Abstract
Background
Prolificacy is the most important trait influencing the reproductive efficiency of pig production systems. The low heritability and sex-limited expression of prolificacy have hindered to some extent the improvement of this trait through artificial selection. Moreover, the relative contributions of additive, dominant and epistatic QTL to the genetic variance of pig prolificacy remain to be defined. In this work, we have undertaken this issue by performing one-dimensional and bi-dimensional genome scans for number of piglets born alive (NBA) and total number of piglets born (TNB) in a three generation Iberian by Meishan F2 intercross.
Results
The one-dimensional genome scan for NBA and TNB revealed the existence of two genome-wide highly significant QTL located on SSC13 (P < 0.001) and SSC17 (P < 0.01) with effects on both traits. This relative paucity of significant results contrasted very strongly with the wide array of highly significant epistatic QTL that emerged in the bi-dimensional genome-wide scan analysis. As much as 18 epistatic QTL were found for NBA (four at P < 0.01 and five at P < 0.05) and TNB (three at P < 0.01 and six at P < 0.05), respectively. These epistatic QTL were distributed in multiple genomic regions, which covered 13 of the 18 pig autosomes, and they had small individual effects that ranged between 3 to 4% of the phenotypic variance. Different patterns of interactions (a × a, a × d, d × a and d × d) were found amongst the epistatic QTL pairs identified in the current work.
Conclusions
The complex inheritance of prolificacy traits in pigs has been evidenced by identifying multiple additive (SSC13 and SSC17), dominant and epistatic QTL in an Iberian × Meishan F2 intercross. Our results demonstrate that a significant fraction of the phenotypic variance of swine prolificacy traits can be attributed to first-order gene-by-gene interactions emphasizing that the phenotypic effects of alleles might be strongly modulated by the genetic background where they segregate.
Background
In the last few years, a major breakthrough in the understanding of the genetic factors that shape complex traits has been the demonstration that, in several species, a non-negligible fraction of the genetic variance is explained by epistatic interactions. The recent identification of multiple epistatic QTL controlling complex traits in mice [1-4], chickens [5,6], and in model organisms such as yeast [7] and Drosophila melanogaster [8,9] has been a major achievement in the understanding of the genetic nature of complex traits. In addition, the discovery that gene expression is modulated, amongst others, by a plethora of regulatory RNAs with diverse functions and properties has added a new and thick layer of complexity in the subsequent identification of the polymorphisms involved in these interactions, since many of them might reside in non-coding regions [10].
In domestic species, traits relying on reproductive physiology, such as prolificacy and fecundity, have a notable impact on the financial outcome of farming enterprises. In pigs, prolificacy is a complex trait that displays a low heritability and strong heterosis [11]. One-dimensional studies have reported the existence of several QTL affecting litter size in pigs [12-16]. However, only one of the reported QTL was significant on a genome-wide level (p < 0.05) [16], and there was a general lack of positional concordance amongst different genome scans [17]. More importantly, these QTL studies exclusively dissected the additive and dominance components of litter size, thus neglecting the analysis of epistatic interactions that, paradoxically, are expected to explain a substantial portion of genetic variation of reproductive traits [18]. In consequence, many unsolved questions concerning to the genetic architecture of pig prolificacy still remain to be answered. Which are the specific contributions of dominance and epistasis in modelling the phenotypic expression of this complex trait? If epistasis is important, which are the dimensions, geometry and intricacy of the network of interacting loci and which types of epistatic interactions are more relevant? In a cross between two inbred mice strains Peripato et al. [3] demonstrated the existence of eight interacting QTL that explain almost 49% of the phenotypic variance of litter size in this cross. These results highlighted the importance of non additive genetic variance as a fundamental component of prolificacy. Nevertheless, laboratory mice strains are usually bred in a very stable environment, where fluctuations are kept to a minimum, and they have been the subject of an intense process of genetic selection without parallel in any other mammal species. Moreover, mice belong to a different superorder (Euarchontoglires) than most of mammalian domestic species (Laurasiatheria), so it is reasonable to expect that in these two distantly related taxonomic groups the biology of reproduction can differ in many instances.
The relevance of the aforementioned questions led us to analyse the genetic architecture of prolificacy traits in pigs. In this way, we have performed an F2 intercross between two distinct European and Asian breeds, the Iberian and Meishan porcine breeds. Chinese Meishan is one of the most prolific pig breeds of the world being an excellent candidate population to perform these kinds of studies [19]. Iberian is an autochthonous Spanish breed with a very low prolificacy [20]. There is a very marked phenotypic difference for prolificacy traits between these two breeds (around 7 piglets per parity), being 14.3 the mean for the number of piglets born alive per parity of the Meishan breed [19] and 7.0 the mean for this trait of the Iberian Guadyerbas strain [20]. Interestingly, the ancestors of these breeds are assumed to have diverged at least 150,000 years before present without subsequent introgressions [21]. In consequence, it is reasonable to expect that these breeds have evolved, since then, by following independent processes of artificial selection and genetic drift, thereby establishing different adaptive epistatic genetic complexes [22]. In the current work, we have performed both a one-dimensional and a bi-dimensional genome-wide scans for prolificacy traits by employing this Iberian by Meishan F2 intercross as a genetic resource. Our main objective was to elucidate if epistasis makes a major contribution in shaping the phenotypic variability of prolificacy in pigs.
Results
Phenotypic data recorded in the F2 sows and linkage map
A description of the data and statistics of phenotypic records of number of piglets born alive (NBA) and total number of piglets born (TNB) in the F2 population is given in Table 1. The phenotypic variance was 10.24 and 9.61 for TNB and NBA, respectively. The linkage map of the 115 markers used in the QTL analyses is shown in Table 2. Marker order and distances as well as average chromosome lengths were in general agreement with other mapping projects and the USDA genome database http://www.animalgenome.org/pig/. Markers provided coverage of the 18 autosomes, with intervals between adjacent markers that were below 20 cM whenever possible. The average marker interval was 17.6 cM (sex-averaged map distance).
Table 1.
Data structure for number of piglets born alive (NBA) and total number of piglets born (TNB) in the Iberian × Meishan F2 experimental population.
| Order of parity | All | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| N of litters | 252 | 225 | 210 | 194 | 881 |
| NBA | 7.9 (3.4)a | 8.3 (3.0) | 8.9 (2.9) | 9.2 (3.0) | 8.5 (3.2) |
| TNB | 8.7 (3.0) | 8.5 (3.1) | 9.3 (2.9) | 9.7 (3.1) | 9.1 (3.1) |
aMean (sd)
Table 2.
Description of the markers employed for linkage analyses
| SSC | Marker | Pos1 | SSC2 | Marker | Pos | SSC | Marker | Pos | SSC | Marker | Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SW1515 | 0 | 5 | SJ024 | 0 | 9 | SW983 | 0 | 13 | S0076 | 0 |
| ESR1 | 10 | SWR453 | 44 | SW21 | 10 | SWR1008 | 26 | ||||
| CGA | 54 | SW2425 | 55 | SW911 | 33 | SW398 | 48 | ||||
| S0113 | 82 | S0005 | 71 | SW2571 | 69 | SW2440 | 69 | ||||
| S0151 | 92 | SW1987 | 80 | SW2093 | 95 | SW769 | 84 | ||||
| SW1828 | 121 | IGF1 | 99 | SW2116 | 130 | 14 | SW857 | 0 | |||
| DBH | 152 | SW378 | 117 | SW1349 | 149 | SW1125 | 19 | ||||
| 2 | IGF2 | 0 | 6 | MC1R | 0 | 10 | S0038 | 0 | SW210 | 37 | |
| S0141 | 35 | SW973 | 22 | SW1894 | 25 | S0007 | 50 | ||||
| SW240 | 49 | SW1057 | 47 | SW2195 | 40 | SW1081 | 61 | ||||
| SW395 | 65 | S0087 | 64 | S0070 | 52 | SW1557 | 81 | ||||
| S0226 | 75 | SW316 | 87 | SW1991 | 66 | SW2515 | 96 | ||||
| S0378 | 94 | S0228 | 104 | SW1626 | 94 | 15 | S0355 | 0 | |||
| S0036 | 140 | SW1881 | 119 | SWR67 | 104 | SW919 | 10 | ||||
| 3 | SW72 | 0 | SW1328 | 153 | 11 | S0385 | 0 | SW1111 | 25 | ||
| S0206 | 16 | SW2419 | 160 | S0182 | 26 | S0149 | 50 | ||||
| S0164 | 33 | 7 | S0025 | 0 | SW2008 | 38 | SW936 | 70 | |||
| S0216 | 64 | TNFB | 64 | S0071 | 56 | SW1119 | 100 | ||||
| S0002 | 88 | S0066 | 87 | SW703 | 85 | 16 | SW742 | 0 | |||
| SW349 | 98 | SW632 | 117 | SW2413 | 100 | PRLR | 20 | ||||
| 4 | SW2403 | 0 | S0212 | 149 | 12 | SW2490 | 0 | SW403 | 30 | ||
| S0301 | 14 | S0101 | 159 | SW2494 | 10 | SW2517 | 60 | ||||
| S0001 | 22 | 8 | SW2410 | 0 | SW1307 | 43 | S0061 | 88 | |||
| SW839 | 45 | SWR1101 | 42 | SW874 | 59 | 17 | SW24 | 0 | |||
| S0214 | 63 | S0017 | 73 | SW1956 | 71 | SW2142 | 14 | ||||
| SW445 | 78 | S0225 | 91 | S0106 | 84 | SW1920 | 30 | ||||
| VCAM1 | 99 | SW61 | 113 | SWR1021 | 100 | S0359 | 43 | ||||
| S0097 | 123 | BMPR1β | 122 | SW2431 | 71 | ||||||
| 18 | SW1023 | 0 | |||||||||
| SW787 | 20 | ||||||||||
| S0120 | 32 | ||||||||||
| SWR414 | 54 |
1Pos: Position of the marker; 2 SSC, Sus Scrofa Chromosome
One dimensional genome scan for TNB and NBA
Results of the whole-genome scan using a single-QTL model for TNB and NBA are summarized in Table 3. Two genome-wide highly significant QTL were identified on SSC13 (P < 0.001) and SSC17 (P < 0.01) at similar positions for both traits. In SSC13, the single QTL for NBA and TNB were found at positions 50 and 55 cM, respectively (Table 3), sharing an overlapping region located between markers SW398 and SW2440. The most likely position for the QTL found on SSC17 was at 22 cM for both traits (Table 3).
Table 3.
Significant single quantitative trait loci (QTL) for number of piglets born alive (NBA) and total number of piglets born (TNB)
| Trait | SSC | Position cM (CI) | LR | Genome-wide significance level (P-value) | a (SE)* | d (SE) |
|---|---|---|---|---|---|---|
| NBA | 13 | 50 (40-59) | 24.61 | < 0.001 | 0.71 (0.18) (Meishan) | 0.69 (0.25) |
| 17 | 22 (11-42) | 22.48 | < 0.01 | 0.73 (0.19) (Iberian) | -0.82 (0.29) | |
| TNB | 13 | 55 (43-64) | 21.93 | < 0.01 | 0.61 (0.18) (Meishan) | 0.89 (0.28) |
| 17 | 22 (12-62) | 21.25 | < 0.01 | 0.68 (0.18) (Iberian) | -0.75 (0.28) |
SSC: Sus Scrofa Chromosome. cM: centimorgan. CI: confidence interval. LR: Likelihood ratio. a: additive effect. d: dominance effect. SE: standard error. * Increase in the number of piglets per copy of either Meishan or Iberian allele.
Highly significant additive and dominance effects were detected for the SSC13 and SSC17 QTL, although the direction of these effects (Iberian vs. Meishan) depended on the chromosome under consideration. For instance, the QTL on SSC13 for NBA increased additively by 0.71 (± 0.18) piglets per copy of the Meishan allele and it had a dominance effect of 0.69 (± 0.25) piglets. Conversely, the Iberian allele was the one associated with an increase in 0.73 (± 0.19) piglets per copy for the QTL on SSC17. Moreover, this QTL on SSC17 displayed a negative dominance effect (-0.82 ± 0.29). We estimated the degree of dominance as the ratio d/a between the estimated dominance (d) and the absolute value of the additive effect (a). Values of d/a larger than unity corresponds to overdominance, while a d/a ratio between 0 and 1 represents partial dominance. In both cases (QTL on SSC13 and SSC17), the estimated d/a values were consistent with a complete dominance situation. Similar values of additive and dominance effects were obtained for the TNB QTL on SSC13 and SSC17 (Table 3), a result that is not surprising since these two traits are highly correlated. Besides, the proportion of phenotypic variance explained by these two single QTL detected on SSC13 and SSC17 ranged from 2% to 3% for both TNB and NBA.
Bi-dimensional genome scans for NBA and TNB
Identification of multiple interacting QTL for NBA and TNB
Results from the bi-dimensional genome scan for NBA are shown in Table 4. Four bi-dimensional genome-wide highly significant (P < 0.01) and five significant (P < 0.05) epistatic interactions between QTL were found. We confirmed that all the observed epistatic interactions were consistently detected across families rather than being the consequence of a single sire effect, a feature that is particularly important when the number of founder males is moderate or even small. The results obtained through the likelihood ratio test were further confirmed by using other approaches. First, a false discovery rate (FDR) was calculated based on the nominal P-values every 30 cM, with a result of 0.018 for a P-value lower than 0.001. Moreover, parametric bootstrapping confirmed the significance of the results.
Table 4.
Results of a bi-dimensional genome scan for number of piglets born alive (NBA)
| SSC1 | Position1 cM (CI) | SSC2 | Position2 cM (CI) | LR | (P-value) | a1 (SE) | d1 (SE) | a2 (SE) | d2 (SE) | a × a (SE) | a × d (SE) | d × a (SE) | d × d (SE) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 153 (146-153) | 6 | 55 (50-60) | 27.34 | <0.05 | 3.34 (0.72) | |||||||
| 1 | 76 (69-84) | 7 | 107 (101-116) | 31.43 | <0.01 | -0.53 (0.18) | 0.73 (0.31) | 0.85 (0.38) | 2.61 (0.57) | 3.83 (0.97) | |||
| 5 | 66 (59-73) | 18 | 11 (2-20) | 27.97 | <0.05 | 0.71 (0.24) | -1.49 (0.40) | 1.75 (0.47) | |||||
| 6 | 59 (54-64) | 7 | 28 (12-37) | 31.25 | <0.01 | 2.88 (0.69) | 6.20 (1.51) | ||||||
| 6 | 4 (1-10) | 14 | 29 (25-36) | 30.27 | <0.01 | 0.61 (0.25) | 1.63 (0.42) | 2.89 (0.74) | |||||
| 8 | 92 (88-94) | 10 | 87 (81-103) | 28.59 | <0.05 | -0.82 (0.32) | -1.50 (0.44) | -2.86 (0.75) | |||||
| 9 | 4 (1-7) | 13 | 73 (66-82) | 30.58 | <0.01 | -0.75 (0.22) | 0.87 (0.35) | -3.31 (0.65) | |||||
| 10 | 99 (89-104) | 15 | 3 (1-8) | 29.41 | <0.05 | 1.25 (0.34) | -1.25 (0.36) | -1.30 (0.62) | |||||
| 12 | 11 (9-18) | 12 | 89 (74-96) | 28.82 | <0.05 | 0.34 (0.17) | 0.90 (0.22) | -1.36 (0.30) | 0.83 (0.39) | 2.45 (0.60) |
SSC1 and Position1: Chromosome and position, in centimorgans (cM), of the first location. SSC2 and Position 2: Chromosome and position, in centimorgans (cM), of the second location. CI: Confidence interval. LR, Likelihood ratio of contrast of the model with and without epistasis. a1 additive effect of the first location. a2 additive effect of the second location d1 dominance effect of the first location. d2 dominance effect of the second location. Epistasis type: a × a: additive by additive effect; a × d: additive by dominance effect; d × a: dominance by additive effect: d × d, dominance by dominance effect. SE: standard error. Only significant values of a, d, and their interactions are shown.
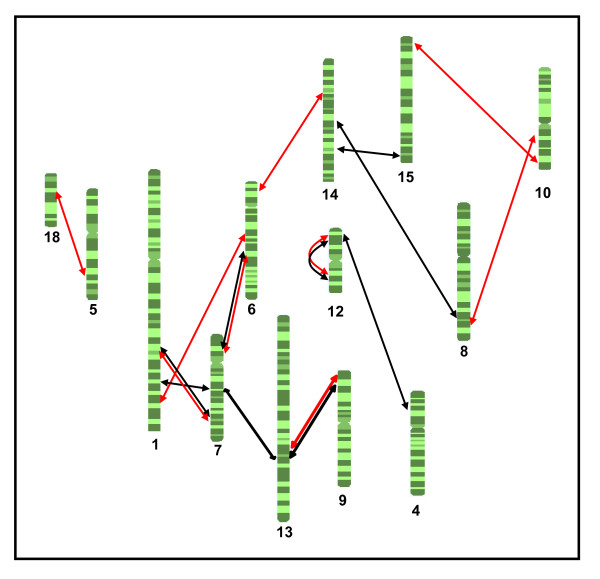
A graphical overview of the epistatic interactions for NBA (red arrows) is shown in Figure 1. As much as twelve of the 18 pig autosomes (1, 5, 6, 7, 8, 9, 10, 12, 13, 14, 15 and 18) were involved in these interactions, forming a complex network with a non-radial geometry. This means that a specific region did not interact simultaneously with multiple loci, but with a very limited number of them (usually interactions were one to one). For example, the SSC12 region located at 11 cM, interacted significantly with another SSC12 region at 89 cM (Figure 1; Table 4). Similarly, two non-overlapping SSC6 QTL regions showed epistatic interactions, one of them with QTL on SSC1 and SSC7 (SSC6, 54-69 cM) and another one with SSC14 (SSC6, 1-10 cM). As shown in Figure 1, other pig chromosomes exhibiting more than one significant interacting QTL were SSC1 (at positions 76 and 153 cM respectively), and SSC7 (at positions 28 cM, and 107 cM). An interesting feature of our analysis was that the highly significant NBA QTL identified in the one-dimensional genome scan (SSC13 at 50 cM and SSC17 at 22 cM) did not show any significant epistatic interaction with other regions across the genome, meaning that its mode of action is purely additive. In contrast, a NBA QTL found on another region of SSC13 (73 cM) had significant epistatic interactions with a QTL located at position 4 cM of SSC9. Similarly, in mice, Peripato et al. [3] identified two significant QTL for litter size in a one-dimensional genome scan (chromosomes 7 and 12) that did not emerge in the bi-dimensional analysis (chromosomes 2, 4, 5, 11, 14, 15 and 18). In the light of these results and ours, we could conclude that there is a low concordance between the QTL identified in one- and bi-dimensional genome scans. This means that, in general, the additive and epistatic components of prolificacy traits encompass different sets of genes.
Figure 1.
Network representation of the epistatic QTL interactions in thirteen pig chromosomes (SSC) for prolificacy traits NBA (red arrows) and TNB (black arrows).
With regard to the bi-dimensional genome-wide scan for TNB, we found three genome-wide highly significant (P < 0.01) and six significant (P < 0.05) epistatic interactions (Table 5; Figure 1). Thirteen out of the 18 pig autosomes were involved in the epistatic QTL interactions for both traits. However, the network of interacting QTL for TNB was not identical to the one reported for NBA. In this sense, only around one third of the QTL epistatic interactions detected in the current study were coincident in both traits. This was an unexpected result since NBA and TNB display a high genetic correlation (rg~ 0.9) and they are expected to share a similar genetic architecture [11]. Moreover, the number of chromosomes displaying multiple interactions was higher for TNB (SSC1, 7, 12, 13 and 14) than for NBA. Notably, four epistatic QTL on SSC7 showed significant interactions with QTL located on SSC6 (60 cM), SSC13 (77 cM) and SSC1 (79 cM and 139 cM, respectively). Other epistatic interactions involved two overlapping regions of SSC13 with one region of SSC7 and one SSC9 QTL, a feature that demonstrates the remarkable complexity and intricacy of these networks. Finally, it is worth mentioning that if we would have assumed a type 1 error α = 0.10, which corresponds to a genome-wide critical value of 25.14 for the LR test, we would have been able to detect 11 and 9 additional epistatic interactions between QTL across the genome for NBA and TNB, respectively (results not shown).
Table 5.
Results of a bi-dimensional genome scan for total number of piglets born (TNB)
| SSC1 | Position1 cM (CI) | SSC2 | Position2 cM (CI) | LR | (P-value) | a1 (SE) | d1 (SE) | a2 (SE) | d2 (SE) | a × a (SE) | a × d (SE) | d × a (SE) | d × d (SE) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 79 (72-84) | 7 | 107 (100-116) | 31.70 | <0.01 | -0.55 (0.17) | -0.68 (0.29) | 0.68 (0.29) | 2.27 (0.49) | 3.29 (0.85) | |||
| 1 | 139 (132-147) | 7 | 89 (84-96) | 28.47 | <0.05 | 0.63 (0.31) | -2.10 (0.54) | 2.08 (0.60) | 2.18 (1.04) | ||||
| 4 | 23 (21-26) | 12 | 5 (1-10) | 27.45 | <0.05 | 0.65 (0.25) | 0.80 (0.33) | -0.92 (0.35) | -2.61 (0.57) | ||||
| 6 | 60 (55-69) | 7 | 24 (12-33) | 35.01 | <0.01 | 2.47 (0.62) | 5.29 (1.30) | ||||||
| 7 | 70 (46-77) | 13 | 77 (73-81) | 28.41 | <0.05 | -1.03 (0.26) | 0.84 (0.28) | 1.08 (0.39) | -1.56 (0.50) | -1.09 (0.46) | -3.14 (0.82) | ||
| 8 | 83 (76-89) | 14 | 58 (54-63) | 29.63 | <0.05 | 2.29 (0.46) | |||||||
| 9 | 4 (1-10) | 13 | 74 (63-81) | 26.99 | <0.05 | -0.70 (0.21) | 0.63 (0.27) | 0.72 (0.34) | -2.98 (0.64) | ||||
| 12 | 11 (8-19) | 12 | 86 (77-93) | 32.10 | <0.01 | 1.01 (0.20) | -1.38 (0.27) | 0.73 (0.34) | 2.00 (0.52) | ||||
| 14 | 89 (80-97) | 15 | 100 (96-101) | 28.30 | <0.05 | 0.79 (0.34) | -1.14 (0.40) | -1.20 (0.41) | -2.62 (0.69) |
SSC1 and Position1: Chromosome and position, in centimorgans (cM), of the first QTL location. SSC2 and Position2: Chromosome and position, of the second QTL location. CI: Confidence interval interval. LR, Likelihood ratio of contrast of the models with and without epistasis. a1: additive effect of the first location. a2: additive effect of the second location. d1: dominance effect of the first location. d2: dominance effect of the second location. a × a: additive by additive effect. a × d: additive by dominance effect. d × a: dominance by additive effect. d × d: dominance by dominance effect. SE: standard error. Only significant values of a, d, and their interactions are shown.
Partition of the phenotypic variance explained by epistatic QTL
We estimated the proportion of the phenotypic variance explained by epistatic genetic components for each TNB and NBA significant epistatic QTL pair. This contribution to the total phenotypic variance ranged from 3.26% (SSC14-SSC15) to 4.04% (SSC12-SSC12) for TNB and from 3.10% (SSC1-SSC6) to 3.62% (SSC9-SSC13) for NBA. The relative contribution of epistasis to the total phenotypic variance was estimated by adding the estimates of the partial epistatic effects of each epistatic interaction. The total phenotypic variance explained by the joint genetic effects of all epistatic QTL pairs for NBA and TNB was 37.6% and 42.4%, respectively. Nevertheless, we would like to mention that this approach might lead to an overestimation of the epistatic contribution because the models employed in our analysis are exclusive [23]. We have calculated the repeatability with a standard animal model, resulting in an estimated value of 0.27 for both TNB and NBA. This value should be considered as the limit of the total variance explained by genetic effects. This result evidences that our estimates of epistatic contributions are clearly overestimated but, in spite of this drawback, we think it is reasonable to assume that a significant proportion of the phenotypic variance of prolificacy is explained by the joint genetic effects of epistatic QTL.
Classification of epistatic effects for NBA and TNB
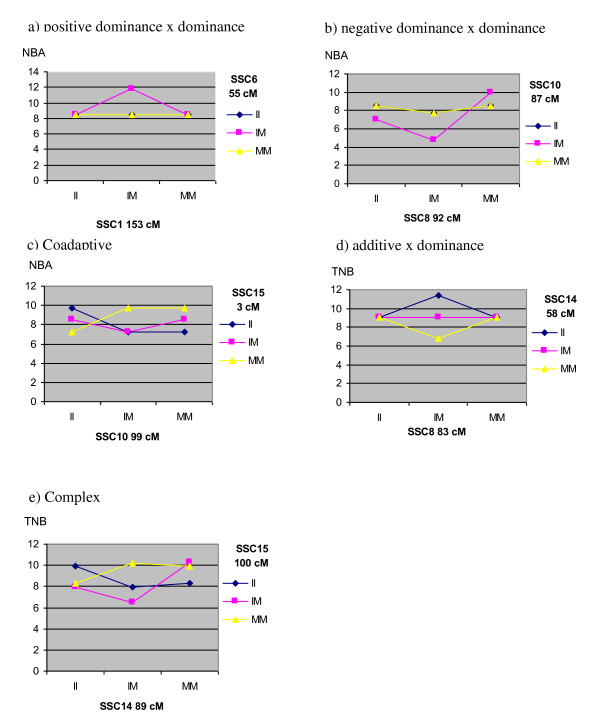
All four forms of epistasis (a × a, a × d, d × a and d × d) were detected among the significant epistatic pairs. In total, 9 a × a, 7 a × d, 11 d × a, and 16 d × d significant interactions were detected (Tables 4 and 5). For the SSC1-SSC6 (NBA) and SSC8-SSC14 (TNB) epistatic pairs, only one type of epistasis was significant (d × d, and a × d, respectively); whereas, the remaining pairs presented two or more types of epistasis, leading to more complex patterns of interactions. We plotted the genotypic values expected for each genotypic class (Figure 2), thus identifying 'dominance-by-dominance' and 'coadaptive' patterns of epistasis as described by Carlborg and Haley [18] and Carlborg et al. [4]. The 'dominance-by-dominance' pattern of epistasis was found in several epistatic QTL pairs and corresponded to interactions were a significant d × d effect was found. In positive d × d interactions (Figure 2a), the genotypic value of double heterozygotes I1M1-I2M2, (being I = Iberian alleles and M = Meishan alleles for loci 1 and 2) was superior to that of simple heterozygotes (I1M1-I2I2, I1M1-M2M2, I1I1-I2M2, and M1M1-I2M2); a pattern that was reversed in negative d × d interactions (Figure 2b).
Figure 2.
Genotypic values expected for each epistatic pattern detected in the experimental F2 Iberian × Meishan intercross. SSC: Sus Scrofa Chromosome. Genotypes are shown as II (Iberian/Iberian homozygote), IM (Iberian/Meishan heterozygote), MM (Meishan/Meishan homozygote).
According to Carlborg and Haley [18], the 'coadaptive' type of interaction occurs when positive a × a interaction effect is significant. This form of epistasis leads to enhanced performance of parental double homozygotes (I1I1-I2I2 and M1M1-M2M2) in comparison to the hybrid double homozygotes (I1I1-M2M2 or M1M1-I2I2). As an example, we found this specific pattern of epistasis in the NBA epistatic QTL pair SSC10-SSC15 (Figure 2c).
Additionally, we found another epistatic pattern where only significant a × d and/or d × a effects were present. These pairs of interacting QTL were included in a category that we named 'additive-by-dominance' epistasis because a × d and d × a interactions showed exactly the same pattern. In positive 'additive-by-dominance' interactions, one locus is overdominant, neutral, or underdominant, depending on the genotype at the second locus. Conversely, the second locus is additive, with the favoured allele being dependent on the genotype at the first locus. In negative 'additive-by-dominance' interactions, the genotypic values of homozygotes at one locus (_._I2I2 and _._M2M2) deviate from what is expectable under an additive model only when the second locus is heterozygous (I1M1-I2I2 and I1M1-M2M2, Figure 2d).
Finally, we considered as a separate category those QTL pairs that could not be classified into any of the preceding epistasis models. The main common feature of QTL pairs gathered in this group was that they all had a significant a × d or d × a epistatic term together with significant a × a and/or d × d terms, leading to 'complex' patterns of interactions (Figure 2e). These complex patterns might be explained by the existence of higher order interactions involving more than two loci or simply because small sample size for certain genotypic classes might lead to inaccurate estimates of the epistatic effects.
Discussion
Identification of genome-wide significant QTL with additive and dominant effects on pig prolificacy
In the single QTL analyses, we found two genome-wide highly significant QTL located on SSC13 and SSC17 affecting NBA and TNB. So far, and to the best of our knowledge, only one study has described a genome-wide significant QTL (P < 0.05) detected at 88 cM on SSC15 for NBA [16]. In the current work, the mode of gene action strongly differed amongst QTL. For instance, the additive effect detected for the SSC13 QTL stemmed from the Meishan breed, and determined an increase of 0.71 and 0.61 piglets for NBA and TNB, respectively. This result is in agreement with the phenotypic differences observed between the purebred parental lines. Conversely, beneficial alleles for the QTL found on SSC17 appeared to derive from the Iberian breed, with increases of 0.73 and 0.68 piglets for NBA and TNB, respectively. These cryptic Iberian QTL alleles, which increase prolificacy, provide a compelling example of the complexity of the genetic architecture of these traits. In addition, significant dominance effects were also detected for the two mentioned QTL; however, the direction of the effects was complete dominance for the QTL on SSC13, and recessivity for the QTL at SSC17.
Our results confirm the existence of several previously reported suggestive QTL for litter size in pigs. De Koning et al. [13] described one suggestive QTL for TNB on SSC17 (43 cM) in an F2 cross between Meishan and commercial Dutch lines. Bidanel et al. [24] reported a QTL for ovulation rate on SSC13 with a similar location (46 cM) and effect (favourable dominant effect of the Meishan allele of 0.7 ova shed). The experiment of Bidanel et al. [24] involved an F2 cross between the same Meishan population employed in our study and a Large White population. Moreover, Cassady et al. [12] described a QTL for number of stillborn piglets on SSC13 (101 cM), although its location is relatively distant to the one reported by us (50-55 cM). Other QTL for litter traits have been described on SSC5, SSC6, SSC7, SSC8, SSC11, SSC14, SSC16 and SSC18 [12-15,25]. However, and as mentioned above, the majority of QTL reported in previous studies none reached the genome-wide significant threshold while two QTL identified in our study did. The most likely explanation for this discrepancy relies on the fact that most of previous experiments involved crosses between the Meishan breed and other standard European pig populations, such as Large White pigs, which have been shown to differ in only three to five piglets born [26]. Moreover, standard European breeds were strongly introgressed with Chinese breeds in the 18-19th centuries and, in consequence, they might not completely fulfil the assumptions and requirements of the F2 intercross design. Conversely, the Meishan and Iberian breeds are highly divergent both at the phenotypic (about 7 piglets per parity) and genetic levels. In this way, phylogenetic studies carried out with mitochondrial DNA have revealed that the Iberian breed has never been introgressed with Asian alleles [21]. Finally, the number of available reproductive records was much larger in our experiment (881 experimental data) than in previous studies (between 200 and 400 experimental data) [12-15,25].
Phenotypic variation of prolificacy traits is affected by a complex network of epistatic QTL
The statistical evidence suggesting that epistasis might be an important component of reproductive traits in mice [3] led us to perform a bi-dimensional genome scan for NBA and TNB. This analysis allowed us to demonstrate that phenotypic variation of these traits can be strongly influenced by a complex network of interacting loci. In this sense, this is the first study that shows genome-wide significant epistatic QTL affecting prolificacy in pigs. After implementing the highly stringent Bonferroni correction for multiple testing, as much as seven QTL interactions remained significant at the 1% bi-dimensional genome-wide level, whereas 11 were significant at the 5% genome-wide level. Rather than identifying one or a few master regions interacting with multiple loci (which would have yielded a star-like geometry of interactions), we found that most chromosomal regions interacted with a single counterpart. However, almost all chromosomes in which significant epistatic QTL were found participated in more than one interaction. Five of the significant epistatic interactions detected had pleiotropic effects on both TNB and NBA traits, whereas five were only related to TNB and three to NBA. These surprising differences for two highly genetically correlated traits [27], might be interpreted in the light of the statistical values of the Likelihood ratio obtained in the contrast between models and the stringent bi-dimensional genome-wide threshold assumed. For instance, the chromosome pair SSC10-SSC15 reached an LR value of 29.41 (p < 0.05) for NBA and an LR value of 22.33 ("no significant") for TNB. Alternatively, there might be a biological meaning behind the specific differences observed in the geometry of the NBA and TNB networks, thus indicating that although similar metabolic and physiological pathways may be implicated in the regulation of both traits, other mechanisms may be operating independently.
Epistatic QTL for pig prolificacy display different types of interactions
This genetic dissection of the epistatic component of prolificacy in pigs was completed with an analysis of the types of interactions detected in the bi-dimensional genome-wide scan. All the epistatic QTL had at least one significant type of interaction (see tables 4 and 5). In total, nine pairs showed additive by additive (a × a) epistasis, which in certain circumstances can have a 'nullification effect' because epistasis might cancel out the effects of individual loci at intermediate frequencies, making difficult to detect them in a conventional one-dimensional genome scan [28]. Furthermore, seven pairs showed additive by dominance (a × d) epistasis, eleven pairs showed dominance by additive (d × a) epistasis, and sixteen pairs showed dominance by dominance (d × d) epistasis. All these forms of epistasis contribute to heterosis [29]. It has been widely supported that heterosis plays an important role in the genetic architecture of reproductive traits [11,12].
Several patterns of interactions among genotypes were identified and classified according to Carlborg and Haley [18] and Carlborg et al. [4]. Eight pairs were classified as 'dominance-by-dominance' and two pairs as 'coadaptive' epistasis. The remaining QTL showed different patterns of epistasis from those previously described in the literature and were, therefore, grouped into two additional categories that we named 'additive-by-dominance' and 'complex' epistasis. 'dominance-by-dominance' epistasis leads to a deviated performance for the double heterozygotes compared to single heterozygotes. Among the d × d interactions found in our study, six had positive sign (Figure 2a) and two had negative sign (Figure 2b). In positive d × d interactions, the genotypic value of the double homozygotes is superior to the simple heterozygotes. Conversely, this pattern is reversed when the sign is negative. Coadaptive epistasis occurs when double homozygotes from the same parental line show enhanced performance [18]. As mentioned above, we only found two QTL pairs which displayed this form of epistasis (Figure 2c). Coadaptive epistasis is fundamental to interpret post-zygotic reproductive isolation [30-32]. In each parental population, selection may have acted leading to fixation of different alleles at the relevant loci regulating prolificacy in a way that statistical epistasis is not apparent in either population. Second-generation hybrids (F2) exhibit combination of alleles at different loci that were not present in any of the parental breeds, leading to the disruption of "co-adapted" gene pools and the appearance of new phenotypes.
The third category included two pairs where only a × d and d × a effects were significant. We called this group 'additive-by-dominance' because the a × d and d × a interactions show the same patterns but with the roles of the loci reversed (Figure 2d). Finally, we found QTL pairs which show 'complex' patterns of interactions characterised by having a significant a × d or d × a term together with significant a × a and/or d × d (Figure 2e). These interactions yielded complex patterns in which the genotypic value for a given genotype at one locus drastically changes depending on the genotype at the second locus.
Conclusions
In summary, the bi-dimensional genome scan of an Iberian × Meishan F2 intercross has allowed to demonstrate that the genetic architecture of pig reproduction is mostly built as a complex network of interacting genes rather than being explained by the sum of the additive effects of a yet to be defined number of loci. Individual epistatic loci have moderate effects on the phenotypic variance of prolificacy and they are distributed in multiple chromosomal locations. Moreover, they display several types of interactions that sometimes cannot be easily ascribed to well defined models, thus suggesting the existence of additional interacting loci. In the next years, the fine mapping and identification of the causal mutations that explain the segregation of epistatic QTL in pigs will be a daunting but fascinating task that will likely unveil many of the secrets that underlie the biological grounds of complex traits.
Methods
Experimental design and phenotypic data
A three-generation F2 intercross between Iberian and Meishan pig breeds was generated to map prolificacy QTL. Eighteen Meishan sows were randomly mated by artificial insemination with three Iberian boars (Guadyerbas line) to produce the F1 progeny in the INRA GEPA experimental unit (Surgères, France). This F1 offspring was purchased and transferred to NOVA GENÈTICA S. A. experimental farm (Lleida, Spain) after weaning at 22-25 days of age. At sexual maturity, eight F1 boars and 97 F1 sows were randomly selected to obtain an F2 progeny. The F1 sows produced only one litter and were slaughtered after weaning. In total, 255 F2 reproductive sows were randomly selected and mated to unrelated boars. They produced a total of 881 litters, i.e. 3.45 parities per F2 sow on average. The number of piglets born alive (NBA) and the total number of piglets born (TNB) were recorded at farrowing. Animals were managed under standard intensive conditions; in all cases, reproduction was carried out by artificial insemination. Protocols were approved by the Ethical and Animal Care Committee at IRTA.
Microsatellite and single nucleotide polymorphism genotyping
DNA was extracted from either frozen blood or tail tissue using commercial protocols (Gentra Systems, Minneapolis). Purebred grandparents, F1 breeding pigs and the 255 F2 sows were genotyped for 115 markers: 109 microsatellites and 6 single nucleotide polymorphisms (SNP). Microsatellite loci were chosen based on their ease of scoring, the absence of null alleles, their genomic location and their informativeness. Microsatellite PCR products were analyzed with the Genescan 3.7 software (Applied Biosystems, Warrington, UK) in an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Single nucleotide polymorphisms markers in DBH, BMPR1β, PRLR and VCAM1 genes were analyzed with the SNapSHOT ddNTP primer extension multiplex kit (Applied Biosystems, Warrington, UK) [33-36]. Moreover, two polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) markers were analysed: the MC1R gene G/A283 SNP was genotyped by PCR-RFLP with NspI [37]. The PvuII estrogen receptor 1 polymorphism was genotyped following Short et al. [38]. Linkage analysis was carried out by using the 'build' option of the CRI-MAP 2.4 program [39].
Statistical analysis
NBA was considered to be the same trait across all parities and the same criterion was applied to TNB. Two statistical models were used to analyze the experimental data. The first model was a one-dimensional QTL mapping performed using a regression approach [40], based on the following mixed model:
| (1) |
where yijk was the ijkth observation for NBA or TNB, Hi was the ith year-season fixed effect, Oj was the jth order of parity fixed effect, uk was the random polygenic effect of the kth individual, pk was the random permanent environmental effects for the kth individual, a was the QTL additive effect, d was the dominance QTL effect and eijkl was the random residual term; ca = pr(QQ) - pr(qq) and cd = 0.5pr(Qq) - 0.5(pr(QQ) + pr(qq)), where pr(QQ) was the probability of being homozygous of Iberian origin, pr(qq) was the probability of being homozygous of Meishan origin and pr(Qq) was the probability of being heterozygous. For computational reasons, heritability (h2) and percentage of permanent environmental effect (p2) were assumed to be known. They were obtained from the posterior mode of a previous Bayesian analysis. Estimates for h2 and p2 were 0.22 and 0.05, respectively. The analyses were performed at every centimorgan along the 2,017 cM of the 18 autosomes, by means of a likelihood ratio test (LR) comparing the models with and without the QTL effects. Nominal P-values were calculated assuming a chi-squared distribution of the LR test. Yet, nominal significance levels cannot be used directly due to the large number of tests performed. Hence, genome-wide significance levels were calculated using a Bonferroni correction and assuming independence between statistical tests every 30 cM. The genome-wide critical values of LR test for level of significance associated with type I errors α = 0.10, 0.05, 0.01 and 0.001 were 13.36, 15.13, 18.27 and 23.22, respectively.
With regard to the two-QTL analyses, two different models were employed. The first model included the effects of two non interacting QTL. The statistical mixed model was:
| (2) |
where a1 and a2 were the additive effects, d1 and d2 were the dominance effects for QTL 1 and 2, respectively. The coefficients ca1, cd1, ca2 and cd2 were calculated as before for locations 1 and 2.
The second model allowed for epistasis, i.e.:
| (3) |
where Ia × a, Ia × d, Id × a and Id × d were the additive × additive, additive × dominance, dominance × additive and dominance × dominance epistatic interaction effects, respectively; ca × a, ca × d, cd × a and cd × d were the regression coefficients calculated following Cockerham's model for epistatic interactions [41,42], i.e.:
 |
The two-QTL analyses were performed using a full bi-dimensional genome scan. LR-tests comparing the models with and without the epistatic interaction effects (model 3 vs model 2) were computed at 1 cM intervals along the 2,017 cM of the 18 autosomes for each of the two QTL, leading to a total of 2,069,595 regression analyses for both NBA and TNB. The values of h2 and p2 used in this analysis were identical to those considered in model 1. The statistical contrast between models for evidence of epistasis was carried out using an LR test with 4 degrees of freedom in the numerator. As before, bi-dimensional genome-wide levels of significance were calculated using a Bonferroni correction assuming statistical independence every 30 cM. The genome-wide critical values of LR test for level of significance associated with type I errors α = 0.10, 0.05, 0.01 and 0.001 were 25.14, 26.68, 30.17 and 35.06 respectively. Confidence intervals for QTL location were calculated using the Likelihood drop method [43]. Finally, we calculated the expected values of the nine genotypic classes using the Cockerham's F2-metric model [42]. In addition, statistical significance was independently assessed by using an approach based on the False Discovery Rate -FDR- [44], that was calculated based on nominal P-values every 30 cM, as well as by employing a parametric bootstrap method [45].
Authors' contributions
JLN conceived and led the project, supervised its execution, participated in the design of the study, drafted and finalized the manuscript. CR participated in the design and coordination of the study, obtained and provided Iberian semen, performs linkage analysis, and helped to revise the manuscript. LV participated in the design of the study, developed the analysis programs, performed the statistical analysis and helped to revise the manuscript. AT has carried out part of the molecular genotyping tasks and has has been involved in drafting and revising critically the manuscript. GM performed part of the molecular genotyping tasks. OR performed part of the molecular genotyping tasks. CB has performed part of the microsatellite genotyping. MA (IRTA) participated in the experimental protocol and data collection. JPB has provided Meishan sows (INRA) to obtain F1 animals, and participated in the discussion of the results and helped to revised the manuscript. MA (UAB) has been involved in drafting the manuscript and revising it critically. CO, was responsible of DNA extraction and the selection of markers to genotype and helped to perform linkage analysis. AS participated in the design and coordination of the study and helped to draft the manuscript.
All authors read and approved the final manuscript.
Contributor Information
José L Noguera, Email: joseluis.noguera@irta.es.
Carmen Rodríguez, Email: valdo@inia.es.
Luis Varona, Email: lvarona@unizar.es.
Anna Tomàs, Email: anna.tomas@cresa.uab.cat.
Gloria Muñoz, Email: gloriammster@gmail.com.
Oscar Ramírez, Email: oscar.ramirez@upf.edu.
Carmen Barragán, Email: barragan@inia.es.
Meritxell Arqué, Email: marqueclemens@gmail.com.
Jean P Bidanel, Email: jean-pierre.bidanel@jouy.inra.fr.
Marcel Amills, Email: marcel.amills@uab.es.
Cristina Ovilo, Email: ovilo@inia.es.
Armand Sánchez, Email: armand.sanchez@uab.es.
Acknowledgements
Financial support was provided by the Spanish Ministerio de Educación y Ciencia (Grant AGL2000-1229-C03). Our sincere acknowledgment to Miguel Angel Toro (INIA) for valuable discussions and useful comments. The authors are indebted to the staff of Nova Genètica, in particular to E. Ramells, F. Marquez, R. Malé, F. Rovira, for cooperating in the experimental protocol, and I. Riart (IRTA) and J. C. Caritez (INRA) for their technical support. The authors gratefully acknowledge the contributions of the INRA (France) and the CIA El Dehesón del Encinar (Spain) for providing the purebred Meishan sows and Iberian boars, respectively.
References
- Brockmann GA, Kratzsch J, Haley CS, Renne U, Schwerin M, Karle S. Single QTL effects, epistasis, and pleiotropy account for two-thirds of the phenotypic F-2 variance of growth and obesity in DU6i × DBA/2 mice. Genome Research. 2000;10:1941–1957. doi: 10.1101/gr.GR1499R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Vaughn TT, Susan L Pletscher, Peripato AC, Adams ES, Erikson CF, King-Ellison KJ. Genetic architecture of adiposity in the cross of LG/J and SM/J inbred mice. Mammalian Genome. 2001;12:3–12. doi: 10.1007/s003350010218. [DOI] [PubMed] [Google Scholar]
- Peripato AC, de Brito RA, Matioli SR, Pletscher LS, Vaughn T, Cheverud JM. Epistasis affecting litter size in mice. Journal of Evolutionary Biology. 2004;3:593–602. doi: 10.1111/j.1420-9101.2004.00702.x. [DOI] [PubMed] [Google Scholar]
- Carlborg O, Brockmann A, Haley CS. Simultaneous mapping of epistatic QTL in DU6i × DBA/2 mice. Mammalian Genome. 2005;16:481–494. doi: 10.1007/s00335-004-2425-4. [DOI] [PubMed] [Google Scholar]
- Carlborg O, Kerje S, Schütz K, Jacobsson L, Jensen P, Andersson L. A global search reveals epistatic interaction between QTL for early growth in the chicken. Genome Research. 2003;13:413–421. doi: 10.1101/gr.528003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlborg O, Burt D, Hocking P, Haley CS. Simultaneous mapping of epistatic QTL in chickens reveals clusters of QTL pairs with similar genetic effects on growth. Genetical Research. 2004;83:197–209. doi: 10.1017/S0016672304006779. [DOI] [PubMed] [Google Scholar]
- Segre D, DeLuna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nature Genetics. 2005;37(1):77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- Fedorowicz GM, Fry DJ, Anholt RR, Mackay TF. Epistatic interactions between smell-impaired loci in Drosophila melanogaster. Genetics. 1998;148:1885–1891. doi: 10.1093/genetics/148.4.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth KL, Marden JH, Clark AG. Mapping determinants of variation in energy metabolism, respiration and flight in Drosophila. Genetics. 2003;165(2):623–635. doi: 10.1093/genetics/165.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L, Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nature Review Genetics. 2004;5:202–212. doi: 10.1038/nrg1294. [DOI] [PubMed] [Google Scholar]
- Rothschild MF, Bidanel JP. In: The Genetics of the Pig. Rothschild MF, Ruvinsky A, editor. Cambridge, UK: CAB International, University Press; 1998. Biology and genetics of reproduction; pp. 313–343. [Google Scholar]
- Cassady JP, Johnson RK, Pomp D, Rohrer GA, Van Vleck LD, Spiegel EK, Gilson KM. Identification of quantitative trait loci affecting reproduction in pigs. Journal of Animal Science. 2001;79:623–633. doi: 10.2527/2001.793623x. [DOI] [PubMed] [Google Scholar]
- De Koning DJ, Ratting AP, Harlizius B, Groenen MAM, Brascamp EW, van Arendonk JAM. Detection and characterization of quantitative trait loci for growth and reproduction traits in pigs. Livestock Production Science. 2001;72:185–198. doi: 10.1016/S0301-6226(01)00226-3. [DOI] [PubMed] [Google Scholar]
- King AH, Jiang ZH, Gibson JP, Haley CS, Archibald AL. Mapping quantitative trait loci affecting female reproductive traits on porcine chromosome 8. Biology of Reproduction. 2003;68(6):2172–2179. doi: 10.1095/biolreprod.102.012955. [DOI] [PubMed] [Google Scholar]
- Tribout T, Iannuccelli T, Druet H, Gilbert J, Riquet R, Gueblez R, Mercat MJ, Bidanel JP, Milan D, le Roy P. Detection of quantitative trait loci for reproduction and production traits in Large White and French Landrace pig populations. Genetics, Selection, Evolution. 2008;40(1):61–78. doi: 10.1186/1297-9686-40-1-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Ren J, Xing Y, Zhang Z, Ma J, Guo Y, Huang L. Quantitative trait loci for litter size and prenatal loss in a White Duroc × Chinese Erhualian resource population. Animal Genetics. 2009;40:963–966. doi: 10.1111/j.1365-2052.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- Spötter A, Distl O. Genetic approaches to the improvement of fertility traits in the pig. Veterinary Journal. 2006;172(2):234–247. doi: 10.1016/j.tvjl.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nature Reviews. 2004;5:619–625. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- Bidanel JP, Caritez JC, Legault C. Estimation of crossbreeding parameters between Large White and Meishan porcine breeds. I. Reproductive performance. Genetics, Selection, Evolution. 1989;21:507–526. doi: 10.1186/1297-9686-21-4-507. [DOI] [Google Scholar]
- Silió L, Rodriguez C, Rodrigáñez J, Toro MA. In: Porcino Ibérico: Aspectos claves. Buxadé C, Daza A, editor. Madrid: Mundiprensa; 2001. La selección de cerdos ibéricos; pp. 125–149. [Google Scholar]
- Alves E, Óvilo C, Rodríguez MC, Silió L. Mitochondrial DNA sequence variation and phylogenetic relationships among Iberian pigs and other domestic and wild pig populations. Animal Genetics. 2003;34(5):319–324. doi: 10.1046/j.1365-2052.2003.01010.x. [DOI] [PubMed] [Google Scholar]
- Fernández A, Toro MA, Rodriguez C, Silió L. Heterosis and epistasis for teat number and fluctuating asymmetry in crosses between Jiaxing and Iberian pigs. Heredity. 2004;93(2):222–227. doi: 10.1038/sj.hdy.6800498. [DOI] [PubMed] [Google Scholar]
- Beavis WD. In: Proceedings of the Forty-Ninth Annual Corn & Sorghum Industry Research Conference: 7-8 December 1994; Chicago. Wilkinson DB, editor. American Seed Trade Association, Washington DC; 1994. The power and deceit of QTL experiments:lessons from comparitive QTL studies; pp. 250–266. [Google Scholar]
- Bidanel JP, Rosendo A, Iannuccelli N, Gilbert H, Caritez JC, Billon Y, Prunier A, Milan D. Detection of quantitative trait loci for teat number and female reproductive traits in Meishan × Large White F2 pigs. Animal. 2008;2:813–820. doi: 10.1017/S1751731108002097. [DOI] [PubMed] [Google Scholar]
- Holl JW, Cassady JP, Pomp D, Johnson RK. A genome scan for quantitative trait loci and imprinted regions affecting reproduction in pigs. Journal of Animal. Science. 2004;82:3421–3429. doi: 10.2527/2004.82123421x. [DOI] [PubMed] [Google Scholar]
- Haley CS, Lee GJ, Ritchie M. Comparative reproductive-performance in Meishan and Large White pigs and their crosses. Animal Science. 1995;60(2):259–267. [Google Scholar]
- Canario L, Roy N, Gruand J, Bidanel JP. Genetic variation of farrowing kinetics traits and relationships with litter size and perinatal mortality in French Large White sows. Journal of Animal Science. 2006;84:1053–1058. doi: 10.2527/jas.2005-775. [DOI] [PubMed] [Google Scholar]
- Routman E, Cheverud JM. Gene effects on a quantitative trait: two-locus epistatic effects measured at microsatellite markers and at estimated QTL. Evolution. 1997;51(5):1654–1662. doi: 10.2307/2411217. [DOI] [PubMed] [Google Scholar]
- Hua JP, Xing YZ, Wu WR, Xu CG, Sun XL, Yu SB, Zhang QF. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. PNAS. 2003;100(5):2574–2579. doi: 10.1073/pnas.0437907100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T Genetics of the evolutionary process 1937New York: Columbia University Press; 17246845 [Google Scholar]
- Muller HJ. In: The new systematics. Huxley J, editor. Oxford: Clarendon; 1940. Bearing of the Drosophila work on systematics; pp. 185–268. [Google Scholar]
- Muller HJ. Isolating mechanisms, evolution and temperature. Biological Symposium. 1942;6:71–125. [Google Scholar]
- Tomàs A, Casellas J, Ramirez O, Pérez-Enciso M, Rodriguez C, Noguera JL, Sánchez A. Polymorphisms of the porcine dopamine beta-hydroxylase gene and their relation to reproduction and piglet survivability in an Iberian × Meishan F2 intercross. Animal Genetics. 2006;37(3):279–282. doi: 10.1111/j.1365-2052.2006.01457.x. [DOI] [PubMed] [Google Scholar]
- Ramirez O, Tomàs A, Casellas J, Blanch M, Noguera JL, Amills M. An Association Analysis Between a Silent C558T Polymorphism at the Pig Vascular Cell Adhesion Molecule 1 Locus and Sow Reproduction and Piglet Survivability Traits. Reproduction Domestic Animals. 2008;43(5):542–546. doi: 10.1111/j.1439-0531.2007.00949.x. [DOI] [PubMed] [Google Scholar]
- Tomàs A, Frigo E, Casellas J, Ramirez O, Ovilo C, Noguera JL, Sánchez A. An association study between polymorphisms of the porcine bone morphogenetic protein receptor type1beta(BMPR1β) and reproductive performance of Iberian × Meishan F2 sows. Animal Genetics. 2006;37(3):297–298. doi: 10.1111/j.1365-2052.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- Tomàs A, Casellas J, Ramirez O, Muñoz G, Noguera JL, Sánchez A. High amino acid variation in the intracellular domain of the pig prolactin receptor (PRLR) and its relation to ovulation rate and piglet survival traits. Journal of Animal Science. 2006;84(8):1991–1998. doi: 10.2527/jas.2005-664. [DOI] [PubMed] [Google Scholar]
- Fernández A, Fabuel E, Alves E, Rodríguez C, Silió L, Ovilo C. DNA test based on coat colour genes for authentication of the raw material of meat products from Iberian pigs. Journal of the Science of Food and Agriculture. 2004;84(14):1855–1860. doi: 10.1002/jsfa.1829. [DOI] [Google Scholar]
- Short TH, Rothschild MF, Southwood OI, McLaren DG, de Vries A, Steen H van der, Eckardt GR, Tuggle CK, Helm J, Vaske DA, Milehan AJ, Plastow GS. Effect of the estrogen receptor locus on reproduction and production traits in four commercial pig lines. Journal of Animal Science. 1997;75(12):3138–3142. doi: 10.2527/1997.75123138x. [DOI] [PubMed] [Google Scholar]
- Green P, Falls K, Crooks S. Documentation of CRI-MAP. http://linkage.rockefeller.edu/soft/crimap/
- Haley CS, Knott SA, Elsen JM. Mapping quantitative trait loci in crosses between outbreed lines using least squares. Genetics. 1994;136:1195–1207. doi: 10.1093/genetics/136.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham CC. An extension of the concept of partitioning hereditary variance for analysis of covariances among relatives when epistasis is present. Genetics. 1954;39:859–882. doi: 10.1093/genetics/39.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C-H, Zeng Z-B. Modeling epistasis of quantitative trait loci using Cockerham's model. Genetics. 2002;160:1243–1261. doi: 10.1093/genetics/160.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- Efron B, Tibshirani R. An Introduction to the Bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]