Abstract
The saePQRS system of Staphylococcus aureus controls the expression of major virulence factors and encodes a histidine kinase (SaeS), a response regulator (SaeR), a membrane protein (SaeQ), and a lipoprotein (SaeP). The widely used strain Newman is characterized by a single amino acid change in the sensory domain of SaeS (Pro18 in strain Newman [SaeSP], compared with Leu18 in other strains [SaeSL]). SaeSP determines activation of the class I sae target genes (coa, fnbA, eap, sib, efb, fib, sae), which are highly expressed in strain Newman. In contrast, class II target genes (hla, hlb, cap) are not sensitive to the SaeS polymorphism. The SaeSL allele (saeSL) is dominant over the SaeSP allele, as shown by single-copy integration of saePQRSL in strain Newman, which results in severe repression of class I target genes. The differential effect on target gene expression is explained by different requirements for SaeR phosphorylation. From an analysis of saeS deletion strains and strains with mutated SaeR phosphorylation sites, we concluded that a high level of SaeR phosphorylation is required for activation of class I target genes. However, a low level of SaeR phosphorylation, which can occur independent of SaeS, is sufficient to activate class II target genes. Using inducible saeRS constructs, we showed that the expression of both types of target genes is independent of the saeRS dosage and that the typical growth phase-dependent gene expression pattern is not driven by SaeRS.
The human pathogen Staphylococcus aureus can cause a wide range of diseases. The versatility of this organism is due to its capacity to produce accessory molecules that mediate specific interactions with the host cells. Most of these factors are tightly regulated by global regulatory loci, such as agr, rot, sigB, and sae, which act as an interactive regulatory network to ensure that there is coordinated temporal expression of virulence factors. Within the network, the sae system appears to be a central downstream regulator. Mutations in sae do not affect the transcription of agr, sigB, sarA, or rot (18, 22, 35). However, at least in some strains, agr activates sae transcription, while sigB and rot repress sae transcription (17, 19, 21, 35, 44). The sae system controls the expression of major virulence genes, including hla (encoding α-hemolysin [Hla]), hlb (encoding β-hemolysin [Hlb]), coa (encoding coagulase [Coa]), eap (encoding extracellular adherence protein [EAP]), and fnbA (encoding fibronectin binding protein A [FnBPA]), probably through direct interaction with these target genes (18, 33, 36, 44, 46, 48, 52).
The sae locus consists of four open reading frames (ORFs), and the products of two of these ORFs (saeR and saeS) show strong sequence homology to bacterial two-component regulators composed of a histidine kinase (HK), SaeS, and a response regulator (RR), SaeR. Two additional ORFs, ORF3 (saeQ) and ORF4 (saeP), which are located upstream of saeRS, are likely to be important for the function of the sae operon (1, 17, 48). saeP encodes a putative lipoprotein, and saeQ encodes a membrane protein with four membrane-spanning stretches. Four overlapping sae-specific transcripts (T1 to T4) have been detected; the T1 message (3.1 kb) initiates upstream of saeP, T2 (2.4 kb) initiates upstream of saeQ, and T3 (2.0 kb) initiates upstream of saeR. T4 (0.7 kb) represents a monocistronic mRNA encompassing only saeP (Fig. 1C). Promoter fusion analysis indicated that the main T2 transcript is generated by endoribonucleolytic processing of the T1 transcript (1, 17). Only two distinct promoter elements (P1 and P3) were detected in the saeRS upstream region. The P3 promoter, which is upstream of saeRS, generates the T3 transcript and is repressed by saeRS. In contrast, the distal P1 promoter is strongly autoregulated (1, 17, 35) and is sensitive to environmental signals, such as pH, H2O2, and α-defensins (1, 17).
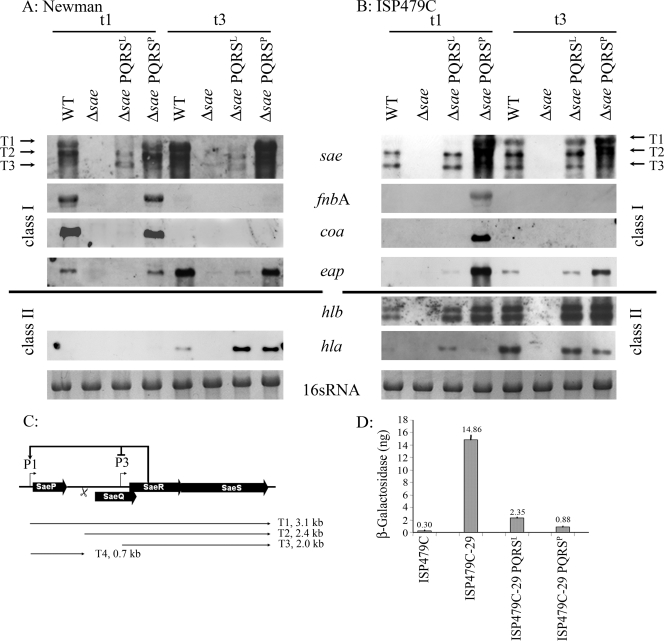
FIG. 1.
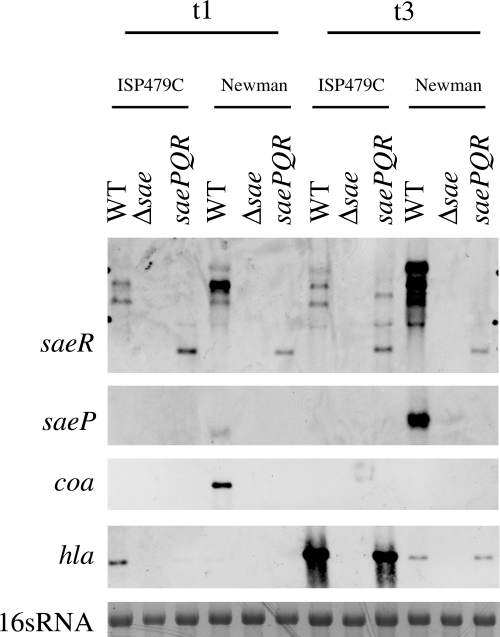
Influence of N-terminal SaeS polymorphism on sae transcription and target gene expression. (A and B) Cross-complementation of sae mutants Newman-29 (A) and ISP479C-29 (B). For single-copy complementation the saePQRS operon from either strain Newman or strain ISP479C was integrated into the lipase gene of sae mutant strains. Bacteria were grown to the exponential (t1) (OD600, 0.5) and postexponential (t3) (OD600, 6) growth phases. RNA was hybridized with digoxigenin-labeled PCR fragments. In strain Newman, the hlb gene is inactivated by phage insertion, and therefore no hlb was detected. The 16S rRNA detected in the ethidium bromide-stained gels was used as a loading control in the lower lanes. WT, wild type. (C) Organization of the sae locus composed of four ORFs. Transcription initiates at the P1 and P3 promoters, generating transcripts T1 and T3, respectively. T2 is generated by processing of T1. P1 is strongly activated and P2 is repressed by SaeR (17). (D) cap promoter fusion assays. cap-lacZ fusions were integrated into strain ISP479C, the sae mutant ISP479C-29, and the complemented sae mutants. ISP479C-29 was complemented by integration of pCWSAE50 (ISP479C-29 PQRSL) and pCWSAE51 (ISP479C-29 PQRSP) into hlb. Bacteria were grown to postexponential growth phase (t3), equal numbers of bacteria were lysed, and promoter activities were expressed in β-galactosidase equivalents (in nanograms). Standard deviations were derived from data for at least two independent cultures.
Two-component systems are the predominant form of signal transduction in bacteria (for a recent review, see reference 15). In response to a specific signal, the HK component uses ATP to autophosphorylate at a conserved histidine residue. The phosphoryl group is then transferred to a conserved aspartate residue in the cognate RR. In most cases, activation of the RR leads to gene activation or repression by interaction with specific binding motifs located upstream of the target genes. In addition, many HKs are predicted to be bifunctional enzymes that also exhibit phosphatase activity towards their cognate RRs (2). Depending on the N-terminal signaling domain of the HK, signaling may occur from the extracytoplasmic space, from the cytoplasm, or from within the membrane. SaeS from S. aureus can be classified as a member of the group of intramembrane sensing HKs (38, 39). These small HKs are characterized by a short intramembrane sensing domain consisting of two transmembrane helices with an extracytoplasmic linker consisting of less than 25 amino acids and are thought to sense their stimulus at or within the membrane. Based on sequence analysis of the conserved domains characterizing HKs (12, 24, 29, 38, 39) or RRs (13, 14, 24), the saeRS system can be considered closely related to the EnvZ/OmpR-like two-component systems.
In S. aureus, regulatory circuits have been analyzed mainly using derivatives of the prototypic strains 8325-4 and Newman. Strain 8325-4 produces high levels of Hla but little clumping factor A, FnBP, or Coa. This is due in part to a mutation in rsbU, which encodes a protein that is important for the activity of alternative sigma factor B (32). Furthermore, due to a mutation in the cap5 gene cluster, strain 8325-4 is not encapsulated (53). In contrast, strain Newman contains a functional rsbU gene and a cap5 operon and is known to produce large amounts of Coa, clumping factor A (55), Eap, and Emp (27). FnBPA, although not functional (25), is also transcribed at high levels (56) in strain Newman. Hla expression in strain Newman is low during growth in vitro, but during infections it can be activated to the level observed in derivatives of strain 8325-4 (22). Overall, strain Newman seems to be more virulent than strain 8325-4 in animal models of infection (K. Ohlsen, personnel communication; our observations). Sequence analysis revealed a single amino acid variation within the first N-terminal transmembrane loop of SaeS of strain Newman compared to SaeS of strain 8325-4 or of other strains (48). Thus, the saeS polymorphism may, at least in part, account for the differences in virulence gene expression between strain Newman and 8325-4 derivatives.
In this study we showed that a single amino acid change in the first transmembrane loop of SaeS with Pro at residue 18 (SacP), as found in strain Newman, is indeed responsible for major peculiarities of this strain. The polymorphism probably results in a high kinase/phosphatase ratio for SaeSP, which is necessary for expression of class I target genes (coa, fnbA, eap, sib, efb) but is not required for expression of class II target genes (hla, hlb, cap). The expression of different target genes is determined by the SaeS polymorphism but is not sensitive to the SaeRS dosage.
MATERIALS AND METHODS
Strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. In vitro growth was performed using CYPG (10 g/liter Casamino Acids, 10 g/liter yeast extract, 5 g/liter NaCl, 0.5% glucose, 0.06 M phosphoglycerate) (43) supplemented with the appropriate antibiotics for strains carrying resistance genes (50 μg/ml kanamycin, 10 μg/ml erythromycin, 5 μg/ml tetracycline, 10 μg/ml chloramphenicol). For RNA isolation and promoter fusion assays bacteria from an overnight culture were diluted to obtain an initial optical density at 600 nm (OD600) of 0.05 in fresh medium without antibiotics and grown with shaking at 37°C to the mid-exponential (t1) (OD600, 0.5), late-exponential (t2) (OD600, 3), or postexponential (t3) (OD600, 6) phase. For analysis of extracellular proteins strains were grown for 2.5 h in basic medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| One Shot TOP10 | Competent E. coli strain for plasmid transformation | Invitrogen, Karlsruhe, Germany |
| CYL316 | RN4220(pYL112Δ19), L54 int gene, r− | 34 |
| CG18 | RN4220(pCG32), φ13 int gene, r− | This study |
| RN4220 | Restriction-deficient S. aureus strain, r− | 31 |
| RN4220-29 | RN4220 ΔsaePQRS::kan | This study |
| RN1 | Strain 8325 lysogenic for three phages | 42 |
| RN1saeN | RN1 with SaeSP from strain Newman | This study |
| Newman | Wild type | 11 |
| NewHG | Newman with SaeSL from strain RN1 | This study |
| ISP479C | Derivative of 8325-4 | 45 |
| Newman-29 | Newman ΔsaePQRS::kan | This study |
| ISP479C-29 | ISP479C ΔsaePQRS::kan | This study |
| Plasmids | ||
| pBT2 | Low-copy-number shuttle vector with Ampr in E. coli and temperature-sensitive replication with Cmr in S. aureus | 7 |
| pCR2.1 | Cloning vector | Invitrogen, Karlsruhe, Germany |
| pMAD | Shuttle vector for gene replacement mutagenesis | 3 |
| pMADsaeSL | pMAD with the saePQRSL operon from strain RN1 | This study |
| pMADsaeSP | pMAD with the saePQRSP operon from strain Newman | This study |
| pEC4 | pBluescript II KS(+) with ermB inserted into ClaI site | 7 |
| pCL84 | Single-copy integration vector, att site for chromosomal integration in geh | 34 |
| pALC2073 | S. aureus vector containing a tetracycline-inducible promoter controlling expression of cloned genes, Cmr | 5 |
| pSK236 | Shuttle vector containing pUC19 cloned into the HindIII site of pC194 | 16 |
| pGEM | Cloning vector | Promega, Mannheim, Germany |
| pCG3 | pCL84 with replacement of tetK by ermB | This study |
| pCG32 | pSK236 with φ13 int gene | This study |
| pCG6 | pCL84 with integrated spoVG-lacZ gene cassette from pKO010 | 17 |
| pCG18 | pCG6 with cap8 promoter | This study |
| pCG33 | hlb integration vector, pCG3 with replacement of L54 attP site with attP of φ13 | This study |
| pCWSAE29 | pMAD construct used to generate ΔsaePQRS::kan | This study |
| pCWSAE7 | pGEM with saePQRSL (nucleotides 1 to 3515), | 48 |
| pCWSAE28 | pCL84 with saePQRSL (nucleotides 1 to 3515), subclone from pCWSAE7 | This study |
| pCWSAE50 | pCG33 with saePQRSL (nucleotides 1 to 3515), subclone of pCWSAE7 | This study |
| pCWSAE32 | pCR2.1 with saePQRSP (nucleotides 1 to 3515) | This study |
| pCWSAE33 | pCL84 with saePQRSP (nucleotides 1 to 3515), subclone from pCWSAE32 | This study |
| pCWSAE51 | pCG33 with saePQRSP (nucleotides 1 to 3515), subclone of pCWSAE32 | This study |
| pCWSAE43 | pALC2073 with saeRSP (nucleotides 1509 to 3449) | This study |
| pCWSAE45 | pALC2073 with saeRSL (nucleotides 1509 to 3449) | This study |
| pCWSAE56 | pCL84 with saePQR(D51N)SL (nucleotides 1 to 3515) | This study |
| pCWSAE54 | pCL84 with saePQR (nucleotides 1 to 3515, Δ2267-3218) | This study |
| pCWSAE59 | pCL84 with saePQR(D51N)SP (nucleotides 1 to 3515) | This study |
| pCWSAE74 | pCR2.1 with saeR(D51N)SL (nucleotides 1509 to 3515), derivative of pCWSAE59 | This study |
| pCWSAE75 | PCR2.1 with saeR(D51N)SP (nucleotides 1509 to 3515), derivative of pCWSAE56 | This study |
| pCWSAE78 | pALC2073 with saeR(D51N)SL (nucleotides 1509 to 3515), subclone from pCWSAE74 | This study |
| pCWSAE79 | pALC2073 with saeR(D51N)SP (nucleotides 1509 to 3515), subclone from pCWSAE75 | This study |
Nucleotide numbers are based on the sae sequence (GenBank accession no. AJ556795).
Replacement of sae.
The sae locus was replaced by a kanamycin resistant cassette. Two fragments flanking the sae locus were amplified from strain ISP479C using oligonucleotides A and B and oligonucleotides C and D (see Table S1 in the supplemental material). Oligonucleotides A and D were modified with a KpnI restriction site. The kanamycin resistance cassette (aphIII) was amplified from pDG782 using oligonucleotides E and F. Reverse oligonucleotide B for amplification of the sae upstream fragment and forward oligonucleotide C for amplification of the sae downstream fragment were synthesized with additional nucleotides complementary to the flanking region of aphIII. All three fragments were annealed without oligonucleotides and amplified by PCR using oligonucleotides A and D to obtain the complete replacement cassettes. The amplicon was restricted with KpnI and cloned into pBT2 (7). To take advantage of blue-white selection, the fusion fragments were subcloned into the EcoRI and SalI sites of pMAD (3), yielding plasmid pCWSAE29. This temperature-sensitive plasmid was electroporated into S. aureus strain RN4220, and sae gene replacement mutants were generated as described previously (3). The mutation was transduced into strain Newman and strain ISP479C using phage φ11 lysates, resulting in sae gene replacement mutants Newman-29 and ISP479C-29. All mutants were verified by Southern hybridization after pulsed-field gel electrophoresis and PCR.
Chromosomal amino acid exchange in SaeS of strain RN1HG and strain Newman.
Amino acid exchange in the products of the native sae operons of strain RN1 and strain Newman was accomplished by homologous recombination with pMAD vectors in which the saePQRS operon was cloned (3). The pMADsaeSP vector included a 3.5-kb PCR fragment amplified from DNA isolated from S. aureus Newman to obtain a chromosomal point mutation in saeS (resulting in a change from SaeS with L18 [SaeSL] to SaeS with P18 [SaeSP]) in strain RN1. Similarly, an amino acid change in SaeS from P18 to L18 in strain Newman was obtained by using pMADsaeSL with a 3.5-kb PCR fragment amplified from strain RN1. Successful mutagenesis of the chromosomal sae operons was verified by DNA sequencing. Strain RN1 with the saeS allele encoding SaeSP (saeSP) was designated RN1saeN, and strain Newman with the saeS allele encoding SaeSL (saeSL) was designated NewHG.
Chromosomal integration of different parts of the sae operon.
Different parts of the sae operon were amplified using high-fidelity polymerases (Phusion HF polymerase; Finnzymes, Espoo, Finland) and oligonucleotides (see Table S1 in the supplemental material) modified with restriction sites. The amplicons were either cloned into pCR2.1 and subcloned or directly cloned into the EcoRI site of the respective vectors. Plasmids were verified by restriction digestion and sequencing of the inserts. The 3.5-kb sae operons of strain ISP479C(pCWSAE7) (48) and strain Newman(pCWSAE32) were subcloned into the single EcoRI site of pCL84 (34), resulting in pCWSAE28 and pCWSAE33, respectively, and alternatively were subcloned into pCG33, resulting in pCWSAE50 and pCWSAE51, respectively. pCG33 is a derivative of pCG3 with a φ13-attP integration site. pCG3 was obtained by replacing the tetK gene of pCL84 with an ermB resistance cassette. The ermB was excised from pEC4 (7) with HindIII and SalI and cloned into the corresponding restriction sites of pCL84. For construction of pCG33 the φ13-attP site was amplified with modified oligonucleotides EcoRIphi13attP-for and Salphi13attP-rev using strain RN1 as the template. The amplicon was cloned into EcoRI-SalI-digested vector pCG3 to replace the L54-specific attP site.
saePQR with an intact terminator region was constructed by strand overlap extension PCR in two steps. Two amplicons were generated using oligonucleotide sae-1U with oligonucleotide 2267hybrid (hybrid reverse oligonucleotide) and oligonucleotide 3218hybrid (hybrid forward oligonucleotide) with oligonucleotide sae-3515L. Diluted amplicons were annealed and amplified by PCR using sae-1U and sae-3515L. The fusion product was restricted with EcoRI and cloned in the corresponding restriction site of the pCL84 vector to obtain pCWSAE54.
The phosphorylation site (D51) in saeR was mutated by strand overlap extension PCR. Two amplicons were generated with overlapping 3′ and 5′ ends with oligonucleotide pairs sae-1U/1680L29 and 1684U31/sae-3515L. Oligonucleotides 1680L29 and 1684U31 are complementary and have a single nucleotide mismatch that changes codon 51 from an aspartate codon (GAT) to an asparagine codon (AAT). DNA from strains Newman and ISP479C were used as templates. Amplicons were annealed without oligonucleotides and amplified using oligonucleotides sae-1U and sae-3515L. The fusion products were restricted with EcoRI and cloned in the corresponding restriction site of pCL84 to obtain pCWSAE56 [saePQR(D51N)SL] and pCWSAE59 [saePQR(D51N)SP].
The pCL84 derivatives were integrated by electroporation into geh of strain CYL316 (34), and the pCG33 derivatives were integrated by electroporation into hlb of strain CG18, which contains the cloned integrase gene of φ13. The φ13 integrase was amplified using modified oligonucleotides listed in Table S1 in the supplemental material, and the amplicons were cloned into the EcoRI and SalI sites of the multicopy vector pSK236, yielding pCG32. RN4220 containing pCG32 was designated strain CG18.
Integrated plasmids were transduced into different strains using φ11 lysates. All transductants were analyzed by PCR and/or Southern hybridization of SmaI-digested DNA separated by pulsed-field gel electrophoresis (PFGE) using probes specific for sae. Due to the single SmaI site in pCL84 and pCG33, integration of the plasmids resulted in digestion of the geh- or hlb-containing SmaI fragment into two bands, one of which reacted with the sae probe in all cointegrants.
Cloning of different parts of the sae operon into pALC2073.
To clone different parts of the sae operon into the anhydrotetracyline (AHT)-inducible vector pALC2073, PCR fragments were generated using oligonucleotides listed in Table S1 in the supplemental material, or fragments were subcloned from the pCL84 derivatives described above. DNA of strain Newman or strain ISP479C was used as the template. For constructs with a mutated saeR gene plasmid pCWSAE56 or pCWSAE59 was used as the template. Amplicons were either cloned into pCR2.1 and subcloned or directly cloned into the EcoRI site of pALC2073. Plasmids and insert orientation were verified by restriction digestion or PCR and sequencing of the inserts. Plasmids were electroporated into strain RN4220 and transduced into different S. aureus strains.
Construction of chromosomally integrated promoter-lacZ fusions.
The cap8 promoter was amplified with the appropriate oligonucleotides (see Table S1 in the supplemental material) with additional EcoRI restriction sites. Amplicons were ligated into the EcoRI site of pCG6 (17) in front of the promoterless lacZ gene, yielding pCG18. Plasmids were verified by restriction digestion, and the correct orientation of the inserts was verified by PCR. Plasmids were electroporated into S. aureus strain CYL316 and transduced into the sae mutant of strain ISP479C, which was complemented by integration of saePQRSP (pCWSAE51) and saePQRSL (pCWSAE50) into hlb. All transductants were verified by PCR.
Promoter activity assays.
Promoter activity assays were performed as described previously (17). Briefly, equal numbers of bacteria were harvested at the appropriate growth phase, resuspended in 1 ml of 0.1 M sodium phosphate buffer (pH 7.4), and lysed with 0.5 ml zirconia-silica beads (diameter, 0.1 mm) in a high-speed homogenizer (Savant Instruments, Farmingdale, NY). β-Galactosidase activity was measured using a FluoReporter galactosidase quantitation kit (Invitrogen, Karlsruhe, Germany). Promoter activities were expressed in ng/ml β-galactosidase based on a standard curve generated by using purified β-galactosidase (Sigma-Aldrich, Munich, Germany). All values are means for at least two independently grown bacterial cultures.
Analysis of extracellular proteins.
The exoproteins of the supernatant were concentrated with StrataClean resin (Stratagene). Proteins were separated according to their molecular weights by one-dimensional PAGE (12%), and after this the gels were stained using 0.1% Coomassie brilliant blue R250, 10% acetic acid, 50% methanol and destained using 10% acetic acid.
RNA isolation and Northern blot hybridization.
RNA isolation and Northern blot analysis were performed as described previously (20). Briefly, approximately 109 S. aureus cells were lysed in 1 ml Trizol reagent (Invitrogen Life Technologies, Karlsruhe, Germany) with 0.5 ml zirconia-silica beads (diameter, 0.1 mm) in a high-speed homogenizer (Savant Instruments, Farmingdale, NY). RNA was isolated as described in the instructions provided by the manufacturer of Trizol. Several digoxigenin-labeled probes for detection of specific transcripts were generated using a DIG PCR labeling kit by following the manufacturer's instructions (Roche Biochemicals, Mannheim, Germany). Oligonucleotides used for probe generation are listed in Table S1 in the supplemental material.
RESULTS
Influence of N-terminal SaeS polymorphism on sae transcription and target gene expression.
The sequences of the products of the saePQRS operons of strain Newman (GenBank accession no. AJ556794) and the 8325-4 derivative ISP479C (GenBank accession no. AJ556795) differ at only a single amino acid residue in SaeS (48). All saeS sequences derived from genome sequencing projects except those for strain Newman were identical to the saeS sequence of strain ISP479C. The mutation in SaeS (Leu18 to Pro18) of strain Newman is located in the first transmembrane loop of the N-terminal signaling domain, indicating that there is a perturbation in the sensing mechanism. Below, SaeSP indicates SaeS from strain Newman, and SaeSL indicates SaeS from strain ISP479C. Previously, we and other workers have shown that strain Newman is characterized by an unusually high level of expression of the sae operon (1, 17, 48). To evaluate whether the sequence polymorphism in SaeS can account for the differential expression of sae, we cloned the saePQRSP and saePQRSL operons into the integration vector pCL84. The vector was integrated into the lipase gene of two sae gene replacement mutants, Newman-29 and ISP479C-29. In these mutants, the whole saePQRS operon was replaced by a kanA resistance cassette. The strains were grown to the exponential (t1) and postexponential (t3) growth phases and analyzed by Northern hybridization. The transcriptional patterns of the three sae-specific transcripts (T1, T2, and T3) were highly dependent on the saeS allele, and the saeS polymorphism fully accounted for the strain-specific pattern of sae transcription (Fig. 1A and B). Integration of saePQRSL led to expression of the three sae transcripts at levels that are comparable to those in ISP479C. On the other hand, integration of saePQRSP led to restoration of the sae expression pattern typical of strain Newman, namely, elevated expression of T1 and T2. The same results were obtained for derivatives of strain Newman (Fig. 1A) and strain ISP479C (Fig. 1B).
Next, we analyzed the effect of the saeS alleles on target gene expression in strain ISP479C and strain Newman. First, prototypic sae target genes (coa, fnbA, and eap), which encode cell-associated proteins with specific binding motifs, were analyzed. In the sae mutant of strain Newman, integration of saePQRSP led to full restoration of coa and fnbA transcription to wild-type levels. In contrast, integration of saePQRSL did not result in detectable transcription of coa and fnbA. In strain ISP479C, the levels of transcription of coa and fnbA were below the detection limit of Northern analysis (Fig. 1B). However, introduction of saePQRSP into the sae mutant of ISP479C resulted in fnbA and coa expression levels comparable to those obtained for strain Newman. Transcription of eap was detected in both wild-type strains, primarily during the postexponential growth phase, and the level of expression in strain Newman was higher than that in strain ISP479C. Again, the higher level of expression of eap in strain Newman could be linked to the sae allele; complementation of the sae mutants with saePQRSP resulted in stronger activation of eap than complementation with saePQRSL. Thus, the high level of transcription of fnbA, coa, and eap in strain Newman compared to the level of transcription in strain ISP479C could clearly be linked to the SaeS polymorphism in the former strain, which seems to be characterized by sae hyperactivity. It is noteworthy that strain ISP479C does not express detectable levels of sigma factor B-activated coa and fnbA due to a mutation in rsbU (6). When strain ISP479r, a derivative of ISP479C with a repaired rsbU gene (49), was analyzed, a low level of sae-dependent expression of coa was detected (data not shown). The sae mutant of ISP479r was negative for coa expression, indicating that saePQRSL can activate coa to some extent, although much less than saePQRSP.
Next, we analyzed genes encoding hemolysins (hla, hlb) that are also known to be strongly sae dependent. In both strains, the sae mutation resulted in downregulation of hla expression, which could be restored by integration of saePQRSP or saePQRSL (Fig. 1A and B). Similarly, hlb expression was restored in the sae mutant of ISP479C by both sae alleles (Fig. 1B). Thus, the regulation of hla and hlb by SaeRS is largely independent of the SaeS genotype.
The capAP operon is one of the few sae target operons shown to be upregulated in an sae mutant. Previously, this was shown only for strain Newman (48). We examined whether this effect is also linked to the SaeSP polymorphism by analyzing complemented sae mutants of ISP479C. We used promoter fusion assays to analyze the role of saePQRS in capAP promoter activity. To this end, the cap promoter region was cloned in front of lacZ and integrated into the chromosomal lipase gene. Cloned saePQRS was integrated into the attB site of the hlb-converting phage φ13. It was shown that sae also results in severe repression of cap promoter activity in strain ISP479C. The repressive effect was independent of the saeS allele since integration of saePQRSP and integration of saePQRSL similarly complemented the mutant phenotype.
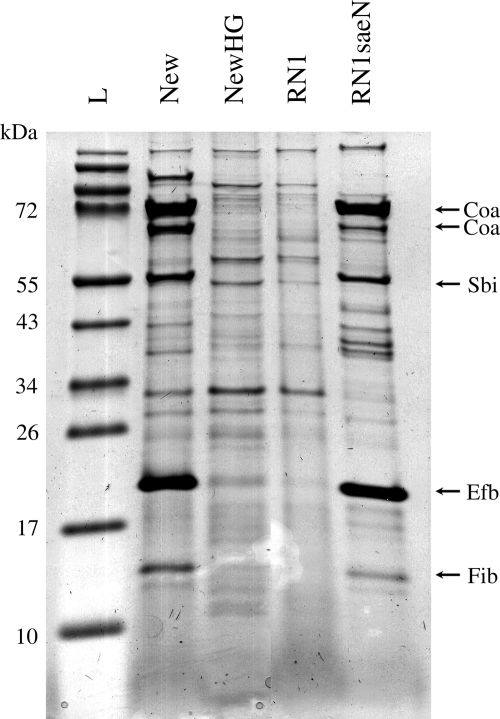
To further show the predominant role of the SaeS polymorphism in target gene repression, we constructed strains in which the native chromosomal saeS was mutated; strain RN1saeN (a derivative of 8325) contains an sae operon with an saeSP allele, whereas strain NewHG (a derivative of strain Newman) contains an saeSL allele. Consistent with the results obtained by cross-complementation (Fig. 1), the gene expression was clearly determined by the SaeS polymorphism (Fig. 2). The extracellular protein profile of strain RN1saeN closely resembles that of strain Newman, namely, high levels of expression of Coa, the IgG binding protein Sbi, and the fibrinogen binding proteins Efb and Fib. This indicates that sbi, efb, and fib also are class I target genes. In contrast, the proteins are clearly downregulated in strain NewHG.
FIG. 2.
Exoprotein pattern of S. aureus strains with mutagenized saeS alleles. Strains were grown for 2.5 h (exponential growth phase), and concentrated proteins from culture supernatants were analyzed by SDS-PAGE. Lane L, ladder; lane New, strain Newman; lane NewHG, strain Newman with saeSL; lane RN1, strain 8325; lane RN1saeN, RN1 with saeSP. The secreted proteins Coa, Sbi, and Efb (MWMN_1069) and the hypothetical fibrinogen-binding MWMN_1066 protein could be identified only in strain Newman and strain RN1 with saeSP.
In summary, the sae-dependent regulation of sae, fnbA, coa, eap, sib, efb, and fib (referred to here as class I target genes) differs markedly from that of hla, hlb, and cap (class II target genes). Class I target genes are highly dependent on SaeSP, whereas class II target genes are independent of the SaeS polymorphism. Below, transcription of coa is used mainly as an example of transcription of the class I target genes and hla is used as a representative of the class II target genes.
Dose-dependent expression of saeRSP and saeRSL using an AHT-inducible promoter system.
It was shown previously using promoter fusion assays that the level of expression of the autoregulated P1 promoter of the sae operon is significantly higher in strain Newman than in other strains (1, 17). As shown in this study, this is solely due to saeSP in strain Newman (Fig. 1). Thus, the concomitant activation of class I target genes may depend on the higher level of transcription of saeRS via SaeSP. To analyze the influence of the saeRS transcript level on target genes, saeRSP and saeRSL were expressed under control of a tetracycline-inducible promoter and introduced into the sae mutants. Low-level saeRS expression was observed in both constructs without induction. However, expression could be clearly enhanced by addition of increasing concentrations of AHT (Fig. 3).
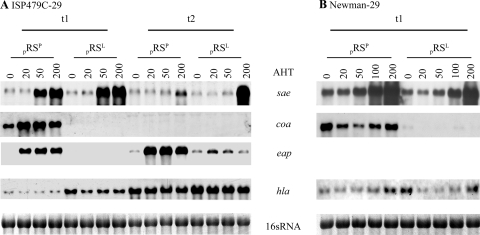
FIG. 3.
Dose-dependent saeRS expression when an AHT-inducible promoter system was used. saeRS from strain Newman (pRSP) or strain ISP479 (pRSL) was cloned behind an AHT-inducible promoter and introduced into the sae mutant ISP479C-29 (A) or Newman-29 (B). Strains were grown to the exponential (t1) (OD600, 0.5) and postexponential (t3) (OD600, 3) growth phases, which was followed by saeRS induction for 1 h with different concentrations of AHT (0, 20, 50, and 200 ng/ml). RNA was hybridized with digoxigenin-labeled PCR fragments. The lane at the bottom contained the 16S rRNA detected in the ethidium bromide-stained gels as a loading control.
hla transcription could be restored in the saePQRS mutant by both saeRS alleles. A higher induction rate was seen with saeRSL than with saeRSP in ISP479C derivatives (Fig. 3A), especially in bacteria from the exponential phase of growth. A similar trend was observed in the single-copy complementation experiments (Fig. 1B). However, different effects of saeRSP and saeRSL on hla transcription were not observed in the analysis of derivatives of strain Newman (Fig. 3B).
When the transcription of coa and eap was analyzed, it was evident that transcription of these genes was driven only by the saeRSP allele. Even a high level of induction of saeRSL did not result in coa expression and yielded only a low level of eap transcription. Similar results were obtained when the same constructs in the saePQRS mutants of strains ISP479C and Newman were analyzed. These results are in line with the finding obtained using the single-copy strains shown in Fig. 1, which indicated that saeSP has a direct effect on class I target genes. It can be concluded that this effect is not due to the higher level of expression of sae in strains harboring saePQRSP, such as strain Newman.
From these experiments, it also became evident that there is little correlation between the level of saeRS and the level of target gene transcription. Increasing the concentration of AHT resulted in a dose-dependent increase in saeRS expression without a concomitant increase in target gene expression. Notably, hla was already induced to maximum levels of expression in cultures without saeRS induction.
We also analyzed whether saeRS is involved in the typical growth phase-dependent expression of the target genes. Thus, saeRS was induced in the exponential and postexponential growth phases by increasing the concentration of AHT for 30 min. Although the AHT induction of saeRS was less prominent in the postexponential phase, it could be clearly shown that the growth phase-dependent expression of the target genes coa and hla is saeRS independent. As in wild-type strains, coa transcription could be induced only in bacteria from the exponential growth phase. In contrast, hla and eap are maximally induced during the postexponential phase. Also, the expression of fnbA, coa, and hla is growth phase dependent in agr mutants. Thus, other regulatory mechanisms besides agr (50, 56) or sae are required for growth phase-dependent regulation of virulence factors.
SaeSL is dominant over SaeSP.
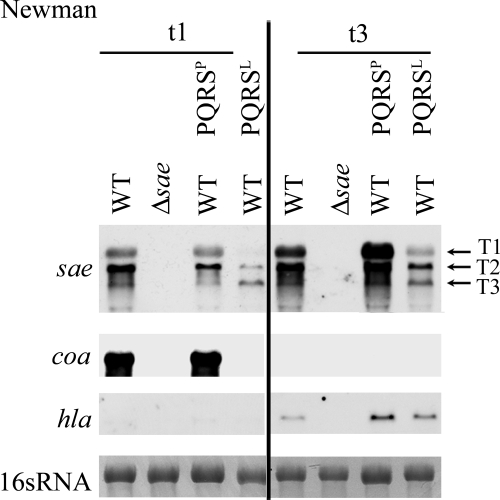
The profound difference in the functionality of the sae regulon between saeSP- and saeSL-expressing strains is presumably due to differences in signal transduction from the membrane-bound SaeS to the response regulator, SaeR, which in turn interacts with the target genes. This assumption is based on the general mechanism elucidated for other two-component systems. Using the SWISS-MODEL Model server (http://swissmodel.expasy.org/), it could be predicted that SaeS belongs to the class of bifunctional HKs that exhibit kinase activity as well as phosphatase activity. One can speculate that the overactive SaeSP protein exhibits constant kinase activity (high levels of phosphorylation of SaeR), whereas in SaeSL the phosphatase activity is dominant. Different target genes may differ in their requirements for the phosphorylation state of SaeR. If the phosphatase activity of SaeSL is dominant, one would expect that SaeSL can titrate the effect of SaeSP. We therefore introduced a copy of saePQRSL into the saePQRSP-containing wild-type Newman strain (Fig. 4). It was clear that in strain Newman just a single copy of saePQRSL resulted in severe inhibition of T1 and T2, as well as repression of coa transcription. As expected, introduction of a second copy of saePQRSP into strain Newman had no effect on either sae or coa transcription. Thus, the effect of SaeSL is dominant over the effect of SaeSP, presumably due to the phosphatase activity of SaeSL.
FIG. 4.
SaeSL is dominant over SaeSP. saePQRS from strain Newman (PQRSP) and saePQRS from strain ISP479C (PQRSL) were integrated into the lipase gene of strain Newman. Bacteria were grown to the exponential (t1) (OD600, 0.5) and late exponential (t2) (OD600, 3) growth phases. RNA was hybridized with digoxigenin-labeled PCR fragments. The lane at the bottom contained the 16S rRNA detected in the ethidium bromide-stained gels as a loading control. WT, wild type.
SaeS-independent activation of the class II target genes.
If SaeSL exhibits mainly phosphatase activity, then the response regulator would be kept mainly in the unphosphorylated state, which in general is thought to have less activity for transcriptional activation of target genes. However, the activity of response regulators may result in activation of some genes without phosphorylation by the cognate histidine kinase (10, 26, 40). We therefore tested whether SaeR can activate target genes without phosphorylation via SaeS. To this end, saeS in the single-copy complementation plasmid was deleted (Fig. 5). The resulting construct contained the native P1 promoter, as well as the native terminator signal downstream of saeS. Especially in strain Newman transcription of the shortened saePQR operon was strongly decreased compared to transcription of the native sae operon in the wild-type strain. The single transcript observed was probably generated by the P3 promoter, which was previously shown not to be autoactivated (17). The main P1 promoter, in contrast, was strongly autoregulated, and the corresponding T1 and T4 (with saeP) transcripts were not detectable in the saeS deletion construct. This indicates that SaeS is required for P1 activation. Additionally, coa expression was undetectable, indicating that SaeS is also needed for the sae-dependent activation of class I target genes. In contrast, hla could be activated without SaeS. These results suggest either that hla can be activated by unphosphorylated SaeR or that SaeR can be activated by other mechanisms (e.g., by cross-activation via other S. aureus HKs).
FIG. 5.
SaeS-independent expression of class II target genes. The saePQR operon (pCWSAE54) was integrated into the lipase gene of the sae mutant Newman-29 or ISP479C-29. Bacteria were grown to the exponential (t1) and postexponential (t3) growth phases. RNA was hybridized with digoxigenin-labeled PCR fragments. The lane at the bottom contained the 16S rRNA detected in the ethidium bromide-stained gels as a loading control. WT, wild type.
Role of phosphorylation of the response regulator SaeR.
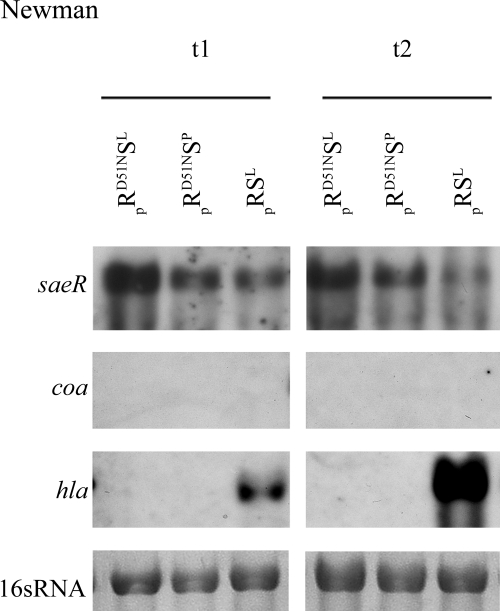
To analyze whether phosphorylation of SaeR is necessary for activation of target genes, the predicted phosphorylation site in SaeR was mutated. Mutation of the conserved Asp was previously shown to lead to complete inactivation of OmpR (28). However, other RRs, such as DegU from Listeria monocytogenes (26) and Bacillus subtilis (10) and AlgR from Pseudomonas aeruginosa (37), were shown to be active in their unphosphorylated forms. Sequence alignment with OmpR class response regulators revealed a conserved Asp residue in SaeR (D51), which is most likely the predicted phosphorylation site. The phosphorylation site was mutated in the AHT-inducible constructs containing saeRSL and saeRSP, and the plasmids were introduced into the saePQRS mutants of strain Newman (Fig. 6) and ISP479C (not shown). The phosphorylation-defective saeR gene is indicated by saeR(D51N). The mutation resulted in complete inactivation of sae regardless of which of the saeS alleles was present. This suggests that at least a low level of SaeR phosphorylation is required for sae activity.
FIG. 6.
Role of the SaeR phosphorylation site. To analyze the requirement for SaeR phosphorylation, mutated saeR(D51N)SL and saeR(D51N)SP were cloned behind an AHT-inducible promoter and introduced into the sae mutant ISP479C-29. Strains containing either mutated saeR(D51N)S or wild-type saeRS were induced with AHT (0.01 μg/ml) and grown to the exponential (t1) (OD600, 0.5) and late exponential (t2) (OD600, 3) growth phases. RNA was hybridized with digoxigenin-labeled PCR fragments. The lane at the bottom contained the 16S rRNA detected in the ethidium bromide-stained gels as a loading control.
DISCUSSION
Sae hyperactivity in strain Newman.
In this study we show that a mutation in the first transmembrane region of SaeS, like that in the widely used strain S. aureus Newman, leads to hyperactivation of the saeRS system and is the reason for the unusual high levels of transcription of some genes, such as fnbA, coa, or eap (sae class I target genes). Additionally, the P1 promoter located upstream of saeP also belongs to this class of target genes and results in strong autoactivation of the T1 and T2 sae transcripts in strain Newman. It was recently shown that the transcription levels of selected sae target genes vary widely between the Newman and Col strains (46). Most of the peculiarities of virulence gene expression in strain Newman are due to the saeS polymorphism. The unusual saeSP allele probably makes this strain relatively virulent in animal models of infection, which is one reason for its wide application in such studies. However, due to the SaeS polymorphism, as well as the recently described truncation of the fibronectin binding proteins (25), results obtained with this strain may not necessarily be applicable to other S. aureus strains. The presence of SaeS in strain Newman results not only in saePQRS overexpression but also in a largely altered response to environmental signaling compared to other strains (1, 17); depending on the stimulus analyzed, either SaeSP is unresponsive (17) or its reaction is opposite that of SaeSL (47).
In contrast to the transcription of class I sae target genes, the transcription of secreted virulence factors, such as the strictly sae-dependent hla and hlb genes, is not elevated in constructs containing SaeSP compared to constructs containing the SaeSL allele. Accordingly, strain Newman does not produce large amounts of α-hemolysin. The level of hla expression seems to be even lower than that in other virulent S. aureus strains, such as the emerging methicillin-resistant strain S. aureus USA300 (8). In addition to the effect on hla and hlb, the repressive effect of saeRS on the expression of the capsular polysaccharide was also shown to be not altered by the SaeS polymorphism (Fig. 1D), placing the capAP gene cluster in class II of the target genes.
Our results are in perfect agreement with the results obtained by Adhikari and Novick (1), who analyzed an saeP::bursa insertion mutant of strain RN6734, a derivative of strain 8325. This mutant curiously resembled strain Newman in the pattern of expression of several virulence factors, such as coa, fnbA, and eap. The original bursa mutation was generated in strain Newman and then transduced into strain RN6734. We assume that upon transduction, the whole saeQRSP sequence from strain Newman replaced the wild-type sequence of strain RN6374 due to the homologous recombination necessary for transduction. Therefore, the phenotype described for the saeP::bursa mutant of stain RN6734 that resembled the Newman strain can be fully explained by the introduction of the SaeSP allele, but it is not related to the bursa insertion.
Role of the transmembrane region of SaeS in the differential activation of target genes.
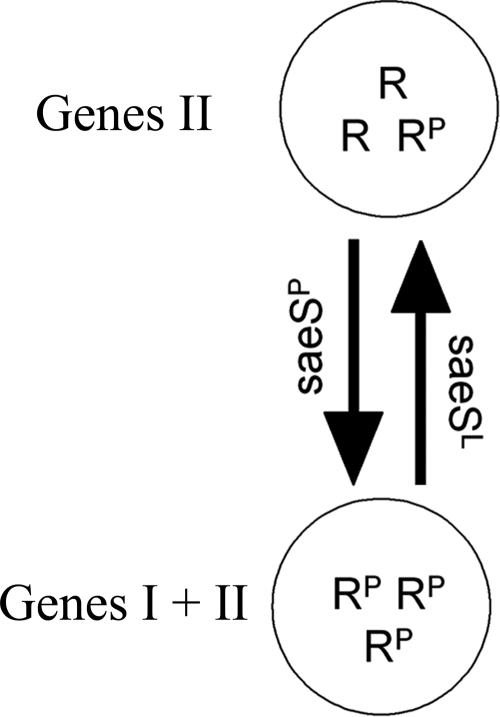
In this study we showed that a single amino acid change in the transmembrane region of SaeS results in severe changes in the expression of class I target genes, whereas class II target genes seem to be regulated equally by SaeSP and SaeSL. This observation can best be explained by differential requirements of the different promoter regions for gene activation via SaeR (Fig. 7). In our working model, the presence of SaeSP results in a shift toward kinase activity and consequently to a high level of SaeR phosphorylation. For SaeSL, the phosphatase activity is dominant, at least under uninduced conditions, and thus only a few SaeR molecules are phosphorylated. The ratio of SaeR-P to SaeR would then determine which genes are activated. This model is based on similarities to the EnvZ/OmpR system of Escherichia coli and the presumption that, depending on the signal, SaeS can exhibit kinase activity and/or phosphatase activity. It could be predicted that SaeS, like EnvZ, is probably a bifunctional HK. This class of HKs is thought to enhance the suppression of signals from noncognate resources due to the phosphatase activity (2). For the prototypic EnvZ/OmpR system in E. coli, it was shown that the ratio of phosphorylated RR to unphosphorylated RR determines which of the target genes are activated. In this model, osmotic signals regulate the level of the phosphorylated RR (OmpR-P) by modulating the ratio of kinase activity to phosphatase activity of EnvZ (57). The target gene ompC requires a high proportion of OmpR to be phosphorylated, while ompF can be activated with just a few OmpR-P molecules. However, the activation of both target genes, ompF and ompC, is dependent on at least a low level of OmpR phosphorylation (28), similar to what was found here for SaeR-mediated gene activation. Mutation of the Asp phosphorylation site in the RR eliminated activation of all target genes in both systems (28) (Fig. 6). Interestingly, deletion of SaeS does not result in complete inactivation of target gene expression. Thus, a low level of SaeR phosphorylation which is independent of SaeS can be assumed. This may occur either through unrelated HKs or through small-molecule phospho donors, such as acetyl phosphate (54).
FIG. 7.
Model for differential regulation of target genes. R, response regulator SaeR; RP, phosphorylated SaeR. The class II target genes (II) include hla, hlb, and cap, and the class I target genes (I) include fnbA, coa, eap, and sae. Class II target genes require only low levels of phosphorylated SaeR. In contrast, class I target genes require high levels of SaeR phosphorylation, like those obtained with SaeSP with strong kinase activity. Under conditions in which the sae system is not fully activated the phosphatase activity of SaeSL results in only a low level of phosphorylated SaeR, which is not sufficient to activate class I target genes.
Role of saeRS in temporal gene expression.
Recently, it was proposed that a different dependence of target genes on the level of RR phosphorylation controls the coordinate multicellular behavior of B. subtilis (30, 51). Target genes involved in swarming motility are activated by very low levels of the phosphorylated RR DegU, whereas genes necessary for complex colony architecture require an intermediate level of DegU phosphorylation. Finally, high levels of DegU-P inhibit both swarming motility and complex colony architecture. Whether a temporal program can also be linked to the different sae target genes remains to be determined. The class II target genes, which in our model require only low levels of phosphorylation (hla, hlb, and cap), are typically expressed in the late growth phase. For class I target genes, there is no correlation with growth phase; coa and fnbA are typically expressed only during the exponential phase, whereas eap is activated in the postexponential phase. This temporal gene expression pattern is clearly determined by additional factors, as shown by the induction of saeRS at different growth phases (Fig. 3). Thus, based on an in vitro analysis, a link to the temporal expression cannot be made easily. However, the gene expression patterns during infection or colonization are markedly different from those under in vitro growth conditions (21-23), and little is known about the gradual expression of sae target genes upon stimulation in vivo. Recently, it was proposed that the sae system may play an important role in environmental sensing after phagocytosis (17), and it was shown that saeRS is important for bacterial escape from the phagocyte through the intracellular activation of toxins (52). One may speculate that the gradual activation of target genes in response to various input signals plays a role after phagocytosis.
Target gene expression is not sensitive to the saeRS dosage.
Using an inducible expression vector system, we showed that the transcription of both types of target genes is largely not sensitive to the level of saeRS transcriptional activation. Induction of a high level of saeRS expression did not result in a significant increase in expression of the target genes. Thus, the different target gene activation of SaeSP and SaeSL was independent of the saeRS dosage. This also suggests that it is the ratio of phosphorylated SaeR to unphosphorylated SaeR mediated by SaeS activity which accounts for the different output, rather than the SaeR concentration. It was also shown that the EnvZ/OmpR system is robust and is only marginally sensitive to the absolute concentrations of EnvZ and/or OmpR (4). However, the robustness seen in the EnvZ/OmpR system is explained by the observation that the translation of OmpR is much more efficient than that of EnvZ, resulting in a >30-fold excess of OmpR over EnvZ (9). Whether there is a similar bias in the translational efficiency of SaeR and SaeS remains to be determined. However, a low ratio of SaeS to SaeR is probable, due to the overlap of the saeR stop and saeS start codons. The same overlap is present in the genetic organization of ompR and envZ and was assumed to cause the high OmpR/EnvZ ratio.
Based on our analysis, the autoregulated P1 promoter of the sae system can be classified as a promoter of the class I target genes. Thus, the saePQRS system, like many other regulatory systems, is positively autoregulated, which seems to contradict the assumption that the system is robust. There would be no point in changing the expression if there were no effect on the output. However, it was recently predicted that positive autoregulation of a two-component system contributes to enhanced output only in the presence of a strong stimulus (41). Since the native signal for SaeS activation is not well defined, we do not know whether our assays were performed under activating conditions. The fact that we could not detect coa and fnbA in strains harboring saePQRS may indicate that the growth conditions used only very weakly stimulated SaeS activation. Thus, under strong stimulating conditions, the function of SaeSL may approximate that of the constitutively activated SaeSP protein. In this way, strain Newman may be a valuable tool for defining the sae regulon by mimicking high-stimulation conditions, which are not well defined yet.
Supplementary Material
Acknowledgments
We thank Vittoria Bisanzio for excellent technical assistance. We are grateful to A. Cheung for providing the vector pALC2073.
This work was supported by grants to C.W. from the Deutsche Forschungsgemeinschaft (grants Wo578/5, SFB766, and TR34) and from the IZKF of Eberhard Karls University, Tübingen, Germany.
Footnotes
Published ahead of print on 20 November 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adhikari, R. P., and R. P. Novick. 2008. Regulatory organization of the staphylococcal sae locus. Microbiology 154:949-959. [DOI] [PubMed] [Google Scholar]
- 2.Alves, R., and M. A. Savageau. 2003. Comparative analysis of prototype two-component systems with either bifunctional or monofunctional sensors: differences in molecular structure and physiological function. Mol. Microbiol. 48:25-51. [DOI] [PubMed] [Google Scholar]
- 3.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batchelor, E., and M. Goulian. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl. Acad. Sci. U. S. A. 100:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman, B. T., N. P. Donegan, T. M. Jarry, M. Palma, and A. L. Cheung. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69:7851-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruckner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 8.Bubeck Wardenburg, J., T. Bae, M. Otto, F. R. Deleo, and O. Schneewind. 2007. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13:1405-1406. [DOI] [PubMed] [Google Scholar]
- 9.Cai, S. J., and M. Inouye. 2002. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 277:24155-24161. [DOI] [PubMed] [Google Scholar]
- 10.Dahl, M. K., T. Msadek, F. Kunst, and G. Rapoport. 1992. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J. Biol. Chem. 267:14509-14514. [PubMed] [Google Scholar]
- 11.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 12.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperin, M. Y. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188:4169-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, R., T. R. Mack, and A. M. Stock. 2007. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem. Sci. 32:225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, R., and A. M. Stock. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaskill, M. E., and S. A. Khan. 1988. Regulation of the enterotoxin B gene in Staphylococcus aureus. J. Biol. Chem. 263:6276-6280. [PubMed] [Google Scholar]
- 17.Geiger, T., C. Goerke, M. Mainiero, D. Kraus, and C. Wolz. 2008. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J. Bacteriol. 190:3419-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 19.Giraudo, A. T., C. Mansilla, A. Chan, C. Raspanti, and R. Nagel. 2003. Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr. Microbiol. 46:246-250. [DOI] [PubMed] [Google Scholar]
- 20.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goerke, C., U. Fluckiger, A. Steinhuber, V. Bisanzio, M. Ulrich, M. Bischoff, J. M. Patti, and C. Wolz. 2005. Role of Staphylococcus aureus global regulators sae and sigmaB in virulence gene expression during device-related infection. Infect. Immun. 73:3415-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439-1447. [DOI] [PubMed] [Google Scholar]
- 23.Goerke, C., and C. Wolz. 2004. Regulatory and genomic plasticity of Staphylococcus aureus during persistent colonization and infection. Int. J. Med. Microbiol. 294:195-202. [DOI] [PubMed] [Google Scholar]
- 24.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41:139-227. [DOI] [PubMed] [Google Scholar]
- 25.Grundmeier, M., M. Hussain, P. Becker, C. Heilmann, G. Peters, and B. Sinha. 2004. Truncation of fibronectin-binding proteins in Staphylococcus aureus strain Newman leads to deficient adherence and host cell invasion due to loss of the cell wall anchor function. Infect. Immun. 72:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gueriri, I., S. Bay, S. Dubrac, C. Cyncynatus, and T. Msadek. 2008. The Pta-AckA pathway controlling acetyl phosphate levels and the phosphorylation state of the DegU orphan response regulator both play a role in regulating Listeria monocytogenes motility and chemotaxis. Mol. Microbiol. 70:1342-1357. [DOI] [PubMed] [Google Scholar]
- 27.Harraghy, N., J. Kormanec, C. Wolz, D. Homerova, C. Goerke, K. Ohlsen, S. Qazi, P. Hill, and M. Herrmann. 2005. sae is essential for expression of the staphylococcal adhesins Eap and Emp. Microbiology 151:1789-1800. [DOI] [PubMed] [Google Scholar]
- 28.Kanamaru, K., H. Aiba, and T. Mizuno. 1990. Transmembrane signal transduction and osmoregulation in Escherichia coli. I. Analysis by site-directed mutagenesis of the amino acid residues involved in phosphotransfer between the two regulatory components, EnvZ and OmpR. J. Biochem. 108:483-487. [DOI] [PubMed] [Google Scholar]
- 29.Kim, D., and S. Forst. 2001. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology 147:1197-1212. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi, K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 66:395-409. [DOI] [PubMed] [Google Scholar]
- 31.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 32.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuroda, H., M. Kuroda, L. Cui, and K. Hiramatsu. 2007. Subinhibitory concentrations of beta-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol. Lett. 268:98-105. [DOI] [PubMed] [Google Scholar]
- 34.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 35.Li, D., and A. Cheung. 2008. Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect. Immun. 76:1068-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang, X., C. Yu, J. Sun, H. Liu, C. Landwehr, D. Holmes, and Y. Ji. 2006. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect. Immun. 74:4655-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, S., U. Selvaraj, D. E. Ohman, R. Quarless, D. J. Hassett, and D. J. Wozniak. 1998. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J. Bacteriol. 180:956-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mascher, T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264:133-144. [DOI] [PubMed] [Google Scholar]
- 39.Mascher, T., J. D. Helmann, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70:910-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsubara, M., and T. Mizuno. 1999. EnvZ-independent phosphotransfer signaling pathway of the OmpR-mediated osmoregulatory expression of OmpC and OmpF in Escherichia coli. Biosci. Biotechnol. Biochem. 63:408-414. [DOI] [PubMed] [Google Scholar]
- 41.Miyashiro, T., and M. Goulian. 2008. High stimulus unmasks positive feedback in an autoregulated bacterial signaling circuit. Proc. Natl. Acad. Sci. U. S. A. 105:17457-17462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 43.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 44.Novick, R. P., and D. Jiang. 2003. The staphylococcal saeRS system coordinates environmental signals with quorum sensing. Microbiology 149:2709-2717. [DOI] [PubMed] [Google Scholar]
- 45.Pattee, P. A. 1981. Distribution of Tn551 insertion sites responsible for auxotrophy on the Staphylococcus aureus chromosome. J. Bacteriol. 145:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogasch, K., V. Ruhmling, J. Pane-Farre, D. Hoper, C. Weinberg, S. Fuchs, M. Schmudde, B. M. Broker, C. Wolz, M. Hecker, and S. Engelmann. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188:7742-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schafer, D., T. T. Lam, T. Geiger, M. Mainiero, S. Engelmann, M. Hussain, A. Bosserhoff, M. Frosch, M. Bischoff, C. Wolz, J. Reidl, and B. Sinha. 25 September 2009. A point mutation in the sensor histidine kinase SaeS of Staphylococcus aureus strain Newman alters response to biocide exposure. J. Bacteriol. doi: 10.1128/JB.00630-09. [DOI] [PMC free article] [PubMed]
- 48.Steinhuber, A., C. Goerke, M. G. Bayer, G. Doring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185:6278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toledo-Arana, A., N. Merino, M. Vergara-Irigaray, M. Debarbouille, J. R. Penades, and I. Lasa. 2005. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 187:5318-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandenesch, F., J. Kornblum, and R. P. Novick. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 173:6313-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhamme, D. T., T. B. Kiley, and N. R. Stanley-Wall. 2007. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol. Microbiol. 65:554-568. [DOI] [PubMed] [Google Scholar]
- 52.Voyich, J. M., C. Vuong, M. Dewald, T. K. Nygaard, S. Kocianova, S. Griffith, J. Jones, C. Iverson, D. E. Sturdevant, K. R. Braughton, A. R. Whitney, M. Otto, and F. R. Deleo. 2009. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J. Infect. Dis. 199:1698-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wann, E. R., B. Dassy, J. M. Fournier, and T. J. Foster. 1999. Genetic analysis of the cap5 locus of Staphylococcus aureus. FEMS Microbiol. Lett. 170:97-103. [DOI] [PubMed] [Google Scholar]
- 54.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolz, C., D. McDevitt, T. J. Foster, and A. L. Cheung. 1996. Influence of agr on fibrinogen binding in Staphylococcus aureus Newman. Infect. Immun. 64:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolz, C., P. Pohlmann-Dietze, A. Steinhuber, Y. T. Chien, A. Manna, W. van Wamel, and A. Cheung. 2000. agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36:230-243. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida, T., S. Phadtare, and M. Inouye. 2007. Functional and structural characterization of EnvZ, an osmosensing histidine kinase of E. coli. Methods Enzymol. 423:184-202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.