Abstract
The σB-dependent general stress response in the common soil bacterium Bacillus subtilis can be elicited by a range of stress factors, such as starvation or an ethanol, salt, or heat shock, via a complex upstream signaling cascade. Additionally, σB can be activated by blue light via the phototropin homologue YtvA, a component of the environmental branch of the signaling cascade. Here we use a reporter-gene fusion to show that σB can also be activated by red light via the energy branch of its upstream signaling cascade. Deletion mutagenesis and homologous overproduction experiments indicate that the RsbP protein (composed of an N-terminal Per-ARNT-Sim [PAS] domain and a C-terminal PP2C-type phosphatase domain) is involved in the red light response. This second light input pathway functions complementarily to YtvA; it shows broader spectral sensitivity but requires higher light intensities. These results are confirmed by transcriptome analyses, which show that both light effects result in upregulation of the σB regulon, with minimal activation of other responses.
When the common soil bacterium Bacillus subtilis is starved or exposed to any of a range of stresses, a complex signal transduction network is activated. This then leads to activation of the alternative RNA polymerase subunit σB, which controls a stress response regulon composed of more than 150 genes (19, 44, 55), of which the ctc gene has generally been accepted as the prototype representative (19). The genes from this σB regulon encode proteins from several different functional categories, including transcriptional regulation, cell protection (like carotenoid biosynthesis and the expression of catalase activity), influx and efflux processes, and carbon metabolism (20).
The σB response can be activated by two separate, but converging, branches of a very complex signal transduction network. These two branches are responsive to different stress signals, as follows: the energy stress branch (elicited by carbon, oxygen, or phosphate limitation) leads to the activation of RsbP, whereas the environmental stress branch (elicited by addition of, e.g., salt or ethanol) activates RsbU. Both RsbP and RsbU are specific phosphatases of the phosphorylated form of RsbV (phosphorylated on a serine residue), which indirectly leads to activation of σB (for reviews, see references 19, 32, and 55). Signaling through the environmental branch involves a large, complex structure, called the stressosome (14, 31), for sensory input. This structure is larger than 1.5 MDa and contains the RsbS, RsbT, and RsbRA proteins and several paralogues of RsbRA (14, 26). Activation of the stressosome leads to RsbT release, which in turn activates RsbU (15).
It has been reported that the σB response in B. subtilis can be activated with blue light via involvement of the phototropin homologue YtvA (4, 17, 49). This protein contains a single LOV (light oxygen voltage) domain (21) that binds a flavin cofactor and shows an authentic photocycle with a very slow thermal recovery rate of its stable ground state (28). It functions in the environmental stress branch of the upstream signal transduction network that regulates activation of σB. It is one of the paralogues of RsbRA, but so far, no evidence has been provided to show that YtvA forms an (integral) part of the stressosome. In this study we show that σB can also be activated with light through the energy stress branch of this cascade, via the RsbP protein. This second light response is preferentially activated by red light. Significantly, transcriptome analyses confirm that YtvA-dependent and -independent light effects upregulate the σB regulon.
MATERIALS AND METHODS
Bacterial strains and genetic manipulation.
The bacterial strains, plasmids, and primers used in this investigation are listed in Table 1. DNA manipulations and molecular genetic techniques were carried out using standard procedures. The strains PB565 (2), PB567 (57), and PB605 (11) were provided by C. Price (University of California at Davis) and were constructed using previously published procedures (Table 1). Escherichia coli MC1061 was used as the intermediate cloning host for plasmids prior to B. subtilis transformation. Strain PB565-605 was obtained by transformation of strain PB605 with chromosomal DNA of strain PB565. Transformants were selected on tryptic soy broth (TSB) agar plates, containing chloramphenicol and erythromycin, after overnight incubation at 37°C.
TABLE 1.
Strains, plasmids, and primers used in this investigation
| Strain, plasmid, or primer | Relevant genotype, characterization, or primer sequencea | Reference, source, or constructionb |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| 168, 1A700 | trpC2 | BGSC; 12 |
| PB198 | amyE::ctc-lacZ trpC2 Cm | 9 |
| PB198/pYtvA | amyE::ctc-lacZ trpC2 Cm pYtvA | 4 |
| PB565 | amyE::ctc-lacZ ΔytvA trpC2 Cm Er | 2 |
| PB565/pRsbP | amyE::ctc-lacZ ΔytvA trpC2 Cm Er pRsbP | This study |
| PB565/pYtvA | amyE::ctc-lacZ ΔytvA trpC2 Cm Er pYtvA | 5 |
| PB605 | amyE::ctc-lacZ trpC2 Cm ΔrsbQ | 11 |
| PB565-605 | amyE::ctc-lacZ ΔytvA trpC2 Cm Er ΔrsbQ | PB565→PB605 |
| PB567 | amyE::ctc-lacZ ΔrsbP trpC2 Cm Sp | 57 |
| E. coli | ||
| MC1061 | F−araD139 (ara-leu)7696 (lac)X74 galU galK hsdR2 mcrA mcrB1 rspL | 13 |
| Plasmids | ||
| pDG148 | Pspac Km Amp | 48 |
| pRsbP | Pspac::rsbP Km Amp | This study |
| pYtvA | Pspac::ytvA | 4 |
| Primers | ||
| His-SalI/FW | 5′-GTCGACAAGGAGGAAGCAGGTATGCATCACCATCACCATCAC-3′ | |
| RsbP/FWPDG148 | 5′-ATGCATCACCATCACCATCACGACAAACAATTGAATGATGCACCATGCGG-3′ | |
| RsbP/RVPDG148 | 5′-GCATGCTTATTTTACATCAACTAATATAAAACATTCGTCATCACTCTTATGATGAGC-3′ |
Underlined bases, restriction enzyme recognition sites; bases in italics, sequences encoding hexahistidine tags.
PB565→PB605, chromosomal DNA of PB565 was used to transform PB605.
Construction of plasmids.
The rsbP gene was amplified by using chromosomal DNA from B. subtilis 168 1A700 and the primers shown in Table 1. A two-step PCR procedure was used to include an N-terminal hexahistidine tag and sites for restriction enzyme digestion. The PCR product was inserted into pCR-Blunt II-TOPO (Invitrogen), in order to obtain large amounts of insert, and subsequently digested with the enzymes PaeI (Fermentas) and SalI (Amersham Biosciences). The resulting restriction fragment was then ligated into the shuttle vector pDG148 (Table 1), thus obtaining pRsbP. In this plasmid, rsbP is under the control of the isopropyl-ß-d-thiogalactopyranoside (IPTG)-inducible spac promoter. This construct is used for low-level overexpression in physiological studies.
Growth conditions and β-galactosidase assays.
The medium used for these experiments was TSB (30 g/liter), supplemented with 0.5% glucose, 5 μg/ml chloramphenicol, 1 μg/ml erythromycin, and 10 μg/ml kanamycin, where applicable. Overnight cultures were grown in this medium in the dark, starting from a single colony of a fresh plate of the same medium. The overnight cultures were diluted and allowed to grow in the dark until they had reached exponential growth phase again, rediluted in an end volume of 10 ml, and distributed over different 100-ml Erlenmeyer flasks to initiate an experiment. The Erlenmeyer flasks were placed in a shaking water bath at 37°C and 250 rpm in the light or in the dark. Dark controls were wrapped tightly in tinfoil. Samples were taken at various time points; for dark controls, they were taken in the dark or with minimal background light. These samples were immediately transferred to an ice/water mixture and subsequently flash frozen with liquid nitrogen for storage at −80°C. β-Galactosidase activities were measured and expressed in Miller units according to reference 25.
Microarray experiments.
For the YtvA-dependent experiment, strain PB565/pYtvA was grown as mentioned above, and IPTG was added as described previously (4) to cells grown exponentially in the dark and in the presence of light. For the YtvA-independent experiment, strain PB565 was grown in the presence and absence of light as described above, until cells reached stationary phase. Samples were taken at different time points and immediately flash frozen with liquid nitrogen and stored at −80°C. For both experiments, white light was used (see below), and two cultures (coming from two independent colonies) were grown in parallel as biological replicas. Microarrays were performed by comparing light and dark samples harvested at 120 min after IPTG induction for the YtvA-dependent experiment and at 390 min after inoculation (∼100 min after the cells reached stationary phase) for the experiment on the YtvA-independent light effect.
RNA isolation.
The RNA isolation protocol was adapted from an extraction method described previously (27). Briefly, cells were disrupted in a 2-ml screw-cap tube in the presence of 100-μm zirconium beads and sodium dodecyl sulfate (SDS)-phenol for 60 s in the Mini-beadbeater-8 (Biospec Products). After chloroform extraction, nucleic acids were precipitated, vacuum dried, and further purified with the ChargeSwitch total RNA cell kit (Invitrogen), according to manufacturer's instructions. RNA purity was verified by its absorption characteristics in the UV region using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA molecular weight (MW) profiles were verified with the 2100 electrophoresis bioanalyzer (Agilent Technologies). The RNA 6000 ladder (Ambion, Inc., Austin TX) consisting of 6 transcripts at a concentration of 20 ng/μl, and ranging from 0.2 to 6.0 kb in length, was used as a marker. In addition, a fast migrating “reference marker” compound was added to all samples for software alignment of the electropherograms within one LabChip run (Agilent Technologies).
Synthesis and labeling of cDNA.
From the isolated RNA, 1 μg was vacuum dried, and cDNA was synthesized by the use of random hexamer pd(N)6 primers (Roche, Mannheim, Germany), Superscript II reverse transcriptase (Life Technologies), and 2 mM dATP, 2 mM dGTP, 2 mM dCTP, 1.2 mM dTTP (Roche), and 0.8 mM aminoallyl dUTP (aa-dUTP) (Sigma). Subsequently, RNA was hydrolyzed in the presence of NaOH, and the cDNA solution was purified using QIAquick spin columns (Qiagen). The cDNA was eluted from the column with 30 μl MilliQ water and vacuum dried. For coupling of the Cy3 dye ester, DNA samples were suspended in 0.1 M Na3CO3 buffer, pH 9.0, at room temperature. The Cy3 ester was added in a final concentration of 0.1 mM (Amersham Pharmacia) and incubated for 1 h at room temperature (RT) in the dark and purified using G50 AutoSeq columns (GE Healthcare). Quality and incorporation of the Cy3 dye were checked with the NanoDrop ND-1000 UV spectrophotometer.
Hybridization.
The NimbleGen hybridization system was used to hybridize the probes to the oligos on the array. First, 4 μl of yeast tRNA was supplemented to each sample prior to the drying step. Dried nucleotides were suspended in 11.5 μl digoxigenin (DIG) Easy Hyb hybridization buffer (Roche) and 0.5 μl of the 50 mM C3-CPK6 48-mer suspension (NimbleGen). The mixture was incubated for 5 min at 95°C, transferred to a 42°C block, and hybridized on 60-mer B. subtilis subsp. subtilis strain NC_000964 microarray slides (Roche NimbleGen, Inc.). Hybridization was performed overnight at 42°C in NimbleGen hybridization system 4 (Roche NimbleGen, Inc.), according to the manufacturer's protocol. Microarray slides were washed for 10 s in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.2% SDS at 37°C, for 10 s in 0.5× SSC at 37°C, and twice for 10 min in 0.2× SSC at room temperature. Slides were dried by the use of a nitrogen flow and scanned with a ScanArray 5000 laser scanner (PerkinElmer Life Sciences). The tagged image file format images were quantified with the NimbleScan software package, version 2.4 (Roche NimbleGen, Inc.).
Microarray data processing.
The raw data was processed using Microsoft Office Excel 2002 Service Pack 2. The intensities of 8,208 spots per microarray, representing 4,104 different genes, were calculated by subtracting the median background (B) from the median signal (S). Values lower than 1 were set to 1. Each value was normalized based on the average signal (S − B) for that channel and array. This average channel signal was determined from spots with signals of at least twice the background level for both channels. Normalized spot intensities from duplicate spots on the array from 2 independent experiments were then averaged, and their light/dark ratios were determined and 2-log transformed.
Microarray data analysis.
Data were analyzed in two independent ways. First, the 2-log ratios of all genes in the data set were analyzed using T-profiler (7), adapted for B. subtilis (51). This tool uses t tests to determine the significance of the total changes in predefined groups of genes. Second, the significance of the changes in spot intensities for each individual gene was determined using Student's t test on the normalized data (before averaging it). For this analysis, all genes for which the average (raw) signal fell below twice the average (raw) background signal were discarded, leaving 3,348 (82%) and 3,285 (80%) of the genes for the YtvA-dependent and the YtvA-independent experiments, respectively. The cutoff for a significant change was determined using a P value of 0.05, corrected for multiple testing with the Bonferroni correction (i.e., 0.05/n, where n is the number of genes tested).
Microarray data accession number.
Microarray data were deposited in GEO with accession number GSE19464 and microarray platform accession number GPL8614.
Illumination.
Without further specification, “light” refers to white light from a compact fluorescent lamp. Moderate white light intensities were set in the range of 25 to 35 microeinstein m−2 s−1. Blue and red light, provided by light-emitting diodes (LEDs) emitting maximally at 470 and 635 nm with a spectral width at half maximum of 35 nm, respectively, were used at a maximal intensity of 20 microeinstein m−2 s−1. Light intensities were measured with a model LI-250 light meter, equipped with a quantum light sensor (LI-COR, Lincoln, NE).
RESULTS
σB activation is enhanced by light in stationary phase in a YtvA-independent manner.
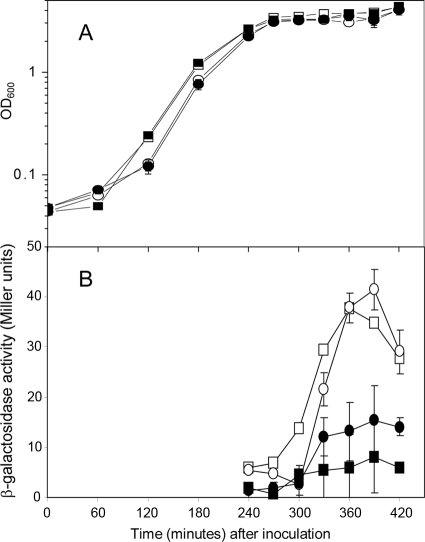
In our previous report on the light activation of σB in B. subtilis via YtvA (4), we observed a significant, light-dependent activation of σB when the cells made the transition from exponential growth into stationary phase in TSB medium. The latter observation was fascinating as well as inexplicable at that moment, as the growth condition was such that ytvA is weakly expressed only (17). One of the possibilities that we proposed in our previous report to explain this observation was remaining activity of YtvA, either through leakage of the spac promoter or a low-level expression from its endogenous promoter. Here we report on our further investigations of σB activation upon entry of B. subtilis into stationary phase by analyzing the σB response (as indicated by the activity of the ctc promoter through β-galactosidase activity) in a strain (PB565) in which the ytvA gene had been deleted. Figure 1A shows that this strain grows slightly slower in a TSB medium containing 0.5% glucose than its corresponding wild-type strain, but the growth yield was not affected by white light of moderate intensity (see Materials and Methods) in either of the strains. When the amount of β-galactosidase was assayed in cells from these cultures (Fig. 1B), it turned out that light elicits a significant increase of the σB response both in the wild-type and in the ΔytvA strain. This light-induced Pctc::lacZ expression coincides with entry of the cells into stationary phase (i.e., at ∼240 min [Fig. 1]) and reaches a maximum after 1 to 2 h, followed by the decrease of β-galactosidase activity.
FIG. 1.
Effect of light on growth and on the expression from the σB-dependent ctc promoter in a wild-type (PB198) and a ΔytvA (PB565) strain of Bacillus subtilis. In both panels, the horizontal axis shows the time after inoculation. Cells were incubated either in the presence of light (open symbols) or in the dark (filled symbols). (A) Growth curves of the two strains (i.e., PB198 [squares] and PB565 [circles]) through exponential growth phase and into stationary phase. The vertical axis shows the optical density at 600 nm (OD600) as an indicator of cell density. (B) Degree of σB activation as indicated by β-galactosidase expression driven by the ctc promoter (and expressed in Miller units) of the wild type and the ΔytvA deletion strain during stationary phase. Symbols have the same definitions as those in panel A. Error bars (shown only when they exceed the size of the symbols) indicate standard deviations calculated from two independent experiments, each with duplicate samples.
The RsbP/Q system is required for the light-dependent enhancement of σB activation.
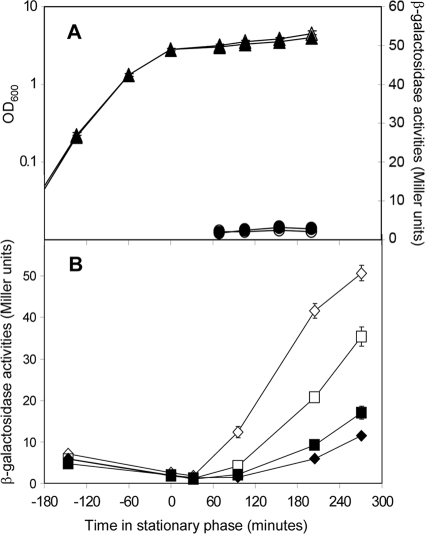
As shown with the experiment in Fig. 1, it was clear that the stationary-phase response cannot be caused by YtvA, so we set up an experiment to identify the potential photoreceptor responsible for this second, “late” light activation of σB. From the fact that this is a stationary-phase response, we assumed that a component from the “energy stress” signaling pathway, rather than the “environmental stress” signaling pathway, would be involved. The former pathway has much fewer components, and one of those few, RsbP, has the typical characteristics of a signal transduction protein: it is composed of a C-terminal PP2C phosphatase domain and an N-terminal Per-ARNT-Sim (PAS) domain, and the latter could well function as a signal input domain to regulate the protein phosphatase activity of RsbP (57). The functioning of RsbP is strictly dependent on RsbQ (11). The latter protein shows sequence homology with hydrolytic enzymes and most probably plays a role in the maturation of RsbP (11, 24). To probe the involvement of RsbP in the light-dependent—but YtvA-independent—activation of σB, we tested the σB response in strain PB565-605, which carries a deletion in both rsbQ and ytvA. In Fig. 2A, it is shown that this strain grows equally well in the absence and presence of light and is completely nonresponsive to visible light in its σB response. To further probe whether RsbP/Q mediates the light-dependent activation of σB upon entry into stationary phase, we constructed an rsbP overexpression plasmid (pRsbP), transformed it into the ytvA deletion strain PB565, and analyzed the σB response of the resulting strain in moderate light, and in the dark, during exponential growth and in stationary phase (Fig. 2B). The results obtained show that the extent of the light-dependent activation of σB in stationary phase in this strain has increased compared to the level of activation in the corresponding wild-type strain. To complete this part of our studies, we analyzed the Pctc::lacZ-dependent transcription level of a strain (PB567) that contains a deletion of the rsbP gene. This experiment revealed that this strain also lacks the ability to activate σB with visible light (data not shown).
FIG. 2.
Role of RsbP/Q in the light-dependent σB activation. The horizontal axes show the time after entry into stationary phase. Cells were incubated either in the presence of light (open symbols) or in the dark (filled symbols). (A) Effect of light on σB activation in a double-deletion ΔytvA ΔrsbQ strain (PB565-605). Growth curves of the PB565-605 strain into stationary phase (triangles) and degree of σB activation, as indicated by β-galactosidase expression from the ctc promoter (in Miller units), upon entry into stationary phase (circles). (B) Effect of RsbP overproduction on the light-dependent activation of the σB-mediated stress response in an ΔytvA background. Strain PB565/pRsbP was grown in TSB medium. At −150 min (relative to the point of entrance into stationary phase), 1 mM IPTG was added to half of the cultures (diamonds; squares, no additions), and cells were incubated in the light or in the dark. Both panels show the results of a typical experiment, in which error bars indicate standard deviations calculated from duplicate samples.
Analysis of the light intensity and color dependence of the two light-responses that activate σB.
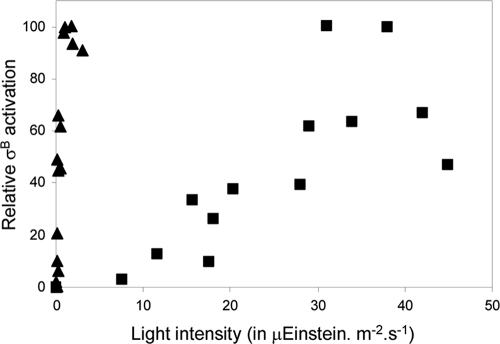
To further discriminate between the two light activation pathways in the upstream signaling cascade of σB from B. subtilis, we analyzed their light intensity dependence. The results, presented in Fig. 3, make it clear that the two responses differ strikingly in this respect, as follows: the YtvA-mediated response (which was measured in the YtvA-overproducing strain PB198/pYtvA because no significant light-induced difference can be observed in a wild-type strain on ctc-driven β-galactosidase expression in the absence of any other stress [4, 49]) saturates at very low light intensities, with a half-saturating intensity of around 1 to 2 microeinstein m−2 s−1. The RsbP/Q-dependent response, in contrast, saturates only at more than 10-fold-higher intensities. Half-saturation of the latter response is observed at approximately 25 microeinstein m−2 s−1, but the accuracy at which these data can be recorded does not allow a clear conclusion as to whether or not a hyperbolic dependency of the magnitude of the σB response on light intensity is observed in this latter response (we cannot exclude that this response has a sigmoidal character). Besides light intensity dependence, the other discriminative characteristic of a photosensory receptor is its spectral sensitivity. We therefore also analyzed the RsbP/Q-dependent σB response with light from two light-emitting diodes, with maximal emissions at 470 and 630 nm, respectively. Table 2 shows that—in contrast to the YtvA-mediated response, which shows no red light sensitivity (4)—the RsbP/Q-mediated response actually is more responsive to red than to blue light. This is surprising, as neither phytochrome- nor retina-based photoreceptor homologues have been identified in the bacilli.
FIG. 3.
Light intensity dependence of the YtvA- and the RsbP-mediated activation of the σB-dependent stress response. The horizontal axis shows the light intensity in units of microeinstein m−2 s−1. The vertical axis shows the relative extent of light-dependent σB activation, expressed in units of β-galactosidase activity (in Miller units) from the ctc promoter. Triangles, percentage of σB activation of the YtvA-overproducing strain PB198/pYtvA, sampled at 2 h after induction with 1 mM IPTG (added at OD600 of 0.6; squares, percentage of σB activation of the PB565 strain at 385 min after inoculation (∼100 min after the cells reached stationary phase). The data shown are the results from a typical experiment; the experiment was performed more than three times.
TABLE 2.
Color dependence of the stationary-phase light-dependent σB activation in PB565 (ΔytvA), measured as the relative level of σB activationa
| Condition | Time (min) after entry into stationary phasec |
|
|---|---|---|
| 30 | 90 | |
| Dark | 20.8 ± 2.0 | 100.0 ± 4.0 |
| Red (635 nm)b | 20.0 ± 0.2 | 179.9 ± 10.0 |
| Blue (470 nm)b | 19.5 ± 2.6 | 120.1 ± 0.8 |
The relative level of activation was calculated according to the following: Miller units of a time point/Miller units of the dark sample at 90 min after entry into stationary phase × 100.
The wavelength of the light emitted by these LEDs was centered at the indicated wavelengths, with an intensity of 20 microeinstein m−2 s−1.
Errors indicate standard deviations calculated from duplicate samples.
Transcriptome analysis of light induction of σB.
So far, experimental evidence on the activation of the general stress response by light has been derived only from the activity of the single promoter of ctc. To investigate this activation at the transcriptome level, we compared light and dark samples, taken during typical experiments for the two light responses, with microarray analyses (see Materials and Methods). The results were analyzed using T-profiler, a tool that identifies significantly regulated gene groups, using all data derived from the microarray experiments (7, 51). Table 3 shows the gene groups that were significantly upregulated in response to visible light in both experiments. Both the YtvA-dependent and the RsbP/Q-dependent responses result in a highly significant upregulation of the σB regulon in response to light, confirming the observations made with the ctc promoter.
TABLE 3.
Significantly upregulated gene groups according to T-profiler
| Gene groupb | Mean 2-log ratioa | t value | E value | No. of genes |
|---|---|---|---|---|
| YtvA-dependent light effect in logarithmic growth phase | ||||
| SigB | 1.459 | 21.44 | <1.0 × 10−15 | 97 |
| SigF | 0.521 | 4.59 | 6.14 × 10−3 | 40 |
| YtvA-independent light effect in stationary growth phase | ||||
| SigB | 0.475 | 6.35 | 2.99 × 10−7 | 97 |
| Xpf_positive | 1.137 | 4.21 | 3.48 × 10−2 | 10 |
Transformation (2-log) of the ratio between spot intensities in light and dark.
See DBTBS (46) for the members of each gene group.
The only other gene group that was significantly upregulated in response to visible light in stationary phase is a group of 10 genes that are regulated by Xpf, a sigma-like transcription factor involved in the regulation of expression from the prophage PBSX (33). In contrast, the YtvA-dependent light response in logarithmic growth phase resulted in a significant upregulation of the sporulation-specific σF regulon, which is normally activated in the prespore during the early stages of sporulation (for a review, see reference 41).
In addition to the T-profiler analysis of gene groups, we also analyzed the data to identify the significantly up- and downregulated individual genes, using a stringent statistical analysis (see Materials and Methods), the result of which is shown in Table 4. Of the genes with significant changes in expression, 12/16 (75%) and 6/13 (46%) genes are known to be part of the σB regulon for the YtvA-dependent and the RsbP/Q-dependent experiments, respectively, confirming the upregulation of the general stress response as seen with T-profiler. One of the genes upregulated by the YtvA-dependent light response (yhcM) is known to be controlled by both σB and σF, which is also in line with the activation of σF shown by T-profiler. The upregulation of the Xpf regulon observed by T-profiler for the RsbP/Q-dependent light response is also confirmed by the analysis of individual genes, as 2/13 of the most significantly affected genes are Xpf regulated (xkdK and xkdJ).
TABLE 4.
Significantly up- or downregulated genes
| Gene | Known control of expression | Comment(s) | 2-Log ratioa | P value | Reference(s) |
|---|---|---|---|---|---|
| YtvA-dependent light effect in logarithmic growth phase | |||||
| yfkM | SigB; Fur (−) | Unknown function; E. coli homologue YhbO protects against multiple stresses | 3.42 | 1.37 × 10−7 | 1, 6, 38, 40, 44 |
| ywzA | SigB | Conserved hypothetical protein; predicted membrane protein | 4.08 | 1.97 × 10−6 | 40, 44 |
| ydaT | SigB | Conserved hypothetical protein; forms an operon with ydaS | 2.87 | 2.39 × 10−6 | 39, 44 |
| yfkJ | SigB; SigA | Protein tyrosine phosphatase involved in ethanol resistance | 2.65 | 4.66 × 10−6 | 34, 44, 46 |
| yflT | SigB | Conserved hypothetical protein | 4.13 | 4.83 × 10−6 | 40, 44 |
| yvgO | SigB | Conserved hypothetical protein; potential lipoprotein | 2.72 | 5.90 × 10−6 | 40, 43 |
| ytxH | SigB; SigH | Conserved hypothetical protein; contains potential transmembrane helix and shows some similarities to desiccation-related proteins in plants | 1.33 | 6.05 × 10−6 | 3, 40, 44, 50 |
| aldY | SigB | NADH-dependent aldehyde dehydrogenase | 2.57 | 6.43 × 10−6 | 39, 44 |
| ydaP | SigB | Putative enzyme with pyruvate as substrate | 3.81 | 8.41 × 10−6 | 38, 40 |
| yhcM | SigB; SigF | No homology to previously reported sequences | 3.09 | 1.20 × 10−5 | 40, 46 |
| ysnF | SigB | Putative protein | 5.70 | 1.21 × 10−5 | 40, 43 |
| yvrE | SigB | Conserved hypothetical protein | 1.09 | 1.38 × 10−5 | 39, 40, 44 |
| glpK | CcpA (−); GlpP (+); SigA | Glycerol kinase; repressed under anaerobic conditions | 2.71 | 1.85 × 10−7 | 30, 46 |
| yuxJ | Putative exporter | 1.79 | 5.99 × 10−7 | ||
| yuaB | AbrB (−); DegU (+) | Small, secreted protein; required for pellicle formation in strain ATCC 6051 | −0.86 | 6.22 × 10−6 | 56 |
| albC | AbrB (−); ResD (+); Rok (−); SigA | Putative transporter involved in subtilosin production | 1.12 | 9.74 × 10−6 | 46 |
| YtvA-independent light effect in stationary growth phase | |||||
| ycbP | SigB | Putative inner integral membrane protein | 1.01 | 6.15 × 10−7 | 40 |
| ylxP | SigB; SigA | Conserved hypothetical protein | 0.52 | 2.97 × 10−6 | 40, 46 |
| ydaS | SigB | Forms an operon with ydaT | 1.02 | 3.01 × 10−6 | 39, 40, 44 |
| yhdN | SigB; YlpC | NAPDH-specific aldo-keto reductase | 0.69 | 5.28 × 10−6 | 16, 38, 40, 46 |
| yoxC | SigB | Conserved hypothetical protein; predicted lipoprotein | 0.85 | 1.35 × 10−5 | 39, 40 |
| csbC | SigB | Sugar transporter | 0.99 | 1.42 × 10−5 | 40, 44 |
| xkdK | Xpf (+) | Conserved hypothetical protein in phage element PBSX | 1.07 | 5.77 × 10−7 | 46 |
| xkdJ | Xpf (+) | Conserved hypothetical protein in phage element PBSX | 1.42 | 1.42 × 10−5 | 46 |
| nhaC | Regulated by pH | Represses the Pho regulon for phosphate starvation | 0.71 | 2.50 × 10−7 | 42, 58 |
| ykoX | Regulated by pH | Putative integral inner membrane protein | −1.00 | 5.86 × 10−7 | 58 |
| yjeA | Regulated by heat shock & pH | Secreted DNase, specific for DNA | −1.09 | 7.36 × 10−6 | 35, 40, 58 |
| ymfJ | Conserved hypothetical protein | −0.53 | 9.71 × 10−6 | ||
| yjlB | Conserved hypothetical protein of the cupin family | 1.00 | 1.42 × 10−5 |
Transformation (2-log) of the ratio between spot intensities in light and dark.
DISCUSSION
B. subtilis is known to possess at least one photoreceptor, YtvA, which is able to activate the general stress response in the presence of blue light (4). In the present work, we have shown that there is another light effect in the energy stress signaling branch of the general stress response that is independent of YtvA. Using microarray experiments, we have confirmed that both light responses result in a significant upregulation of the entire σB regulon (Tables 3 and 4). The observed level of upregulation of this regulon for the YtvA-dependent response is higher than the level of upregulation due to the RsbP/Q-dependent response, which corresponds well with the observed differences between light and dark in our reporter enzyme assays (compare Fig. 1B in this article with Fig. 1D in reference 4). To our knowledge, this is the first microarray data that shows an effect of visible light on a chemotropic bacterium.
In the RsbP/Q-dependent light response in stationary phase, one also observes upregulation of the phage-related regulon of Xpf. Transcription of Xpf itself is controlled by σB (44), but during exponential growth, this is insufficient to lead to induction of its regulon of late-stage PBSX genes (Table 3) (44). The other known induction mechanism of PBSX is the DNA damage-induced SOS response, mediated by RecA and LexA (18, 47). However, the complete absence of induction of the rest of the SOS response regulon suggests that in addition to σB, there must be an alternate, unknown regulatory mechanism for the Xpf regulon in stationary phase.
On the photobiology of B. subtilis.
With the characterization of the RsbP/Q-dependent response as the (chronologically) second light response of B. subtilis, two separate light activation pathways of σB have now been described. Therefore, it is relevant to discuss the photobiology of this spore-forming common soil bacterium. In this respect, it is important to note that, just like in the atmosphere and also in a highly (Raleigh) scattering environment like the soil, red light has a higher penetration depth than blue light (29). Nevertheless, the activation of σB in B. subtilis via RsbP/Q requires approximately 10-fold-higher light intensity than the YtvA-mediated response. This may be due to an intrinsically lower stability of the signaling state formed in the putative photoreceptor involved in the RsbP-dependent response than that of the corresponding state in YtvA (which in vitro has a very low rate of thermal relaxation [<10−3 s−1] [28]).
Which of these two pathways is most important for the protective physiology of B. subtilis depends on many different factors, including the physiological conditions and growth phase of the cells and the ambient light climate. Under conditions of exponential growth, YtvA will only be able to activate σB if the cells have previously been exposed to a significant “environmental stress,” induced by, e.g., the addition of salt (4, 49). Transition of the cells to stationary phase, induced by, e.g., depletion of free energy, activates RsbP (57). This, however, leads to maximal stimulation of σB only when visible light is available. Although the necessary light intensity is higher than that required for activation of YtvA (Fig. 3), it is equivalent to only ∼1% of the light intensity at noon on a sunny summer day at moderate latitude (52).
The σB response has been characterized as a mechanism for general but low-level protection against a wide range of stresses. Therefore, this response may show cross talk to many other signal transduction pathways (19). As no other photoreceptors, beyond YtvA and the one involved in the RsbP/Q-mediated light response, have so far been identified in this organism (see also reference 54), it can now be proposed that this cross talk includes the sporulation phosphorelay. It has previously been shown that visible light can delay sporulation in B. subtilis by several hours (45). In this study, we observed a YtvA- and light-dependent induction of the sporulation sigma factor σF (Tables 3 and 4). A limited overlap exists between the regulons of σB and σF, which may explain this observation. Moreover, a decrease in the intracellular GTP concentration has been identified as a potential signal for sporulation (37), and interestingly, the function of YtvA in vivo is abolished when the GTP-binding site of this protein is interrupted (5). These mechanisms could jointly serve as a very primitive type of “circadian clock” to stimulate the cells to sporulate during the night. Moreover, we have observed that cross talk from the σB pathway also affects biofilm formation (M. Ávila-Pérez, unpublished results).
Systems biology of B. subtilis.
The transition from exponential growth phase to stationary phase in B. subtilis is an extremely complex biological process, regulated through a signal transduction network of which the output characteristics are far beyond intuitive reasoning. Parts of this network have been subject to very detailed mathematical modeling (22, 23), but its systems-level understanding is not yet within sight. A major difficulty in the further characterization of this network is the absence of well-defined stimuli for activation. The discovery of the two pathways for light activation of σB has considerably alleviated this situation. Accordingly, light activation of the σB response has already revealed that current models for the σB response to salt and ethanol stress may overemphasize the role of RsbX as a feedback loop in this response, because we recently observed that a persistent light stimulus also evokes a persistent σB response (M. Ávila-Pérez, unpublished results) rather than a transient one (8). Furthermore, the involvement of stressosomes in the activation of σB through the “environmental” branch brings up many questions regarding the involvement of cooperativity, stochastics, and cytoplasmic diffusion, etc., in signal transduction that are ideally suited for a systems biology approach. This can now be studied by comparing the responses to low-intensity blue light and moderate-intensity red light.
Molecular basis of the red light response.
The present work does not provide enough data to allow for a conclusion about the mechanism by which light is sensed in the RsbP/Q-dependent response. Since no upstream components of the energy stress pathway are known, we think that RsbP is responsible for the second (red) light effect. Our working hypothesis for the light-sensing mechanism in RsbP is that light absorption by an unidentified chromophore leads to a structural change in its PAS domain, which then activates phosphatase activity in its C-terminal PP2C-type phosphatase domain. We consider it unlikely that the RsbP/Q-mediated response activates σB via a photosensitizer for the following reasons: first, because of the still relatively low light intensity at which saturation is observed of the response that it generates; second, because of the high efficiency of red versus blue light in generating the response; and finally, because of the absence of an induction of oxidative stress responses in the microarray data (Tables 3 and 4). Furthermore, if so, one would have to postulate that synthesis of the putative photosensitizing pigment is dependent on functional RsbP/Q. There is no evidence available that would support this latter notion. Accordingly, the mechanism of primary photochemistry in the RsbP-mediated response must be different from the photochemistry in all six hereto well-characterized families of photosensory receptors (53), as well as from the photochemistry involved in phototaxis in hemH mutants of E. coli (59).
It has not escaped our attention that other examples of the involvement of as yet uncharacterized photoreceptors, responsible for a red light-dependent phenotype, are present in the literature. Light regulation of motility in Agrobacterium tumefaciens (36) and Acinetobacter baylyi (M. Bitrian and B. C. Nudel, unpublished experiments) are recent examples. In this respect, it is relevant to mention that RsbP and RsbQ form a bicistronic operon, not only in B. subtilis (11) but in many other organisms as well, be it that the phosphatase domain can be substituted by other output modules like histidine protein kinases or guanylate cyclases (10). RsbP/Q may therefore turn out to be the prototype of a new family of photosensory proteins.
Acknowledgments
We thank C. Price (UC Davis, CA) for generously providing strains and stimulating discussions. Furthermore, we thank Huub C. F. Hoefsloot for help with the statistical analysis of the microarray data, Jos C. Arents for help with experiments, Michel Ossendrijver for the skillful contribution regarding RNA extractions and microarray analyses.
This work was supported by the Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research (NWO) through a grant to K.J.H.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Abdallah, J., T. Caldas, F. Kthiri, R. Kern, and G. Richarme. 2007. YhbO protects cells against multiple stresses. J. Bacteriol. 189:9140-9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbar, S., T. A. Gaidenko, C. M. Kang, M. O'Reilly, K. M. Devine, and C. W. Price. 2001. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor sigma(B) of Bacillus subtilis. J. Bacteriol. 183:1329-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ávila-Pérez, M., K. J. Hellingwerf, and R. Kort. 2006. Blue light activates the sigmaB-dependent stress response of Bacillus subtilis via YtvA. J. Bacteriol. 188:6411-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ávila-Pérez, M. E., J. Vreede, Y. Tang, O. Bende, A. Losi, W. Gärtner, and K. J. Hellingwerf. 2009. In vivo mutational analysis of the Bacillus subtilis lov-domain containing protein YtvA: mechanism of light-activation of the general stress response. J. Biol. Chem. 284:24958-24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 7.Boorsma, A., B. C. Foat, D. Vis, F. Klis, and H. J. Bussemaker. 2005. T-profiler: scoring the activity of predefined groups of genes using gene expression data. Nucleic Acids Res. 33:W592-W595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the sigma B transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boylan, S. A., A. Rutherford, S. M. Thomas, and C. W. Price. 1992. Activation of Bacillus subtilis transcription factor sigma B by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brody, M. S., V. Stewart, and C. W. Price. 2009. Bypass suppression analysis maps the signalling pathway within a multidomain protein: the RsbP energy stress phosphatase 2C from Bacillus subtilis. Mol. Microbiol. 72:1221-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brody, M. S., K. Vijay, and C. W. Price. 2001. Catalytic function of an alpha/beta hydrolase is required for energy stress activation of the sigma(B) transcription factor in Bacillus subtilis. J. Bacteriol. 183:6422-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burkholder, P. R., and N. H. Giles. 1947. Induced biochemical mutations in Bacillus subtilis. Am. J. Bot. 34:345. [PubMed] [Google Scholar]
- 13.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 14.Delumeau, O., C. C. Chen, J. W. Murray, M. D. Yudkin, and R. J. Lewis. 2006. High-molecular-weight complexes of RsbR and paralogues in the environmental signaling pathway of Bacillus subtilis. J. Bacteriol. 188:7885-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delumeau, O., S. Dutta, M. Brigulla, G. Kuhnke, S. W. Hardwick, U. Volker, M. D. Yudkin, and R. J. Lewis. 2004. Functional and structural characterization of RsbU, a stress signaling protein phosphatase 2C. J. Biol. Chem. 279:40927-40937. [DOI] [PubMed] [Google Scholar]
- 16.Ehrensberger, A. H., and D. K. Wilson. 2004. Structural and catalytic diversity in the two family 11 aldo-keto reductases. J. Mol. Biol. 337:661-673. [DOI] [PubMed] [Google Scholar]
- 17.Gaidenko, T. A., T. J. Kim, A. L. Weigel, M. S. Brody, and C. W. Price. 2006. The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J. Bacteriol. 188:6387-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goranov, A. I., E. Kuester-Schoeck, J. D. Wang, and A. D. Grossman. 2006. Characterization of the global transcriptional responses to different types of DNA damage and disruption of replication in Bacillus subtilis. J. Bacteriol. 188:5595-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecker, M., J. Pane-Farre, and U. Volker. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61:215-236. [DOI] [PubMed] [Google Scholar]
- 20.Hoper, D., U. Volker, and M. Hecker. 2005. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J. Bacteriol. 187:2810-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huala, E., P. W. Oeller, E. Liscum, I. S. Han, E. Larsen, and W. R. Briggs. 1997. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278:2120-2123. [DOI] [PubMed] [Google Scholar]
- 22.Iber, D., J. Clarkson, M. D. Yudkin, and I. D. Campbell. 2006. The mechanism of cell differentiation in Bacillus subtilis. Nature 441:371-374. [DOI] [PubMed] [Google Scholar]
- 23.Igoshin, O. A., C. W. Price, and M. A. Savageau. 2006. Signalling network with a bistable hysteretic switch controls developmental activation of the sigma transcription factor in Bacillus subtilis. Mol. Microbiol. 61:165-184. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko, T., N. Tanaka, and T. Kumasaka. 2005. Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis. Protein Sci. 14:558-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenney, T. J., and C. P. Moran, Jr. 1987. Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J. Bacteriol. 169:3329-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, T. J., T. A. Gaidenko, and C. W. Price. 2004. A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J. Mol. Biol. 341:135-150. [DOI] [PubMed] [Google Scholar]
- 27.Kort, R., B. J. Keijser, M. P. Caspers, F. H. Schuren, and R. Montijn. 2008. Transcriptional activity around bacterial cell death reveals molecular biomarkers for cell viability. BMC Genomics 9:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Losi, A., E. Polverini, B. Quest, and W. Gartner. 2002. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys. J. 82:2627-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandoli, D. F., G. A. Ford, L. J. Waldron, J. A. Nemson, and W. R. Briggs. 1990. Some spectral properties of several soil types: implications for photomorphogenesis. Plant Cell Environ. 13:287-294. [Google Scholar]
- 30.Marino, M., T. Hoffmann, R. Schmid, H. Mobitz, and D. Jahn. 2000. Changes in protein synthesis during the adaptation of Bacillus subtilis to anaerobic growth conditions. Microbiology 146(Pt. 1):97-105. [DOI] [PubMed] [Google Scholar]
- 31.Marles-Wright, J., T. Grant, O. Delumeau, G. van Duinen, S. J. Firbank, P. J. Lewis, J. W. Murray, J. A. Newman, M. B. Quin, P. R. Race, A. Rohou, W. Tichelaar, M. van Heel, and R. J. Lewis. 2008. Molecular architecture of the “stressosome,” a signal integration and transduction hub. Science 322:92-96. [DOI] [PubMed] [Google Scholar]
- 32.Marles-Wright, J., and R. J. Lewis. 2007. Stress responses of bacteria. Curr. Opin. Struct. Biol. 17:755-760. [DOI] [PubMed] [Google Scholar]
- 33.McDonnell, G. E., H. Wood, K. M. Devine, and D. J. McConnell. 1994. Genetic control of bacterial suicide: regulation of the induction of PBSX in Bacillus subtilis. J. Bacteriol. 176:5820-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musumeci, L., C. Bongiorni, L. Tautz, R. A. Edwards, A. Osterman, M. Perego, T. Mustelin, and N. Bottini. 2005. Low-molecular-weight protein tyrosine phosphatases of Bacillus subtilis. J. Bacteriol. 187:4945-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng, K. L., C. C. Lam, Z. Fu, Y. F. Han, K. W. Tsim, and W. K. Wong. 2007. Cloning and characterization of the yjeA gene, encoding a novel deoxyribonuclease, from Bacillus subtilis. J. Biochem. 142:647-654. [DOI] [PubMed] [Google Scholar]
- 36.Oberpichler, I., R. Rosen, A. Rasouly, M. Vugman, E. Z. Ron, and T. Lamparter. 2008. Light affects motility and infectivity of Agrobacterium tumefaciens. Environ. Microbiol. 10:2020-2029. [DOI] [PubMed] [Google Scholar]
- 37.Ochi, K., J. Kandala, and E. Freese. 1982. Evidence that Bacillus subtilis sporulation induced by the stringent response is caused by the decrease in GTP or GDP. J. Bacteriol. 151:1062-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersohn, A., H. Antelmann, U. Gerth, and M. Hecker. 1999. Identification and transcriptional analysis of new members of the sigmaB regulon in Bacillus subtilis. Microbiology 145(Pt. 4):869-880. [DOI] [PubMed] [Google Scholar]
- 39.Petersohn, A., J. Bernhardt, U. Gerth, D. Hoper, T. Koburger, U. Volker, and M. Hecker. 1999. Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 181:5718-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piggot, P. J., and D. W. Hilbert. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579-586. [DOI] [PubMed] [Google Scholar]
- 42.Pragai, Z., C. Eschevins, S. Bron, and C. R. Harwood. 2001. Bacillus subtilis NhaC, an Na+/H+ antiporter, influences expression of the phoPR operon and production of alkaline phosphatases. J. Bacteriol. 183:2505-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pragai, Z., and C. R. Harwood. 2002. Regulatory interactions between the Pho and sigma(B)-dependent general stress regulons of Bacillus subtilis. Microbiology 148:1593-1602. [DOI] [PubMed] [Google Scholar]
- 44.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 45.Propst-Ricciuti, C., and L. B. Lubin. 1976. Light-induced inhibition of sporulation in Bacillus licheniformis. J. Bacteriol. 128:506-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sierro, N., Y. Makita, M. de Hoon, and K. Nakai. 2008. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 36:D93-D96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simmons, L. A., A. I. Goranov, H. Kobayashi, B. W. Davies, D. S. Yuan, A. D. Grossman, and G. C. Walker. 2009. Comparison of responses to double-strand breaks between Escherichia coli and Bacillus subtilis reveals different requirements for SOS induction. J. Bacteriol. 191:1152-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52:697-704. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki, N., N. Takaya, T. Hoshino, and A. Nakamura. 2007. Enhancement of a sigma(B)-dependent stress response in Bacillus subtilis by light via YtvA photoreceptor. J. Gen. Appl. Microbiol. 53:81-88. [DOI] [PubMed] [Google Scholar]
- 50.Tam le, T., H. Antelmann, C. Eymann, D. Albrecht, J. Bernhardt, and M. Hecker. 2006. Proteome signatures for stress and starvation in Bacillus subtilis as revealed by a 2-D gel image color coding approach. Proteomics 6:4565-4585. [DOI] [PubMed] [Google Scholar]
- 51.Ter Beek, A., B. J. Keijser, A. Boorsma, A. Zakrzewska, R. Orij, G. J. Smits, and S. Brul. 2008. Transcriptome analysis of sorbic acid-stressed Bacillus subtilis reveals a nutrient limitation response and indicates plasma membrane remodeling. J. Bacteriol. 190:1751-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorington, L. 1985. Spectral, irradiance, and temporal aspects of natural and artificial light. Ann. N. Y. Acad. Sci. 53:28-54. [DOI] [PubMed] [Google Scholar]
- 53.van der Horst, M. A., and K. J. Hellingwerf. 2004. Photoreceptor proteins, “star actors of modern times”: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc. Chem. Res. 37:13-20. [DOI] [PubMed] [Google Scholar]
- 54.van der Horst, M. A., J. Key, and K. J. Hellingwerf. 2007. Photosensing in chemotrophic, non-phototrophic bacteria: let there be light sensing too. Trends Microbiol. 15:554-562. [DOI] [PubMed] [Google Scholar]
- 55.van Schaik, W., and T. Abee. 2005. The role of sigmaB in the stress response of Gram-positive bacteria—targets for food preservation and safety. Curr. Opin. Biotechnol. 16:218-224. [DOI] [PubMed] [Google Scholar]
- 56.Verhamme, D. T., E. J. Murray, and N. R. Stanley-Wall. 2009. DegU and Spo0A jointly control transcription of two loci required for complex colony development by Bacillus subtilis. J. Bacteriol. 191:100-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 58.Wilks, J. C., R. D. Kitko, S. H. Cleeton, G. E. Lee, C. S. Ugwu, B. D. Jones, S. S. BonDurant, and J. L. Slonczewski. 2009. Acid and base stress and transcriptomic responses in Bacillus subtilis. Appl. Environ. Microbiol. 75:981-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, H., H. Inokuchi, and J. Adler. 1995. Phototaxis away from blue light by an Escherichia coli mutant accumulating protoporphyrin IX. Proc. Natl. Acad. Sci. U. S. A. 92:7332-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]