). They hypothesize that the helical phase clash introduced by the stutter contributes to oppositional stability of the flanking coiled-coil structures. Indeed, coiled-coils can contain subdomains of relatively high and low stability (40, 45), and stutters cause localized distortion in the coiled-coil structure (19, 46, 66). According to the dynamic bundle model, strong helix packing within the HAMP domain (kinase-off signaling state) leads to relaxed helix packing in the adaptation region, whereas relaxed helix packing within the HAMP domain (kinase-on signaling state) leads to strong helix packing in the adaptation region (84). The helix-packing mode within the adaptation region, in turn, is hypothesized to be antisymmetrically coupled with that of the protein interaction region that controls CheA autokinase activity (68).
ERT signature function.
In human guanylyl cyclase 2D, certain missense substitutions for residue Arg-838 in the ERT signature cause a severe retinopathy, Leber congenital amaurosis (29). Molecular dynamics simulations provide a hypothetical structure for this S helix and suggest a central role for residue Arg-838 in forming salt bridges with nearby Glu residues (59). These bridges stabilize the local interface and thereby constrain mobility in the distal portion of the coiled-coil. The resulting interchain electrostatic repulsion involving these and other S-helix Glu residues serves to repel the two helical strands, resulting in a destabilized “splayed” structure immediately distal to the ERT signature (59). This specific mechanism likely is not general, as these Glu residues are either not strongly conserved in receptor guanylyl cyclase sequences (16) or in S-helix sequences generally (8). Nevertheless, it illustrates a critical function for the ERT signature in control of distal helix packing.
A dynamic bundle in NarX?
The dynamic bundle model provides a useful framework for interpreting effects of alterations in the NarX HAMP-S-helix element. Evidence suggests that signal-ligand binding to the NarX periplasmic domain corresponds to the ligand-free signaling state of MCPs (Tar or Tsr) (9, 24, 73) (for commentary, see reference 32). Thus, nitrate binding would effect the HAMP-relaxed, S-helix-strong packing conformations analogous to the MCP kinase-on output response.
For NarX, it can be imagined that a structural perturbation transmitted across the stutter residue controls formation or geometry of interactions with ERT signature residues, thereby influencing the distal coiled-coil structure. Support for this notion comes from the mutant phenotypes reported here (Fig. 4 and 5). Two of the three heptad deletions that uniquely conferred the impaired induction phenotype, Δ(V234-G240) and Δ(K237-H243), affect the ERT signature in the S-helix core (Fig. 2B). Likewise, the only Ala substitution conferring an impaired induction phenotype, L241A, is at the end of the S-helix conserved core just distal to the ERT signature. These alterations, by abolishing ERT function, would trap the distal portion of the S helix in the stabilized conformation, thereby preventing output activation. In contrast, heptad deletions and missense substitutions proximal to the ERT signature almost exclusively yielded constitutive or elevated basal phenotypes.
FIG. 4.
Positive-function phenotypes conferred by HAMP and S-helix heptad scan deletions. Expression of the Φ(narG-lacZ) reporter is normalized to the induced wild-type value, which corresponds to about 2,000 Miller units. Hatched bars show data from cultures grown in the absence of nitrate (uninduced). Filled bars show data from cultures grown in the presence of nitrate (induced). Data for mutants indicated with asterisks are from reference 10.
Heptad deletions through the S-helix distal portion yielded the range of phenotypes (Fig. 4). Thus, even if the above model is correct, the mechanism by which signal is propagated to control transmitter output activity is unclear. The NarX HAMP-S-helix element connects to the output transmitter module through the intervening central module (Fig. 1), a variant of the conserved GAF domain found in a variety of signaling proteins (11) (C. E. Noriega, M. J. Gray, J. A. Appleman, H.-C. Wu, L.-L. Chen, and V. Stewart, unpublished data). We hypothesize that the distal portion of the NarX S helix comprises the initial α-helix for this GAF-like domain (Fig. 2A). Thus, signal-responsive modulation of the S-helix conserved core structure might control how the distal portion interacts with the remainder of the GAF-like domain.
HAMP control of transmitter activity.
As described above, a heptad stutter at the HD2 distal boundary seems critical for HAMP-mediated signaling in two distinct cases. To search for HAMP-related stutters in other contexts, we examined sensors for which the HAMP domain and transmitter module are adjacent or nearly so. Below we first summarize transmitter structure and dynamics and then consider three different examples of conserved stutters, suggesting a broad role for this discontinuity in HAMP-mediated signaling.
The homodimeric transmitter module comprises two domains connected by a flexible tether. The dimerization and histidyl phosphotransfer (DHp) domain is an antiparallel four-helix bundle that contains the phosphoaccepting His residue, and the catalytic and ATP-binding (CA) domain embodies the autokinase activity (reviewed in references 31 and 35). The DHp domain governs receiver domain phosphorylation (phosphotransfer activity) and dephosphorylation (phosphatase activity). Therefore, transmitter activity reflects an equilibrium between two distinct active conformations, autokinase and phosphotransfer or phosphatase.
X-ray structures reveal a dynamic lateral interface between the DHp and CA domains (2, 14, 21, 51). Thus, it is hypothesized that regulatory signal influences the DHp antiparallel four-helix bundle structure in order to modulate this interface, which must be destabilized for autokinase action (allowing the CA domain to interact with the phospho-accepting His residue) yet stabilized for phosphotransfer or phosphatase activity (sequestering the CA domain away from the His residue) (2, 21, 51).
These alternate conformations appear to result from rotation between α-helices in the DHp domain antiparallel four-helix bundle. This is obviously congruent with the model, suggested by NMR structure analysis of the Af1503 HAMP domain, that the HD1 and HD2 α-helices rotate relative to one another (42). Confoundingly, however, HAMP domain rotation seems incompatible with the ligand-responsive piston-type motion exhibited by MCP transmembrane signaling modules (67) (reviewed in reference 33). Analogous piston-type signaling motions likely operate in many two-component sensors, including EnvZ, NarX, and CpxA (9, 24, 72, 74) (for commentary, see reference 32).
The dynamic bundle hypothesis provides a mechanism through which a TM2 piston-type motion results in destabilization (i.e., helical rotation) within a distal helical element (84). If this hypothesis applies broadly to two-component sensors, one expects to find appropriately positioned heptad stutters in different classes of HAMP-containing sensors. Three such examples follow.
(i) EnvZ-type sensors.
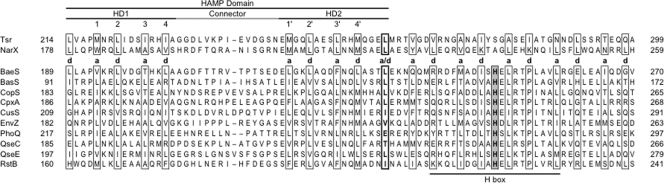
The well-studied EnvZ sensor represents a common structure, with an MCP-like transmembrane signaling module and HAMP domain linked directly to a transmitter module (72). Among the 15 HAMP-containing sensors encoded by E. coli K-12 (10), 10 conform to this architecture. (Four others, NarX, NarQ, BarA, and TorS, are described above or below, whereas the fifth, RcsC, is not considered here.) Sequence from the HAMP domain through the first helix of the DHp domain for these 10 sensors is presented in Fig. 6.
FIG. 6.
EnvZ-type sensor HAMP domain extensions. Sequences from E. coli K-12 Tsr, NarX, and 10 HAMP-containing sensors are shown. The HAMP domain and H-box motif (79) are indicated. Flanking numbers denote positions of the terminal residues within the overall sequence. Numbers within the HD1 and HD2 helices indicate interactions within the HAMP domain (42). Residues at heptad positions a and d are enclosed within boxes, residues at the stutter position a/d are enclosed within a thickly outlined box, and DHp domain invariant His residues are enclosed within a shaded box. Alternate designations are as follows: CopS also called YedV, CusS also called YbcZ, QseC also called YgiY, and QseE also called YfhK.
All 10 of these EnvZ-type sensors are in the closely related HPK 1, HPK 2, and HPK 3 sequence subfamilies (79), the DHp domains for which are annotated in databases under the HisKA designation (e.g., pfam00512) (34). Available structures for this DHp domain class reveal an antiparallel four-helix bundle, rather than a coiled-coil (14, 21, 51, 71). Nevertheless, the proximal (H-box) helix does exhibit a heptad-like repeat pattern that reflects hydrophobic packing interactions in the domain interior (51), and a computational study concluded that this helix forms a coiled-coil at least through the conserved Pro residue (60). Moreover, the structure of the Bacillus subtilis DesK sensor DHp domain reveals an antiparallel four-helix coiled-coil (2), although DesK is in the distinct HPK 7 sequence subfamily, the DHp domain for which is annotated as HisKA_3 (pfam07730).
Remarkably, for all 10 EnvZ-type sensors, the junction between the HAMP HD2 helix and the DHp helix exhibits the identical stutter pattern exhibited by MCPs and Sln1p-NarX sensors (Fig. 6). Thus, it is easy to envision the dynamic bundle model in operation, with control exerted directly over alternate conformations within the DHp domain.
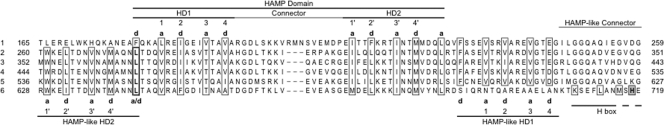
(ii) S-helix sensors.
Like NarX, many sensors have intervening domains between the HAMP-S-helix element and the transmitter module (8). However, the HAMP, S-helix, and transmitter sequences are contiguous in the Sln1p sensor. We extracted a somewhat arbitrary selection of protein sequences with similar architecture from the data set provided in reference 8, taking care to include representatives in which the S helix appeared to have insertions or deletions. Examples are presented in Fig. 7. For most, the 40-residue S-helix segment overlaps the HAMP or DHp sequence or both. Nevertheless, almost all have a separate S-helix conserved core sequence. We aligned the S-helix portions according to their ERT signature elements (as defined by reference 8) for ease of comparison. Three distinct classes are apparent.
FIG. 7.
S-helix-containing sensors. Sequences from representative examples are shown. The HAMP domain HD2 element, S-helix conserved core, and the initial portion of the H-box motif (79) are indicated. Flanking numbers denote positions of the terminal residues within the overall sequence. Numbers within the HD2 helix indicate interactions within the HAMP domain (42). Residues at heptad positions a and d are enclosed within boxes, residues at stutter positions a/d are enclosed within a thickly outlined box, residues in the S-helix ERT signature are in bold typeface, and DHp domain invariant His residues are enclosed within shaded boxes. Sequence gaps introduced to maximize alignment are indicated by dots, and those corresponding to heptad position a or d are indicated. Sequences are denoted by gene name or by locus tag as appropriate. The named proteins are Sln1p (S. cerevisiae), BarA and TorS (E. coli K-12), TorS (Shewanella oneidensis MR-1), and PgtB (S. enterica serovar Typhimurium LT2). Anonymous protein locus tags are indicated as follows: PA for Pseudomonas aeruginosa PAO; VC and VCA for Vibrio cholerae N16961 chromosomes I and II, respectively; ebA for Aromatoleum aromaticum EbN1; lpg for Legionella pneumophila Philadelphia 1; and Npun for Nostoc punctiforme PCC 73102.
One class, exemplified by S. cerevisiae Sln1p and E. coli BarA, displays a HAMP-S-helix stutter identical to those in MCPs and NarX (compare Fig. 2A and 7) and a second stutter at the S-helix-DHp interface. The intervening sequences exhibit uninterrupted heptad repeat patterns, but their length varies, as does the position of the ERT signature with respect to upstream and downstream HAMP and DHp elements.
A second class, exemplified by E. coli TorS, also has the S-helix-DHp stutter, but the HAMP-S-helix interface exhibits an uninterrupted heptad repeat pattern. Again, the length of the intervening sequence varies, as does the position of the ERT signature. Finally, the third class, exemplified by Salmonella enterica serovar Typhimurium PgtB, exhibits a variety of nonstutter heptad discontinuities at both interfaces.
Thus, although variations of the dynamic bundle hypothesis may apply to many or most of these HAMP-S-helix-DHp sensors, specific details likely span a range of possibilities. Nevertheless, the overall conservation of the S-helix conserved core, along with relatively uninterrupted heptad repeat patterns for most, suggests that signaling functions through generally similar mechanisms for this broad sensor class.
(iii) Tandem HAMP domains in Nik-1.
Finally, alert to HAMP domain coiled-coil extensions, we encountered the Neurospora crassa Nik-1 (Os-1) sensor, for which orthologs are present in many species of ascomycetes (22). The Nik-1 amino terminus, which has no obvious transmembrane segments, contains six tandem HAMP domains (22). Alex et al. (3) described this sequence as five tandem repeats of a 90-residue sequence followed by a sixth truncated repeat.
All six tandem HAMP domains display the defining HD1-connector-HD2 architecture, although the 17-residue connector for the first domain is slightly longer than usual (Fig. 8). Each of the first five HD2 elements is followed by an in-phase heptad repeat sequence corresponding to two turns of a coiled-coil, and each of the last five HD1 elements is preceded by a two-turn contiguous heptad repeat joined by a stutter.
FIG. 8.
Nik-1 tandem HAMP domains. Sequence from the amino-terminal portion of Neurospora crassa Nik-1 is shown. The HAMP and HAMP-like domains and the initial portion of the H-box motif (79) are indicated. Flanking numbers denote positions of the terminal residues within the overall sequence. Numbers within the HD1 and HD2 helices indicate interactions within the HAMP domain (42). Residues at heptad positions a and d are enclosed within boxes, residues at the stutter position a/d are enclosed within a thickly outlined box, and DHp domain invariant His residues are enclosed within a shaded box.
Remarkably, these apparent coiled-coil extensions exhibit a HAMP-like architecture, with a pair of 15-residue amphipathic α-helixes joined by a 15-residue connector (Fig. 8). However, they differ from canonical HAMP domains, most obviously by the conserved acidic residue at position 4 in the HD1-like element, and by the absence of the conserved Glu at the beginning of the HD2-like element.
Thus, the Nik-1 amino terminus contains a tandem array of alternating HAMP and HAMP-like elements in which each HAMP plus HAMP-like element pair comprises one of the 90-residue repeats (3). This structure provides a striking example of HAMP domain coiled-coil extensions that likely are essential for proper signaling and further emphasizes the functional importance of stutter discontinuities at boundaries between HAMP domains and their adjacent coiled-coil extensions.
Conclusions.
The HAMP domain NMR structure suggests a helix rotation model for signal transmission (42), which logically implies that distal helical elements also function through rotation (2, 21, 54). Distal helices or coiled-coils seemingly could act passively to convey the rotary motion from upstream to downstream. In the case of S-helix function, however, the opposite phenotypes (constitutive versus impaired induction) conferred by different heptad deletions, together with dependence on the conserved heptad stutter, imply an active, dynamic role for different portions of the S-helix coiled-coil. Indeed, replacing the S helix with a heterologous coiled-coil restored function but not regulation to the Sln1p sensor (69). Thus, we favor models such as the dynamic bundle hypothesis to guide future analysis of S-helix structure and function.
The NarX sensor represents only one of several cases where a HAMP domain coiled-coil extension mediates intramolecular signaling. Although such extensions are S-helix elements in some sensors (8), other examples abound as noted above. Moreover, coiled-coils mediate intramolecular signaling in other proteins that contain no HAMP domain (18, 54). Therefore, coiled-coils likely conduct signal-responsive conformational changes through a variety of mechanisms.
Acknowledgments
We thank Michael Gray and Alex Appleman for their contributions to this work and Chet Price for valuable critique of the manuscript.
This study was supported by Public Health Service grant GM036877 from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 4 December 2009.
REFERENCES
- 1.Adamson, J. G., N. E. Zhou, and R. S. Hodges. 1993. Structure, function and application of the coiled-coil protein folding motif. Curr. Opin. Biotechnol. 4:428-437. [DOI] [PubMed] [Google Scholar]
- 2.Albanesi, D., M. Martin, F. Trajtenberg, M. C. Mansilla, A. Haouz, P. M. Alzari, D. de Mendoza, and A. Buschiazzo. 2009. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc. Natl. Acad. Sci. U. S. A. 106:16185-16190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alex, L. A., K. A. Borkovich, and M. I. Simon. 1996. Hyphal development in Neurospora crassa: involvement of a two-component histidine kinase. Proc. Natl. Acad. Sci. U. S. A. 93:3416-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander, R. P., and I. B. Zhulin. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 104:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ames, P., and J. S. Parkinson. 1988. Transmembrane signaling by bacterial chemoreceptors: E. coli transducers with locked signal output. Cell 55:817-826. [DOI] [PubMed] [Google Scholar]
- 7.Ames, P., Q. Zhou, and J. S. Parkinson. 2008. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 190:6676-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anantharaman, V., S. Balaji, and L. Aravind. 2006. The signaling helix: a common functional theme in diverse signaling proteins. Biol. Direct 1:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Appleman, J. A., L.-L. Chen, and V. Stewart. 2003. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J. Bacteriol. 185:4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Appleman, J. A., and V. Stewart. 2003. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 185:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravind, L., and C. P. Ponting. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22:458-459. [DOI] [PubMed] [Google Scholar]
- 12.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 13.Balbás, P., X. Soberón, E. Merino, M. Zurita, H. Lomeli, F. Valle, N. Flores, and F. Bolivar. 1986. Plasmid vector pBR322 and its special-purpose derivatives-a review. Gene 50:3-40. [DOI] [PubMed] [Google Scholar]
- 14.Bick, M. J., V. Lamour, K. R. Rajashankar, Y. Gordiyenko, C. V. Robinson, and S. A. Darst. 2009. How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda. J. Mol. Biol. 386:163-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biegert, A., C. Mayer, M. Remmert, J. Söding, and A. N. Lupas. 2006. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34:W335-W339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biswas, K. H., A. R. Shenoy, A. Dutta, and S. S. Visweswariah. 2009. The evolution of guanylyl cyclases as multidomain proteins: conserved features of kinase-cyclase domain fusions. J. Mol. Evol. 68:587-602. [DOI] [PubMed] [Google Scholar]
- 17.Bornhorst, J. A., and J. J. Falke. 2003. Quantitative analysis of aspartate receptor signaling complex reveals that the homogeneous two-state model is inadequate: development of a heterogeneous two-state model. J. Mol. Biol. 326:1597-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brody, M. S., V. Stewart, and C. W. Price. 2009. Bypass suppression analysis maps the signalling pathway within a multidomain protein: the RsbP energy stress phosphatase 2C from Bacillus subtilis. Mol. Microbiol. 72:1221-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown, J. H., C. Cohen, and D. A. D. Parry. 1996. Heptad breaks in α-helical coiled coils: stutters and stammers. Proteins 26:134-145. [DOI] [PubMed] [Google Scholar]
- 20.Butler, S. L., and J. J. Falke. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 37:10746-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casino, P., V. Rubio, and A. Marina. 2009. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139:325-336. [DOI] [PubMed] [Google Scholar]
- 22.Catlett, N. L., O. C. Yoder, and B. G. Turgeon. 2003. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell 2:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chervitz, S. A., and J. J. Falke. 1995. Lock on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J. Biol. Chem. 270:24043-24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheung, J., and W. A. Hendrickson. 2009. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure 17:190-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang, R. C., R. Cavicchioli, and R. P. Gunsalus. 1997. ‘Locked-on’ and ‘locked-off’ signal transduction mutations in the periplasmic domain of the Escherichia coli NarQ and NarX sensors affect nitrate- and nitrite-dependent regulation by NarL and NarP. Mol. Microbiol. 24:1049-1060. [DOI] [PubMed] [Google Scholar]
- 26.Collins, L. A., S. M. Egan, and V. Stewart. 1992. Mutational analysis reveals functional similarity between NARX, a nitrate sensor in Escherichia coli K-12, and the methyl-accepting chemotaxis proteins. J. Bacteriol. 174:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crick, F. H. C. 1953. The packing of α-helices: simple coiled-coils. Acta Crystallogr. 6:689-697. [Google Scholar]
- 28.Cunningham, B. C., and J. A. Wells. 1989. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244:1081-1085. [DOI] [PubMed] [Google Scholar]
- 29.den Hollander, A. I., R. Roepman, R. K. Koenekoop, and F. P. Cremers. 2008. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog. Retin. Eye Res. 27:391-419. [DOI] [PubMed] [Google Scholar]
- 30.Doebber, M., E. Bordignon, J. P. Klare, J. Holterhues, S. Martell, N. Mennes, L. Li, M. Engelhard, and H. J. Steinhoff. 2008. Salt-driven equilibrium between two conformations in the HAMP domain from Natronomonas pharaonis: the language of signal transfer? J. Biol. Chem. 283:28691-28701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutta, R., L. Qin, and M. Inouye. 1999. Histidine kinases: diversity of domain organization. Mol. Microbiol. 34:633-640. [DOI] [PubMed] [Google Scholar]
- 32.Falke, J. J., and A. H. Erbse. 2009. The piston rises again. Structure 17:1149-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao, R., and A. M. Stock. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray, M. J. 2002. M.S. thesis. University of California, Davis, CA.
- 37.Gruber, M., J. Söding, and A. N. Lupas. 2006. Comparative analysis of coiled-coil prediction methods. J. Struct. Biol. 155:140-145. [DOI] [PubMed] [Google Scholar]
- 38.Harbury, P. B., T. Zhang, P. S. Kim, and T. Alber. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401-1407. [DOI] [PubMed] [Google Scholar]
- 39.Hazelbauer, G. L., J. J. Falke, and J. S. Parkinson. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33:9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodges, R. S., J. Mills, S. McReynolds, J. P. Kirwan, B. Tripet, and D. Osguthorpe. 2009. Identification of a unique “stability control region” that controls protein stability of tropomyosin: a two-stranded alpha-helical coiled-coil. J. Mol. Biol. 392:747-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsing, W., F. D. Russo, K. K. Bernd, and T. J. Silhavy. 1998. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J. Bacteriol. 180:4538-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hulko, M., F. Berndt, M. Gruber, J. U. Linder, V. Truffault, A. Schultz, J. Martin, J. E. Schultz, A. N. Lupas, and M. Coles. 2006. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126:929-940. [DOI] [PubMed] [Google Scholar]
- 43.Inouye, M. 2006. Signaling by transmembrane proteins shifts gears. Cell 126:829-831. [DOI] [PubMed] [Google Scholar]
- 44.Liu, J. D., and J. S. Parkinson. 1989. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J. Bacteriol. 171:1254-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, S. M., and R. S. Hodges. 2004. Defining the minimum size of a hydrophobic cluster in two-stranded alpha-helical coiled-coils: effects on protein stability. Protein Sci. 13:714-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lupas, A., S. Muller, K. Goldie, A. M. Engel, A. Engel, and W. Baumeister. 1995. Model structure of the Ompα rod, a parallel four-stranded coiled coil from the hyperthermophilic eubacterium Thermotoga maritima. J. Mol. Biol. 248:180-189. [DOI] [PubMed] [Google Scholar]
- 47.Lupas, A. N., and M. Gruber. 2005. The structure of α-helical coiled coils. Adv. Protein Chem. 70:37-78. [DOI] [PubMed] [Google Scholar]
- 48.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria. A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 49.Manson, M. D. 2008. The tie that binds the dynamic duo: the connector between AS1 and AS2 in the HAMP domain of the Escherichia coli Tsr chemoreceptor. J. Bacteriol. 190:6544-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manson, M. D. 2009. A mutational wrench in the HAMP gearbox. Mol. Microbiol. 73:742-746. [DOI] [PubMed] [Google Scholar]
- 51.Marina, A., C. D. Waldburger, and W. A. Hendrickson. 2005. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 24:4247-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLachlan, A. D., M. Stewart, and L. B. Smillie. 1975. Sequence repeats in α-tropomyosin. J. Mol. Biol. 98:281-291. [DOI] [PubMed] [Google Scholar]
- 53.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 54.Möglich, A., R. A. Ayers, and K. Moffat. 2009. Design and signaling mechanism of light-regulated histidine kinases. J. Mol. Biol. 385:1433-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 56.Pauling, L., R. B. Corey, and H. R. Branson. 1951. The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. U. S. A. 37:205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potter, L. R. 2005. Domain analysis of human transmembrane guanylyl cyclase receptors: implications for regulation. Front. Biosci. 10:1205-1220. [DOI] [PubMed] [Google Scholar]
- 58.Rabin, R. S., and V. Stewart. 1992. Either of two functionally redundant sensor proteins, NarX and NarQ, is sufficient for nitrate regulation in Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 89:8419-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramamurthy, V., C. Tucker, S. E. Wilkie, V. Daggett, D. M. Hunt, and J. B. Hurley. 2001. Interactions within the coiled-coil domain of RetGC-1 guanylyl cyclase are optimized for regulation rather than for high affinity. J. Biol. Chem. 276:26218-26229. [DOI] [PubMed] [Google Scholar]
- 60.Singh, M., B. Berger, P. S. Kim, J. M. Berger, and A. G. Cochran. 1998. Computational learning reveals coiled coil-like motifs in histidine kinase linker domains. Proc. Natl. Acad. Sci. U. S. A. 95:2738-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sodek, J., R. S. Hodges, L. B. Smillie, and L. Jurasek. 1972. Amino-acid sequence of rabbit skeletal tropomyosin and its coiled-coil structure. Proc. Natl. Acad. Sci. U. S. A. 69:3800-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stewart, V. 2003. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem. Soc. Trans. 31:1-10. [DOI] [PubMed] [Google Scholar]
- 63.Stewart, V., and J. Parales, Jr. 1988. Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J. Bacteriol. 170:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 65.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis, p. 25-51. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, DC.
- 66.Strelkov, S. V., and P. Burkhard. 2002. Analysis of α-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 137:54-64. [DOI] [PubMed] [Google Scholar]
- 67.Swain, K. E., and J. J. Falke. 2007. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry 46:13684-13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swain, K. E., M. A. Gonzalez, and J. J. Falke. 2009. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry 48:9266-9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tao, W., C. L. Malone, A. D. Ault, R. J. Deschenes, and J. S. Fassler. 2002. A cytoplasmic coiled-coil domain is required for histidine kinase activity of the yeast osmosensor, SLN1. Mol. Microbiol. 43:459-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tomenius, H., A. K. Pernestig, C. F. Mendez-Catala, D. Georgellis, S. Normark, and O. Melefors. 2005. Genetic and functional characterization of the Escherichia coli BarA-UvrY two-component system: point mutations in the HAMP linker of the BarA sensor give a dominant-negative phenotype. J. Bacteriol. 187:7317-7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tomomori, C., T. Tanaka, R. Dutta, H. Park, S. K. Saha, Y. Zhu, R. Ishima, D. Liu, K. I. Tong, H. Kurokawa, H. Qian, M. Inouye, and M. Ikura. 1999. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat. Struct. Biol. 6:729-734. [DOI] [PubMed] [Google Scholar]
- 72.Utsumi, R., R. E. Brissette, A. Rampersaud, S. A. Forst, K. Oosawa, and M. Inouye. 1989. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science 245:1246-1249. [DOI] [PubMed] [Google Scholar]
- 73.Ward, S. M., A. F. Bormans, and M. D. Manson. 2006. Mutationally altered signal output in the Nart (NarX-Tar) hybrid chemoreceptor. J. Bacteriol. 188:3944-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ward, S. M., A. Delgado, R. P. Gunsalus, and M. D. Manson. 2002. A NarX-Tar chimera mediates repellent chemotaxis to nitrate and nitrite. Mol. Microbiol. 44:709-719. [DOI] [PubMed] [Google Scholar]
- 75.Watts, K. J., M. S. Johnson, and B. L. Taylor. 2008. Structure-function relationships in the HAMP and proximal signaling domains of the aerotaxis receptor Aer. J. Bacteriol. 190:2118-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams, S. B., and V. Stewart. 1997. Nitrate- and nitrite-sensing protein NarX of Escherichia coli K-12: mutational analysis of the amino-terminal tail and first transmembrane segment. J. Bacteriol. 179:721-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams, S. B., and V. Stewart. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 33:1093-1102. [DOI] [PubMed] [Google Scholar]
- 78.Winston, S. E., R. Mehan, and J. J. Falke. 2005. Evidence that the adaptation region of the aspartate receptor is a dynamic four-helix bundle: cysteine and disulfide scanning studies. Biochemistry 44:12655-12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wolanin, P. M., P. A. Thomason, and J. B. Stock. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 3:reviews3013.1-3013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woolfson, D. N. 2005. The design of coiled-coil structures and assemblies. Adv. Protein Chem. 70:79-112. [DOI] [PubMed] [Google Scholar]
- 81.Xu, Q., W. P. Black, E. M. Mauriello, D. R. Zusman, and Z. Yang. 2007. Chemotaxis mediated by NarX-FrzCD chimeras and nonadapting repellent responses in Myxococcus xanthus. Mol. Microbiol. 66:1370-1381. [DOI] [PubMed] [Google Scholar]
- 82.Xu, Q., W. P. Black, S. M. Ward, and Z. Yang. 2005. Nitrate-dependent activation of the Dif signaling pathway of Myxococcus xanthus mediated by a NarX-DifA interspecies chimera. J. Bacteriol. 187:6410-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yanofsky, C., V. Horn, and D. Thorpe. 1964. Protein structure relationships revealed by mutational analysis. Science 146:1593-1594. [DOI] [PubMed] [Google Scholar]
- 84.Zhou, Q., P. Ames, and J. S. Parkinson. 2009. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol. Microbiol. 73:801-814. [DOI] [PMC free article] [PubMed] [Google Scholar]