Abstract
The integrase of the temperate bacteriophage mv4 catalyzes site-specific recombination between the phage attP site and the host attB site during Lactobacillus delbrueckii lysogenization. The mv4 prophage is excised during the induction of lytic growth. Excisive site-specific recombination between the attR and attL sites is also catalyzed by the phage-encoded recombinase, but the directionality of the recombination is determined by a second phage-encoded protein, the recombination directionality factor (RDF). We have identified and functionally characterized the RDF involved in site-specific excision of the prophage genome. The mv4 RDF, mv4Xis, is encoded by the second gene of the early lytic operon. It is a basic protein of 56 amino acids. Electrophoretic mobility shift assays demonstrated that mv4Xis binds specifically to the attP and attR sites via two DNA-binding sites, introducing a bend into the DNA. In vitro experiments and in vivo recombination assays with plasmids in Escherichia coli and Lactobacillus plantarum demonstrated that mv4Xis is absolutely required for inter- or intramolecular recombination between the attR and attL sites. In contrast to the well-known phage site-specific recombination systems, the integrative recombination between the attP and attB sites seems not to be inhibited by the presence of mv4Xis.
During the establishment of lysogeny, many temperate bacteriophages integrate their genomes into the bacterial host chromosome in a site-specific manner: recombination between the attachment sites attB and attP, located on the bacterial and phage genomes, respectively, is catalyzed by a phage-encoded recombinase (integrase) (14, 42) and leads to formation of the hybrid attachment sites attL and attR at the junctions between the phage and bacterial genomes. Prophage excision, which occurs during the induction of lytic growth, involves site-specific recombination between the attachment junctions attL and attR and requires at least one phage-encoded recombination directionality factor (RDF) or excise, in addition to integrase (32, 43).
In the model Escherichia coli phage λ and its close relatives, the recombination directionality is controlled directly by the RDF Xis. The gene xis is located upstream from the int gene encoding the integrase. Xis is required for the excision reaction but inhibits integrative recombination (2, 41). Xis determines the directionality of recombination by influencing the formation of specific protein-DNA architectures (30). To exert its effect, Xis binds cooperatively with Int and FIS to specific sites, thereby inducing and stabilizing a DNA bend that alters the intasome structure formed during recombination (1, 15, 44, 53).
In bacteriophages of the P2 family, site-specific recombination-related genes are organized differently, even though the directionality of recombination seems to be controlled as in phage λ. The P2 genetic switch region contains two overlapping face-to-face promoters, pc and pe (50). The leftward promoter, pc, produces the lysogenic transcript, including the C repressor and P2 integrase, whereas the rightward promoter, pe, produces the lytic transcript. The first protein encoded by the lytic operon, Cox, is a λ Cro-like repressor of the promoter pc controlling lysogeny (50). However, Cox is a multifunctional protein that also acts as a recombination directionality factor (57).
Site-specific recombination has been well studied for bacteriophages of Gram-positive bacteria, essentially for bacteriophages of Actinomycetales, such as Mycobacterium phage L5 (36, 46-48) and Streptomyces phage φC31 (52, 55), but also for bacteriophages of lactic acid bacteria (3, 10, 11, 22, 49). However, only few RDFs have been characterized to date, for example, those of mycobacteriophage L5 (34, 36) and of lactococcal phage TP901-1 (9).
The site-specific integration system of bacteriophage mv4 has been characterized previously (6, 22). The mv4 integrase belongs to the tyrosine recombinase family and has no absolute requirement for accessory factors, unlike most of the λ Int family recombinases (4). The mv4 integrase also functions in a wide range of Gram-positive bacteria and in Escherichia coli (6). Furthermore, a site-specific integrative vector derived from mv4 is stably maintained in the chromosome of several lactobacilli in the absence of selection, suggesting that mv4Int cannot catalyze excisive recombination on its own (6). The minimal 234-bp attP site of mv4 contains five putative integrase arm-binding sites and has a 17-bp core sequence in common with the attB site (4). During recombination, strand exchange occurs inside the core sequence. The attB site is located at the 3′ end of a tRNAser gene (22). The minimal size of the functional attB site is 16 bp, and no symmetric inverted repeats have been found in the core sequence (5).
In this study, we identified the RDF gene mv4xis as the second gene of the mv4 early lytic operon. This location is unusual, being reported here for the first time in a temperate bacteriophage harboring a tyrosine recombinase. mv4Xis is a 56-residue basic peptide that acts by binding to two sites within attR or attP DNA, thereby bending the DNA. The presence of mv4Xis is absolutely required, together with mv4Int, for excisive recombination between the attL and attR attachment sites. However, in contrast to results for the well-studied systems (λ, L5, HP1, and P2 phages), preliminary results suggest that mv4Xis does not inhibit integrative recombination between the attP and attB sites.
MATERIALS AND METHODS
Bacterial strains, bacteriophage, and growth conditions.
The bacterial strains, bacteriophage, and plasmids used in this study are listed in Table 1. Lactobacillus plantarum strains were grown at 37°C in MRS medium (19) or on MRS medium solidified with 1.5% (wt/vol) agar. Erythromycin and chloramphenicol were used at concentrations of 5 μg/ml and 10 μg/ml, respectively. E. coli was grown at 37°C in Luria-Bertani broth (Difco Laboratories, Detroit, MI) or on LB broth solidified with 1.5% (wt/vol) agar. Antibiotics were used at the following concentrations: 100 μg/ml for ampicillin, 150 μg/ml for erythromycin, 10 μg/ml for chloramphenicol, and 65 μg/ml for spectinomycin.
TABLE 1.
Bacteriophage, bacterial strains, and plasmids used in this study
| Strain/phage/plasmid | Relevant characteristic(s) | Reference/source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169endA1 λ−hsdR17(rK− mK+) deoRthi-1supE44recA1 gyrA96 relA1 | Life Technologies |
| TG2 | supEhsdΔ5thi Δ(lac-proAB) Δ(srl-recA)306::Tn10 (Tetr) F′ [traD36proAB+lacIqlacZΔM15] | 51 |
| BL21(DE3) | F− omp T hsdSΒ(rΒ− mΒ−) gal dcm (DE3) | Novagen |
| L. plantarum | ||
| LP80 | Lactobacillus plantarum DSM 4229 | 29 |
| LP80-3 | LP80::pMC1 integrant | 22 |
| Bacteriophage mv4 | Temperate phage isolated from the L. delbrueckii LT4 strain | 16 |
| Plasmids | ||
| pRC1 | ermAM+ (Emr), 3.5-kb cloning vector, ColE1 replicon | 33 |
| pMC1 | Emr, 5.15 kb, pRC1 with mv4 attP-int region | 22 |
| pRC10 | Emr, 3.85 kb, pRC1 with mv4 attP region | 4 |
| pRCattR | Emr, 5.2 kb, pRC1 with attR region cloned at the SalI and XbaI sites | This work |
| pRCattRDR2 | Emr, 5.2 kb, pRC1 with a deletion derivative of the attR region cloned at the BamHI and KpnI sites | This work |
| pRCattL | Emr, 4 kb, pRC1 with attL region cloned at the SalI and XbaI sites | This work |
| pMPM-A3 | bla+ (Ampr), 3.4-kb cloning vector, p15A replicon | 40 |
| pA3int | Ampr, 4.75 kb, pMPM-A3 with mv4 int gene | 4 |
| pNZ8037 | cat+ (Cmr), 3-kb expression vector, pSH71 replicon | 20 |
| pNZ57 | Cmr, 3.65 kb, pNZ8037 with mv4 ORF-57 cloned at the SmaI site | This work |
| pNZ109 | Cmr, 3.6 kb, pNZ8037 with mv4 ORF-109 cloned at the BamHI and SmaI sites | This work |
| pNZ179 | Cmr, 3.6 kb, pNZ8037 with mv4 ORF-179 cloned at the SmaI and PstI sites | This work |
| pNZxis | Cmr, 3.2 kb, pNZ8037 with mv4 ORF-56 cloned at the SmaI site | This work |
| pNZxish | Cmr, 3.2 kb, pNZ8037 with mv4 ORF-56 His tagged at the C terminus cloned at the BamHI and EcoRI sites | This work |
| pNZ9530 | Emr, 7 kb, pIL252 carrying nisRK (pAMβ1 replicon) | 31 |
| pAM239 | aad9 (Spcr), 4.3-kb cloning vector, pSC101 replicon | D. Gil |
| pAMattB | Spcr, 4.6 kb, pAM239 with the attB region | 4 |
| pAMattR | Spcr, 4.6 kb, pAM239 with the attR region cloned at the BamHI and SmaI sites | This work |
| pET15b | bla+ (Ampr), 5.7-kb expression vector, pBR322 replicon | Novagen |
| pETxish | Ampr, 5.9 kb, pET15b with mv4 ORF-56 His tagged at the C terminus cloned at the NcoI and XhoI sites | This work |
| pCointRL | Emr, Spcr, 8.45 kb, cointegrate between pAMattB and pRC10 bearing the attR and attL sites | This work |
DNA techniques.
DNA techniques were performed essentially as described by Sambrook et al. (51). Restriction enzymes, the Klenow fragment of DNA polymerase I, Taq polymerase, T4 polynucleotide kinase, and T4 DNA ligase were purchased from either Roche Molecular Biochemicals (Mannheim, Germany) or New England Biolabs (Beverly, MA) and used as recommended by the suppliers.
E. coli and L. plantarum were electrotransformed using a gene pulser device (Bio-Rad, Richmond, CA) according to the manufacturer's recommendations and according to the method of Auvray et al. (6), respectively.
E. coli plasmid DNA was isolated using a Qiaprep spin kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. Total DNA was obtained from L. plantarum and E. coli as previously described (22).
Plasmid construction.
The plasmids used in this work are listed in Table 1 and the primers (Eurogentec, Seraing, Belgium) in Table 2. The 1,757-bp SalI-XbaI fragment (containing the attR region) of pBSattR (22) was ligated to the 3,438-bp SalI-XbaI fragment of pRC1 to give pRCattR. The 590-bp SalI-XbaI fragment (containing the attL region) of pBSattL (22) was ligated to the 3,438-bp SalI-XbaI fragment of pRC1 to give pRCattL. The 278-bp BamHI PCR fragment, amplified from pRCattR using the DR9 and attb primers, was ligated to the 4,330-bp BamHI-SmaI fragment of pAM239 to give pAMattR. The 636-bp PCR fragment, amplified from mv4 DNA using the F3 and F4 primers, was ligated to the 3,016-bp SmaI fragment of pNZ8037 to give pNZ57. The 588-bp BamHI-XmnI PCR fragment, amplified from mv4 DNA using the C1 and BX1 primers, was ligated to the 3,010-bp BamHI-XmnI fragment of pNZ8037 to give pNZ109. The 216-bp BamHI-EcoRI PCR fragment, amplified from pNZxis (pNZxis was constructed by inserting a 224-bp SmaI-XmnI fragment from mv4 DNA containing the xis gene into the SmaI site of pNZ8037) using the Xis1 and XisHis primers, was ligated to the 3,001-bp BamHI-EcoRI fragment of pNZ8037 to give pNZxish. The 243-bp NcoI-XhoI PCR fragment, amplified from pNZxis using the Xis1 and XisHis primers, was ligated to the 5,643-bp NcoI-XhoI fragment of pET15b to give pETxish. pCointRL is the cointegrate resulting from site-specific recombination between pRC10 (attP) and pAMattB in the E. coli TG2 strain in the presence of pA3int. pCointRL was purified from agarose gel and introduced by transformation into the TG2 strain for amplification.
TABLE 2.
List of primers used in this study
| Primer | Sequence (5′ → 3′)a | Restriction site(s) |
|---|---|---|
| DR2 | GGGGTACCAGAAACGCTTTTTAGC | KpnI |
| DR3 | ATGCCATCTATTAACTAGCT | |
| DR4 | AACTGCAGTGAAACGCATGGAAACA | PstI |
| DR9 | CGGGATCCAGTTCTAAATCAACTAGATTTTTAACT | BamHI |
| BX1 | GCATCTGGATCTTAT | |
| C1 | GCATCTGGATCTTATAATTA | |
| F3 | TCACCCGGGCAAGATCATCG | SmaI |
| F4 | TTTGACTGCAGGATTACGGC | PstI |
| attb | CATTTGATTTAGATGTCCTT | |
| L1 | CACCATCTTAAAAATAACTT | |
| Xis1 | TCACCATGGTACTGCACTGGATCCC | NcoI, BamHI |
| XisHis | ACTCGAGTCTAGAATTCCTAGTGATGGTGATGGTGATGTTCCTCCTTAAA | XhoI, XbaI, EcoRI |
| X1mut | AAACGCAGCAACCCGACCAGA | |
| X2mut | AACCCTCGCAACCCGTGGTTG | |
| X2A | CCAGAAACATGGTTGAAAGAACC | |
| X2B | ATGTTTCTGGAGGGTTCTGG | |
| Uni | GTAAAACGACGGCCAGT | |
| Rev | GGAAACAGCTATGACCATG |
The restriction sites contained in the primers are underlined. The mutations are in bold and underlined (for X1mut and X2mut). The Xis binding site sequences are in bold (for X2A and X2B).
Construction of the attR site deletion derivative.
The mv4 attR DR2 DNA segment was amplified by PCR, using DR2 and Uni oligonucleotides and pRCattR as the template. The amplified fragment was cut with BamHI and KpnI and purified by polyacrylamide gel electrophoresis, and the 1,655-bp BamHI-KpnI fragment was ligated to the 3,461-bp BamHI-KpnI fragment of pRC1 to give pRCattRDR2.
Protein production and preparation.
Cell extracts containing soluble mv4Xis were prepared from L. plantarum LP80 cells carrying pNZxish and pNZ9530 (nisRK). Control crude extract without mv4Xis was prepared from L. plantarum LP80 cells carrying pNZ8037 (empty expression vector) and pNZ9530. Overnight cultures were diluted 1/50 in MRS and grown at 37°C. When the cultures reached an optical density at 600 nm (OD600) of 0.2, nisin was added to a final concentration of 25 ng/ml. The cells were collected by centrifugation 5 h after nisin addition; washed three times with 50 mM Tris-HCl, pH 7.5, 10% glycerol; and incubated in a 1/20 volume of lysis buffer (50 mM Tris-HCl, pH 7.5, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mg/ml lysozyme) overnight at 4°C. The cells were sonicated 10 times, for 30 s each, on ice and stored for 1 h at 4°C with 1% Triton X-100. Cell debris was removed by centrifugation at 3,000 × g for 1 h at 4°C. E. coli preparations containing soluble mv4Int were obtained as previously described (4). The protein concentrations of extracts were determined by the Bradford assay (8).
Gel electrophoretic mobility shift assay (GEMSA).
The primers were end labeled using polynucleotide kinase and [γ-32P]ATP (Amersham) (51) before amplification of the 242-bp DR9-DR3 (attP), 281-bp DR9-attb (attR), 244-bp L1-DR3 (attL), 283-bp L1-attb (attB), 207-bp DR4-DR3 (attPD4), 187-bp DR2-DR3 (attPD2), 197-bp X1mut-DR3 (attPX1mut), and 137-bp X2mut-DR3 (attPX2mut) DNA fragments with pRC10, pRCattR, pRCattL, or pAMattB as the template. The 187-bp DR2-DR3 fragment was digested with BstBI or BstXI, and the resulting 149-bp (attPBB) or 123-bp (attPBX) fragment, respectively, was purified from the acrylamide gel. DNA binding reactions were performed with a reaction volume of 20 μl containing 30 mM Tris-HCl, pH 7.5, 5 mM NaCl, 90 mM KCl, 5 mM EDTA, 1% glycerol, 2 μg poly(dI-dC), 105 cpm of probe, and 10 μg of protein for 10 min at room temperature before electrophoresis. Protein-DNA complexes were separated by electrophoresis with 5% nondenaturing polyacrylamide gels. The gels were run in TBE buffer (0.1 M Tris-HCl, pH 8, 0.1 M borate, 0.7 mM EDTA). The radioactive compounds were detected with a Fuji BAS1000 bioimaging analyzer system (Fuji Photo Film Co., Tokyo, Japan) and analyzed with Tina version 2.07c software (Raytest Isotopenmeβgeräte GmbH, Straubenhardt, Germany) and/or by autoradiography after the gels were placed against BioMax MR film (Kodak) at room temperature.
Bending.
The 552-bp Uni-Rev, 182-bp X2A-Rev, and 216-bp Uni-X2B DNA fragments were radioactively labeled during amplification, using 20 μCi [α-32P]dATP per reaction mixture and pRCattRDR2 as the template. The 181-bp AccI-BstBI, 182-bp KpnI-MslI, 180-bp SpeI-XhoI, 181-bp AccI-XbaI, and 185-bp HindIII-SacI fragments were purified from acrylamide gels after digestion of the 552-bp Uni-Rev fragment with the corresponding restriction enzymes. The 180-bp SfcI-X2B fragment was purified from an acrylamide gel after digestion of the 216-bp Uni-X2B fragment with SfcI. DNA-protein complex analysis by electrophoresis was carried out as described above for GEMSA.
Preparation of radioactive substrates.
The 283-bp L1-attb (attB), 281-bp DR9-attb (attR), 244-bp L1-DR3 (attL), and 242-bp DR9-DR3 (attP) DNA fragments were radioactively labeled during amplification, using 20 μCi [α-32P]dATP per reaction mixture.
In vitro excisive recombination assays.
attL × attR intermolecular recombination reaction mixtures (20 μl), containing 200 ng supercoiled plasmid carrying the att site to be tested and a linear radiolabeled att fragment (105 cpm) in 30 mM Tris-HCl, pH 7.5, 5 mM NaCl, 90 mM KCl, 5 mM EDTA, 1% glycerol, and 4 mM spermidine, were incubated for 90 min at 42°C in the presence or absence of Int cell extracts (3 μg protein equivalent) and with Xis or control cell extracts (3 μg or 6 μg protein equivalent, respectively). The reactions were stopped by adding 0.1% sodium dodecyl sulfate (SDS) and dye. For time course experiments, aliquots were removed at various time points and the reaction was stopped. Samples were analyzed as previously described (4).
In vivo site-specific recombination test with E. coli.
E. coli recA TG2 cells carrying pA3int and (i) pRC10 and pAMattB, (ii) pCointRL, or (iii) pRCattL and pAMattR were transformed with pNZ8037 or pNZxish. Cells were cultured in the presence of appropriate antibiotics for about 25 generations, and the plasmids were then extracted. After restriction digestion, the plasmids were analyzed by agarose gel electrophoresis with ethidium bromide staining.
Nucleotide sequence accession numbers.
The sequence data have been submitted to the GenBank database under accession numbers AF182207 (nucleotides 5392 to 5222) and ABG46345 (Xis).
RESULTS
Identification of the mv4 RDF gene.
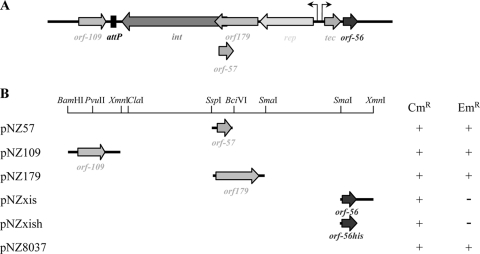
We previously showed that phage mv4 integrates its genome into the Lactobacillus delbrueckii chromosome by site-specific recombination at the 3′ end of a tRNAser gene (22). Prophage mv4 excision seems to involve site-specific recombination between the attR and attL sites, because after prophage induction by mitomycin C treatment of the lysogenic strain, the cured host strain contained an intact tRNAser gene (22). Owing to the localization of the RDF gene close to the integrase gene in mobile genetic elements (35), we tried to identify the gene encoding the phage factor promoting excisive recombination by testing various open reading frames (ORFs) located in the close vicinity of the integrase gene: ORF-179, ORF-109, and ORF-57 (Fig. 1). We investigated the possibility that one of these ORFs was able to induce the excision of pMC1 (integrative vector containing the mv4 integrase gene and the attP site; Emr) from the chromosome of L. plantarum LP80-3. The three mv4 ORFs were independently cloned into the expression vector pNZ8037 (Cmr), which was then introduced into LP80-3, and Cmr transformants were selected. We then screened these transformants for erythromycin sensitivity (Ems), indicating that the integrated plasmid had been excised and lost, since pMC1 is not replicative in L. plantarum. All of the colonies tested (>400) remained Emr. The mv4 RDF gene was therefore not located in the close vicinity of the integrase gene.
FIG. 1.
Identification of phage mv4 RDF gene. (A) Genetic organization of the lysogeny region containing the characterized rep, tec, and int genes, the attP site, and ORFs of unknown functions. (B) Series of chloramphenicol-resistant (Cmr) recombinant plasmids, in which mv4 ORFs are cloned into the expression vector pNZ8037. The cloned gene present in each plasmid is shown, with an arrow indicating its orientation relative to the promoter. Each plasmid was introduced into an L. plantarum strain containing an erythromycin-resistant (Emr) chromosomal integrated vector (pMC1), and chloramphenicol-resistant (Cmr) transformants were screened for loss of erythromycin resistance (−), i.e., loss of the integrated vector pMC1, as indicated.
In the early lytic operon, just downstream from the tec gene, there is a small ORF encoding a putative basic (pI 9.69) protein containing 56 amino acids (Fig. 1A) (17). Comparison of the sequence of this protein with sequences in protein databases revealed no significant amino acid identity to any other known RDF. Nevertheless, this ORF displayed weak similarity to the RDF of the SGI1 genomic island from Salmonella enterica serovar Typhimurium DT104 (37% identity over a 53-residue segment) (21). Predictions of the secondary structure of ORF-56 by PsiPred (http://bioinf.cs.ucl.ac.uk/psipred/) identified a helix-turn-helix (HTH) motif within the N-terminal domain of the protein (data not shown). CLUSTAL X (1.81) analysis of ORF-56, comparing this protein with the proteins from the RDF database (http://www.pitt.edu/∼gfh/download.html) (35), showed that ORF-56 could be aligned with the SLP1 family, which contains SGI1-Xis (data not shown).
The same test used for ORF-179, ORF-57, and ORF-109 was used to determine whether ORF-56 could induce the excision of pMC1 from the L. plantarum LP80-3 chromosome. We found that 100% of the Cmr transformants (>400) containing the expression plasmid pNZxis had excised pMC1 (Ems colonies) (Fig. 1B). We carried out the same experiment with the expression vector pNZxish, in which the ORF-56-encoded protein was C-terminally His tagged. Identical results were obtained, demonstrating that the His-tagged protein functioned similarly to the wild-type protein (Fig. 1B). No pMC1 excision event was detected in the absence of mv4Xis, even if integrant strains were cultured over more than 150 generations in the absence of selective pressure (6).
In vivo, the presence of ORF-56 stimulates the excision of the pMC1 vector that had integrated into the chromosome of a heterologous Gram-positive strain, L. plantarum, and this ORF seems to encode a factor that promotes excisive recombination, possibly corresponding to the mv4 RDF, mv4Xis.
mv4Xis is absolutely required for excisive recombination.
We investigated the site-specific excision reaction mediated by mv4Xis by developing an in vitro excision assay.
The mv4Xis protein was overproduced in L. plantarum by use of a nisin-controlled expression system (45). Following the induction of L. plantarum strains containing pNZxish, we observed the synthesis of a 7-kDa protein, which is absent in induced cells containing the empty expression vector pNZ8037 (data not shown). This protein was produced in relatively small amounts but was completely soluble. The presence of the His-tagged mv4Xis protein was confirmed by Western blotting with anti-His antibodies (data not shown). In contrast, when the mv4Xis protein was overproduced in E. coli by use of the T7 expression system (pETxish), large amounts of totally insoluble protein were detected for each induction condition, medium, and temperature tested. For these reasons, the L. plantarum mv4Xis cell extract was used in the in vitro assay.
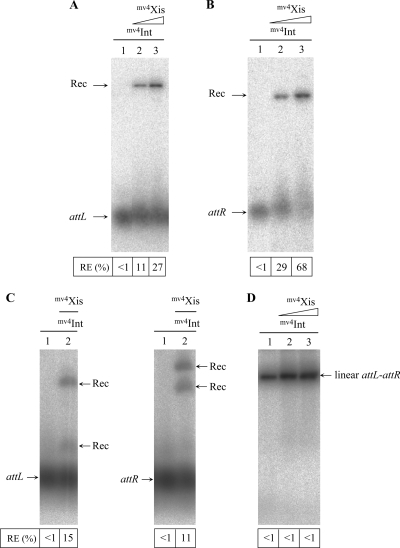
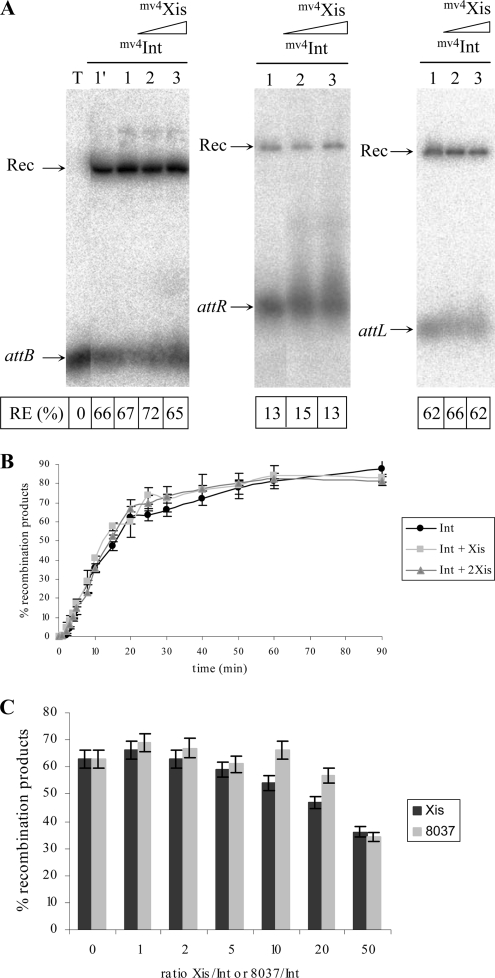
The in vitro recombination between the attR and attL sites was monitored by the detection of a radiolabeled linear recombination product resulting from the insertion of a supercoiled attR- or attL-containing plasmid into a radiolabeled 283-bp or 244-bp DNA fragment containing attL or attR, respectively. The recombination product, Rec, was detected only in the presence of both mv4Int and mv4Xis; no Rec was detected in reaction mixtures containing only mv4Int (Fig. 2A and B). The excisive recombination efficiency was Xis dose dependent between 0 and 6 μg of Xis crude extract present in the reaction mixture (Fig. 2A and B). The recombination between attL on a plasmid and attR on a linear fragment is more efficient (by a factor of two to three) than the reverse situation (Fig. 2A and B). We suggest that the assembly of the higher-order nucleoprotein complex on the attL site is favored by this site being supercoiled and/or that the excisive synapsis with the attR site is more efficiently constituted. In contrast to the integrative recombination (4), the recombination between the attL and attR sites on separate linear fragments is effective and is about half as efficient as the recombination between one site on a plasmid and the other on a linear fragment (Fig. 2C).
FIG. 2.
Role of mv4Xis in in vitro attR-attL recombination. Samples were incubated at 42°C for 90 min, analyzed by electrophoresis with a 1% agarose gel, and visualized using a PhosphorImager. The intensities of the bands corresponding to the radioactive recombination product (Rec) and the att substrates (attL and attR) were quantified for each sample (as described in reference 4). The recombination efficiency (RE) is indicated under the gels. (A) In vitro recombination assays between the attR site-containing plasmid and the linear attL site. Each reaction mixture contained 200 ng of the supercoiled attR plasmid (lanes 1 to 3), the labeled 283-bp linear attL fragment, 3 μg of mv4Int-enriched cell extract (lane 1), and 3 μg (lane 2) or 6 μg (lane 3) of mv4Xis-enriched cell extract. (B) Recombination between the attL site-containing plasmid and the linear attR site. Each reaction mixture contained 200 ng of the supercoiled attL plasmid, the labeled 281-bp linear attR fragment, 3 μg of mv4Int-enriched cell extract (lane 1), and 3 μg (lane 2) or 6 μg (lane 3) of mv4Xis-enriched cell extract. (C) In vitro recombination assays between attR and attL sites on linear fragments. Each reaction mixture contained 100 ng of the 2,144-bp linear attR fragment (or the 4,028-bp linear attL fragment), the labeled 283-bp linear attL fragment (or the 281-bp linear attR fragment), 3 μg of mv4Int-enriched cell extract (lane 1), and 15 μg of mv4Xis-enriched cell extract (lane 2). (D) In vitro recombination assays between the attR and attL sites on a linear fragment. Each reaction mixture contained the labeled 4,134-bp linear pCointRL fragment, 3 μg of mv4Int-enriched cell extract (lane 1), and 6 μg (lane 2) or 15 μg (lane 3) of mv4Xis-enriched cell extract.
We also assayed in vitro excision activity by monitoring recombination between the attR and attL sites present on the same circular DNA molecule, the cointegrate pCointRL, by detecting the two DNA products, the attP-containing plasmid (pRC10) and the attB-containing plasmid (pAMattB). In the presence of mv4Xis, the resolution of pCointRL into the two plasmids pRC10 and pAMattB was observed (data not shown). On the contrary, the recombination between the attL and attR sites on the same linear fragment (pCointRL linearized) is inefficient in the presence or absence of mv4Xis (Fig. 2D).
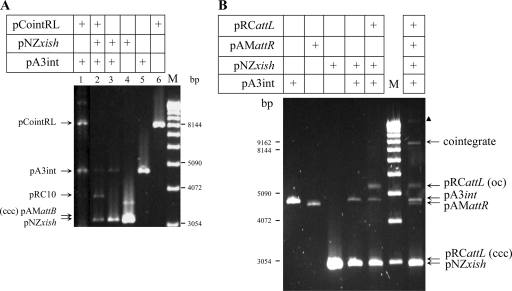
In vivo, in a heterologous Gram-negative host, E. coli, in an mv4Int-producing strain, the plasmid pCointRL carrying attR and attL on the same circular DNA molecule was stably maintained in the absence of mv4Xis (Fig. 3A, lane 1). In the presence of mv4Xis, the resolution of pCointRL into two plasmids, the attP-containing plasmid (pRC10) and the attB-containing plasmid (pAMattB; not visible because of low copy number), was strongly stimulated and the cointegrate pCointRL was no longer detectable (Fig. 3A, lane 2).
FIG. 3.
Role of mv4Xis in in vivo attR-attL recombination. (A) In E. coli, the recombination between the attR and attL sites placed on pCointRL is visualized by the resolution of pCointRL and the formation of the resulting plasmids pRC10 and pAMattB in the presence of Int (pA3int) and Xis (pNZxish). Plasmids isolated from the E. coli strains were digested with EcoRI, which linearizes pA3int, pNZxish, pRC10, and pCointRL but not pAMattB. The resulting fragments were separated by electrophoresis on a 0.8% agarose gel, which was stained with ethidium bromide. The arrows indicate the locations of the different plasmids. pAMattB was not detectable because of the low copy number. Lane M, 1-kb DNA ladder (Invitrogen). (B) In E. coli, the intermolecular site-specific recombination between the attR and attL sites placed on pAMattR and pRCattL, respectively, was visualized by the formation of a cointegrate plasmid carrying the resulting attB and attP sites. Plasmids isolated from the E. coli strains were digested with BstBI, which linearizes pAMattR, pA3int, pNZxish, and the cointegrate but not pRCattL. The resulting fragments were analyzed as described for panel A. The black triangle indicates the possible multimeric form of pRCattL or an additional recombination product (cointegrate + pRCattL). Lane M, 1-kb DNA ladder (Invitrogen). oc, open circular; ccc, covalently closed circular.
In vivo, in E. coli, the recombination between two plasmids carrying attR (pAMattR) and attL (pRCattL) is characterized by the formation of a cointegrate carrying the two resulting attP and attB sites. The cointegrate was detected only in E. coli strains containing both pNZxish and pA3int simultaneously (Fig. 3B), whereas no cointegrate was present in strains expressing only the int gene (data not shown).
In conclusion, mv4Xis is required for the recombination between the attL and attR sites. These experiments were carried out in vivo with heterologous Gram-negative or Gram-positive hosts (E. coli or L. plantarum) or in vitro with protein preparations from these hosts, demonstrating that, as integrative recombination (4), excisive recombination either does not require any L. delbrueckii-specific accessory factor or requires a universally conserved or substituted factor.
mv4Xis binds specifically to the attR and attP sites.
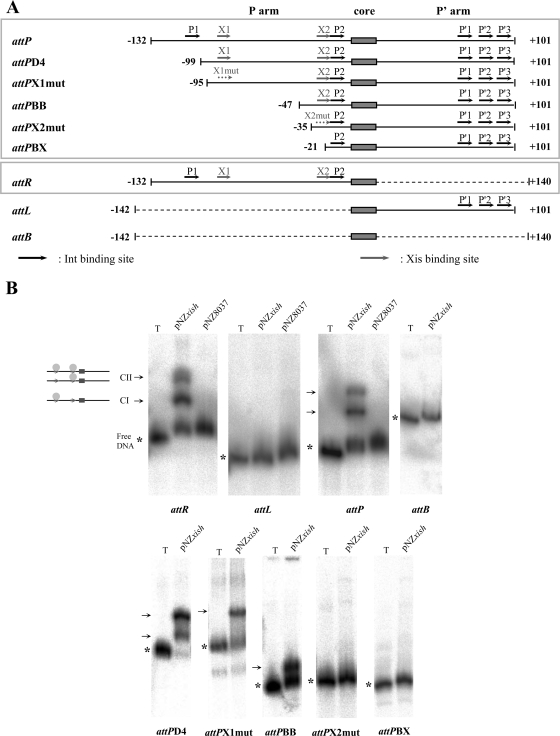
Generally, RDFs control recombination by binding to the attachment sites. mv4Xis probably affects mv4 recombination in a similar way, because it contains a putative DNA-binding domain (the HTH motif). To demonstrate this property, we investigated the ability of mv4Xis to specifically retard the migration of substrates for excisive recombination, attL and attR, and substrates for integrative recombination, attB and attP, on gel electrophoresis.
Following the incubation of mv4Xis-enriched cell extract with attR or attL DNA fragments, two retarded complexes were detected with attR and none with attL (Fig. 4B). When this experiment was carried out with attB and attP, we observed two retarded complexes with attP and none with attB. No complex was formed with any of these sites following incubation with control cell extract (pNZ8037) (Fig. 4B). These results indicated that mv4Xis binds to the P arm and not to the P′ arm of the attachment sites.
FIG. 4.
mv4Xis binds to attR and attP DNA. (A) Schematic representation of attR, attL, attB, and deletion derivatives of the attP sites used as radiolabeled fragments for the experiment shown in panel B. The solid horizontal lines designate the attP-derived DNA and the dashed lines the attB-derived DNA. The numbers at both ends indicate the first and last bases of the segment according to the attP region (4). The 17-bp core sequence is indicated by the shaded box. (B) Gel electrophoresis of protein-DNA complexes. Linear radiolabeled fragments (indicated under the gels and diagrammed in panel A) were incubated with 10 μg of an mv4Xis-enriched cell extract (pNZxish) or with 10 μg of a control cell extract (pNZ8037). The protein-DNA complexes were analyzed by electrophoresis with a 5% polyacrylamide gel. In lane T, only the radiolabeled fragment was included in the reaction mixture, without cell extract. The linear radiolabeled fragments are indicated with an asterisk (*). The complexes are indicated with arrows, and the drawings to the left of the complexes represent the different configurations of mv4Xis binding.
mv4Xis binds to two distinct regions in the P arm of the attR and attP sites.
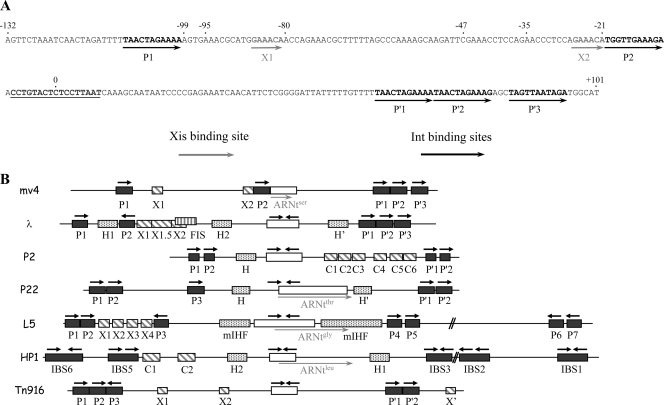
The presence of two retarded complexes suggests that two Xis binding sites may be available in the attP and attR sites. Deletion analysis of the attP substrate by electrophoretic mobility shift assays in the presence of mv4Xis identified two independent regions likely to contain such binding sites (Fig. 4, compare attPD4, attPBB, and attPBX). Analysis of the sequences of these two regions, extending from positions −99 to −80 and from positions −47 to −21, respectively, led to the identification of a 5′-GAAACA sequence present in both regions (Fig. 5A). These two 6-bp sequences, designated X1 and X2, are separated by 54 bp and are in direct orientation. They are located on the same side of the DNA, separated by six turns of the DNA helix, between the P1 and P2 putative Int-binding sites. The X1 binding site is 13 bp away from the P1 site, and the X2 site is adjacent to the P2 site (Fig. 5A).
FIG. 5.
Organization of the attP binding sites. (A) Structure and nucleotide sequence of the mv4 attP region. The numbers above the sequence correspond to the boundaries of the corresponding attP sites presented in Fig. 4A (except 0, which corresponds to the central base of the core region). The core sequence is underlined. The black arrows indicate the 11-bp consensus sequence 5′-TRRYTRRWARR-3′, which is represented five times in attP, and the gray arrows indicate the 6-bp sequence of the Xis-binding site. (B) Comparison of the attP site organizations of λ, P2, P22, L5, HP1, and mv4 phages and the Tn916 transposon (according to references 18, 25, 36, 39, 44, 48, and 57). Integrase binding sites are represented by black squares, IHF binding sites by gray plotted squares, Xis binding sites by gray diagonally striped squares, and the FIS binding site by a gray vertically striped square. The gray arrows represent the 3′ end of the tRNA gene. The core site is represented by a white square, and the black arrows indicate the orientations of the Int arm-type and core-type binding sites.
We investigated whether this motif constituted the binding site for mv4Xis by introducing multiple mutations into X1 or X2 to yield attP sites with potentially defective Xis-binding sites (X1mut and X2mut, AACCCG [where the mutated residues are underlined]). The effects of these changes were evaluated by electrophoretic mobility shift assays with mv4Xis (Fig. 4). When the attP substrate containing one of the two mutated binding sites (attPX1mut) was incubated with an mv4Xis-enriched cell extract, only one complex was detected. This is consistent with there being only one site available, X2 (Fig. 4). When the mutated X2 binding site was tested with the truncated attP site attPX2mut (Fig. 4A), no retarded complex was detected (Fig. 4B).
The same experiments were performed with the equivalent attR derivatives, and the results confirmed those obtained with the attP site and its derivatives (data not shown).
mv4Xis bends the P-arm DNA.
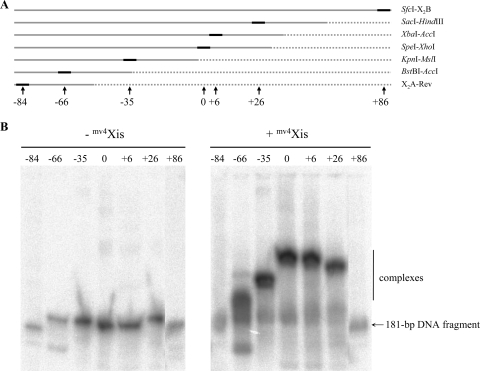
Evidence that mv4Xis induces a bend in the DNA at the P arm was obtained by analyzing the relative mobilities of mv4Xis-DNA complexes with a set of attR substrates circularly permuted for the X2 site (Fig. 6A). mv4Xis formed detectable retarded complexes with five of the substrates, and the relative mobilities of the protein-DNA complexes are consistent with a Xis-induced DNA bend (Fig. 6B). mv4Xis did not form complexes with two of the DNA substrates, generated with restriction enzymes cutting close to the X2 site (Fig. 6A). The approximate bending angle was estimated at 90° (54).
FIG. 6.
mv4Xis bends the P-arm DNA. (A) Seven circularly permuted DNA fragments containing the X2 binding site of mv4Xis were generated by cleavage of pRCattRDR2 with different enzymes, generating 181-bp fragments. The short black line represents the X2 binding site, and the long gray line represents the circularly permuted DNA fragments. The numbers under the arrows indicate the relative locations of the X2 binding site with respect to the center of the fragment (0). (B) These labeled fragments were incubated with the mv4Xis-enriched cell extract (+mv4Xis) or with the control cell extract (−mv4Xis), and the DNA-protein complex mobility was analyzed by gel electrophoresis.
mv4Xis does not inhibit Int-mediated integrative recombination.
We investigated the role of mv4Xis in integrative site-specific recombination between the attP site and the attB site but also between the attP site and the attR or the attL site, which can be secondary integration sites. We previously set up an in vitro integration reaction in which mv4Int efficiently catalyzes recombination between the attP and attB attachment sites (4).
Comparisons of the in vitro integrative recombination efficiencies of reactions between an attP-containing plasmid and the radiolabeled linear fragments containing the attB, attL, or attR site showed that the attL site was essentially equivalent to attB in recombination with attP (Fig. 7A). Conversely, the recombination efficiency of the reaction between the attP and attR sites was about a quarter as efficient as that between the attP and attL sites (Fig. 7A). In experiments carried out with the attL or attR site on a plasmid and the attP site on a linear radiolabeled fragment, the recombination efficiencies were similar to those previously obtained with the reverse locations (data not shown).
FIG. 7.
Role of mv4Xis in in vitro integrative recombination. The samples and the recombination efficiencies (RE) were analyzed as described in the legend for Fig. 2. (A) In vitro recombination assays between linear radiolabeled fragments containing the attB, attL, or attR site and the attP-containing plasmid. Each reaction mixture contains the linear attB, attL, or attR fragment, 200 ng of the supercoiled attP plasmid, 3 μg of mv4Int-enriched cell extract (lanes 1′ and 1 to 3), and 3 μg (lane 2) or 6 μg (lane 3) of mv4Xis-enriched cell extract or 6 μg of control cell extract (pNZ8037) (lane 1′). The recombination efficiencies are indicated under the gels. (B) Kinetics of attP × attB recombination in the presence of Int or Int and Xis. The reactions were realized as described for panel A. At the indicated times, aliquots were removed, and the reactions were stopped by adding 0.1% SDS. The recombination efficiencies are plotted on the graph. (C) In vitro attP × attB recombination assays in the presence of Int (1 μg in each reaction mixture) and various amounts of the control cell extract (8037) or the mv4Xis-enriched cell extract (Xis). The ratios of Xis/Int or 8037/Int are shown below the graph. The reactions were realized as described for panel A, and the recombination efficiencies are plotted on the graph. Error bars indicate standard deviations.
Whatever the substrate used for recombination with the attP site, the integrative recombination efficiency is not affected by the presence of mv4Xis (Fig. 7A).
Similarly, no marked effect on the time course of the integrative recombination reaction was observed in the presence of mv4Int and two different concentrations of mv4Xis compared to the time course of the reaction in the presence of mv4Int alone (Fig. 7B). In addition, the intermolecular recombination efficiency between the attP-containing plasmid and the radiolabeled linear fragment containing attB was analyzed in the presence of various ratios of mv4Xis/mv4Int (up to 50). Compared to the recombination efficiency in the presence of various ratios of control cell extract/mv4Int (up to 50), no significant inhibition was observed (Fig. 7C).
In the in vitro test used here, in the presence of both mv4Int and mv4Xis, the recombination reaction remains unidirectional, i.e., integrative, since the resulting attL and attR sites neoformed by the attP-attB recombination reaction are present on a linear DNA fragment and the intramolecular attL-attR recombination is not detectable in vitro in this case (Fig. 2D). Therefore, the results observed are only due to the effect of mv4Xis on the sole integrative recombination.
DISCUSSION
We have demonstrated that mv4 ORF-56 encodes the phage RDF, a 56-amino-acid protein required for efficient site-specific excisive recombination. The amino acid sequence of mv4 RDF does not resemble that of the phage lambda excisionase but is distantly related to the RDF of the L5-SAM-SPL1 family, especially to the SGI1 and SLP1 RDF (35). Otherwise, mv4Xis shares several common characteristics with the other RDF, such as a small size and a basic pI (35). mv4Xis appears to have a function similar to that of the lambda Xis, in that it is required for excisive recombination, but mv4Xis differs from the lambda Xis in that it seems not to inhibit integrative recombination. This is the main difference between this system and other previously described excisive recombination systems involving the RDF (2, 12, 24, 36).
The mv4 RDF is encoded by the second gene of the early lytic operon, downstream from the Ptec promoter, whereas the int gene is the third gene of the lysogenic operon transcribed from Prep (17). To our knowledge, this is the first time that such a position has been reported for an RDF gene in a nondefective temperate bacteriophage harboring tyrosine recombinase. Usually, for the majority of the phages, particularly for the lambdoid phages, the xis gene is in the immediate vicinity of the int gene and is often located in the same operon (35). In nonlambdoid bacteriophages of the P2 family and in phage HP1, the gene encoding the RDF, cox, is the first gene of the early lytic operon (24, 57), but in this case, Cox has an additional function, regulating the activity of the promoters involved in the genetic switch between lysis and lysogeny (26, 50). The two functions of the Cox protein are carried out by two distinct proteins, Tec and Xis, in phage mv4 (17).
The presence of the xis gene in the early lytic operon may reflect the regulation of recombination via the regulation of xis and int expression. Indeed, the recombination directionality factor, antagonizing the integration process, should be expressed only during induction of the lytic cycle and would therefore need to be tightly regulated. The mv4 int and xis gene localization implies that the expression of the two genes is not coregulated during the mv4 phage cycle. In the lysogenic strain, the int gene is transcribed at a low level from the Prep promoter, whereas the early lytic operon with the xis gene is totally repressed. During prophage induction, the transcription of the lysogenic operon with the int gene is shut off and the lytic operon is transcribed from the Ptec promoter at a high level, leading to extensive synthesis of Xis (17). In P2-like phages and in phage TP901-1, where the overall genetic organization of the genetic switch resembles that described for mv4 (17, 37, 38, 50, 57), the regulation of the site-specific recombination genes is achieved in a similar way. In the lambdoid phages, the int and xis genes are present in the same operon. When both the integrase and the RDF are required, the transcription of the two genes is initiated from the major early PL promoter (23). During lysogenization or in a lysogenic strain, int transcription is initiated at a promoter, PI, located in the xis gene (28).
We investigated the role of mv4Xis in excisive recombination. In the presence of mv4Int, we observed an absolute requirement of mv4Xis for recombination between attR and attL to occur both in vivo and in vitro. We also noted a significant influence of the supercoiling state of the recombination substrates on the excisive reaction. Although the linear attL and attR substrates proved to be totally competent for recombination, the reaction efficiency was strongly enhanced with at least one supercoiled substrate, regardless of which one was provided in circular form. This contrasts with the integrative reaction, in which the superhelicity of the attP site is required and linear substrates are not efficient (4).
As was shown for the integrative reaction with mv4Int (4), the excisive reaction seems independent from host accessory factors, like IHF or FIS for the lambda recombination system, since the experiments were performed in vivo with various heterologous bacterial hosts (E. coli and L. plantarum) or in vitro with cell extracts of these hosts. However, we cannot exclude the requirement of at least one accessory factor universally present in these strains. The independence of host accessory factors in the site-specific recombination reaction is unusual.
We have shown that mv4Xis binds to and bends attR and attP DNA. The two Xis-binding sites are located on the P arm of attR or attP between the putative mv4Int-binding sites P1 and P2, without overlapping these sites (Fig. 5A). In the P arms of the various phages analyzed, the number of the P-arm binding sites of the integrase is relatively constant (2 or 3), but between the Int arm-binding sites and the core sequence, multiple binding sites for host and phage DNA bending proteins are present (Fig. 5B). Compared to other phage systems (Fig. 5B), the recombination protein binding site organization on the P arm is unique for mv4 due to the absence of host or phage DNA bending protein binding sites between the Int arm-binding sites and the core sequence, suggesting a specific, less complex P-arm folding mechanism. The binding of mv4Xis to X1 and X2 might bend the DNA P arm by about 180° to bring the P1-arm site close to the core sequence, thereby facilitating the bridging of these two sites (P1-arm site-core sequence) by a bivalent DNA-binding integrase monomer. Based on these observations, mv4Xis may play at least an architectural role, influencing excisive intasome assembly on attR. This situation evokes that of Xis-L5. In L5 directionality control, Xis-L5 has only an architectural role, with the direction of recombination controlled exclusively by means of the binding and bending of DNA (36). Xis-L5 promotes excision by facilitating the formation of the attR intasome, in which Xis-L5 indirectly enhances Int-L5 occupancy of the P1/P2 arm-type sites by assisting in the formation of intramolecular bridges between the P1/P2 sites and the core (36). In phage λ, Xis controls the directionality of recombination by stimulating the excisive reaction while simultaneously inhibiting the integrative recombination. These dual and opposing effects are realized in two ways. First, the cooperative binding of three Xis monomers to the P arm forms a micronucleoprotein filament (1), which substantially bends attR DNA and alters its trajectory within the excisive intasome, thereby stabilizing the synaptic complex in which attL and attR are bridged by bivalent DNA-binding Int proteins (30). Second, Xis cooperatively recruits an integrase monomer to the P2-arm site through protein-protein interactions at its C terminus (13, 56) and therefore inhibits the formation of a proficient integrative intasome on attP. Due to the close proximity of the P2-arm site and the X2 mv4Xis binding site in the mv4 P arm, we cannot exclude interactions between mv4Xis and mv4Int in order to facilitate the formation of the excisive synapsis.
We have shown that mv4Xis does not inhibit integrative recombination between the attP site and the attB, attL, or attR site under our experimental conditions (Fig. 7). The lack of inhibition by mv4Xis of the integration reaction would be an unprecedented behavior among characterized recombination directionality factors. Indeed, in all of the site-specific recombination systems in which RDFs have been described, even if the RDF genes are expressed from the lytic operon, as with P2-like phages, recombination between attP and attB has always been reported to be inhibited, in various ways, by the RDF (7, 24, 27, 36, 41, 57). For P2 (57) or λ (2, 41) phage, the RDF concentration completely inhibiting the integrative recombination is the same as that necessary for maximal stimulation of excision. For the two phages, the integrative recombination inhibitory effect is observed with a ratio of Xis/Int of about 0.5 to 1 in the in vitro reaction. The in vitro mv4 experiments used to test the inhibition of the integrative recombination were realized under the same conditions. Moreover, we have tested mv4Xis/mv4Int ratios up to 50 without inhibition of the integrative recombination.
The formation of the integrative intasome on the mv4 attP site seems to be unaffected by mv4Xis, as are the attp intrasome interactions with the attB, attR, and attL sites. The higher-order nucleoprotein complex assembly on the P arm in the attP intasome probably involves a bridge to be formed between the P1-arm site and the core region by an mv4Int monomer, and this structure may be stabilized by protein-protein interactions with the three mv4Int monomers assembled on the P′ arm to get a functional intasome. However, this structure may be unstable when formed on the attR site, because of the absence of the P′ arm. The binding of mv4Xis to the P arm and the subsequent DNA bending might then be required for the stabilization of this attR intasome and its assembly into the excisive attL-attR synapsis. The P′-arm nucleoprotein complex on attL, the attL intasome, seems to be stable independently of the presence of the P arm or mv4Xis. Thus, mv4Xis may play a purely architectural role, allowing the higher-order nucleoprotein complex to assemble on attR and facilitating the formation of the excisive synapsis. This function seems to have no inhibitory effect on integrative synapsis assembly involving the attP and attB (or attL or attR) sites.
During phage infection or prophage induction, inhibition of the integrative reaction is not mandatory to avoid phage (re)integration events and to get an efficient lytic cycle. Due to the specific genetic organization of the recombination system in mv4, in both cases, there is a Ptec-driven burst of transcription of the early lytic operon, which leads to overproduction of mv4Xis and Tec, whereas at the same time the lysogenic operon expression is very low, due to nonactivation by Rep or repression by Tec, conditions allowing the presence of a basal level of integrase sufficient to catalyze excision.
Acknowledgments
We thank F. Auvray for his participation in the identification of the mv4 RDF.
We are very grateful to our colleagues in the laboratory, P. Le Bourgeois, M. L. Daveran-Mingot, D. Passerini, and S. Nolivos, for advice and helpful discussions.
This work was supported by grants from the Centre National de la Recherche Scientifique (CNRS-UMR5100) and from the EC Biotech II program (BIO4-CT98-0406).
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Abbani, M. A., C. V. Papagiannis, M. D. Sam, D. Cascio, R. C. Johnson, and R. T. Clubb. 2007. Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly. Proc. Natl. Acad. Sci. U. S. A. 104:2109-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abremski, K., and S. Gottesman. 1982. Purification of the bacteriophage lambda xis gene product required for lambda excisive recombination. J. Biol. Chem. 257:9658-9662. [PubMed] [Google Scholar]
- 3.Alvarez, M. A., M. Herrero, and J. E. Suarez. 1998. The site-specific recombination system of the Lactobacillus species bacteriophage A2 integrates in gram-positive and gram-negative bacteria. Virology 250:185-193. [DOI] [PubMed] [Google Scholar]
- 4.Auvray, F., M. Coddeville, G. Espagno, and P. Ritzenthaler. 1999. Integrative recombination of Lactobacillus delbrueckii bacteriophage mv4: functional analysis of the reaction and structure of the attP site. Mol. Gen. Genet. 262:355-366. [DOI] [PubMed] [Google Scholar]
- 5.Auvray, F., M. Coddeville, R. C. Ordonez, and P. Ritzenthaler. 1999. Unusual structure of the attB site of the site-specific recombination system of Lactobacillus delbrueckii bacteriophage mv4. J. Bacteriol. 181:7385-7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auvray, F., M. Coddeville, P. Ritzenthaler, and L. Dupont. 1997. Plasmid integration in a wide range of bacteria mediated by the integrase of Lactobacillus delbrueckii bacteriophage mv4. J. Bacteriol. 179:1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibb, L. A., M. I. Hancox, and G. F. Hatfull. 2005. Integration and excision by the large serine recombinase phiRv1 integrase. Mol. Microbiol. 55:1896-1910. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Breuner, A., L. Brondsted, and K. Hammer. 1999. Novel organization of genes involved in prophage excision identified in the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 181:7291-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brussow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 11.Bruttin, A., F. Desiere, S. Lucchini, S. Foley, and H. Brussow. 1997. Characterization of the lysogeny DNA module from the temperate Streptococcus thermophilus bacteriophage phi Sfi21. Virology 233:136-148. [DOI] [PubMed] [Google Scholar]
- 12.Burrus, V., and M. K. Waldor. 2003. Control of SXT integration and excision. J. Bacteriol. 185:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bushman, W., S. Yin, L. L. Thio, and A. Landy. 1984. Determinants of directionality in lambda site-specific recombination. Cell 39:699-706. [DOI] [PubMed] [Google Scholar]
- 14.Campbell, A. 1962. Episomes. Adv. Genet. 11:101-145. [Google Scholar]
- 15.Cho, E. H., R. I. Gumport, and J. F. Gardner. 2002. Interactions between integrase and excisionase in the phage lambda excisive nucleoprotein complex. J. Bacteriol. 184:5200-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cluzel, P. J., M. Veaux, M. Rousseau, and J. P. Accolas. 1987. Evidence for temperate bacteriophages in two strains of Lactobacillus bulgaricus. J. Dairy Res. 54:397-405. [DOI] [PubMed] [Google Scholar]
- 17.Coddeville, M., F. Auvray, M. Mikkonen, and P. Ritzenthaler. 2007. Single independent operator sites are involved in the genetic switch of the Lactobacillus delbrueckii bacteriophage mv4. Virology 364:256-268. [DOI] [PubMed] [Google Scholar]
- 18.Connolly, K. M., M. Iwahara, and R. T. Clubb. 2002. Xis protein binding to the left arm stimulates excision of conjugative transposon Tn916. J. Bacteriol. 184:2088-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeMan, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for cultivation of lactobacilli. J. Appl. Microbiol. 23:130-135. [Google Scholar]
- 20.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doublet, B., D. Boyd, M. R. Mulvey, and A. Cloeckaert. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55:1911-1924. [DOI] [PubMed] [Google Scholar]
- 22.Dupont, L., B. Boizet-Bonhoure, M. Coddeville, F. Auvray, and P. Ritzenthaler. 1995. Characterization of genetic elements required for site-specific integration of Lactobacillus delbrueckii subsp. bulgaricus bacteriophage mv4 and construction of an integration-proficient vector for Lactobacillus plantarum. J. Bacteriol. 177:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Echols, H., and G. Guarneros. 1983. Control of integration and excision, p. 75-92. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Esposito, D., and J. J. Scocca. 1997. Purification and characterization of HP1 Cox and definition of its role in controlling the direction of site-specific recombination. J. Biol. Chem. 272:8660-8670. [DOI] [PubMed] [Google Scholar]
- 25.Esposito, D., J. S. Thrower, and J. J. Scocca. 2001. Protein and DNA requirements of the bacteriophage HP1 recombination system: a model for intasome formation. Nucleic Acids Res. 29:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esposito, D., J. C. Wilson, and J. J. Scocca. 1997. Reciprocal regulation of the early promoter region of bacteriophage HP1 by the Cox and Cl proteins. Virology 234:267-276. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh, P., L. R. Wasil, and G. F. Hatfull. 2006. Control of phage Bxb1 excision by a novel recombination directionality factor. PLoS Biol. 4:e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guarneros, G. 1988. Retroregulation of bacteriophage lambda int gene expression. Curr. Top. Microbiol. Immunol. 136:1-19. [DOI] [PubMed] [Google Scholar]
- 29.Josson, K., T. Scheirlinck, F. Michiels, C. Platteeuw, P. Stanssens, H. Joos, P. Dhaese, M. Zabeau, and J. Mahillon. 1989. Characterization of a gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid 21:9-20. [DOI] [PubMed] [Google Scholar]
- 30.Kim, S., and A. Landy. 1992. Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR. Science 256:198-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleerebezem, M., M. M. Beerthuyzen, E. E. Vaughan, W. M. de Vos, and O. P. Kuipers. 1997. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landy, A. 1989. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu. Rev. Biochem. 58:913-949. [DOI] [PubMed] [Google Scholar]
- 33.Le Bourgeois, P., M. Lautier, M. Mata, and P. Ritzenthaler. 1992. New tools for the physical and genetic mapping of Lactococcus strains. Gene 111:109-114. [DOI] [PubMed] [Google Scholar]
- 34.Lewis, J. A., and G. F. Hatfull. 2000. Identification and characterization of mycobacteriophage L5 excisionase. Mol. Microbiol. 35:350-360. [DOI] [PubMed] [Google Scholar]
- 35.Lewis, J. A., and G. F. Hatfull. 2001. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 29:2205-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis, J. A., and G. F. Hatfull. 2003. Control of directionality in L5 integrase-mediated site-specific recombination. J. Mol. Biol. 326:805-821. [DOI] [PubMed] [Google Scholar]
- 37.Madsen, P. L., and K. Hammer. 1998. Temporal transcription of the lactococcal temperate phage TP901-1 and DNA sequence of the early promoter region. Microbiology 144:2203-2215. [DOI] [PubMed] [Google Scholar]
- 38.Madsen, P. L., A. H. Johansen, K. Hammer, and L. Brondsted. 1999. The genetic switch regulating activity of early promoters of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 181:7430-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattis, A. N., R. I. Gumport, and J. F. Gardner. 2008. Purification and characterization of bacteriophage P22 Xis protein. J. Bacteriol. 190:5781-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer, M. P. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163:41-46. [DOI] [PubMed] [Google Scholar]
- 41.Moitoso de Vargas, L., and A. Landy. 1991. A switch in the formation of alternative DNA loops modulates lambda site-specific recombination. Proc. Natl. Acad. Sci. U. S. A. 88:588-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nash, H. A. 1974. Purification of bacteriophage lambda Int protein. Nature 247:543-545. [DOI] [PubMed] [Google Scholar]
- 43.Nash, H. A., and C. A. Robertson. 1981. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J. Biol. Chem. 256:9246-9253. [PubMed] [Google Scholar]
- 44.Papagiannis, C. V., M. D. Sam, M. A. Abbani, D. Yoo, D. Cascio, R. T. Clubb, and R. C. Johnson. 2007. Fis targets assembly of the Xis nucleoprotein filament to promote excisive recombination by phage lambda. J. Mol. Biol. 367:328-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavan, S., P. Hols, J. Delcour, M. C. Geoffroy, C. Grangette, M. Kleerebezem, and A. Mercenier. 2000. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl. Environ. Microbiol. 66:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pena, C. E., J. M. Kahlenberg, and G. F. Hatfull. 1998. The role of supercoiling in mycobacteriophage L5 integrative recombination. Nucleic Acids Res. 26:4012-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pena, C. E., J. M. Kahlenberg, and G. F. Hatfull. 1999. Protein-DNA complexes in mycobacteriophage L5 integrative recombination. J. Bacteriol. 181:454-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pena, C. E., M. H. Lee, M. L. Pedulla, and G. F. Hatfull. 1997. Characterization of the mycobacteriophage L5 attachment site, attP. J. Mol. Biol. 266:76-92. [DOI] [PubMed] [Google Scholar]
- 49.Raya, R. R., C. Fremaux, G. L. De Antoni, and T. R. Klaenhammer. 1992. Site-specific integration of the temperate bacteriophage phi adh into the Lactobacillus gasseri chromosome and molecular characterization of the phage (attP) and bacterial (attB) attachment sites. J. Bacteriol. 174:5584-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saha, S., E. Haggard-Ljungquist, and K. Nordstrom. 1987. The cox protein of bacteriophage P2 inhibits the formation of the repressor protein and autoregulates the early operon. EMBO J. 6:3191-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Smith, M. C., R. Till, K. Brady, P. Soultanas, H. Thorpe, and M. C. Smith. 2004. Synapsis and DNA cleavage in phiC31 integrase-mediated site-specific recombination. Nucleic Acids Res. 32:2607-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swalla, B. M., E. H. Cho, R. I. Gumport, and J. F. Gardner. 2003. The molecular basis of co-operative DNA binding between lambda integrase and excisionase. Mol. Microbiol. 50:89-99. [DOI] [PubMed] [Google Scholar]
- 54.Thompson, J. F., and A. Landy. 1988. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 16:9687-9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thorpe, H. M., S. E. Wilson, and M. C. Smith. 2000. Control of directionality in the site-specific recombination system of the Streptomyces phage phiC31. Mol. Microbiol. 38:232-241. [DOI] [PubMed] [Google Scholar]
- 56.Wu, Z., R. I. Gumport, and J. F. Gardner. 1998. Defining the structural and functional roles of the carboxyl region of the bacteriophage lambda excisionase (Xis) protein. J. Mol. Biol. 281:651-661. [DOI] [PubMed] [Google Scholar]
- 57.Yu, A., and E. Haggard-Ljungquist. 1993. The Cox protein is a modulator of directionality in bacteriophage P2 site-specific recombination. J. Bacteriol. 175:7848-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]