Abstract
In its natural habitats, Bacillus subtilis is exposed to changing osmolarity, necessitating adaptive stress responses. Transcriptomic and proteomic approaches can provide a picture of the dynamic changes occurring in salt-stressed B. subtilis cultures because these studies provide an unbiased view of cells coping with high salinity. We applied whole-genome microarray technology and metabolic labeling, combined with state-of-the-art proteomic techniques, to provide a global and time-resolved picture of the physiological response of B. subtilis cells exposed to a severe and sudden osmotic upshift. This combined experimental approach provided quantitative data for 3,961 mRNA transcription profiles, 590 expression profiles of proteins detected in the cytosol, and 383 expression profiles of proteins detected in the membrane fraction. Our study uncovered a well-coordinated induction of gene expression subsequent to an osmotic upshift that involves large parts of the SigB, SigW, SigM, and SigX regulons. Additionally osmotic upregulation of a large number of genes that do not belong to these regulons was observed. In total, osmotic upregulation of about 500 B. subtilis genes was detected. Our data provide an unprecedented rich basis for further in-depth investigation of the physiological and genetic responses of B. subtilis to hyperosmotic stress.
Bacillus subtilis, a metabolically versatile Gram-positive bacterium, is able to adapt efficiently to a wide variety of stress conditions such as heat stress or phosphate deprivation. It is frequently exposed to nutrient limitations, and in response to such growth-restricting conditions, B. subtilis mounts a complex developmental program that results in the formation of a highly stress-resistant spore (14). In its natural habitat, the upper layers of soil, B. subtilis is often challenged with sudden and often long-lasting changes in osmolality due to flooding and drying of the soil. These extreme conditions threaten the cell with rupture or dehydration and necessitate active countermeasures to ensure survival and growth (7).
After sudden osmotic upshift, B. subtilis rapidly accumulates K+ from the environment via two different K+ uptake systems (KtrAB and KtrCD) (26). This initial phase of osmotic adaptation is followed by the accumulation of compatible solutes thereby permitting a reduction in the cellular K+ level (65). Compatible solutes are either endogenously synthesized by B. subtilis or accumulated from exogenous sources (7). They counteract the deleterious effects of a hypertonic environment on cellular water content and cell physiology and allow B. subtilis to proliferate under a wide range of external osmolalities. Proline, the major compatible solute produced by B. subtilis (65), is synthesized via a dedicated osmostress-responsive pathway that comprises at least two paralogue enzymes of the vegetative proline biosynthetic pathway (4, 28). These paralogue proline biosynthetic enzymes are ProH, a pyrroline-5-carboxylate reductase, and ProJ, a glutamate 5-kinase. Additionally, YerD, a putative ferredoxin-dependent glutamate synthase, might be involved in providing the precursor for proline biosynthesis subsequent to a severe osmotic upshift (28). B. subtilis possesses five osmotically regulated transport systems (OpuA to OpuE) for the acquisition of a broad spectrum of compatible solutes from environmental sources, and the uptake of these compounds provides a considerable degree of osmostress resistance (7).
Sudden osmotic down-shocks trigger a rapid influx of water, driving up turgor, which threatens the integrity of the cell. Like many other microorganisms, B. subtilis expells compatible solutes and ions via mechanosensitive channels (MscL and YkuT) to reduce the osmotic potential of its cytoplasm and thereby counteracts the influx of water (24, 64).
Under severe hyperosmotic conditions that no longer allow growth and cell division, B. subtilis activates the σB-controlled general stress response which provides a nonspecific and preemptive multiple stress resistance to the cells (21). Environmental and cellular stress conditions are detected by a complex set of sensor and regulatory proteins that are organized in the stressosome (43). Signal output from the stressosome activates the alternative transcription factor σB, which in turn triggers the coordinated transcription of about 150 genes comprising a general stress regulon (51, 52). Thirty-seven general stress proteins have an important function in cellular protection against a severe salt shock since the disruption of their structural genes causes a salt-sensitive phenotype. Such a phenotype is also exhibited by a sigB mutant and by the disruption of yerD. Among the σB-controlled genes are those coding for the import systems for the compatible solutes glycine betaine and proline, OpuD and OpuE (28). Transcription of both the opuE and opuD genes is under simultaneous control of σB and the vegetative sigma factor σA, thereby linking the nonspecific general stress response with the salt-specific adaptive response of B. subtilis (7, 21).
Two extracytoplasmic function (ECF) sigma factor regulons, the σW and σM regulons, are also induced subsequent to a hyperosmotic shock. Both the σW and σM regulons are typically induced by cell envelope stress, and σM is essential for growth and cellular survival in high-salt concentrations (29, 51). Severe salt stress has pleiotropic effects on the physiology of B. subtilis: the composition of the cytoplasmic membrane is altered (40, 41), cell wall properties are adjusted (42), and swarming capability of the cells is largely impaired (57).
The physiological response of B. subtilis to changing osmolality has been analyzed in substantial detail at the level of single genes or proteins (7), but the potential of large-scale transcriptomics and proteomics approaches has not yet been fully explored. Such studies are well suited to provide a global view on the cellular responses to a particular environmental challenge. Hoffmann et al. used classical two-dimensional gel electrophoresis (2-DE) to compare protein expression patterns of B. subtilis cultures growing under low- and high-salinity conditions (25), and Höper et al. performed a detailed analysis of the protein synthesis pattern along the growth curve of severely salt-stressed B. subtilis using 2-DE in combination with dual-channel imaging and gel warping (27). A transcriptomics approach analyzing mRNA changes on a time-resolved scale following a hyperosmotic shock was performed by Steil et al., who focused on transcriptional changes that are taking place in cells of a ΔsigB mutant strain that were either continuously cultured at high salinity or subjected to an osmotic upshift (57).
Many membrane proteins, especially transporters of compatible solutes and ions, play a crucial role in adaptation of B. subtilis to salt stress. However, no study has yet addressed the changes in the membrane proteome of salt-stressed B. subtilis cells in a quantitative manner. Furthermore, quantitative studies investigating salt-induced changes in a prokaryotic membrane proteome are generally rare (6, 22, 31, 67, 68). For these reasons, we designed an experimental strategy that combined quantitative shotgun proteomics for the analysis of both the cytosolic and the cytoplasmic membrane proteome with whole-genome microarray data to profile in a time-dependent fashion the physiological adaptation of B. subtilis to a sudden and strong osmotic upshift.
MATERIALS AND METHODS
Growth conditions and preparation of the crude membrane fraction.
The B. subtilis 168 (trpC2) wild-type strain (2) was grown aerobically at 37°C and 180 rpm in Belitsky minimal medium (58) without glutamate to avoid its usage as precursor for biosynthesis of the compatible solute proline. Exponentially growing cells (optical density at 500 nm [OD500] of 0.4) were challenged with 6% (wt/vol) NaCl, and samples were taken before and 10, 30, 60, and 120 min after the onset of stress. Cells were harvested by centrifugation (8,000 × g for 10 min at 4°C), and cell pellets were washed twice with TBS (50 mM Tris, 150 mM NaCl, pH 7.5). Cell lysis was performed in a French press (minicell; SLM Aminco, Rochester, NY). Cell debris was removed by centrifugation (20,000 × g for 20 min at 4°C), and the protein concentration of the supernatant was determined (NanoRotiquant; Roth, Germany).
The above-described growth experiment was performed three times in 14N-labeled medium and three times in 15N-labeled medium for relative quantification of proteins on a proteome-wide level using either normal or 15N-labeled ammonium sulfate as a supplement. 15N reagents were purchased from Eurisotope (Saint Aubin Cedex, France). Equal amounts of protein from each 14N-labeled sample time point were mixed with the respective pooled reference proteins containing equal amounts of 15N-labeled samples from all five time points. The mixed 14N/15N-labeled samples were subjected to ultracentrifugation (100,000 × g for 60 min at 4°C). The resulting pellet was designated as the crude membrane fraction, and an aliquot with a protein content of 50 mg was used as the starting material for subsequent membrane preparations. The supernatant was designated as the cytosolic fraction and analyzed further. A schematic representation of the workflow for the proteomics experiment is given in Fig. S1 in file S1 in the supplemental material.
Membrane purification and SDS-PAGE of membrane and cytosolic proteins.
The membrane preparation was performed exactly as described previously (19). Briefly, to remove contaminating cytosolic proteins, the crude membrane fraction was subjected to a two-step purification procedure using a strongly alkaline carbonate buffer followed by a high-salt buffer. The purified membrane fraction was solubilized using SDS-PAGE sample buffer, and proteins from both subcellular fractions were separated via SDS-PAGE using a 10% (wt/vol) acrylamide minigel for membrane proteins and a 12% (wt/vol) minigel for cytosolic proteins. A gel lane was cut into 12 equidistant pieces, and the tryptic in-gel digestion and the following peptide elution for liquid chromatography-tandem mass spectrometry (LC-MS/MS) were carried out as described previously (15).
Nano-LC-MS/MS analysis.
The nano-LC-MS/MS analysis of peptides derived from tryptic in-gel digestion was performed on a linear trap quadrupole (LTQ) Oribtrap (Thermo Fisher Scientific, Waltham, MA) equipped with a nanoACQUITY UPLC (Waters, Milford, MA). Peptides were loaded onto a trapping column (nanoAcquity Symmetry UPLC column, C18, 5 μm, 180 μm by 20 mm; Waters) at a flow rate of 10 μl/min and washed for 3 min with 99% buffer A. Peptides were then eluted and separated via an analytical column (nanoAcquity BEH130 UPLC column, C18, 1.7 μm, 100 μm by 100 mm; Waters) with a decreasing buffer gradient (from 99% buffer A to 60% buffer B (0.1% acetic acid, 90% acetonitrile, distilled water [dH2O]) in a time frame of 80 min. The mass spectrometric analysis started with a full survey scan in the Orbitrap (m/z 300 to 2,000, resolution of 60,000) followed by collision-induced dissociation and acquisition of MS/MS spectra of the four most abundant precursor ions in the LTQ. Precursors were dynamically excluded for 30 s, and unassigned charge states as well as singly charged ions were rejected.
Database search and filtering of data sets.
Sequest peak list files (*.dta) were generated from the raw instrument data (*.raw) using Extract_msn with default settings implemented in BioworksBrowser 3.3.1 SP1. The database search was performed with SEQUEST (version 27, rev. 12) (13), and the result files (*.out) were combined and filtered with DTASelect 1.9 and Contrast (59). The searched database contained the target sequences, which include the complete proteome set of B. subtilis extracted from UniProtKB (release 12.7) (63) and a set of common laboratory contaminants as well as a decoy database. The precursor tolerance was set to 10 ppm, and tolerance for fragment ions was set to 1 atomic mass unit (amu). In these searches, we only allowed fully tryptic peptides, two missed cleavage sites, and methionine oxidation (+15.9949) as differential modification. Only b- and y-ion series were included in the database search. All searches were performed for either light or heavy (with 15N as fixed modification) peptides using the same set of parameters. Peptide filter criteria were adjusted by keeping the empirically determined false-positive rate (FPR) (50) for each set of combined 14N and 15N search results on a protein level below 3% and on a peptide level below 1%. The following filter criteria were sufficiently stringent: minimum XCorr scores were set to 2.2, 2.8 and 3.5 for +2, +3 and +4 charged ions, respectively, and a ΔCN value of 0.1 was demanded.
Quantification, normalization, and filtering of proteomic data.
The quantification of metabolically labeled peptides was performed using the quantification program Census (49). MS1 files were generated from the raw instrument data with the RawExtractor program (version 1.9.3) and SEQUEST search results of heavy and light peptides were combined with Contrast and filtered with DTASelect 1.9, applying the above-mentioned filter criteria. Both MS1 files and the combined search results were passed to Census, and quantification was performed using the default setting for high-resolution mass spectrometry data. Quantified peptides were extracted from Census, using a default filter and a stringent determinant score of 0.8. Singleton peptides (i.e., peptides with one isotopomer below the detection threshold) as well as proteins with only one quantified unique peptide were discarded. Protein ratios were calculated from peptide ratios using the weighted average approach implemented in Census. Weighted average protein ratios were log2 transformed and median centered. The final data set retained only those proteins that were quantified with at least two 14N- or 15N-labeled peptides per sample and that had at least two expression values from separate biological replicates at the initial reference time point. Expression profiles from different replicates were averaged (see Tables S1 and S2 in file S2 in the supplemental material).
Transcriptome analysis.
Cell harvesting and preparation of total RNA were performed as described previously (16). RNA samples were DNase treated and purified using the RNeasy minikit (Qiagen, Hilden, Germany). The quality of RNA preparations was controlled using the Agilent 2100 Bioanalyzer according to the manufacturer's instructions (Agilent Technologies, Waldbronn, Germany). Generation of the Cy3/Cy5-labeled cDNAs and hybridization to whole-genome DNA microarrays (Eurogentec, Cologne, Germany) containing DNA fragments originating from 4,022 B. subtilis genes as duplicate spots were performed as described by Jürgen et al. (35). The slides were scanned with a ScanArray Express scanner (PerkinElmer Life and Analytical Sciences, Monza, Italy), and signal intensities of the individual spots were quantified with the ScanArray Express image analysis software (PerkinElmer Life and Analytical Sciences). Three hybridizations with mRNA from three biological replicates were performed with the samples labeled with Cy5 and a pooled reference labeled with Cy3, resulting in six measurements per gene. Genes were considered significantly expressed if 70% of the feature pixels had intensities more than 2 standard deviations above the background pixel intensity in four of the six measurements. Intensity-dependent (Lowess) normalization was performed using the GeneSpring software (Agilent Technologies), and the ratios of duplicate spots were averaged for further analyses, resulting in three biologically independent expression values per time point. The expression profiles from three biological replicates were averaged, and incomplete expression profiles discarded. The mRNA expression data are available as supplemental material (see Table S3 in file S2 in the supplemental material).
Statistical, clustering, and functional annotation analyses.
The TIGR Multiexperiment Viewer belonging to the TM4 software suite (53) was used for one-way analysis of variance (ANOVA), figure of merit (FOM) analyses, and K-medians clustering (KMC) of both the transcriptomic and the proteomic data sets. The confidence level for the ANOVA results was set to 99.9% (P < 10−3) for transcriptomic data and to 95% (P < 0.05) for proteomic data. The KMC was only performed on statistically significant mRNA and protein expression profiles. The number of clusters for the KMC approach was derived using the figure of merit implementation of the TM4 software suite (53). Briefly, a figure of merit is an estimate of the predictive power of a clustering algorithm. Since the FOM and KMC calculations were both performed in the Euclidean space, the average expression profiles were standardized in order to have a mean expression value of zero and standard deviation of 1. This simple standardization ensures that similar expression profiles are close in Euclidean space, thereby clustering together. The averaged and standardized mRNA and protein expression profiles were subjected to KMC using 11 clusters for transcriptomic data and nine clusters for proteomic data. Euclidean distance was used as metric, and a maximum of 50 iterations was allowed. The Gene Functional Classification tool provided by DAVID (Database for Annotation, Visualization and Integrated Discovery) bioinformatics resources was used to derive the first biologically meaningful interpretations of gene or protein clusters (11, 30), and for all DAVID tools, default parameters were employed. Lists of regulon members were compiled for the σB, σM, σW, and σX regulons using the DBTBS database (55) and additional data from genome-wide studies (8, 12, 33, 51, 52).
Membrane protein and signal peptide prediction.
Four algorithms were used to predict integral membrane proteins (IMPs) and transmembrane helices (TMH): TMHMM2.0 (39, 56), SOSUI1.1 (23, 45, 46), HMMTOP2.0 (61, 62), and Phobius (37, 38). Signal peptides were predicted using LipoP1.0 (34), SignalP3.0 (5, 47), and Phobius. Prediction results were combined in the following way. (i) Integral membrane proteins were considered if at least two of all four algorithms (TMHMM2.0, SOSUI, HMMTOP, and Phobius) predicted transmembrane helices and (in order to discriminate between N-terminal TMHs and signal peptides) if Phobius predicted at least one TMH or no signal peptide sequence. (ii) Lipoproteins were considered if LipoP1.0 predicts a signal peptidase II cleavage site and Phobius a signal peptide. (iii) Finally, secreted proteins were considered if SignalP3.0 as well as Phobius predicted a signal peptide and if the protein was not assigned as a lipoprotein using the programs mentioned in parameter ii. This cellular localization prediction of proteins resulted in the assignment of 1,027 integral membrane proteins, 99 lipoproteins, and 274 Sec-type secreted proteins in the complete proteome set of B. subtilis.
Microarray data accession number.
The microarray data are accessible through GEO Series accession no. GSE18345 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE18345).
RESULTS AND DISCUSSION
Aiming for a comprehensive picture of the transcriptional response of B. subtilis to a sudden and strong increase in medium osmolality, we monitored the mRNA expression profile using whole-genome microarrays. We complemented this data set with a 14N/15N metabolic labeling strategy in order to track changes in the relative protein levels of the cytosol and the cytoplasmic membrane proteome.
Bacterial cultures were propagated in a minimal medium to early exponential phase (OD500 of 0.4) and were then exposed to a strong osmotic upshift by the addition of 6% (wt/vol) NaCl. This sudden increase in salinity resulted in a growth arrest for 60 to 90 min, after which growth was resumed at a strongly reduced rate (27, 60). Samples for the proteomic as well as for the transcriptomic experiments were taken along the growth curve before and 10, 30, 60, and 120 min after the addition of NaCl.
Global changes on transcriptome and proteome level.
Overall, in our proteome study we could identify 2,169 proteins with at least one unique peptide (FPR, 11.2% on protein level) and 1,710 proteins with at least two unique peptides (FPR, 0.6%) along the growth curve. The proteins identified in our study represent 53% and 42% of the theoretical protein complement of B. subtilis, respectively. Moreover, the protein identifications are based on 14,334 and 13,875 unique peptide identifications (FPR, 1.9% and 0.1%, respectively, on the peptide level). With an average of 6.6 unique peptides or about 26% average sequence coverage per protein identification, this large data set represents a rich resource for further analyses (targeted analyses and/or reanalyses) of changes in the B. subtilis proteome under salt stress conditions, but this data set (for details, see files S1 and S3 in the supplemental material) might prove useful also for studies not connected with the cellular osmotic stress response.
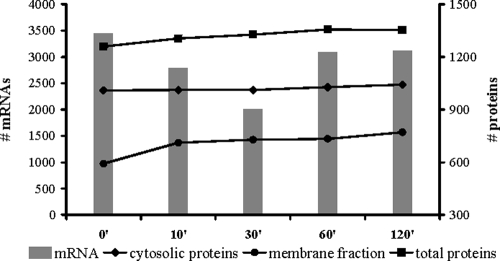
To monitor global changes in gene transcription subsequent to an osmotic upshift, microarrays of the entire gene inventory of B. subtilis were used to screen the transcription profile of the osmotically challenged cells at various times. The results of these experiments were then compared to changes observed in the proteome profile of these cells (Fig. 1). Immediately after the hyperosmotic shock with 6% (wt/vol) NaCl, the B. subtilis cells ceased growth, and concomitantly the number of significantly expressed mRNA species decreased dramatically from 3,447 in exponentially growing cells down to 2,007 mRNA species 30 min after the cells were exposed to salt stress. In contrast, the total number of identified proteins remained nearly unaffected by the salt treatment of the B. subtilis cultures. Only the proteins identified in the membrane fraction showed a substantial increase (20%) within the first 10 min after salt stress. Of the 218 proteins identified, only a subset (30%) can be considered as truly associated with the cytoplasmic membrane either as integral membrane proteins, lipoprotein, or secreted or peripheral membrane protein. The remaining 70% of the proteins detected in the membrane fraction are of cytosolic origin. A functional annotation analysis of these proteins using the DAVID Gene Functional Classification tool revealed five overrepresented groups among these cytosolic proteins: ribosomal proteins, proteins involved in histidine and arginine biosynthesis, aminoacyl-tRNA synthetases, and glycolytic enzymes. It is certainly possible that these proteins end up in the membrane fraction due to their physiological roles during salt stress adaptation. However, one should keep in mind that denatured proteins or proteins subjected to degradation might appear in the membrane fraction as well.
FIG. 1.
Numbers of significantly expressed transcripts and reliably identified proteins along the growth curve. Microarrays were screened for significantly expressed genes using pixel statistics as described in Materials and Methods, and the corresponding numbers of protein identifications along the growth curve are based on at least two unique peptide identifications from all three biological replicates.
In contrast, more than 70% of the 98 proteins identified in growing cells—but not in early salt-exposed cells—are associated with the cytoplasmic membrane. This finding may point to either a general protection mechanism against hyperosmotic shock or the vulnerability of membrane proteins to high salt concentrations.
K-medians clustering of significantly regulated mRNA and proteins.
The stringent filtering procedures of the transcriptome and proteome data set resulted in a comprehensive data set of 3,961 mRNA expression profiles as well as expression profiles for 590 proteins identified in the cytosolic fraction and 383 proteins identified in the membrane fraction. Among these, 165 proteins could be quantified in either subcellular fraction. Within these data sets, the one-way ANOVA identifies 949 mRNAs with significant changes at P < 10−3, as well as 29 proteins in the cytosol and 59 proteins in the membrane fraction with significant changes at P < 0.05. Among the 59 significant proteins of the membrane fraction were, with overlapping predictions, 17 integral membrane proteins, 6 lipoproteins, 5 secreted proteins, and 33 proteins that were either peripheral membrane proteins or cytosolic contaminants. The difference between significantly regulated mRNAs and proteins illustrates nicely the different induction behavior and turnover rates of bacterial mRNAs (with half-lives between seconds and minutes) (20) and proteins (half-lives of minutes to hours).
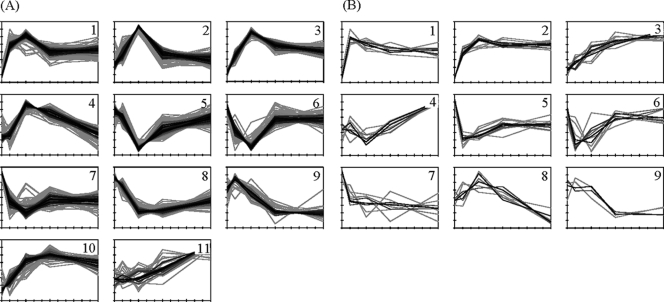
KMC was applied to both the statistically significant transcriptome and proteome data sets in order to group mRNAs and proteins with a similar temporal expression pattern in response to the imposed osmotic upshift. The results are displayed in Fig. 2. The corresponding functionally enriched gene and protein groups are summarized in Table 1, and detailed information is available as supplemental material (see files S4A and S4B in the supplemental material). Overall, mRNA and protein clusters are in good accordance. In particular, the temporary downregulation of most vegetative functions due to the growth arrest of the osmotically challenged B. subtilis cells is well reflected by both the mRNA and the protein complement. However, most changes in response to the osmotic upshift at the mRNA level were observed within the first 30 to 60 min, whereas the corresponding changes in the protein pattern occur at a far slower rate. The changes in the proteome of the salt-stressed cells were probably not complete at the last time point (120 min) analyzed in our experiments.
FIG. 2.
Results of the K-medians clustering of the transcriptomic (A) and proteomic (B) expression profiles. The cytosolic and the membrane proteomes were clustered together. The expression profiles are color-coded, corresponding to their similarity (correlation coefficient) to the cluster median. The KMC results as well as the respective functional analysis data are available in files S4A and S4B in the supplemental material.
TABLE 1.
Summary of functional annotation enrichment analyses for the KMC results of the transcriptomic and proteomic data
| Cluster | No. of members | Enriched functional group(s) | DAVID functional annotation clustering result(s) (P value)a | Quantile (%) | Fold change (vs. 0 min) at: |
|||
|---|---|---|---|---|---|---|---|---|
| 10 min | 30 min | 60 min | 120 min | |||||
| Transcriptome | ||||||||
| 1 | 127 | This cluster contains 54 genes previously assigned to the σB-dependent general stress regulon. Of these, 46 show a minimum 8-fold induction 30 min after osmotic upshift. | Stress response (0.035), CBS domain (0.041) | 75 | 10.9 | 19.2 | 6.0 | 7.2 |
| 50 | 5.2 | 10.5 | 3.3 | 3.9 | ||||
| 25 | 3.0 | 4.3 | 2.2 | 2.2 | ||||
| 2 | 104 | This cluster contains 83 genes of unknown function. The remaining 21 genes are functionally very divergent. 47 genes encode IMPs, lipoproteins, or secreted proteins. | Membrane protein (0.026), DNA binding (0.031) | 75 | 2.0 | 6.1 | 1.7 | 1.5 |
| 50 | 1.5 | 3.6 | 1.4 | 1.2 | ||||
| 25 | 1.1 | 2.5 | 1.1 | −1.1 | ||||
| 3 | 109 | This cluster contains 23 σB-dependent genes and 27 genes involved in transport processes (e.g., glnT, gntP, mntACDH, mrpBCDFG, and opu transporters). | Transporter (5.9E−4), lipid-anchored proteins/ABC transporter (0.006), amino/organic acid transport (0.009), tellurium resistance protein (0.013), stress response (0.014) | 75 | 3.2 | 10.5 | 5.0 | 3.8 |
| 50 | 1.9 | 6.1 | 3.3 | 2.5 | ||||
| 25 | 1.6 | 3.9 | 2.3 | 2.0 | ||||
| 4 | 74 | This cluster contains 53 genes of unknown function. The remaining 21 genes are functionally very divergent. This cluster also includes 26 genes encoding IMPs. | Membrane protein (0.046) | 75 | 1.2 | 3.7 | 2.9 | 1.4 |
| 50 | 1.1 | 2.4 | 2.0 | 1.1 | ||||
| 25 | −1.1 | 1.8 | 1.4 | −1.2 | ||||
| 5 | 114 | This cluster shows strong enrichment of genes of primary metabolism (TCA cycle, metabolism of amino acids, nucleotides, lipids, and coenzymes). | Amino acid biosynthesis (1.5E−5), primary metabolism/biosynthesis (1.6E−5), ligase (i.e., aminoacyl-tRNA synthtases), lysine biosynthesis (0.012), arginine biosynthesis (0.013), coenzyme/prosthetic group biosynthesis (0.029), TCA cycle (0.034), nucleotide-binding (0.040) | 75 | 1.0 | −2.2 | −1.3 | 1.0 |
| 50 | −1.2 | −3.0 | −1.7 | −1.2 | ||||
| 25 | −1.5 | −4.6 | −2.2 | −1.6 | ||||
| 6 | 99 | This cluster shows enrichment of genes of the primary metabolism (oxidative phosphorylation, glycolysis, amino acid, nucleotide, lipid, and coenzyme metabolism). | SAM and 4Fe-4S enzymes (0.029), metal binding (0.033), (phospho)transferase (0.048) | 75 | −1.7 | −2.6 | −1.1 | −1.2 |
| 50 | −2.1 | −3.5 | −1.4 | −1.4 | ||||
| 25 | −3.2 | −5.5 | −1.8 | −1.6 | ||||
| 7 | 99 | This cluster comprises 61 genes of unknown function and several small groups with divergent functional annotations. | 2-component system (0.060)* | 75 | −2.1 | −2.6 | −1.9 | −2.0 |
| 50 | −2.6 | −3.5 | −2.5 | −2.4 | ||||
| 25 | −3.4 | −5.2 | −3.1 | −3.1 | ||||
| 8 | 105 | Chemotaxis and motility genes are strongly enriched in this cluster. Moreover, this cluster contains genes involved in a broad range of transport processes and amino acid metabolism. | Motility (7.6E−7), chemotaxis (0.003), membrane protein (0.007), methyl-accepting transducer (0.012), amino acid biosynthesis (0.017) | 75 | −1.1 | −2.9 | −2.8 | −1.9 |
| 50 | −1.4 | −3.7 | −3.9 | −2.5 | ||||
| 25 | −1.8 | −5.8 | −6.2 | −3.5 | ||||
| 9 | 32 | This cluster comprises 18 genes coding for the phage-like element PBSX. | LysM repeat-containing proteins/cell wall (0.016) | 75 | 1.4 | 1.1 | −1.4 | −1.5 |
| 50 | 1.3 | −1.1 | −1.6 | −1.7 | ||||
| 25 | 1.2 | −1.4 | −2.1 | −2.0 | ||||
| 10 | 58 | This cluster contains no prevalent functional gene group(s), but many small groups. Among these are genes for proline biosynthesis and Opu transporters. | Proline/glutamine family biosynthesis (0.055)* | 75 | 1.6 | 3.2 | 4.8 | 3.0 |
| 50 | 1.3 | 2.3 | 3.1 | 2.1 | ||||
| 25 | 1.1 | 1.8 | 2.3 | 1.6 | ||||
| 11 | 28 | This cluster contains no prevalent functional gene group(s). One small group is related to sporulation and germination (5 genes). | Sporulation/germination (0.083)* | 75 | 1.3 | 1.5 | 1.9 | 4.0 |
| 50 | 1.1 | 0.0 | 1.5 | 2.5 | ||||
| 25 | −1.1 | −1.2 | 1.3 | 2.3 | ||||
| Proteome | ||||||||
| 1 | 10 | All proteins are non-membrane proteins of the primary metabolism detected in the membrane fraction. | Biosynthetic process (0.182)†, ATP binding (0.184)† | 75 | 2.3 | 2.2 | 1.9 | 1.9 |
| 50 | 2.0 | 1.9 | 1.6 | 1.6 | ||||
| 25 | 1.9 | 1.7 | 1.5 | 1.5 | ||||
| 2 | 15 | Peripheral membrane proteins as well as ribosomal proteins dominate this cluster. In addition, three (putative) detoxification proteins (SodA, YaaN, YceC) grouped in this cluster were detected in the membrane fraction. | Ribosomal proteins (0.080) | 75 | 2.1 | 2.6 | 2.9 | 2.7 |
| 50 | 1.9 | 2.3 | 2.3 | 2.1 | ||||
| 25 | 1.6 | 2.2 | 1.9 | 1.9 | ||||
| 3 | 12 | Membrane proteins involved in osmostress resistance (Opu proteins, YtxH, and YuaG) are enriched in this cluster. | Glycine betaine/proline transport (0.036) | 75 | 1.2 | 1.9 | 2.8 | 3.1 |
| 50 | 1.1 | 1.6 | 2.1 | 2.4 | ||||
| 25 | −1.0 | 1.3 | 1.7 | 2.0 | ||||
| 4 | 8 | Cytosolic proteins with only very weak significant changes are comprised in this cluster. The assignment of any biological significance to this cluster is therefore difficult or questionable. | Oxidoreductase (0.020) | 75 | 1.1 | −1.0 | 1.1 | 1.4 |
| 50 | 0.0 | −1.1 | 1.1 | 1.3 | ||||
| 25 | −1.0 | −1.1 | −1.0 | 1.2 | ||||
| 5 | 13 | 12 proteins of this cluster are known/predicted to be IMPs, lipoproteins, or secreted proteins. All proteins of this cluster were detected in the membrane fraction. | Membrane protein (0.038) | 75 | −1.4 | −1.3 | −1.2 | −1.2 |
| 50 | −1.6 | −1.5 | −1.2 | −1.3 | ||||
| 25 | −1.7 | −1.6 | −1.4 | −1.4 | ||||
| 6 | 10 | 7 proteins of this cluster are known to be IMPs or lipoproteins. These proteins were detected in the membrane fraction. | Membrane protein (0.402)† | 75 | −1.3 | −1.3 | 1.0 | 0.5 |
| 50 | −1.9 | −1.6 | −1.1 | −1.1 | ||||
| 25 | −2.7 | −3.3 | −1.1 | −1.3 | ||||
| 7 | 9 | 8 proteins of this cluster are IMPs, lipoproteins, or known to be cell surface associated, mostly detected in the membrane fraction. Two cell wall proteins, PonA and PbpC, also fall within this group. | Cell envelope biogenesis/developmental process (0.192)† | 75 | −1.3 | −1.3 | −1.4 | −1.3 |
| 50 | −1.6 | −1.5 | −1.8 | −1.8 | ||||
| 25 | −1.6 | −1.9 | −2.2 | −2.0 | ||||
| 8 | 7 | All proteins are of cytosolic origin and show only very weak significant changes. The inference of biological significance is therefore difficult or questionable. | Primary metabolic process (0.206)† | 75 | 1.1 | 1.2 | 1.1 | −1.1 |
| 50 | 1.0 | 1.1 | 1.0 | −1.1 | ||||
| 25 | 0.0 | 1.1 | 1.0 | −1.1 | ||||
| 9 | 4 | 2 proteins of this very small cluster are involved in motility (Hag and FlgG), show a ≥2-fold downregulation, and were detected in the membrane fraction. | No terms clustered | 75 | 1.2 | −0.4 | −1.5 | −1.2 |
| 50 | 0.0 | −1.1 | −1.8 | −1.6 | ||||
| 25 | −1.1 | −1.2 | −2.1 | −2.1 | ||||
Different significance thresholds for the DAVID Functional Annotation Clustering approach were applied for the transcriptomic data and the proteomic data. Since the number of genes per cluster is reasonably high for the transcriptomic data, a P value of ≤0.05 was applied. Because the number of proteins per cluster is, with a few exceptions, rather low, a less stringent P value (≤0.1) was chosen as a significance threshold. The P value is given in parentheses. *, no significant (P ≤ 0.05) functional annotation cluster (the next best hit is displayed); †, no significant (P ≤ 0.1) functional annotation cluster (the next best hit is displayed).
General and cell envelope stress response.
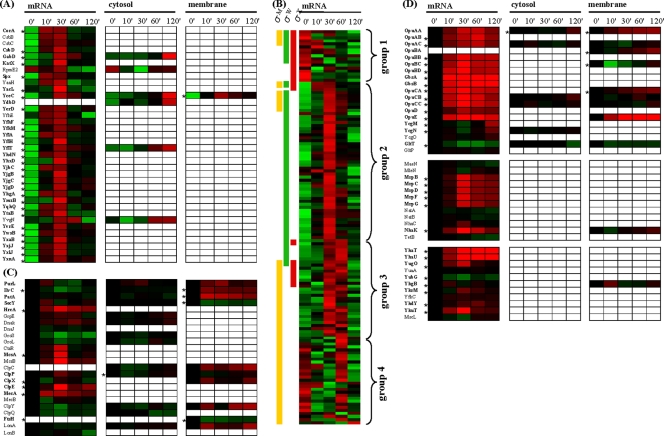
The initial response of B. subtilis to a sudden and strong osmotic upshift is characterized by a strong and immediate activation of the σB regulon, leading to synthesis and accumulation of proteins conferring a broad stress resistance to B. subtilis cells (21). The σB regulon comprises at least 37 proteins that are critical for providing osmostress resistance, as judged by the salt-sensitive phenotype of their structural genes (28). Almost all of these σB-dependent salt stress protective proteins are significantly upregulated either early at the mRNA level or in later adaptation stages of the osmotically challenged cells on the protein level (Fig. 3A). Only a small number of these proteins are of known function. However, their immediate and strong induction subsequent to the osmotic up-shock and, at later times, their accumulation in osmotically stressed cells strongly suggest a physiologically important function for this group of proteins within the acclimatization process of B. subtilis to high-salinity surroundings.
FIG. 3.
General, cell envelope, and specific osmostress responses. Expression profiles shown in panels A and B are based on standardized log2 ratio values, and expression profiles in panels C and D are log2 ratio based. The minimum and maximum displayed log2 ratios are ±3 for the transcriptomics data and ±2 for the proteomics data. Genes/proteins exhibiting significant changes are asterisked. (A) The 35 proteins given here belong to the σB-dependent general stress regulon and were already shown to be indispensable for the adaptation to high salinity (28). (B) Displayed are 131 mRNAs which are presumed or shown to be under the control of either σM, σW, or σX. In order to reveal overall expression patterns, the mRNA expression profiles were sorted according to their ECF membership and their expression profile. (C) Expression profiles of chaperones and ATP-dependent proteases as well as regulator and adapter proteins involved in protein quality control are shown. (D) Specific osmostress response proteins. The upper group contains proteins involved in uptake and synthesis of compatible solutes, whereas both lower groups comprise systems (presumably) involved in Na+ extrusion (middle group) and K+ extrusion (lower group).
In addition to the σB regulon, three large ECF regulons (σM, σW, and σX), which specifically govern the physiological response to cell envelope stress, are affected by a suddenly increase in the external osmolality. The σM and σW regulons are known to be involved in salt stress adaptation (29, 51). A compiled set of 132 known or probable members of the σM, σW, and σX regulons was sorted according to expression patterns and regulon memberships (Fig. 3B; see Table S1 in file S5 in the supplemental material). This classification revealed four groups of genes with similar expression profiles, each of which was associated with one prevalent ECF transcription factor. The color-coded expression pattern of these genes indicates a sequential activation of the general and cell-surface stress regulons. The σB regulon is activated immediately after the shift to high salinity, and the onset of σB-controlled gene expression is followed by the induction of the σW regulon, which shows maximum activation after 30 min. The σM regulon is the last to be induced, with maximum induction 60 min after the salt shock. We also observed partial repression of the σX regulon. Our data clearly show that there is a sequential activation of the transcription of the σB, σW, and σM regulons, but one should keep in mind that there is evidence for functional and regulatory redundancy of the ECFs (44).
We observed that the differential activation of the above-discussed stress regulons at the mRNA level by salt stress was not reflected one-to-one at the protein level. Fifty-four expression profiles were acquired for 43 proteins whose structural genes are under the (partially overlapping) control of σM, σW, and σX (see Table S2 in file S5 in the supplemental material). Remarkably, among this set of proteins, only six proteins showed significant changes in response to salt stress. The penicillin-binding protein PonA and the membrane-localized protease FtsH are under the control of σM, and both proteins exhibited a significant downregulation in the membrane fraction. In contrast, three “Y” proteins (YaaN, YuaG, and YuaI) under the control of σW show significant upregulation. In addition, YceC, which is a member of the detoxification operon yceCDEFGH and under the control of σA, σM, and σW is significantly upregulated in the membrane fraction. Proteins under the control of σX (DltD, LytR, PbpX, YabP, YwbL, and YwbM) are mostly integral membrane proteins, lipoproteins, or secreted proteins and were detected in our experiments in the membrane fraction. Each of these six proteins exhibits a nonsignificant downregulation in the salt-stressed cells.
Overall, expression profiles of proteins and mRNAs correspond reasonably well, but some proteins exhibit regulation patterns that are opposed to their observed profile at the mRNA level. Several explanations for this unexpected behavior are conceivable. Since many ECF-controlled genes encode for proteins that are known to be secreted or localized at the cellular surface, a plausible explanation is that the respective proteins are subject to a localization shift upon stimulation (e.g., secretion). However, targeted protein degradation and mRNA-related processes are possible explanations. In any case, further experimental evidence is required to elucidate the physiological significance of the observed counterintuitive changes in the context of hyperosmotic stress.
Protein quality control.
In B. subtilis, the functional integrity of proteins is ensured through protein quality control mechanisms that include the action of molecular chaperones, protein-folding catalysts, and ATP-dependent proteases. As mentioned above, shortly after the shift to hyperosmotic conditions numerous cytosolic proteins with predicted vegetative functions were observed to be enriched in the membrane fraction. A representative set of these types of proteins (PurL, phosphoribosylformylglycinamidine synthase II involved in purine biosynthesis; IlvC, ketol-acid reductoisomerase involved in branched-chain amino acid synthesis; and PatA, an aminotransferase involved in lysine biosynthesis) is shown in Fig. 3C. We also found that numerous integral membrane proteins or peripheral membrane proteins (e.g., ATP-binding proteins of ABC transporters) showed a clear downregulation in the membrane fraction after hyperosmotic shock. These proteins are represented in Fig. 3C by SecY, which possesses 10 transmembrane helices and belongs to the Sec protein secretion system. Both facts indicate the involvement of protein quality control mechanisms in response to the osmotic upshift, either in the refolding or degradation of unfolded vegetative proteins or in the (targeted) degradation of membrane proteins. Figure 3C shows expression profiles of some quality control proteins. Significant induction on mRNA level could be observed for hrcA, mcsA, clpP, clpX, clpE, and mecA. On the protein level, it is interesting to note that an enrichment of chaperones (ClpC, ClpX, and ClpY) is observed in the membrane fraction concomitantly with their depletion in the cytosolic fraction. In addition, the ATP-dependent LonA protease was upregulated in the membrane fraction and hence could carry out degradation of misfolded proteins, while the membrane-anchored protease FtsH, which degrades both cytosolic and membrane proteins (54), was itself subject to degradation. The issue of whether the observed enrichment of vegetative proteins in the membrane fraction corresponds to a real physiologically relevant change in protein localization during salt stress needs further careful studies. However, our data hint that the sudden imposition of hyperosmotic conditions might induce denaturation and misfolding of both cytosolic and membrane proteins in B. subtilis.
Osmospecific response.
The stress response of B. subtilis to sudden increases in osmolality comprises not only σB-controlled general stress proteins, but also many salt-specific stress proteins. The transcription of the genes encoding this latter group of proteins appears to be mostly independently from alternative sigma factors. Many of the salt-specific proteins are of known function. These proteins display a significant upregulation at either the mRNA or protein level (Fig. 3D). In particular, all Opu transporters (OpuA to OpuE) for compatible solute acquisition are immediately upregulated at the mRNA level, with maximum induction after 30 min, and the corresponding transport proteins accumulate to significant levels in the cytoplasmic membrane after 60 or 120 min. It has to be noted in this context that the specific substrates of these osmoprotectant uptake systems (7) were not present in the growth medium. Consequently, their induction reflects their osmotic control. When B. subtilis uses proline as a nutrient rather than as an osmoprotectant, this amino acid is imported via the high-affinity proline uptake system YcgO and degraded by the YcgM and YcgN enzymes to glutamate (7). Expression of the ycgMNO operon is induced by low levels of extracellular proline (S. Moses and E. Bremer, unpublished data). We observed a significant upregulation of the transcription of the ycgMNO operon in the salt-stressed cells in the late adaptation phase; this apparent osmotic induction of the ycgMNO operon probably reflects the leakage of proline from the osmotically stressed and hence proline-producing cells and the subsequent induction of ycgMNO expression (S. Moses and E. Bremer; unpublished data). The H+/glutamate symporter GltP did not show a significant change in mRNA level, whereas the H+ and Na+/glutamate symporter GltT was downregulated at both the mRNA and the protein level, consistent with the cellular need for a tightly controlled low intracellular Na+ level. Twenty-nine possible candidates for the active Na+ extrusion systems were extracted from UniProtKB using the GO annotation “sodium ion transport” (GO:0006814). Among these, two Na+/H+ antiporters were found to be significantly upregulated in the osmotically challenged B. subtilis cells: (i) the mrpABCEDEFG operon, involved in pH homeostasis (32), and the Mrp complex, which probably constitutes the major Na+ extrusion system operating in B. subtilis; and (ii) NhaK, an Na+, K+, Li+, and Rb+/H+ antiporter (18). Disruption of the mrpABCEDEFG operon causes a strong sensitivity toward Na+ (36), and upregulation of nhaK expression in response to either NaCl or KCl has already been reported (18).
When B. subtilis is exposed to a sudden shift from hyper- to hypo-osmotic conditions, the cells have to jettison very rapidly those ions and organic solutes that were accumulated under high-osmolality growth conditions in order to reduce water entry (7). Expulsion of these solutes by B. subtilis is accomplished via the transient opening of mechanosensitive channels (24). A single channel of large conductivity (MscL type) and three channel proteins of small conductivity (MscS types YhdY, YkfC, and YkuT) are present in B. subtilis (24, 64). We observed a minor reduction in the mRNA level for the mscL gene, but found a significant induction on the mRNA level for the yhdY and ykuT genes in our transcriptome study. An upregulation of the yhdY and ykuT genes at high salinity has been observed previously (64). The transient opening of mechanosensitive channels in B. subtilis serves as a safety valve for osmotically down-shocked cells. The observed induction of yhdY and ykuT might thus be interpreted as a preemptive measure of high-salinity-challenged B. subtilis cells for the next osmotic down-shock. It also should be noted that the ykuT gene is part of the σB regulon.
The initial response of B. subtilis to an osmotic upshift is the massive accumulation of K+ ions (26, 65). The K+ content of the cells is reduced again when compatible solutes are accumulated in the second adaptation phase to the high-salinity challenge (65). To search for B. subtilis proteins potentially involved in the extrusion of K+, a list of all proteins with the GO annotation “potassium ion transport” (GO:0006813) was extracted from the UniProtKB database. Among the nine proteins retrieved by this search, the K+ export system YhaTU (17) and the putative K+ channel protein YugO were significantly induced at the mRNA level. These observations suggest that both the YhaTU and the YugO systems might function in a concerted fashion in the export of K+ during the prolonged adaptation process of B. subtilis to high-salinity surroundings.
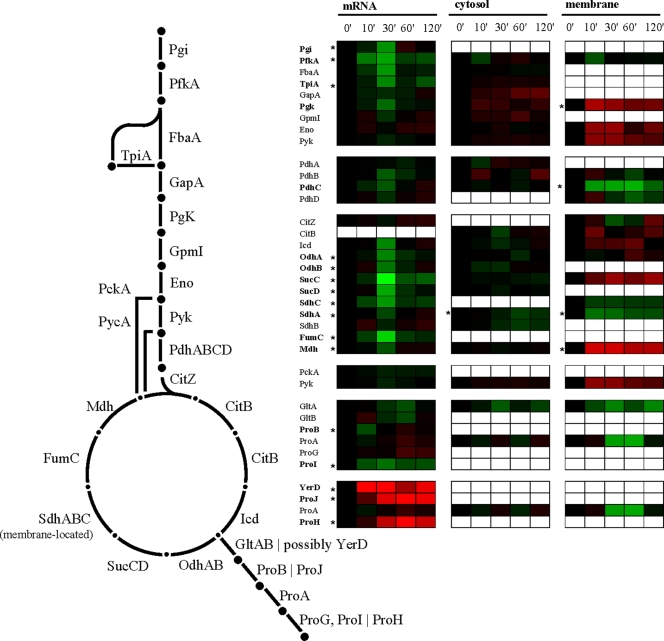
After the initial uptake of K+, B. subtilis starts to synthesize large amounts of the compatible solute proline, which is accumulated to maintain turgor and general cellular functionality (26, 65). Two routes for proline biosynthesis from its precursor glutamate operate in B. subtilis (4): ProB, ProG, and ProI catalyze anabolic proline biosynthesis, and at least two paralogue proteins (ProH and ProJ) are presumed to be responsible for high-level proline synthesis under hyperosmotic conditions (4, 7, 28). The ProA enzyme is used by both proline biosynthetic routes (4, 7). Consistent with the cellular role for these two proline biosynthetic routes, we found that the genes involved in the anabolic proline biosynthesis were repressed, presumably due to the growth arrest, but those used for the osmoadaptive proline production (proHJ) were induced (Fig. 4).
FIG. 4.
Glycolysis, TCA cycle, and vegetative as well as salt-specific proline biosynthesis. Expression profiles are displayed on the basis of log2 ratios. The minimum and maximum displayed log2 ratios are ±3 for the transcriptomics data and ±2 for the proteomics data. Genes/proteins exhibiting significant changes are asterisked.
The large amounts of proline produced under osmotic stress conditions (65) require a continuous supply of the precursor molecules 2-oxoglutarate and glutamate. Recently, Höper et al. (27) observed in a comprehensive proteome study of salt-shocked B. subtilis cells that the enzymes of the tricarboxylic acid (TCA) cycle leading to the synthesis of 2-oxoglutarate remained present in the salt-stressed cells, whereas the synthesis and amounts of those enzymes that participate in the TCA cycle reactions from 2-oxoglutarate to oxaloacetate were reduced. In this way, the osmotically challenged B. subtilis cell ensures an adequate supply of 2-oxoglutarate that then can be converted via the GOGAT (glutamine-2-oxoglutarate aminotransferase) enzyme into glutamate, the immediate precursor for the biosynthesis of proline. Our transcriptome and proteome data support the findings of Höper et al. (27).
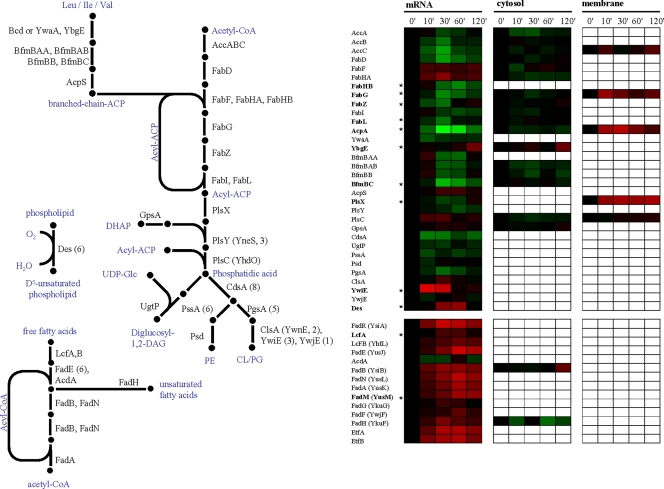
The adaptation of B. subtilis to high salinity is also accompanied by rearrangements in either the composition or structure of the cell envelope. In particular, the lipid and fatty acid composition of the cytoplasmic membrane is affected (42). Lopez et al. found an increase in the content of cardiolipin at the expense of phosphatidylglycerol, an increase in saturated straight-chain fatty acids, and a decrease in the level of isobranched fatty acids, as well as an increase in unsaturated fatty acids. These changes are thought to lead to a higher salt resistance of the B. subtilis cell (41). Due to the salinity-induced growth arrest observed in our experiments, most enzymes involved in fatty acid and lipid synthesis are repressed on the mRNA level and are downregulated on the protein level (Fig. 5). In particular, the significant repression of the genes encoding for the fatty acid synthesis proteins AcpA, FabG, FabZ, and FabL strongly indicates a reduced need for membrane building blocks in these almost nongrowing cells.
FIG. 5.
Fatty acid and lipid biosynthesis and fatty acid degradation. Expression profiles are displayed on the basis of log2 ratios. The minimum and maximum displayed log2 ratios are ±3 for the transcriptomics data and ±2 for the proteomics data. Genes/proteins exhibiting significant changes are asterisked. PE, phosphatidylethanolamine; CL/PG, cardiolipin/phosphatidylglycerol.
In contrast to the above-described findings, we observed an induction for a few genes and proteins involved in fatty acid synthesis. The initial condensation step of the fatty acid synthesis in B. subtilis is carried out by two β-ketoacyl acyl carrier protein (ACP) synthases, FabHA and FabHB, which can use either acetyl coenzyme A (CoA) or branched acyl-CoA primers (9). A third β-ketoacyl-ACP synthase, FabF, carries out chain elongation. Interestingly, in the course of cellular adaptation of the B. subtilis cells to high salinity, we found that the transcription of the fabHA and fabHB genes was reciprocally regulated. The fabHB gene was repressed, and, interestingly, the encoded enzyme has the higher specific activity for branched-chain primers (9). This result is in good agreement with the decreased need for isobranched fatty acids under salt stress conditions (41). More detailed studies on the apparent different roles for the FabHA and FabHB enzymes during salt stress adaptation might thus be rewarding.
B. subtilis has three (putative) cardiolipin synthase genes, clsA (formerly known as ywnE), ywiE, and ywjE. In our data, transcription of the clsA gene as well as the ywjE gene is only marginally affected by salt stress, but the σB-controlled ywiE gene is induced more than 8-fold. Nevertheless, ywiE and ywjE are not essential during hyperosmotic conditions, whereas clsA is required for increased osmotolerance in B. subtilis (28, 40). A third distinct biochemical change in the membrane composition is the increase in unsaturated fatty acids, which is catalyzed by a membrane-located Δ5-desaturase encoded by the cold-inducible des gene (1). The expression of the des gene is regulated via the DesKR two-component regulatory system responsive to membrane lipid fluidity (10). After a 60-min imposition of hyperosmotic stress, we observed a 3-fold induction of the des gene. This is not only in good agreement with the observation made by Lopez et al. (40) on membrane fatty acid and lipid composition under hyperosmotic conditions, but it also suggests that the DesKR two-component system senses changes in membrane fluidity after the exposure of B. subtilis cells to NaCl in the growth medium.
We found that the anabolic reactions of the fatty acid and lipid metabolism are downregulated, likely a consequence of the growth arrest. However, the expression of genes coding for proteins involved in degradation of free fatty acids via β-oxidation show global upregulation (Fig. 5). These genes are mostly controlled by the FadR repressor, which is negatively regulated by long-chain acyl-CoAs. It is worth notice that both end products from straight-chain as well as branched-chain acyl-CoA degradation end up in the TCA cycle either as acetyl-CoA or succinyl-CoA and could in principle complement the anaplerotic reactions catalyzed by the PckA and PycA enzymes.
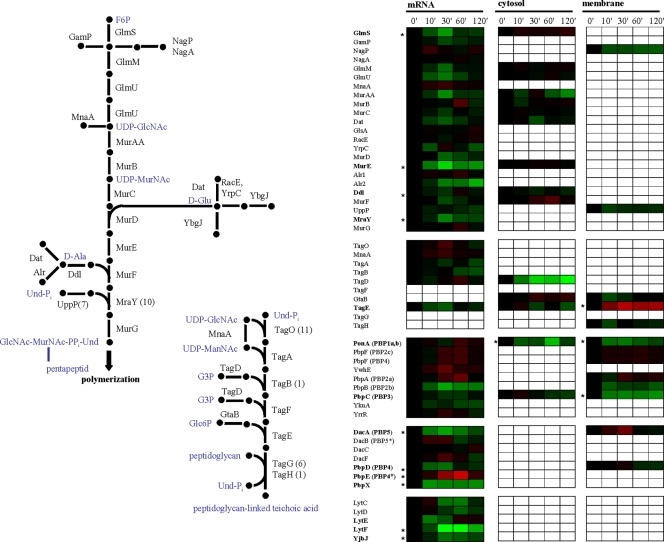
Along with the composition of the cytoplasmic membrane, exposure of B. subtilis to high salinity also influences the properties of the cell wall (42). Given the growth arrest that the B. subtilis cells experienced in our salt stress experiment, nearly all enzymes involved in the biosynthesis of the cell wall and its precursors are downregulated on both the mRNA and protein levels (Fig. 6). Few genes and enzymes show the opposite regulation. Among this group of salt-stress-induced genes is penicillin-binding protein Pbp4* (pbpE); upregulation of pbpE expression has already been reported, and the disruption of pbpE results in a salt-sensitive phenotype (48). Expression of pbpE is controlled by the alternative sigma factor σW, and Pbp4* functions as an endopeptidase that is involved in the cleavage of peptidoglycan cross-links. Hence, Pbp4* could play an important role in the cell wall rearrangements that take place during osmoadaptation of B. subtilis (48).
FIG. 6.
Biosynthesis of peptidoglycan and teichoic acids, different groups of penicillin-binding proteins, and vegetative autolysins. Expression profiles are displayed on the basis of log2 ratios. The minimum and maximum displayed log2 ratios are ±3 for the transcriptomics data and ±2 for the proteomics data. Genes/proteins exhibiting significant changes are asterisked.
Concluding remarks.
The strategy we employed for the study of severely salt-stressed B. subtilis cells examines in parallel and in a time-resolved manner the quantitative changes of the transcriptome, the cytosolic proteome, and the membrane proteome. This genome-wide approach covers not only the whole mRNA complement of B. subtilis cells, but also the two largest subproteomes. A higher proteome coverage of salt-stressed B. subtilis can be achievable in future studies by employing tools that specifically target the most hydrophobic complement of the proteome (66), the secretome (3), and the cell wall proteome of B. subtilis (Hempel et al., unpublished data).
The data presented here provide a comprehensive picture of the dynamic changes in gene expression and the concomitant alterations in the level of proteins that occur in B. subtilis cells exposed to a severe salt shock. Our study complements previous genetic and physiological studies on the osmotic stress response of B. subtilis. In combination with other genome-wide transcriptional profiling (57) and proteomic studies (27) of salt-stressed B. subtilis cells, our data provide an unprecedented rich basis for further in-depth investigation of the physiological and genetic responses of B. subtilis to hyperosmotic stress.
Supplementary Material
Acknowledgments
We are indebted to Sylvia Melzer for valuable contributions to bioinformatics aspects of this work. We thank Vickie Koogle for kindly editing the manuscript and Katrin Harder, Anja Hoffmann, and Julia Kretschmer for excellent technical assistance.
This work was supported by grants from the Bundesministerium für Bildung und Forschung (0313812C and 03ZIK011), the Deutsche Forschungsgemeinschaft (SFB/TR34), and the European Union (BaSysBio).
Footnotes
Published ahead of print on 30 November 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aguilar, P. S., J. E. Cronan, Jr., and D. de Mendoza. 1998. A Bacillus subtilis gene induced by cold shock encodes a membrane phospholipid desaturase. J. Bacteriol. 180:2194-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484-1502. [DOI] [PubMed] [Google Scholar]
- 4.Belitsky, B. R., J. Brill, E. Bremer, and A. L. Sonenshein. 2001. Multiple genes for the last step of proline biosynthesis in Bacillus subtilis. J. Bacteriol. 183:4389-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Bidle, K. A., P. A. Kirkland, J. L. Nannen, and J. A. Maupin-Furlow. 2008. Proteomic analysis of Haloferax volcanii reveals salinity-mediated regulation of the stress response protein PspA. Microbiology 154:1436-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bremer, E. 2002. Adaptation to changing osmolarity, p. 385-391. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 8.Cao, M., and J. D. Helmann. 2004. The Bacillus subtilis extracytoplasmic-function sigmaX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 186:1136-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, K. H., R. J. Heath, and C. O. Rock. 2000. beta-ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J. Bacteriol. 182:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cybulski, L. E., D. Albanesi, M. C. Mansilla, S. Altabe, P. S. Aguilar, and D. de Mendoza. 2002. Mechanism of membrane fluidity optimization: isothermal control of the Bacillus subtilis acyl-lipid desaturase. Mol. Microbiol. 45:1379-1388. [DOI] [PubMed] [Google Scholar]
- 11.Dennis, G., Jr., B. T. Sherman, D. A. Hosack, J. Yang, W. Gao, H. C. Lane, and R. A. Lempicki. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:P3. [PubMed] [Google Scholar]
- 12.Eiamphungporn, W., and J. D. Helmann. 2008. The Bacillus subtilis sigmaM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67:830-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976. [DOI] [PubMed] [Google Scholar]
- 14.Errington, J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117-126. [DOI] [PubMed] [Google Scholar]
- 15.Eymann, C., A. Dreisbach, D. Albrecht, J. Bernhardt, D. Becher, S. Gentner, T. Tam le, K. Büttner, G. Buurman, C. Scharf, S. Venz, U. Völker, and M. Hecker. 2004. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 4:2849-2876. [DOI] [PubMed] [Google Scholar]
- 16.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujisawa, M., M. Ito, and T. A. Krulwich. 2007. Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity. Proc. Natl. Acad. Sci. U. S. A. 104:13289-13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujisawa, M., A. Kusumoto, Y. Wada, T. Tsuchiya, and M. Ito. 2005. NhaK, a novel monovalent cation/H+ antiporter of Bacillus subtilis. Arch. Microbiol. 183:411-420. [DOI] [PubMed] [Google Scholar]
- 19.Hahne, H., S. Wolff, M. Hecker, and D. Becher. 2008. From complementarity to comprehensiveness—targeting the membrane proteome of growing Bacillus subtilis by divergent approaches. Proteomics 8:4123-4136. [DOI] [PubMed] [Google Scholar]
- 20.Hambraeus, G., C. von Wachenfeldt, and L. Hederstedt. 2003. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics 269:706-714. [DOI] [PubMed] [Google Scholar]
- 21.Hecker, M., and U. Völker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 22.Herranen, M., N. Battchikova, P. Zhang, A. Graf, S. Sirpio, V. Paakkarinen, and E. M. Aro. 2004. Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803. Plant Physiol. 134:470-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann, T., C. Boiangiu, S. Moses, and E. Bremer. 2008. Responses of Bacillus subtilis to hypotonic challenges: physiological contributions of mechanosensitive channels to cellular survival. Appl. Environ. Microbiol. 74:2454-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann, T., A. Schutz, M. Brosius, A. Völker, U. Völker, and E. Bremer. 2002. High-salinity-induced iron limitation in Bacillus subtilis. J. Bacteriol. 184:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holtmann, G., E. P. Bakker, N. Uozumi, and E. Bremer. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 185:1289-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Höper, D., J. Bernhardt, and M. Hecker. 2006. Salt stress adaptation of Bacillus subtilis: a physiological proteomics approach. Proteomics 6:1550-1562. [DOI] [PubMed] [Google Scholar]
- 28.Höper, D., U. Völker, and M. Hecker. 2005. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J. Bacteriol. 187:2810-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horsburgh, M. J., and A. Moir. 1999. Sigma M, an ECF RNA polymerase sigma factor of Bacillus subtilis 168, is essential for growth and survival in high concentrations of salt. Mol. Microbiol. 32:41-50. [DOI] [PubMed] [Google Scholar]
- 30.Huang, D. W., B. T. Sherman, Q. Tan, J. Kir, D. Liu, D. Bryant, Y. Guo, R. Stephens, M. W. Baseler, H. C. Lane, and R. A. Lempicki. 2007. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35:W169-W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, F., S. Fulda, M. Hagemann, and B. Norling. 2006. Proteomic screening of salt-stress-induced changes in plasma membranes of Synechocystis sp. strain PCC 6803. Proteomics 6:910-920. [DOI] [PubMed] [Google Scholar]
- 32.Ito, M., A. A. Guffanti, B. Oudega, and T. A. Krulwich. 1999. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 181:2394-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jervis, A. J., P. D. Thackray, C. W. Houston, M. J. Horsburgh, and A. Moir. 2007. SigM-responsive genes of Bacillus subtilis and their promoters. J. Bacteriol. 189:4534-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jürgen, B., S. Tobisch, M. Wumpelmann, D. Gordes, A. Koch, K. Thurow, D. Albrecht, M. Hecker, and T. Schweder. 2005. Global expression profiling of Bacillus subtilis cells during industrial-close fed-batch fermentations with different nitrogen sources. Biotechnol. Bioeng. 92:277-298. [DOI] [PubMed] [Google Scholar]
- 36.Kajiyama, Y., M. Otagiri, J. Sekiguchi, S. Kosono, and T. Kudo. 2007. Complex formation by the mrpABCDEFG gene products, which constitute a principal Na+/H+ antiporter in Bacillus subtilis. J. Bacteriol. 189:7511-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kall, L., A. Krogh, and E. L. Sonnhammer. 2007. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 35:W429-W432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kall, L., A. Krogh, and E. L. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027-1036. [DOI] [PubMed] [Google Scholar]
- 39.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 40.Lopez, C. S., A. F. Alice, H. Heras, E. A. Rivas, and C. Sanchez-Rivas. 2006. Role of anionic phospholipids in the adaptation of Bacillus subtilis to high salinity. Microbiology 152:605-616. [DOI] [PubMed] [Google Scholar]
- 41.Lopez, C. S., H. Heras, H. Garda, S. Ruzal, C. Sanchez-Rivas, and E. Rivas. 2000. Biochemical and biophysical studies of Bacillus subtilis envelopes under hyperosmotic stress. Int. J. Food Microbiol. 55:137-142. [DOI] [PubMed] [Google Scholar]
- 42.Lopez, C. S., H. Heras, S. M. Ruzal, C. Sanchez-Rivas, and E. A. Rivas. 1998. Variations of the envelope composition of Bacillus subtilis during growth in hyperosmotic medium. Curr. Microbiol. 36:55-61. [DOI] [PubMed] [Google Scholar]
- 43.Marles-Wright, J., T. Grant, O. Delumeau, G. van Duinen, S. J. Firbank, P. J. Lewis, J. W. Murray, J. A. Newman, M. B. Quin, P. R. Race, A. Rohou, W. Tichelaar, M. van Heel, and R. J. Lewis. 2008. Molecular architecture of the “stressosome,” a signal integration and transduction hub. Science 322:92-96. [DOI] [PubMed] [Google Scholar]
- 44.Mascher, T., A. B. Hachmann, and J. D. Helmann. 2007. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors. J. Bacteriol. 189:6919-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitaku, S., and T. Hirokawa. 1999. Physicochemical factors for discriminating between soluble and membrane proteins: hydrophobicity of helical segments and protein length. Protein Eng. 12:953-957. [DOI] [PubMed] [Google Scholar]
- 46.Mitaku, S., T. Hirokawa, and T. Tsuji. 2002. Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics 18:608-616. [DOI] [PubMed] [Google Scholar]
- 47.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 48.Palomino, M. M., C. Sanchez-Rivas, and S. M. Ruzal. 2009. High salt stress in Bacillus subtilis: involvement of PBP4* as a peptidoglycan hydrolase. Res. Microbiol. 160:117-124. [DOI] [PubMed] [Google Scholar]
- 49.Park, S. K., J. D. Venable, T. Xu, and J. R. Yates III. 2008. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng, J., J. E. Elias, C. C. Thoreen, L. J. Licklider, and S. P. Gygi. 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2:43-50. [DOI] [PubMed] [Google Scholar]
- 51.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 53.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 54.Schumann, W. 1999. FtsH—a single-chain charonin? FEMS Microbiol. Rev. 23:1-11. [DOI] [PubMed] [Google Scholar]
- 55.Sierro, N., Y. Makita, M. de Hoon, and K. Nakai. 2008. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 36:D93-D96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 57.Steil, L., T. Hoffmann, I. Budde, U. Völker, and E. Bremer. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stülke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139:2041-2045. [DOI] [PubMed] [Google Scholar]
- 59.Tabb, D. L., W. H. McDonald, and J. R. Yates III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tam, L. T., H. Antelmann, C. Eymann, D. Albrecht, J. Bernhardt, and M. Hecker. 2006. Proteome signatures for stress and starvation in Bacillus subtilis as revealed by a 2-D gel image color coding approach. Proteomics 6:4565-4585. [DOI] [PubMed] [Google Scholar]
- 61.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 62.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 63.UniProt Consortium. 2007. The universal protein resource (UniProt). Nucleic Acids Res. 35:D193-D197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wahome, P. G., and P. Setlow. 2008. Growth, osmotic downshock resistance and differentiation of Bacillus subtilis strains lacking mechanosensitive channels. Arch. Microbiol. 189:49-58. [DOI] [PubMed] [Google Scholar]
- 65.Whatmore, A. M., J. A. Chudek, and R. H. Reed. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136:2527-2535. [DOI] [PubMed] [Google Scholar]
- 66.Wolff, S., H. Hahne, M. Hecker, and D. Becher. 2008. Complementary analysis of the vegetative membrane proteome of the human pathogen Staphylococcus aureus. Mol. Cell. Proteomics 7:1460-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu, C., H. Ren, S. Wang, and X. Peng. 2004. Proteomic analysis of salt-sensitive outer membrane proteins of Vibrio parahaemolyticus. Res. Microbiol. 155:835-842. [DOI] [PubMed] [Google Scholar]
- 68.Xu, C., S. Wang, H. Ren, X. Lin, L. Wu, and X. Peng. 2005. Proteomic analysis on the expression of outer membrane proteins of Vibrio alginolyticus at different sodium concentrations. Proteomics 5:3142-3152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.