Abstract
The 3.1-Mb genome of an outbreak methicillin-resistant Staphylococcus aureus (MRSA) strain (TW20) contains evidence of recently acquired DNA, including two large regions (635 kb and 127 kb). The strain is resistant to a wide range of antibiotics, antiseptics, and heavy metals due to resistance genes encoded on mobile genetic elements and also mutations in housekeeping genes.
A 2-year outbreak of a highly transmissible methicillin-resistant Staphylococcus aureus (MRSA) strain (designated TW) in an intensive care unit (ICU) in London was recently reported (12). Acquisition of TW MRSA was four times more likely to be associated with bacteremia than was acquisition of other commonly found MRSA strains [>95% epidemic (E)MRSA-15 or EMRSA-16]. TW MRSA was also significantly more frequently isolated from vascular access device cultures but less frequently from carriage sites (anterior nares, axilla, and perineum), suggesting that TW differs in its colonization capacity from other MRSA strains. TW was initially defined by its extended antibiotic resistance pattern, being resistant to penicillin, methicillin, erythromycin, ciprofloxacin, gentamicin, neomycin, trimethoprim, and tetracycline (12). TW also had elevated minimum bactericidal concentrations (MBCs) for chlorhexidine and was resistant to a chlorhexidine-based antiseptic protocol effective against other MRSA strains in the ICU (6). TW20 (strain 0582) was a representative bacteremic isolate cultured on 21 October 2003 (12).
Multilocus sequence typing (MLST) identified TW20 as sequence type 239 (ST239), an international health care-associated (HA) MRSA lineage prevalent in Asia (19, 38), South America (2, 37), and Eastern Europe (5, 33), which includes EMRSA-1, -4, -7, and -11 and the Brazilian, Portuguese, Hungarian, and Viennese clones (24). To investigate the genetic basis for increased resistance and transmissibility, the TW20 genome was completely sequenced, assembled, and finished and annotated as described previously (16, 25). The final finished genome (10) assembly contained 64,087 capillary reads, giving an average coverage of 13.3. At 3,075,806 bp, the TW20 genome is the largest S. aureus genome sequenced thus far. It consists of a single chromosome of 3,043,210 bp in size (Fig. 1) and 2 plasmids (pTW20_1 and pTW20_2), of 29,585 bp and 3,011 bp.
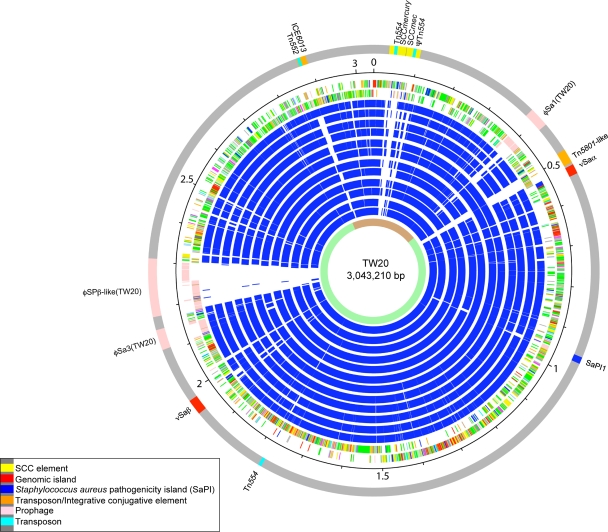
FIG. 1.
Schematic circular diagram of the S. aureus TW20 chromosome. Key for the circular diagram (outer to inner): outer colored segments on the gray outer ring represent genomic islands and horizontally acquired DNA (see the key in the figure); scale (in Mb); annotated CDSs colored according to predicted function are shown on a pair of concentric circles, representing both coding strands; S. aureus reciprocal Fasta matches shared with the S. aureus strains: MRSA252, (accession number BX571856) (16), MSSA476 (accession number BX571857) (16), MW2 (accession number BA000033) (4), N315 (accession number BA000018) (20), Mu50 (accession number BA000017) (20), Mu3 (accession number AP009324) (23), COL (accession number CP000046) (13), NCTC8325 (accession number CP000253) (14), USA3000 FPR3757 (accession number CP000255) (11), JH9 (accession number CP000703) (22), Newman (accession number AP009351) (3), and RF122 (accession number AJ938182) (15); regions of the chromosome derived from a CC8 ancestor (light green) or the CC30 ancestor (brown). Color coding for TW20 CDS functions: dark blue, pathogenicity/adaptation; black, energy metabolism; red, information transfer; dark green, surface associated; cyan, degradation of large molecules; magenta, degradation of small molecules; yellow, central/intermediary metabolism; pale green, unknown; pale blue, regulators; orange, conserved hypothetical; brown, pseudogenes; pink, phage and IS elements; gray, miscellaneous.
TW20 belongs to clonal complex 8 (CC8), which contains strains NCTC8325 (ST8) (14), Newman (ST8) (3), USA300 (ST8) (11), and COL (ST250) (13). Comparative genomic analysis with these strains by reciprocal Fasta analysis (36) revealed that between 83.7 and 82.7% of protein coding sequences (CDSs) in the TW20 chromosome have reciprocal matches with CC8 members. The highest numbers of matches in any sequenced S. aureus strain, however, was with MRSA 252 (85.9% of CDSs). MRSA 252 (ST36) belongs to CC30 and is a representative of EMRSA-16 that has been a dominant MRSA clone in United Kingdom hospitals for more than 10 years (16). In comparison to CC8, most of the additional matches to MRSA 252 are to CDSs in horizontally acquired mobile genetic elements (MGEs) rather than to orthologous CDSs. A significant component of the S. aureus genome is derived from MGEs that contribute to the accessory genome (21). In the TW20 genome, 16.2% of the CDSs (12.6% of the total genomic DNA) are found in MGEs (Fig. 1). Both TW20 and MRSA 252 are representatives of successful hospital-associated MRSA lineages and have large accessory genomes that contain many of the CDSs associated with drug resistance.
Methicillin resistance is conferred by a mecA gene on a type III staphylococcal cassette chromosome (SCC) mec element (SCCmecIII). TW20 has a composite SCC region of two SCC elements, SCCmercury and SCCmecIII, identical in structure to the type III SCCmec region found by Ito et al. (18) in an isolate from New Zealand in 1985. The SCCmecIII region is present in a part of the chromosome hypothesized to have been transferred from CC30 into a CC8 background as part of a large block of DNA (26). The approximate boundaries of the recombination were identified from pairwise comparisons of the TW20 chromosome with MRSA 252 (CC30) and USA300 TCH1516 (CC8). A marked shift in DNA percent identity of approximately 1 percentage point was observed across the approximate recombination breakpoints (data not shown), demonstrating that 635 kb (∼20.6% of the TW20 chromosome; SATW20_26800 to SATW20_03960) may have been transferred from a CC30 donor. This transfer event also contributes to the high level of reciprocal Fasta matches between TW20 and MRSA252 (ST36).
The origins of SCCmecIII in the TW20 genome are unclear, since SCCmecIII has not been found in the CC30 lineage. Each of the SCC elements contains further MGEs: SCCmercury contains Tn554, encoding a streptomycin 3′-adenylyltransferase and an erythromycin resistance protein, ErmA1, and SCCmec contains an integrated plasmid, pT181, and ΨTn554, containing cadmium resistance CDSs. In addition to Tn554 and ΨTn554 in the SCCmec region, the TW20 chromosome contains an additional Tn554 and a Tn552 transposon, encoding the β-lactamase BlaZ, within an integrative conjugative element (ICE) (31).
Further resistance determinants are found on plasmid pTW20_1. Importantly, it carries a gene encoding an antiseptic resistance protein, QacA, that confers resistance to antiseptics such as cationic biocides, quaternary ammonium salts, and diamidines via an export-mediated mechanism (29). In addition, part of the plasmid is highly similar (98 to 100% DNA identity) to the mer operon of the SCCmercury region found on the chromosome (Fig. 2). pTW20_1 also contains a homologue of the gene encoding the cadmium-transporting ATPase CadA, found in ΨTn554 of SCCmec. This region in pTW20_1 is bordered by IS431 elements, as it is in the chromosomal copy of SCCmercury. Notably, upstream of the SCCmercury mer operon, there is a CDS that encodes a putative NADH-binding protein. A fragment homologous to the 3′ end of this CDS is also present on pTW20_1 upstream of the mer operon and is truncated by an IS431 element. The absence of the 5′ region of this CDS on pTW20_1 suggests that this region, including the mer operon, may have arisen on the plasmid by recombination between chromosomal and plasmid IS431 elements. It is therefore possible that IS431-mediated recombination plays a role in the evolution of the SCC region.
FIG. 2.
Comparative analysis of the TW20 plasmid pTW20_1 with the mer operon of the TW20 SCC region. Pairwise comparisons of the TW20 SCC region containing the mer operon from the TW20 chromosome (top) with the TW20 plasmid pTW20_1 (bottom) using the Artemis Comparison Tool (ACT) (9) are shown. The colored bars separating each sequence (red and blue) represent matches identified by BlastN (1); red lines link matches in the same orientation, and blue lines link matches in the reverse orientation. CDSs associated with metal and drug resistance are marked, as are IS431 elements. Colored bars at the top of the figure indicate parts of the sequence found in the SCCmercury (blue) and SCCmec (green) elements, including ΨTn554 (yellow), that make up this region.
Two other drug resistance genes, encoding a tetracycline resistance protein, TetM, and a trimethoprim-resistant dihydrofolate reductase, DfrG, are found in a 31.3-kb region (Tn5801-like), similar to transposons/ICE found in the genomes of S. aureus strains Mu50 (20) and Mu3 (23) and Streptococcus agalactiae strain COH1 (35). In comparison to the Tn5801 elements in Mu50 and Mu3, the TW20 element contains an additional four CDSs, including dfrG, in the central region of the element.
There are three prophage within the TW20 genome, two of which are similar to those previously found in sequenced S. aureus genomes: φSa1(TW20) is 43.3 kb in size, is integrated within the 5′ region of a lipase gene, and does not carry CDSs with homology to known virulence factors; φSa3(TW20) is 44.7 kb in size, is integrated in the phospholipase C gene, and carries the staphylococcal complement inhibitor SCIN (28), staphylokinase (30), and enterotoxin A (7) genes associated with virulence. Genes for two other enterotoxins, enterotoxins K and Q, are carried on a Staphylococcus aureus pathogenicity island (SaPI), SaPI1.
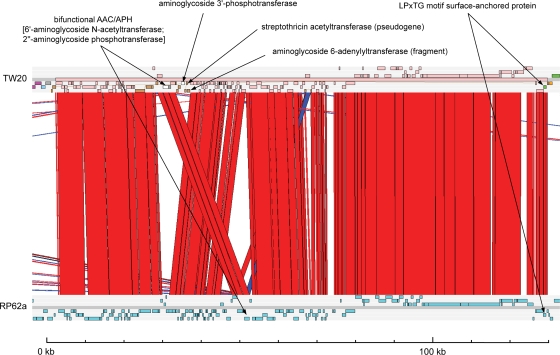
At 127.2 kb, the third prophage, φSPβ-like(TW20), is markedly larger than the other two and does not display similarity with other S. aureus prophage. φSPβ-like(TW20) exhibits extended similarity with the φSPß-like region in the Staphylococcus epidermidis RP62a genome (13) (Fig. 3). Comparison of the two sequences reveals a region of sequence divergence and rearrangement in the center of the prophage. In φSPβ-like(TW20), this region contains CDSs associated with aminoglycoside resistance and streptothricin resistance (Fig. 3). In addition, φSPβ-like(TW20) contains a CDS that may have a role in promoting persistence of TW20 in the hospital setting. S. aureus possesses many surface-anchored proteins with the LPxTG motif, which bind host molecules (27). SATW20_21850 encodes an LPxTG motif surface-anchored protein that does not have orthologs in any of the genomes of the other sequenced S. aureus strains currently available. A highly similar CDS (95.1% amino acid identity), sesI (8), is present in the S. epidermidis φSPβ-like region (Fig. 3). A recent study by Söderquist et al. found that sesI was absent from normal S. epidermidis flora of healthy individuals without any health care association but was found in approximately 50% of clinical isolates causing invasive infections, leading them to suggest that this gene was a potential marker of invasive capacity (32). The presence of an LPxTG motif surface-anchored protein on an MGE in TW20 suggests that this strain has augmented its array of this family of functionally important proteins through a recent acquisition event and therefore this LPxTG motif surface-anchored protein may not be widely distributed in related strains. Genome sequencing of a global collection of ST239 strains revealed only 7% (3/42) of isolates were positive for orthologs of this CDS (14a). Work is under way to survey the wider distribution of this gene in the S. aureus population and investigate the function of the encoded protein.
FIG. 3.
Comparative analysis of φSPβ-like(TW20) prophage with the S. epidermidis RP62a φSPß-like prophage. Pairwise BlastN comparison of the S. aureus TW20 prophage φSPβ-like(TW20) region from the TW20 chromosome (top) with the S. epidermidis RP62a φSPß-like prophage region from the RP62a chromosome (bottom) (13) displayed in ACT is shown. The extent of the φSPβ-like(TW20) prophage in the TW20 sequence, which extends from SATW20_20290 to SATW20_21850, is marked by the pink horizontal bar.
Evidence of adaptation to survive in a health care environment is also found in the core genome. Several housekeeping genes have alleles associated with antibiotic resistance. The TW20 DNA gyrase subunit A (GyrA) contains a leucine residue at position 84. The more-widespread residue in S. aureus GyrA proteins is serine, suggesting this is the plesiomorphic amino acid at this position. In vitro studies have demonstrated that substitution of Ser84Leu generates resistance to quinolones in S. aureus (34). TW20 exhibits low-level resistance to mupirocin. The TW20 isoleucyl-tRNA synthetase contains a phenylalanine residue at position 588. The substitution of Val588Phe has been shown to confer chromosomal low-level mupirocin resistance in S. aureus without significantly affecting fitness (17).
In conclusion, genomic analysis of TW20 provides evidence of its adaptation to survive in a health care setting through acquisition of drug and antiseptic resistance genes carried on MGEs, large chromosomal insertions, and point mutations in housekeeping genes. The large size of the TW20 genome reflects the ability of the ST239 lineage to undergo prolonged and continuing evolution to adapt to the hospital environment. Further studies are under way to elucidate the components of the genome that promote transmission and interaction with the host.
Nucleotide sequence accession numbers.
The sequence and annotation of the TW20 genome have been deposited in the EMBL database under the accession numbers FN433596, FN433597, and FN433598.
Acknowledgments
The Sanger Institute is core funded by the Wellcome Trust. Funding for the sequencing of the TW20 genome was provided by Guy's and St. Thomas' Charity. J.D.E. receives funding from the Department of Health via the NIHR comprehensive Biomedical Research Centre award to Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London.
We thank the Sanger Institute's Pathogen Production Group for shotgun and finishing sequencing and the core Informatics Group for support.
Footnotes
Published ahead of print on 30 November 2009.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amaral, M. M., L. R. Coelho, R. P. Flores, R. R. Souza, M. C. Silva-Carvalho, L. A. Teixeira, B. T. Ferreira-Carvalho, and A. M. Figueiredo. 2005. The predominant variant of the Brazilian epidemic clonal complex of methicillin-resistant Staphylococcus aureus has an enhanced ability to produce biofilm and to adhere to and invade airway epithelial cells. J. Infect. Dis. 192:801-810. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., T. Bae, O. Schneewind, F. Takeuchi, and K. Hiramatsu. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190:300-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 5.Bartels, M. D., A. Nanuashvili, K. Boye, S. M. Rohde, N. Jashiashvili, N. A. Faria, M. Kereselidze, S. Kharebava, and H. Westh. 2008. Methicillin-resistant Staphylococcus aureus in hospitals in Tbilisi, the Republic of Georgia, are variants of the Brazilian clone. Eur. J. Clin. Microbiol. Infect. Dis. 27:757-760. [DOI] [PubMed] [Google Scholar]
- 6.Batra, R., B. S. Cooper, C. Whiteley, A. Patel, D. Wyncoll, and J. D. Edgeworth. The TW variant of MRSA sequence type 239 is not controlled by a surface antiseptic protocol effective at preventing transmission of endemic MRSA on an intensive care unit. Clin. Infect. Dis., in press.
- 7.Betley, M. J., and J. J. Mekalanos. 1988. Nucleotide sequence of the type A staphylococcal enterotoxin gene. J. Bacteriol. 170:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowden, M. G., W. Chen, J. Singvall, Y. Xu, S. J. Peacock, V. Valtulina, P. Speziale, and M. Hook. 2005. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 151:1453-1464. [DOI] [PubMed] [Google Scholar]
- 9.Carver, T. J., K. Rutherford, M. Berriman, M. A. Rajandream, B. Barrell, and J. Parkhill. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 10.Chain, P. S., D. V. Grafham, R. S. Fulton, M. G. Fitzgerald, J. Hostetler, D. Muzny, J. Ali, B. Birren, D. C. Bruce, C. Buhay, J. R. Cole, Y. Ding, S. Dugan, D. Field, G. M. Garrity, R. Gibbs, T. Graves, C. S. Han, S. H. Harrison, S. Highlander, P. Hugenholtz, H. M. Khouri, C. D. Kodira, E. Kolker, N. C. Kyrpides, D. Lang, A. Lapidus, S. A. Malfatti, V. Markowitz, T. Metha, K. E. Nelson, J. Parkhill, S. Pitluck, X. Qin, T. D. Read, J. Schmutz, S. Sozhamannan, P. Sterk, R. L. Strausberg, G. Sutton, N. R. Thomson, J. M. Tiedje, G. Weinstock, A. Wollam, and J. C. Detter. 2009. Genomics. Genome project standards in a new era of sequencing. Science 326:236-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 12.Edgeworth, J. D., G. Yadegarfar, S. Pathak, R. Batra, J. D. Cockfield, D. Wyncoll, R. Beale, and J. A. Lindsay. 2007. An outbreak in an intensive care unit of a strain of methicillin-resistant Staphylococcus aureus sequence type 239 associated with an increased rate of vascular access device-related bacteremia. Clin. Infect. Dis. 44:493-501. [DOI] [PubMed] [Google Scholar]
- 13.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. DeBoy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. X. Jiang, H. Y. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillaspy, A. F., V. Worrell, J. Orvis, B. A. Roe, D. W. Dyer, and J. J. Iandolo. 2006. The Staphylococcus aureus NCTC 8325 genome, p. 381-412. In V. Fischetti, R. Novick, J. Ferretti, D. Portnoy, and J. Rood (ed.), Gram positive pathogens. ASM Press, Washington, DC.
- 14a.Harris, S., E. J. Feil, M. T. G. Holden, M. A. Quail, E. K. Nickerson, N. Chantratita, S. Gardete, A. Tavares, N. Day, J. A. Lindsay, J. D. Edgeworth, H. de Lencastre, J. Parkhill, S. J. Peacock, and S. D. Bentley. Evolution of MRSA during hospital transmission and intercontinental spread. Science, in press. [DOI] [PMC free article] [PubMed]
- 15.Herron-Olson, L., J. R. Fitzgerald, J. M. Musser, and V. Kapur. 2007. Molecular correlates of host specialization in Staphylococcus aureus. PLoS One 2:e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holden, M. T. G., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. J. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurdle, J. G., A. J. O'Neill, and I. Chopra. 2004. The isoleucyl-tRNA synthetase mutation V588F conferring mupirocin resistance in glycopeptide-intermediate Staphylococcus aureus is not associated with a significant fitness burden. J. Antimicrob. Chemother. 53:102-104. [DOI] [PubMed] [Google Scholar]
- 18.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko, K. S., J. Y. Lee, J. Y. Suh, W. S. Oh, K. R. Peck, N. Y. Lee, and J. H. Song. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Z. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Q. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 21.Lindsay, J. A., and M. T. G. Holden. 2004. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 12:378-385. [DOI] [PubMed] [Google Scholar]
- 22.Mwangi, M. M., S. W. Wu, Y. Zhou, K. Sieradzki, H. de Lencastre, P. Richardson, D. Bruce, E. Rubin, E. Myers, E. D. Siggia, and A. Tomasz. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 104:9451-9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neoh, H. M., L. Cui, H. Yuzawa, F. Takeuchi, M. Matsuo, and K. Hiramatsu. 2008. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 52:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 25.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 26.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roche, F. M., R. Massey, S. J. Peacock, N. P. J. Day, L. Visai, P. Speziale, A. Lam, M. Pallen, and T. J. Foster. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149:643-654. [DOI] [PubMed] [Google Scholar]
- 28.Rooijakkers, S. H., M. Ruyken, A. Roos, M. R. Daha, J. S. Presanis, R. B. Sim, W. J. van Wamel, K. P. van Kessel, and J. A. van Strijp. 2005. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 6:920-927. [DOI] [PubMed] [Google Scholar]
- 29.Rouch, D. A., D. S. Cram, D. DiBerardino, T. G. Littlejohn, and R. A. Skurray. 1990. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol. Microbiol. 4:2051-2062. [DOI] [PubMed] [Google Scholar]
- 30.Sako, T., and N. Tsuchida. 1983. Nucleotide sequence of the staphylokinase gene from Staphylococcus aureus. Nucleic Acids Res. 11:7679-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth, D. S., and D. A. Robinson. 2009. Integrative and sequence characteristics of a novel genetic element, ICE6013, in Staphylococcus aureus. J. Bacteriol. 191:5964-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soderquist, B., M. Andersson, M. Nilsson, A. Nilsdotter-Augustinsson, L. Persson, O. Friberg, and S. Jacobsson. 2009. Staphylococcus epidermidis surface protein I (SesI): a marker of the invasive capacity of S. epidermidis? J. Med. Microbiol. 58:1395-1397. [DOI] [PubMed] [Google Scholar]
- 33.Szczepanik, A., M. Koziol-Montewka, Z. Al-Doori, D. Morrison, and D. Kaczor. 2007. Spread of a single multiresistant methicillin-resistant Staphylococcus aureus clone carrying a variant of staphylococcal cassette chromosome mec type III isolated in a university hospital. Eur. J. Clin. Microbiol. Infect. Dis. 26:29-35. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka, M., T. Wang, Y. Onodera, Y. Uchida, and K. Sato. 2000. Mechanism of quinolone resistance in Staphylococcus aureus. J. Infect. Chemother. 6:131-139. [DOI] [PubMed] [Google Scholar]
- 35.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U. S. A. 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson, N. R., S. Howard, B. W. Wren, M. T. Holden, L. Crossman, G. L. Challis, C. Churcher, K. Mungall, K. Brooks, T. Chillingworth, T. Feltwell, Z. Abdellah, H. Hauser, K. Jagels, M. Maddison, S. Moule, M. Sanders, S. Whitehead, M. A. Quail, G. Dougan, J. Parkhill, and M. B. Prentice. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vivoni, A. M., B. A. Diep, A. C. de Gouveia Magalhaes, K. R. Santos, L. W. Riley, G. F. Sensabaugh, and B. M. Moreira. 2006. Clonal composition of Staphylococcus aureus isolates at a Brazilian university hospital: identification of international circulating lineages. J. Clin. Microbiol. 44:1686-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, B. L., G. Zhang, H. F. Ye, E. J. Feil, G. R. Chen, X. M. Zhou, X. M. Zhan, S. M. Chen, and W. B. Pan. 2009. Predominance of the Hungarian clone (ST 239-III) among hospital-acquired meticillin-resistant Staphylococcus aureus isolates recovered throughout mainland China. J. Hosp. Infect. 71:245-255. [DOI] [PubMed] [Google Scholar]