Abstract
The damX gene product of Salmonella enterica serovar Typhimurium is a protein located in the inner membrane. DamX migrates as a 70-kDa protein in SDS-PAGE even though the predicted protein size is 46 kDa. Synthesis of DamX protein occurs in both exponential- and stationary-phase cultures. Disruption of damX causes severe sensitivity to bile. Lack of the outer membrane protein AsmA suppresses bile sensitivity in Salmonella damX mutants.
The damX locus, also known as Urf74.3, is an open reading frame (ORF) located in an operon that also contains the aroB, aroK, rpe, gph, and dam genes (11, 12). The region is highly conserved in Escherichia coli and Salmonella enterica (2, 13). The known genes contained in the operon are functionally heterogeneous: aroB and aroK are involved in aromatic amino acid metabolism; dam encodes DNA adenine methyl transferase; trpS is the gene for tryptophan aminoacyl-tRNA synthetase; and the rpe and gph genes are involved in carbohydrate metabolism (8, 11). The product of the E. coli damX (Urf74.3) gene has previously been described as a 70-kDa protein (8, 11), and two recent studies have shown that DamX accumulates in the E. coli septal ring (1, 6). Below, we show that the damX gene product of Salmonella enterica serovar Typhimurium is an inner membrane protein whose absence causes bile sensitivity. The study has been carried out with strain ATCC 14028. However, the nucleotide sequences of the damX-dam chromosomal region are 100% identical in strains LT2 (13) and SL1344 (ftp://ftp.sanger.ac.uk/pub/pathogens/Salmonella).
DamX is an inner membrane protein.
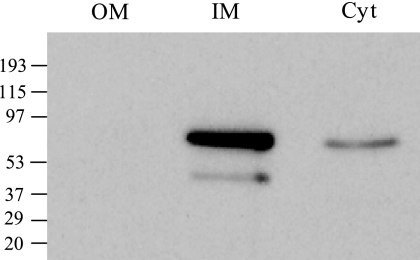
In silico analysis of the DamX secondary structure with the TMpred algorithm (http://www.ch.embnet.org/software/TMPRED_form.html) predicted that DamX has one transmembrane segment. To detect DamX by Western immunoblot analysis, we constructed a DamX protein derivative tagged with a 3× FLAG epitope (24). The primers used for introduction of the 3× FLAG epitope were 5′ GTG GGC AAA ACC GTT GCA TCA GGT TCA GGC CGA TCT GAA AGA CTA CAA AGA CCA TGA CGG 3′ and 5′ GTC AGC GAC ACT CAC AGG CAA TTG CAG GAC ACA GCC TGG ACA TAT GAA TAT CCT CCT TAG 3′. Protein purification, cell fractionation (cytosol, inner membrane, and outer membrane), and Western analysis of the resolved protein extracts were carried out as described elsewhere (17). DamX migrated as a 70-kDa protein under denaturing conditions, as previously described for E. coli (11). DamX was found in the S. enterica serovar Typhimurium inner membrane (Fig. 1) and was detected in protein preparations from both exponential- and stationary-phase cultures (Fig. 2).
FIG. 1.
Distribution of DamX protein tagged with a 3× FLAG epitope in subcellular fractions of S. enterica serovar Typhimurium. Anti-FLAG western hybridization is shown for three fractions: the outer membrane (OM), the inner membrane (IM), and the cytosol (Cyt). The volumes loaded for all fractions were normalized to the same number of bacteria (5 × 107 CFU). Prestaining molecular mass standards in kDa are indicated.
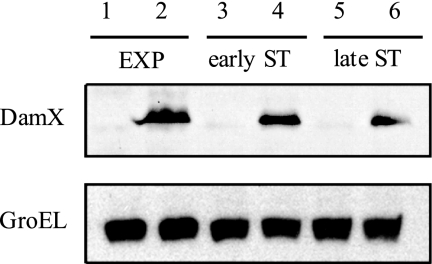
FIG. 2.
Levels of DamX protein in S. enterica cultures grown in LB medium at 37°C with shaking. Protein preparations were made from cultures in exponential phase (EXP; optical density at 660 nm [OD660] = 0.2), early stationary phase (early ST; OD660 = 2.5), and late stationary phase (late ST; OD660 = 3.4). All samples were prepared from 4 × 108 cells. Levels of the heat shock protein GroEL were monitored as internal loading controls. The strains were ATCC 14028 (damX+; lanes 1, 3, and 5) and SV5448 (damX+::3×FLAG; lanes 2, 4, and 6).
Construction of damX deletion mutants.
The S. enterica damX gene was disrupted by lambda Red recombination (4). The ΔdamX1 deletion was generated with primers 5′ ATG GAT GAA TTC AAA CCA GAA GAC GAG CTG AAA CCC GAT CGT GTA GGC TGG AGC TGC TTC 3′ and 5′ AGC GAC ACT CAC AGG CAA TTG CAG GAC ACA GCC TGG ATT ACA TAT GAA TAT CCT CCT TAG 3′. This deletion eliminated the entire damX ORF except 40 bp in the 5′ region. The ΔdamX2 deletion was generated with primers 5′ AGA CGA GCT GAA ACC CGA TCC CAG CGA TCG TCG TAC TGG TAT TCC GGG GAT CCG TCG ACC 3′ and 5′ TCG TGG TGG TCG CTT TCG GCG TCG CGG CTG GCG CTG CCG GGT GTA GGC TGG AGC TGC TTC 3′. The ΔdamX2 deletion eliminated 795 bp, thus leaving about one-third of the damX coding sequence (423 bp) at the 3′ end as well as 60 bp at the 5′ end. The external primers for deletion verification by the PCR were 5′ GGA CTA TTT GCC GCA CAT GC 3′ and 5′ AGG TCG CTC TTG ATA TCG GC 3′.
Consequences of damX disruption.
Both the ΔdamX1 and the ΔdamX2 alleles conferred sensitivity to sodium deoxycholate (DOC) (Table 1) but not to sodium dodecyl sulfate, crystal violet, or malachite green (data not shown). Because dam mutants are extremely sensitive to bile (21) and the damX gene lies immediately upstream of dam (11), these experiments did not ascertain whether the ΔdamX1 and ΔdamX2 deletions caused bile sensitivity on their own or by a polar effect on dam gene expression. To clarify this point, two kinds of experiments were carried out: (i) tests for SOS induction using the cea::lac fusion of plasmid pGE108 (23) as a reporter (18, 19) and (ii) analysis of expression of a translational std::lac fusion, which is extremely sensitive to Dam methylation (7). The main conclusion from these experiments, whose results are summarized in Table 2, was that the ΔdamX1 deletion caused a certain degree of polarity on dam gene expression, reflected in its ability to cause moderate induction of std::lac expression. The polarity was mild, however, as indicated by the fact that neither ΔdamX1 nor ΔdamX2 caused significant SOS induction (Table 2).
TABLE 1.
MICs of sodium deoxycholate in S. enterica strains carrying dam and damX deletion alleles
| Strain | Genotype | MIC of DOC (%)a |
|---|---|---|
| ATCC 14028 | wt | 6.0 |
| SV4536 | Δdam-230 | 0.2 |
| SV5116 | ΔdamX1 | 0.4 |
| SV5307 | ΔdamX2 | 0.4 |
| SV5441 | DUP [ΔdamX2/ΔdamX2] | 0.2 |
| SV5443 | DUP [ΔdamX2/dam+] | 4.0 |
| SV5538 | ΔdamX2 ΔasmA::Kmr | 6.0 |
| SV5539 | ΔdamX2 ΔmutH::Kmr | 0.4 |
MICs are medians of results from >5 independent determinations. Overnight cultures in LB were diluted 1:50 in fresh LB and incubated at 37°C with shaking until late exponential phase (OD660 = 0.6 to 0.8). Cultures were then diluted to obtain a concentration of 3,000 cells/ml. Aliquots were transferred to microtiter plates whose wells had previously been filled with aqueous solutions of DOC (0.2%, 0.4%, 0.6%, 0.8%, 1%, 2%, 3%, 4%, 5%, 6%, and 7%). After overnight incubation at 37°C, bacterial growth was visually monitored.
TABLE 2.
Effects of dam and damX mutations on the expression of cea::lac and stdA::lac fusions
| Genotypea | β-Galactosidase activityb |
|
|---|---|---|
| cea::lacZ | sdtA::lacZ | |
| Wild type | 54.75 ± 2 | <1 |
| ΔdamX1 | 57.49 ± 2 | 15 ± 1 |
| ΔdamX2 | 55.36 ± 3 | <1 |
| Δdam-230 | 727.38 ± 63 | 1273 ± 88 |
The strains were SV4933 (ATCC 14028/pGE108), SV5119 (ΔdamX1/pGE108), SV5488 (ΔdamX2/pGE108), SV4930 (Δdam-230 /pGE108), SV5206 (stdA::lacZ), SV5287 (stdA::lacZ ΔdamX1), SV5486 (stdA::lacZ ΔdamX2), and SV5208 (stdA::lacZ Δdam-230). Strains SV4930 and SV4933 were described in reference 18. Strains SV5206 and SV5208 were described in reference 7.
Strains were grown in LB at 37°C with shaking. Aliquots for β-galactosidase analysis were extracted at an OD660 of 0.8. β-Galactosidase activities were assayed using the CHCl3-sodium dodecyl sulfate permeabilization procedure (14). Data are averages and standard deviations from 3 experiments.
Examination of the nucleotide sequence in the S. enterica damX-dam region provided an explanation for the partial polarity of the ΔdamX1 deletion. The E. coli dam gene is transcribed from five promoters, and one of them (p4) is located within the damX coding region (10). In silico analysis of the Salmonella damX ORF unambiguously predicts the existence of nearly canonical −10 and −35 promoter modules in a region which has been eliminated by the ΔdamX1 deletion but not by ΔdamX2 (see Fig. S1 in the supplemental material). A polar effect on dam expression may likewise explain SOS induction by transposon insertions in damX (15) as well as the invasion defect of Salmonella enterica serovar Typhi damX mutants (9).
A nonpolar damX deletion causes bile sensitivity on its own.
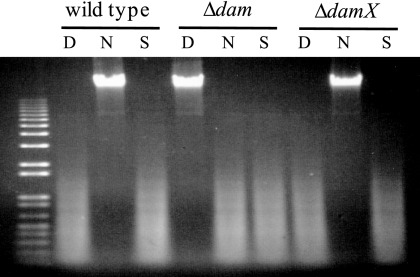
Lack of polarity of the ΔdamX2 deletion on the downstream dam gene was confirmed by digestion of genomic DNA preparations with restriction endonucleases DpnI, Sau3AI, and NdeII. All these enzymes recognize the sequence 5′ GATC 3′. NdeII activity is blocked by Dam methylation, while DpnI cuts methylated DNA only, and Sau3AI cuts both methylated and unmethylated DNA. The DNA restriction pattern in the ΔdamX2 mutant was similar to that of the wild type (dam+) strain, thus ruling out polarity (Fig. 3).
FIG. 3.
Digestion of genomic DNAs isolated from the wild type, a dam mutant (SV4536), and a mutant carrying the ΔdamX2 allele (SV5307) by restriction enzymes DpnI (D), NdeII (N), and Sau3AI (S).
Evidence that lack of DamX causes bile sensitivity on its own (and not by polarity on dam) was also obtained by complementation analysis using a chromosomal duplication (3). A damX+/ΔdamX2 merodiploid was 10-fold more resistant to DOC than the ΔdamX2 mutant and 20-fold more resistant than a ΔdamX2/ΔdamX2 merodiploid (Table 1).
Lack of AsmA suppresses bile sensitivity in damX mutants.
Previous studies have shown that bile sensitivity of S. enterica dam mutants is suppressed by two classes of recessive mutations: (i) in the mismatch repair genes mutH, mutL, and mutS (18) and (ii) in the asmA gene (17). The MIC analyses represented in Table 1 indicated that lack of AsmA suppresses DOC sensitivity in a damX background, while inactivation of the MutHLS system does not. This is a relevant difference between dam and damX mutants despite the fact that both mutant types are bile sensitive. Lack of AsmA has previously been shown to activate the Mar regulon, which may in turn activate efflux pumps and other protective devices (17). As a consequence, asmA mutations suppress bile sensitivity in genetic backgrounds as diverse as dam, phoP, and wec (17). This study adds damX to the list.
Relevance of envelope integrity for bile resistance.
The importance of the cell envelope as a barrier for bile salts is illustrated by the variety of envelope alterations that cause bile sensitivity. The current list includes mutations that alter the outer membrane (17, 20), the lipopolysaccharide (5, 16), and the enterobacterial common antigen (22). It is thus interesting that DamX, an inner membrane protein, is likewise required for bile resistance. The high sensitivity of damX mutants to DOC suggests that lack of DamX may cause a major change in envelope structure or function. Unlike S. enterica dam mutants (21), however, S. enterica damX mutants do not show envelope instability (see Fig. S2 in the supplemental material). The relevance of DamX as an envelope component is supported by two recent studies indicating that E. coli DamX may participate in cell division (1, 6). However, the observation that DamX is produced by both dividing and nondividing Salmonella cells (Fig. 2) suggests that DamX may be a constant component of the S. enterica inner membrane.
Supplementary Material
Acknowledgments
This study was supported by collaborative grants BIO2007-67457-CO2 and CSD2008-00013 from the Spanish Ministry of Science and Innovation (MCINN) and the European Regional Fund. J.L.-G. holds an FPU fellowship from the MCINN. F.G.-Q. holds an FPI fellowship from the MCINN. Nancy Cheng's stay at the University of Seville was supported by a Weissman Fellowship from Harvard University, Cambridge, MA.
We are grateful to Anabel Prieto, Yuri Orlov, and Ignacio Cota for discussions and to Modesto Carballo of the Servicio de Biología (CITIUS, Universidad de Sevilla) for help in experiments performed at the facility.
Footnotes
Published ahead of print on 30 November 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arends, S. J. R., K. Williams, R. J. Scott, S. Rolong, D. L. Popham, and D. S. Weiss. 30 October 2009. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD and RipA. J. Bacteriol. doi: 10.1128/JB.01244-09. [DOI] [PMC free article] [PubMed]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 3.Camacho, E. M., and J. Casadesus. 2001. Genetic mapping by duplication segregation in Salmonella enterica. Genetics 157:491-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 90:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Froelich, J. M., K. Tran, and D. Wall. 2006. A pmrA constitutive mutant sensitizes Escherichia coli to deoxycholic acid. J. Bacteriol. 188:1180-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerding, M. A., B. Liu, F. Bendezú, C. Hale, T. G. Berhanrdt, and P. A. de Boer. 14 August 2009. Self-enhanced accumulation of FtsN at division sites, and roles for other proteins with a SPOR domain (DamX, DedD, and RipA) in Escherichia coli cell constriction. J. Bacteriol. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed]
- 7.Jakomin, M., D. Chessa, A. J. Baumler, and J. Casadesus. 2008. Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA, and HdfR. J. Bacteriol. 190:7406-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonczyk, P., R. Hines, and D. W. Smith. 1989. The Escherichia coli dam gene is expressed as a distal gene of a new operon. Mol. Gen. Genet. 217:85-96. [DOI] [PubMed] [Google Scholar]
- 9.Leclerc, G. J., C. Tartera, and E. S. Metcalf. 1998. Environmental regulation of Salmonella typhi invasion-defective mutants. Infect. Immun. 66:682-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lobner-Olesen, A., E. Boye, and M. G. Marinus. 1992. Expression of the Escherichia coli dam gene. Mol. Microbiol. 6:1841-1851. [DOI] [PubMed] [Google Scholar]
- 11.Lyngstadaas, A., A. Lobner-Olesen, and E. Boye. 1995. Characterization of three genes in the dam-containing operon of Escherichia coli. Mol. Gen. Genet. 247:546-554. [DOI] [PubMed] [Google Scholar]
- 12.Lyngstadaas, A., A. Lobner-Olesen, E. Grelland, and E. Boye. 1999. The gene for 2-phosphoglycolate phosphatase (gph) in Escherichia coli is located in the same operon as dam and at least five other diverse genes. Biochim. Biophys. Acta 1472:376-384. [DOI] [PubMed] [Google Scholar]
- 13.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.O'Reilly, E. K., and K. N. Kreuzer. 2004. Isolation of SOS constitutive mutants of Escherichia coli. J. Bacteriol. 186:7149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picken, R. N., and I. R. Beacham. 1977. Bacteriophage-resistant mutants of Escherichia coli K12. Location of receptors within the lipopolysaccharide. J. Gen. Microbiol. 102:305-318. [DOI] [PubMed] [Google Scholar]
- 17.Prieto, A. I., S. B. Hernandez, I. Cota, M. G. Pucciarelli, Y. Orlov, F. Ramos-Morales, F. Garcia-del Portillo, and J. Casadesus. 2009. Roles of the outer membrane protein AsmA of Salmonella enterica in the control of marRAB expression and invasion of epithelial cells. J. Bacteriol. 191:3615-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prieto, A. I., F. Ramos-Morales, and J. Casadesus. 2004. Bile-induced DNA damage in Salmonella enterica. Genetics 168:1787-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prieto, A. I., F. Ramos-Morales, and J. Casadesus. 2006. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics 174:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prouty, A. M., J. C. van Velkinburgh, and J. S. Gunn. 2002. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J. Bacteriol. 184:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pucciarelli, M. G., A. I. Prieto, J. Casadesus, and F. García-del-Portillo. 2002. Envelope instability in DNA adenine methylase mutants of Salmonella enterica. Microbiology 148:1171-1182. [DOI] [PubMed] [Google Scholar]
- 22.Ramos-Morales, F., A. I. Prieto, C. R. Beuzon, D. W. Holden, and J. Casadesus. 2003. Role for Salmonella enterica enterobacterial common antigen in bile resistance and virulence. J. Bacteriol. 185:5328-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salles, B., J. M. Weiseman, and G. Weinstock. 1987. Temporal control of colicin E1 induction. J. Bacteriol. 169:5028-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uzzau, S., N. Figueroa-Bossi, S. Rubino, and L. Bossi. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 98:15264-15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.