FYCO1 recognition of LC3 on autophagosomes facilitates microtubule-mediated cytosolic transport of this degradative organelle.
Abstract
Autophagy is the main eukaryotic degradation pathway for long-lived proteins, protein aggregates, and cytosolic organelles. Although the protein machinery involved in the biogenesis of autophagic vesicles is well described, very little is known about the mechanism of cytosolic transport of autophagosomes. In this study, we have identified an adaptor protein complex, formed by the two autophagic membrane-associated proteins LC3 and Rab7 and the novel FYVE and coiled-coil (CC) domain–containing protein FYCO1, that promotes microtubule (MT) plus end–directed transport of autophagic vesicles. We have characterized the LC3-, Rab7-, and phosphatidylinositol-3-phosphate–binding domains in FYCO1 and mapped part of the CC region essential for MT plus end–directed transport. We also propose a mechanism for selective autophagosomal membrane recruitment of FYCO1.
Introduction
The term autophagy is commonly used to describe several distinct cellular processes during which cytosolic/nucleosolic proteins or organelles are transported into the lysosomes for degradation. Macroautophagy (hereafter autophagy) is considered the major pathway for delivery of cytosolic cargo to lysosomes. During autophagy, part of the cytosol is sequestered into a double-membrane vesicle, called an autophagosome, which subsequently fuses with endosomes and lysosomes. This way, the cytosolic cargo is delivered for degradation by lysosomal proteases (Mizushima, 2007; Xie and Klionsky, 2007).
Genetic screens in yeast identified a group of ∼30 autophagy-related proteins (Atg) essential for autophagosome biogenesis (Tsukada and Ohsumi, 1993; Thumm et al., 1994). The majority of these proteins participate in the initial stages of autophagosome formation. Currently, very little is known about the protein machinery involved in intracellular transport of autophagosomes and their docking and fusion with other membranous compartments. In mammals, Rab7, the core class C Vps-tethering complex and UVRAG are important for fusion of autophagosomes with lysosomes (Gutierrez et al., 2004; Jäger et al., 2004; Liang et al., 2008). Several other Rab family members, including Rab5 (Ravikumar et al., 2008), Rab11 (Fader et al., 2008), Rab24 (Munafó and Colombo, 2002), Rab32 (Hirota and Tanaka, 2009), and Rab33B (Itoh et al., 2008), are suggested to be involved in autophagy. Except for the Atg16L–Rab33 complex, the autophagy-specific effectors of these small GTPases are currently unknown. Multiple studies on the importance of microtubules (MTs) for mammalian autophagy have been published in recent years (Webb et al., 2004; Köchl et al., 2006), but the protein machinery involved in MT-dependent transport of autophagosomes has not been characterized yet.
The lipid phosphatidylinositol-3-phosphate (PI3P) is also essential for autophagosome biogenesis (Seglen and Gordon, 1982; Blommaart et al., 1997). Although the exact function or functions of PI3P in autophagy are still unclear, it is generally accepted that PI3P-enriched membranes recruit and activate effector proteins containing FYVE (Fab1, YOTB/ZK632.12, Vac1, and EEA1) or PX (Phox) PI3P-binding domains (Gaullier et al., 1998; Song et al., 2001).
ATG8/MAP1-LC3/GABARAP is a family of small globular proteins containing a C-terminal ubiquitin-like domain and a short N-terminal arm formed by two amphipathic α-helices (Paz et al., 2000; Sugawara et al., 2004). All members of the family are membrane associated via a phosphatidylethanolamine lipid anchor attached to a C-terminal glycine (Kirisako et al., 2000; Kabeya et al., 2004). In yeast, Atg8 localizes to phagophores and autophagosomes, where it participates in membrane expansion (Kirisako et al., 1999; Xie et al., 2008).
In this paper, we identify FYCO1 (FYVE and coiled-coil [CC] domain containing 1) as a novel LC3-, Rab7-, and PI3P-interacting protein. The LC3–FYCO1 interaction is mediated by an LC3-interacting region (LIR) motif adjacent to the FYVE domain of FYCO1. We demonstrate that FYCO1 dimerizes via the CC region, interacts with PI3P via its FYVE domain, and forms a complex with Rab7 via a part of the CC region located in front of the FYVE domain. Overexpression of FYCO1 redistributes LC3- and Rab7-positive structures to the cell periphery in an MT-dependent manner. This effect is mediated by the central part of the CC region and suggests a role for FYCO1 in MT plus end–directed transport of autophagic vesicles.
Results
FYCO1 is a novel LC3-interacting protein
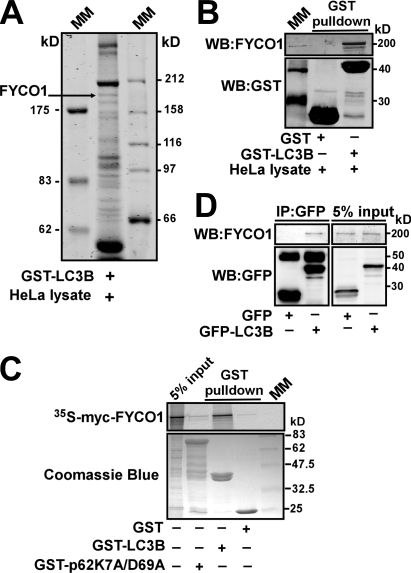
To identify new interaction partners of LC3B, we performed affinity purification using GST-LC3B as an affinity ligand. HeLa cell lysate from ∼108 cells was incubated in a batch mode with GST-LC3B bound to glutathione–Sepharose. Bound proteins were eluted and resolved on SDS-PAGE (Fig. 1 A). Protein bands were subjected to trypsin digestion, and the corresponding proteins were identified by mass spectrometry. One of the proteins identified by this approach was FYCO1, a novel protein with uncharacterized function (Fig. 1 A). The identity of FYCO1 and the specificity of its interaction with LC3B were confirmed by GST pull-down and coimmunoprecipitation experiments (Fig. 1, B–D).
Figure 1.
FYCO1 is a novel LC3B-interacting protein. (A and B) Endogenous FYCO1 binds to GST-LC3B. GST-LC3B bound to glutathione–Sepharose beads was incubated with HeLa cell lysate, and copurified proteins were identified by Coomassie blue staining and mass spectrometry (A) or immunoblotting with anti-FYCO1 antibody (B). (C) GST-LC3B but not GST-p62 or GST alone binds to FYCO1 in a GST pull-down assay. GST, GST-LC3B, or GST-p62 bound to glutathione–Sepharose beads was incubated with [35S]methionine-labeled myc-FYCO1. Formed protein complexes were isolated and visualized by autoradiography (top) or Coomassie blue staining (bottom). (D) GFP-LC3B immunoprecipitates endogenous FYCO1 from HeLa cell lysate. HeLa cells expressing GFP or GFP-LC3B were lysed and processed as in B. IP, immunoprecipitation; MM, molecular mass; WB, Western blot.
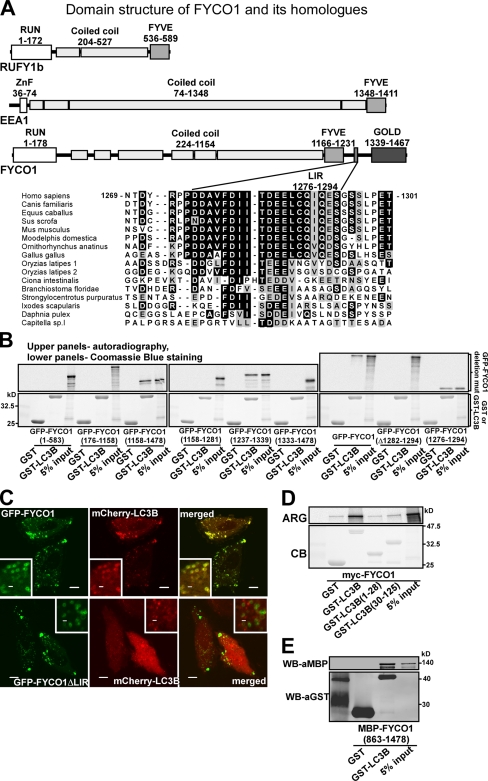
FYCO1 is a novel protein encoded by a gene located at chromosome 3 common eliminated region 1 (C3CER1; 3p21.3; Kiss et al., 2002). Analyses using the Conserved Domain Database (Marchler-Bauer et al., 2009) and COILS web server (Lupas et al., 1991) predict FYCO1 to be a long CC protein similar to two families of Rab effector proteins: RUN and FYVE domain–containing proteins (RUFY1–4) and EEA1 (Fig. 2 A; Rose et al., 2005). Similar to these proteins, FYCO1 contains a central, 850-aa-long CC region flanked by globular domains. The globular domain at the C-terminal end of the CC is common for all three protein groups and is a PI3P lipid-binding FYVE domain (Gaullier et al., 1998). Similar to RUFY1–4 proteins, FYCO1 contains an N-terminal RUN domain, which is an α-helical protein–protein interaction domain shown to bind to the small GTPases of Rab and Rap families (Callebaut et al., 2001; Recacha et al., 2009). Unique for FYCO1 compared with RUFYs and EEA1 is the presence of a 100-aa-long unstructured region C-terminally to the FYVE domain, followed by a globular GOLD (Golgi dynamics) domain (Fig. 2 A and Fig. S2 C).
Figure 2.
The interaction between FYCO1 and LC3B is mediated by a short conserved acidic motif C-terminally from the FYVE domain in FYCO1 and both N- and C-terminal domains of LC3B. (A) Schematic structure of FYCO1 and its homologous proteins RUFY1b and EEA1. In the multialignment sequence, identity is indicated by black highlighting, and residues highlighted in gray indicate substitutions to chemically similar amino acids in more than 50% of the compared sequences. (B) The region of FYCO1 between aa 1,276 and 1,294 is essential and sufficient for the interaction with LC3B. GST or GST-LC3B were incubated with [S35]methionine-labeled deletion mutants of FYCO1 and processed as in Fig. 1 C. (C) LIR is essential for colocalization of FYCO1 with LC3B in HeLa cells. HeLa cells expressing the indicated constructs were imaged by confocal microscopy. Insets show an enlarged field of interest. (D) Both N- and C-terminal domains of LC3B are needed for efficient interaction with FYCO1. GST, GST-LC3B, or its deletion mutants were incubated with [S35]methionine-labeled myc-FYCO1 and processed as in Fig. 1 C. ARG, autoradiography; CB, Coomassie blue. (E) The interaction between FYCO1 and LC3B is direct. GST or GST-LC3B was incubated with recombinant MBP-FYCO1863–1,478. Protein complexes were isolated and visualized by immunoblotting with anti-GST or anti-MBP antibodies. WB, Western blot. Bars: (C) 10 µm; (C, insets) 1 µm.
Using GST pull-down assays, we mapped a 19-aa-long conserved LIR at aa 1,276–1,294 of FYCO1 as essential and sufficient for binding to LC3B (Fig. 2 B and Fig. S1). Deletion of LIR (Δ1,282–1,294) resulted in loss of colocalization between FYCO1 and LC3B (Fig. 2 C). Similar to binding of other LIR-containing proteins, both N- and C-terminal domains of LC3B were needed for efficient binding to FYCO1 (Fig. 2 D). Using GST pull-down with recombinant GST-LC3B and a maltose-binding protein (MBP) fusion of the C-terminal half of FYCO1, we confirmed a direct interaction between the two proteins (Fig. 2 E).
FYCO1 localizes to autophagosomes, endosomes, and lysosomes
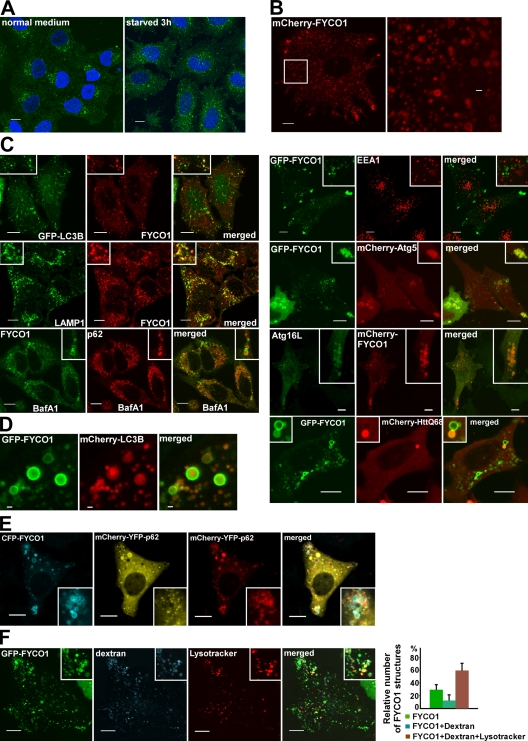
Immunostaining showed that endogenous FYCO1 is localized to punctuated structures concentrated in the juxtanuclear region of the cytoplasm of HeLa cells (Fig. 3 A, left). Interestingly, FYCO1 puncta redistributed to other parts of the cytoplasm upon starvation of HeLa cells in Hanks’ buffer saline for 3 h (Fig. 3 A, right). mCherry-FYCO1, stably expressed in HeLa cells, decorated cytosolic puncta (0.3–0.8 µm in diameter) and the rim of cytosolic vesicles (0.8–2 µm in diameter), with partial enrichment of these structures in the periphery of the cell (Fig. 3 B). Costaining and cotransfection experiments identified a partial colocalization of FYCO1 with LC3B, LAMP1, ATG5, and ATG16 (Fig. 3 C). We also observed accumulation of mCherry-tagged p62 and HuntingtinQ68 inside FYCO1-decorated vesicles in untreated cells as well as endogenous p62 inside FYCO1 vesicles after bafilomycin A1 (BafA1) treatment (Fig. 3 C). No colocalization could be seen between FYCO1 and EEA1 or p40phox (Fig. 3 C), which are two markers of early endosomes. These data suggest that FYCO1 decorates autophagosomes or autophagosome-derived structures. Interestingly, FYCO1-positive structures larger than 0.8 µm were always seen as vesicles, with FYCO1 present on the rim but not inside the vesicles, independent of whether acid-sensitive GFP or acid-stable mCherry was used as a fusion tag (Fig. 3 B). In contrast, LC3 was easily detected inside FYCO1 vesicles on structures resembling autophagic bodies (Fig. 3 D). This suggests that FYCO1 localizes to the external but not to the internal membrane of autophagosomes, and upon autophagosome/late endosome (LE)/lysosome fusion, it stays on the external surface of autolysosomes.
Figure 3.
FYCO1 decorates cytosolic vesicles and cytosolic punctuated structures containing the protein markers of autophagosomes/LEs/lysosomes. (A) Endogenous FYCO1 localizes to the cytosolic punctuated structures. HeLa cells cultured in complete growth medium (left) or in HBSS for 3 h (right) were fixed and stained with anti-FYCO1 antibody (green) and Draq5 (blue). (B) Overexpressed mCherry-FYCO1 localizes to the cytosolic punctuated structures and cytosolic vesicles in stably transfected HeLa cells. (C) FYCO1 partially colocalizes with LC3B, LAMP1, ATG5, ATG16L, p62, and HuntingtinQ64 and does not colocalize with EEA1. HeLa cells or mouse embryonic fibroblasts (for ATG16L staining) were transfected with the indicated vectors or stained with the indicated antibodies and imaged 24 h after transfection. For p62 staining, HeLa cells were incubated with 0.2 µM BafA1 for 12 h before fixation. (D) FYCO1 colocalizes with LC3B on the rim but not inside of the vesical structures. HeLa cells cotransfected with GFP-FYCO1, mCherry-LC3B, and myc-p62 were imaged by confocal microscopy 48 h after transfection. (E and F) FYCO1-decorated structures are heterogenous in their luminal content. (E) HeLa cells transfected with CFP-FYCO1 and mChery-YFP-p62 were imaged 48 h after transfection. (F) HeLa cells transfected with GFP-FYCO1 were labeled with LysoTracker red for 60 min and Alexa Fluor 647–dextran (10,000 D) for 4 h in normal growth medium to stain late endocytic compartments (left). Relative number of FYCO1 structures ± SD colocalized with LysoTracker red or Alexa Fluor 647–dextran (10,000 D; right). (B, C, E, and F) Insets show an enlarged field of interest. Bars: (A, B [left], C, E, and F) 10 µm; (B [right] and D) 1 µm.
Measurements of pH status of FYCO1-decorated structures with pH-sensitive tandem fluorescent tag mCherry-YFP (Pankiv et al., 2007) fused to the selective autophagic substrate p62 or labeling with LysoTracker red showed that the majority of FYCO1-positive structures were acidic. (Fig. 3, E and F). Around 60% of them were LysoTracker and dextran (10,000 D) positive, suggesting that they represent the fusion products of late endocytic compartments with autophagosomes (Fig. 3 F).
FYCO1 colocalizes with Rab7 and is located to the limiting membrane of the autophagosomes and LEs
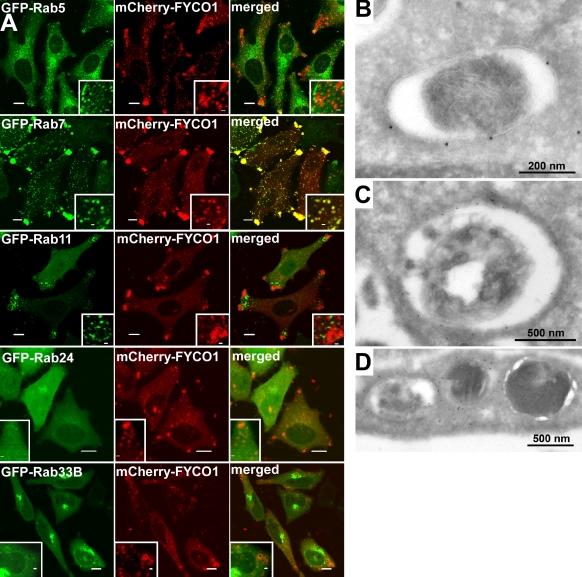
Because FYCO1 is homologous to two groups of Rab effector proteins and resides on the surface of autophagosomes/autolysosomes, we tested whether it colocalizes with Rab family members implicated in autophagy. We found that only Rab7 and not other Rab family proteins (Rab5, -11, -24, and -33B) strongly colocalized with FYCO1 when expressed as wild-type proteins in HeLa cells (Fig. 4 A).
Figure 4.
FYCO1 colocalizes with Rab7 and decorates the limiting membranes of autophagosomes, LEs, and lysosomes. (A) HeLa cells transfected with the indicated constructs were imaged by confocal microscopy 24 h after transfection. Insets show an enlarged field of interest. (B–D) Immunoelectron micrographs of HeLa cells transfected with GFP-FYCO1 and stained with anti-GFP polyclonal antibody (10-nm gold particle). Bars: (A) 10 µm; (A, insets) 1 µm.
To confirm the nature of FYCO1-decorated compartments, we studied the subcellular distribution of GFP-tagged FYCO1 in HeLa cells by immunoelectron microscopy. FYCO1 was detected on single- and double-membrane vesicles 0.2–1.5 µm in diameter with or without luminal content (Fig. 4, B–D). Autophagosome-like double-membrane vesicles were labeled with FYCO1 on external but not on internal membranes (Fig. 4 B), confirming the results obtained by immunofluorescence microscopy.
Membrane recruitment of FYCO1 requires both the FYVE domain and dimerization via its CC domain
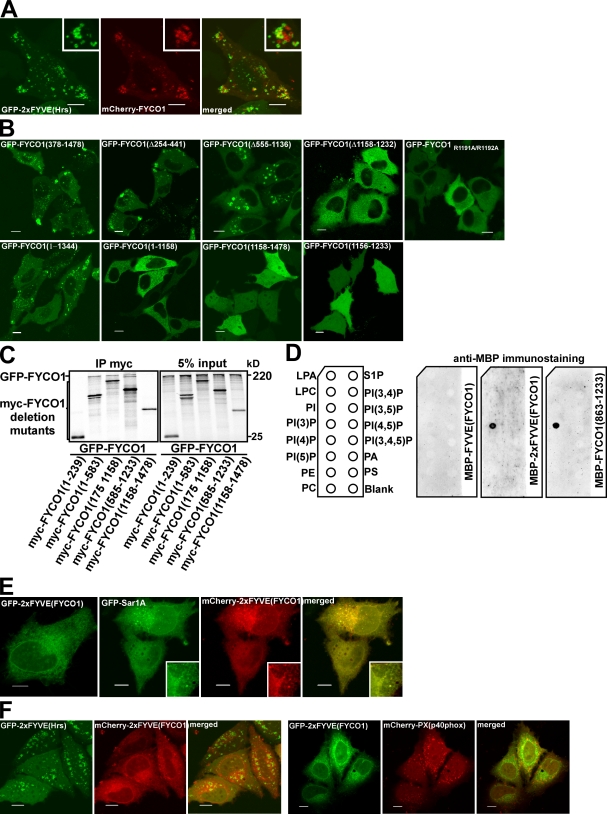
Despite the presence of a predicted PI3P-binding FYVE domain in FYCO1, we observed very little or no colocalization between FYCO1 and two protein probes commonly used as PI3P sensors, a tandem fusion of two FYVE domains from Hrs (2xFYVEHrs) and the PX domain from p40phox (PXp40phox; Fig. 5 A), suggesting that other parts of FYCO1 contribute to membrane recruitment. To map the region involved in membrane recruitment, we analyzed the subcellular distribution of deletion mutants of FYCO1. Only constructs lacking the FYVE domain (Δ1,158–1,232) but not those lacking RUN or GOLD domains, LIR, or the N-terminal part of the CC domain were deficient in vesicle recruitment (Fig. 5 B and Fig. S1). Mutation of the PI3P-binding motif RRHHCR to AAHHCR in the FYVE domain confirmed the importance of the PI3P–FYVE domain interaction for the membrane recruitment of FYCO1 (Fig. 5 B). Interestingly, neither the FYVE domain alone nor the C-terminal fragment of the protein containing FYVE, LIR, and GOLD domains were recruited to intracellular membranes (Fig. 5 B), indicating that the FYVE domain is essential but not sufficient for membrane targeting of FYCO1.
Figure 5.
FYCO1 can dimerize and is recruited to intracellular membranes in an FYVE domain–dependent manner. (A) FYCO1 shows only weak or no colocalization with a PI3P probe 2xFYVEHrs. HeLa cells were imaged 24 h after transfection with the indicated constructs. (B) The FYVE domain of FYCO1 is essential but not sufficient for its recruitment to the cytosolic vesicles. HeLa cells were imaged 24 h after transfection with the indicated constructs. (C) FYCO1 can self-interact in a CC-dependent manner. Full-length GFP-FYCO1 was cotranscribed and cotranslated with the indicated deletion mutants of myc-tagged FYCO1 in rabbit reticulocyte lysate. S35-labeled FYCO1 complexes were immunoprecipitated with anti-myc antibody, separated by SDS-PAGE, and visualized by autoradiography. IP, immunoprecipitation. (D) Dimers but not a monomer of the FYVE domain from FYCO1 can bind PI3P in protein–lipid overlay assay. PIP Strips were incubated with 1-µg/ml solutions of MBP-FYVEFYCO1, MBP-2xFYVEFYCO1, or MBP-FYCO1863–1,233 for 1 h, and bound proteins were detected by immunostaining with anti-MBP antibody. LPA, lysophosphatic acid; LPC, lysophosphocholine; PI, phosphatidylinositol; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine-1-phosphate; PA, phosphatidic acid; PS, phosphatidylserine. (E) The construct containing a tandem fusion of two FYVE domains from FYCO1 (2xFYVEFYCO1) localizes to the nuclear envelope and ER in HeLa cells. HeLa cells were imaged 24 h after transfection with the indicated constructs. (A and E) Insets show an enlarged field of interest. (F) 2xFYVEFYCO1 does not colocalize with 2xFYVEHrs (left) and PXp40phox (right). HeLa cells were imaged 24 h after transfection with the indicated constructs. Bars, 10 µm.
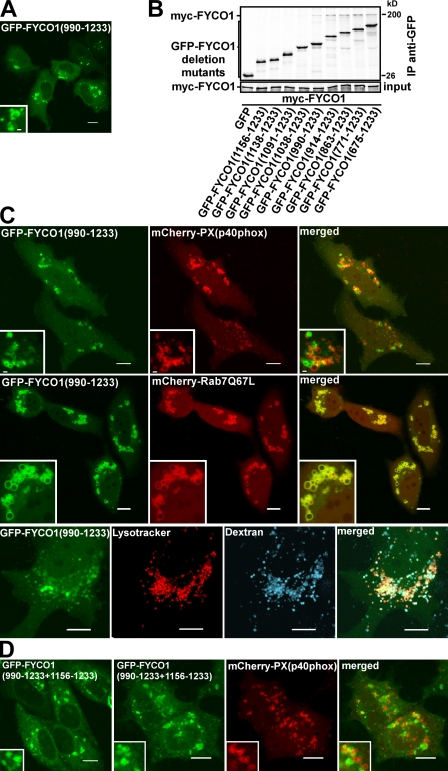
Several reported FYVE domain–containing proteins function as homodimers or contain tandem FYVE domains (Hayakawa et al., 2004; Kutateladze, 2006). Because FYCO1 can self-interact in a CC-dependent manner (Fig. 5 C), we decided to test whether the FYVE domain from FYCO1 also has to be part of a dimer for efficient membrane recruitment. An in vitro protein–lipid overlay assay demonstrated that recombinant MBP-2xFYVEFYCO1 (tandem fusion of two FYVE domains from FYCO1, similar to the 2xFYVE probe from Hrs; Fig. S3; Gillooly et al., 2000) and MBP-FYCO1863–1,233 (single FYVE domain with dimerizing CC) but not MBP-FYCO11,156–1,233 (single FYVE domain) bound to PI3P (Fig. 5 D). Surprisingly, we failed to detect binding of MBP-FYCO1863–1,478 (C-terminal fragment of FYCO1) to any of the lipids by this assay (Fig. S2 A), suggesting that part of the protein C-terminally to the FYVE domain (unstructured region or GOLD domain) has an inhibitory effect on the binding of FYCO1 to lipids. Incubation of MBP-FYCO1863–1,478 with recombinant GST-LC3B partially rescued the PI3P-binding activity of FYCO1 (Fig. S2 A). Interestingly, in cells, the 2xFYVEFYCO1 construct showed a strong staining of the ER and nuclear envelope, as revealed by its colocalization with the ER-resident small GTPase Sar1A (Fig. 5 E). Although 2xFYVEFYCO1 had similar domain borders and the same interdomain linker sequence (Fig. S3 A), it showed no colocalization with 2xFYVEHrs (Fig. 5 F, left). Neither did it colocalize with the PXp40phox probe (Fig. 5 F, right). Inhibition of PI3P synthesis with wortmannin resulted in the partial release of 2xFYVEFYCO1 from the membranes of the nuclear envelope and ER (Fig. S4 A). The retention of 2xFYVEFYCO1 on some membranes was possibly caused by the highly hydrophobic nature of the membrane insertion loop (MIL) in the FYVE domain of FYCO1. Indeed, similar to the RRHHCR motif, the FYCO1-specific MIL was essential for the membrane recruitment of 2xFYVEFYCO1 and could not be substituted with the MIL from EEA1 (Fig. S4 B). Surprisingly, the 2xFYVEFYCO1 construct containing a C-terminal end of FYCO1 after the second FYVE domain (FYCO11,156–1,233+1,156–1,478; Fig. S1) was only weakly recruited to the ER and could be seen together with LC3 in cytosolic punctuated structures (Fig. S4 C). In contrast, FYCO1 deletion constructs containing a dimerizing CC and a single FYVE domain showed a different pattern of subcellular distribution. The FYVE domain alone (FYCO11,156–1,233) as well as the FYVE domain with a short CC (FYCO11,138–1,233 and FYCO11,091–1,233) were diffusely distributed throughout the cytosol and nucleus without recruitment to membranes (Fig. 5 B and see Fig. 7 A, bottom). FYCO11,038–1,233 was recruited to perinuclear vesicles in 30% of transfected cells (see Fig. 7 A, top and middle), whereas FYCO1990–1,233, FYCO1914–1,233, and deletion mutants with a longer CC could be detected on cytosolic vesicles in all transfected cells (Fig. 6 A and see Fig. 8 E). These data correlate well with the in vitro dimerization properties of deletion mutants of FYCO1. FYCO1990–1,233 is the smallest FYVE domain–containing construct that efficiently interacted with full-length FYCO1 in in vitro immunoprecipitation assays (Fig. 6 B), suggesting that the dimer of the FYVE domain from FYCO1 but not a monomer is recruited to cellular membranes. Despite the recruitment to vesicles, FYCO1990–1,233 showed very little or no colocalization with 2xFYVEHrs or PXp40phox constructs (Fig. 6 C, top). At the same time, FYCO1990–1,233 staining completely overlapped with Rab7 (Fig. 6 C, middle) and colocalized with LysoTracker and dextran (Fig. 6 C, bottom), indicating a lysosomal/late endosomal localization of this construct. To confirm that the CC in front of the FYVE domain is responsible for recruitment to LEs/lysosomes, we generated a chimeric 2xFYVEFYCO1 construct containing a CC region of different length in front of the first FYVE domain. As expected, FYCO1990–1,233+1,156–1,233 showed a mixed ER and late endosomal/lysosomal localization but colocalized only weakly with PXp40phox (Fig. 6 D).
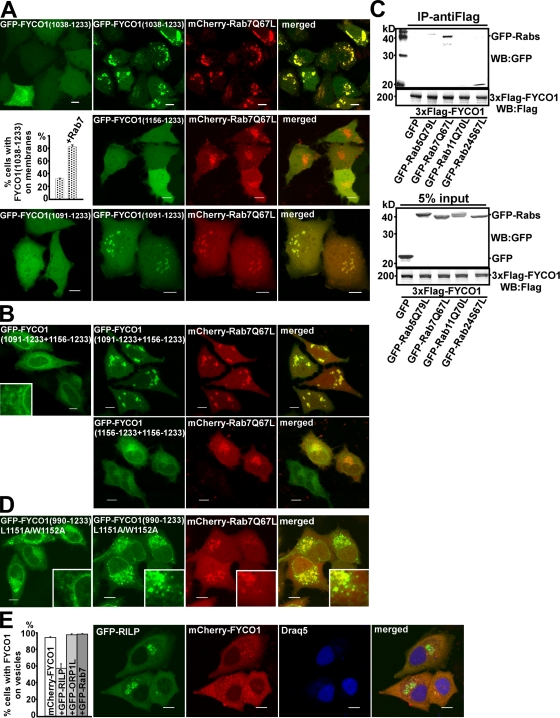
Figure 7.
Rab7 forms a complex with FYCO1 and induces the recruitment of FYCO1 to intracellular membranes. (A) Rab7Q67L recruits FYCO11,038–1,233 and FYCO11,091–1,233 but not FYCO11,156–1,233 to the cytosolic vesicles. HeLa cells transfected with GFP-FYCO11,038–1,233 (top), GFP-FYCO11,091–1,233 (bottom), or GFP-FYCO11,156–1,233 (middle right) with or without mCherry-Rab7Q67L were imaged 24 h after transfection. The graph shows the relative number of cells ± SD with GFP-FYCO11,038–1,233 recruited to intracellular membranes without and with cotransfected mCherry-Rab7Q67L. (B) Rab7Q67L recruits the 2xFYVEFYCO1 construct containing a short CC region in front of the first FYVE domain (FYCO11,091–1,233+1,156–1,233) but not a 2xFYVE alone to the perinuclear vesicles. HeLa cells transfected with GFP-FYCO11,091–1,233+1,156–1,233 or GFP-FYCO11,156–1,233+1,156–1,233 with or without mCherry-Rab7Q67L were imaged 24 h after transfection. (C) Rab7 coimmunoprecipitates with FYCO1 from HEK293 cell lysate. HEK293 cells stably transfected with 3xFlag-FYCO1 were cotransfected with GTP-locked mutants of Rab5, -7, -11, and -24 and lysed 24 h after transfection, and protein complexes were immunoprecipitated with anti–Flag M2 agarose affinity gel. Bound proteins were eluted with Flag peptide, resolved by SDS-PAGE, and visualized by immunoblotting with anti-GFP and anti-Flag antibody. IP, immunoprecipitation; WB, Western blot. (D) Leucine-1151 and tryptophan-1152 are important for the recruitment of FYCO1 to Rab7-containing vesicles. HeLa cells transfected with GFP-FYCO1990–1,233L1151A/W1152A with or without mCherry-Rab7Q67L were imaged 24 h after transfection. (B and D) Insets show an enlarged field of interest. (E) RILP but not Rab7 or ORP1L competes with FYCO1 for recruitment to intracellular vesicles. (right) HeLa cells cotransfected with GFP-RILP together with mCherry-FYCO1 and stained with Draq5 were imaged 24 h after transfection. (left) Relative number of HeLa cells cotransfected with mCherry-FYCO1 and the indicated constructs, in which FYCO1 is recruited to the intracellular vesicles. Error bars represent SD. Bars, 10 µm.
Figure 6.
The FYVE domain from FYCO1 containing the dimerizing CC is recruited to late endosomal/lysosomal structures. (A) FYCO1990–1,233 is the smallest FYVE domain–containing deletion mutant of FYCO1 consistently recruited to cytosolic vesicles. HeLa cells transiently transfected with FYCO1990–1,233 were imaged 24 h after transfection. (B) FYCO1990–1,233 is the smallest FYVE domain–containing deletion mutant of FYCO1 that can efficiently interact with the full-length FYCO1. Full-length myc-FYCO1 was cotranscribed and cotranslated with the indicated deletion mutants of GFP-tagged FYCO1 in rabbit reticulocyte lysate. S35-labeled FYCO1 complexes were immunoprecipitated with anti-myc antibody, separated by SDS-PAGE, and visualized by autoradiography. IP, immunoprecipitation. (C) FYCO1990–1,233 strongly colocalizes with Rab7, LysoTracker red, and Alexa Fluor 647–dextran (10,000 D) but only weakly with PXp40phox. HeLa cells transfected with the indicated constructs or labeled with LysoTracker red for 60 min and Alexa Fluor 647–dextran (10,000 D) for 4 h were imaged 24 h after transfection. (D) FYCO1 construct containing a CC region followed by a tandem repeat of two FYVE domains from FYCO1 (FYCO1990–1,233+1,156–1,233) localizes to the ER and perinuclear vesicles and only weakly colocalizes with PXp40phox. HeLa cells were imaged 24 h after transfection with the indicated constructs. (A, C, and D) Insets show an enlarged field of interest. Bars, 10 µm.
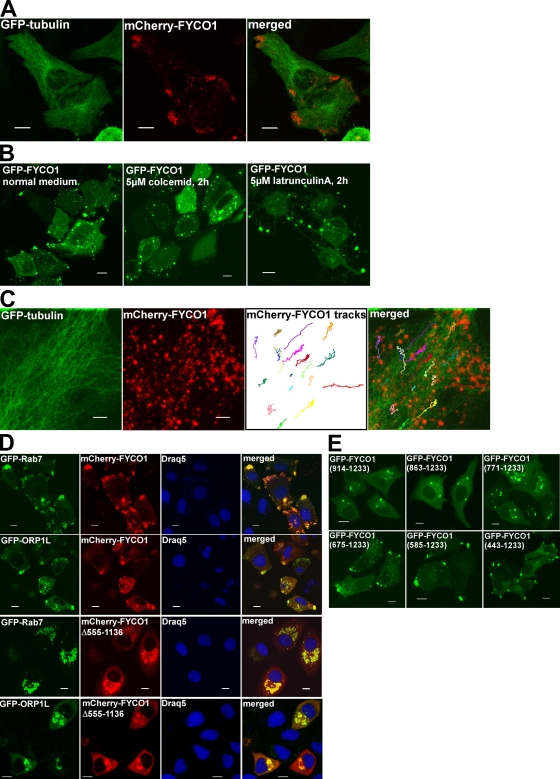
Figure 8.
FYCO1 redistributes Rab7- and ORP1L-decorated vesicles to the MT plus end. (A) Overexpressed FYCO1 is enriched in the cell periphery, close to the MT plus end. HeLa cells were imaged 48 h after transfection with mCherry-FYCO1 and GFP-tubulin. (B) Peripheral enrichment of FYCO1 is dependent on intact MTs. HeLa cells were transfected with GFP-FYCO1 and cultured in normal medium (left) or treated with 5 µM colcemid (middle) or 5 µM latrunculin A (right) for 2 h. (C) FYCO1-decorated vesicles move along the MT tracks. HeLa cells transfected with mCherry-FYCO1 and GFP-tubulin were live imaged by confocal microscopy 48 h after transfection. Movements of FYCO1-decorated structures were tracked with ImageJ MTrackJ plug-in (National Institutes of Health) and presented as 8-min-long tracks in the overlay image. (D) Full-length FYCO1 but not the FYCO1 lacking aa 555–1,136 can redistribute Rab7 and ORP1L to the cell periphery. HeLa cells were transfected with the indicated constructs and stained with DRAQ5. (E) The region of FYCO1 between aa 675 and 771 is essential for the MT plus end translocation of FYCO1-decorated structures. HeLa cells were imaged 24 h after transfection with the indicated constructs. Bars: (A, B, D, and E) 10 µm; (C) 5 µm.
There are several possible explanations for the difference in subcellular distribution of 2xFYVEFYCO1 compared with FYCO1990–1,233. Although both probes bound only to PI3P in the in vitro assay, we cannot exclude the possibility that they have different lipid-binding preferences in vivo. Alternatively, the CC region in front of the FYVE domain could be involved in protein–protein interactions that would target FYCO1 to specific membrane subdomains. Interestingly, both EEA1 and RUFY1 can bind Rab5 or -4 with their CC regions in front of the FYVE domains (Simonsen et al., 1998; Cormont et al., 2001). Multiple sequence alignment of FYCO1 from different species revealed that amino acid conservation in a region preceding the FYVE domain (aa 1,120–1,165) is much higher than in other parts of the CC and comparable to the conservation of the FYVE domain itself (Fig. S3 B), suggesting its important functional role.
Rab7 forms a complex with FYCO1 and induces the recruitment of FYCO1 to intracellular membranes
Because Rab7 strongly colocalizes with FYCO1990–1,233 and full-length FYCO1, we tested whether it can recruit the FYVE domain from FYCO1 onto LEs/lysosomes in a CC-dependent manner. Although FYCO11,038–1,233 was recruited to membranes only in 30% of transfected cells, it colocalized with Rab7Q67L (GTP-locked mutant of Rab7)-decorated vesicles in 80% of cotransfected cells (Fig. 7 A, top and graph). Moreover, although the FYCO11,091–1,233 deletion mutant displayed only diffuse nuclear–cytosolic distribution when expressed alone, it was readily recruited to vesicles upon cotransfection with Rab7Q67L (Fig. 7 A, bottom). No such effect was observed with the FYCO11,156–1,233 construct lacking the CC domain (Fig. 7 A, middle). Similarly, the FYCO11,091–1,233+1,156–1,233 chimeric protein containing a 65-aa-long CC in front of the first FYVE domain of tandem repeat of two FYVE domains displayed an ER/nuclear envelope staining pattern when expressed alone and was redistributed to Rab7Q67L-decorated vesicles upon cotransfection (Fig. 7 B). To confirm that such redistribution is mediated by a CC region and is not FYVE domain specific, we generated chimeric constructs containing a CC region from FYCO1 fused to a tandem of FYVE domains from Hrs (FYCO11–1,160 + 2xFYVEHrs and FYCO1990–1,160 + 2xFYVEHrs). These two fusion proteins but not 2xFYVEHrs were recruited to Rab7Q67L-decorated vesicles (Fig. S5). These data show that Rab7 can indeed recruit the FYVE domain from FYCO1 to late endosomal/lysosomal compartments in a CC-dependent manner. Consistently, Rab7 and FYCO1 were in the same complex and coimmunoprecipitated together from HEK293 cells lysate (Fig. 7 C). FYCO1 interacted preferentially with the GTP-locked Rab7Q67L mutant and not with the GDP-locked Rab7T22N mutant (Fig. S5 A). Moreover, mutation of conserved L1151 and W1152 in the CC region preceding the FYVE domain to alanines led to the dramatic redistribution of the FYCO1990–1,233 construct to the nuclear envelope and ER in a pattern similar to the subcellular distribution of 2xFYVEFYCO1 and strongly reduced its colocalization with Rab7 (Fig. 7 D).
RILP (Rab-interacting lysosomal protein) and ORP1L (oxysterol-binding protein–related protein 1 long) are two well-known Rab7 effector proteins. They can bind simultaneously to Rab7 and regulate the MT minus end–directed transport of LEs/lysosomes (Johansson et al., 2007). Interestingly, overexpression of RILP but not ORP1L or Rab7 resulted in a strong decrease in the number of FYCO1-decorated vesicles in cells and a decrease in the number of cells with FYCO1 vesicles (Fig. 7 E), suggesting a competition between RILP and FYCO1 for binding to Rab7.
FYCO1 redistributes Rab7- and ORP1L-decorated vesicles to the MT plus end
Interestingly, although endogenous FYCO1 was stained in punctuated structures in the juxtanuclear region or throughout the cytosol, overexpressed FYCO1 was enriched in cytosolic dots and vesicles at the cell periphery, close to the MT plus end (Fig. 8 A). This localization of overexpressed protein was dependent on intact MTs. Treatment of cells with the MT-depolymerizing agents colcemid and vinblastine led to the redistribution of FYCO1 vesicles from the cellular periphery to clusters throughout the cytosol (Fig. 8 B), whereas actin-depolymerizing agents latrunculin A and cytochalasin B preserved FYCO1 vesicles in distinct peripheral cellular protrusions (Fig. 8 B). Movement of FYCO1 structures along the MT tracks was also detected by live cell imaging of HeLa cells transfected with GFP-tubulin and mCherry-FYCO1 (Fig. 8 C and Video 1). Overexpressed FYCO1 was not only enriched close to the MT plus ends itself, but it also redistributed LC3B (Fig. 2 C), Rab7, and ORP1L to this location (Fig. 8 D). There are several possible explanations for this phenotype. Overexpressed FYCO1 can compete with endogenous RILP for binding to Rab7, uncoupling the MT minus end–directed transport. Alternatively, FYCO1 can actively participate in anterograde MT transport by linking Rab7 to kinesin motor proteins. The first hypothesis implies that any deletion mutant of FYCO1 that colocalizes with Rab7 should promote the redistribution of Rab7 vesicles to MT plus ends. This was not the case for small constructs such as FYCO1990–1,233 (Fig. 6 A) and FYCO1914–1,233 (Fig. 8 E), which, together with Rab7, decorated perinuclear vesicles (Fig. 6 C). To map the region of FYCO1 important for the MT plus end transport, we analyzed the subcellular distribution of deletion mutants of FYCO1. RUN and GOLD domains as well as the N-terminal part of the CC were not essential for the FYCO1 overexpression phenotype. A FYCO1 mutant lacking the CC domain between aa 555 and 1,136 was localized to perinuclear vesicles instead of cellular periphery (Fig. 5 B) and was not able to redistribute Rab7 or ORP1L to a peripheral location (Fig. 8 D). To increase the mapping accuracy, we compared the subcellular distribution of FYCO1 deletion mutants containing only a CC domain of different length, followed by the FYVE domain. Although FYCO1914–1,233, FYCO1863–1,233, and FYCO1771–1,233 were localized to the perinuclear vesicles, similar to FYCO1Δ555–1,136 (Fig. 8 E), extension of the CC from aa 771 to 675 in the FYCO1675–1,233 construct resulted in the dramatic redistribution of the FYCO1 deletion mutant to the cell periphery (Fig. 8 E). A similar phenotype of peripherally enriched FYCO1 vesicles was also observed with FYCO1586–1,233 and FYCO1483–1,233 constructs (Fig. 8 E). Based on this data, we can conclude that the region of FYCO1 between aa 675 and 771 contains a determinant, likely a kinesin-binding site, for MT plus end targeting of FYCO1.
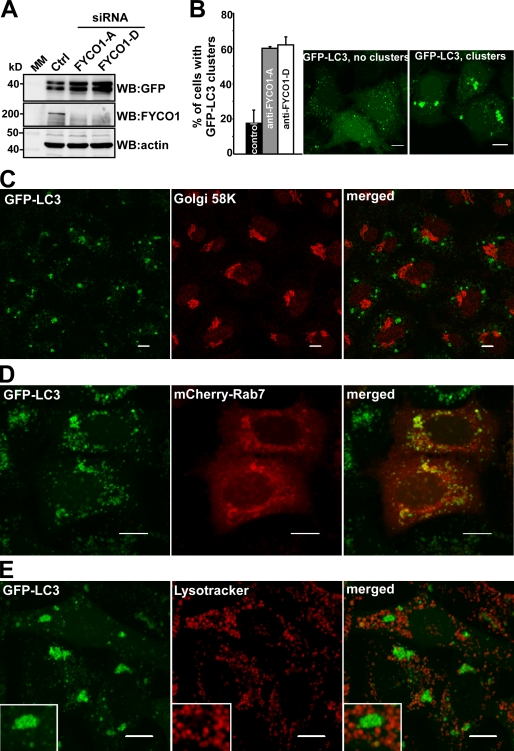
Depletion of FYCO1 leads to the accumulation of perinuclear clustering of autophagosomes
To further elucidate the function of FYCO1, we tested the effect of its siRNA-mediated knockdown in HeLa cells stably transfected with GFP-LC3. Depletion of endogenous FYCO1 with two different siRNAs resulted in the accumulation of GFP-LC3 (Fig. 9 A), primarily in the form of perinuclear and peri-Golgi clusters of GFP-LC3–positive vesicles and membranes (Fig. 9, B and C). These vesicles colocalized with mCherry-Rab7 (Fig. 9 D) but did not stain positive with LysoTracker (Fig. 9 E), suggesting that they represent early autophagosomes or phagophores.
Figure 9.
Depletion of FYCO1 leads to the accumulation of perinuclear clusters of autophagosomes. (A) Western blot (WB) of cell lysates from HeLa cells stably expressing GFP-LC3 48 h after transient transfection with siRNA against FYCO1 (siRNAs: A, Applied Biosystems; D, Thermo Fisher Scientific). MM, molecular mass. (B) Representative images of HeLa cells with or without the perinuclear clusters of GFP-LC3–positive vesicles (right) and quantification of the number of cells with this phenotype 48 h after transient transfection with siRNAs against FYCO1 (left). Error bars represent SDs based on three independent experiments. (C, D, and E) The perinuclear clusters of GFP-LC3–positive vesicles colocalize with mCherry-Rab7 but not with Golgi protein marker 58K or LysoTracker red. GFP-LC3 HeLa cells were transiently transfected with siRNA against FYCO1 (Thermo Fisher Scientific) 48 h before imaging or cotransfected with mCherry-Rab7. Cells were either fixed and stained with anti-Golgi 58K antibody or incubated with 50 nM LysoTracker red for 1 h. Insets show an enlarged field of interest. Bars, 10 µm.
Discussion
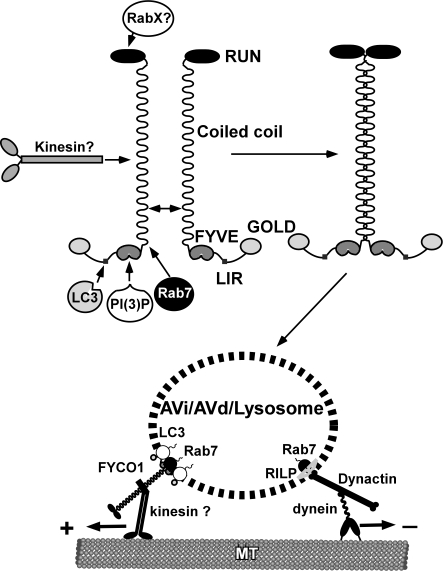
The evolutionary expansion of ATG8 family proteins in mammals and plants implies a more versatile and diverse functions for these proteins in higher eukaryotes compared with yeast. The growing number of identified interaction partners (Wang et al., 1999; Okazaki et al., 2000; Sagiv et al., 2000; Kim et al., 2002; Mansuy et al., 2004; Mohrlüder et al., 2007a,b; Pankiv et al., 2007; Cook et al., 2008; Kirkin et al., 2009; Nowak et al., 2009; Schwarten et al., 2009; Zhang et al., 2009) suggests that similar to Rabs, ATG8 family proteins may function as adaptors, linking autophagosomal membranes to downstream effector proteins. Our results suggest that FYCO1, the novel interaction partner of LC3 identified in this study, functions as an effector of both LC3 and Rab7. It resides on the external surface of autophagosomes/LEs/lysosomes and promotes MT plus end–directed transport of these membranous compartments (Fig. 10). FYCO1 contains an N-terminal RUN domain, a central CC region, and a C-terminal cluster of globular domains. The RUN domain of FYCO1 is likely to bind Rab or Rap family GTPases (Kukimoto-Niino et al., 2006; Recacha et al., 2009). The central CC region has several important functions. First of all, if fully extended, it spatially separates N- and C-terminal globular domains of FYCO1 a distance of ∼120–130 nm (predicted length of the CC of 850 nm). This enables FYCO1 to interact with two spatially distant protein–protein or protein–lipid complexes and connect these complexes or associated membranous compartments into a single functional unit. Second, the CC region is responsible for the dimerization of FYCO1. Finally, the CC of FYCO1 has several protein-binding regions. The central part between aa 675 and 771 contains a determinant, likely a kinesin-binding site, for the targeting of FYCO1 to the MT plus end. The C-terminal part of the CC is responsible for the colocalization of FYCO1 with Rab7.
Figure 10.
Proposed model of FYCO1 function in the MT-dependent transport of autophagic vesicles.
The subcellular distribution of the 2xFYVEFYCO1 and FYCO1990–1,233L1151A/W1152A constructs suggests that the FYVE domain of FYCO1 has intrinsic affinity for ER membranes. Recent studies show that in addition to PI3P binding, several other protein–lipid interactions may determine the subcellular localization of the FYVE domains (Dumas et al., 2001; Kutateladze et al., 2004).
In addition to lipid binding, the subcellular distribution of full-length FYCO1 is fine-tuned by protein–protein interactions mediated by regions adjacent to the FYVE domain. C-terminally to the FYVE domain, an ∼100-aa-long acidic region lacking regular secondary structure connects the globular FYVE and GOLD domains and possibly forms a flexible acidic loop (Fig. S2 C). Because this region of FYCO1 showed an inhibitory effect on in vitro PI3P binding and reduced the recruitment of FYVE domain–containing mutants to intracellular membranes (Fig. S2 B), we speculate that it can fold onto the basic lipid interaction surface of the FYVE domain and maintain FYCO1 in a lipid-unbound state. Interestingly, the middle part of this unstructured region contains an LIR and can bind LC3. Recombinant LC3B partially rescues the PI3P-binding properties of FYCO1863–1,478 in vitro, suggesting that the binding of LC3 to LIR can unfold the acidic loop from the lipid-binding surface and prime FYCO1 for the membrane binding (Fig. S2 D). This would ensure binding of FYCO1 only to membranes containing LC3 on their surface.
The second protein–protein interaction determining the subcellular localization of FYCO1 is mediated by Rab7 and the conserved part of the CC region of FYCO1 directly adjacent to the N-terminal end of the FYVE domain. We suggest that, similar to RILP-Rab7 and GCC185-Rab6 structures (Wu et al., 2005; Burguete et al., 2008), FYCO1 and Rab7 may form a heterotetrameric complex with the central parallel CC of FYCO1 binding two molecules of Rab7.
Cytosolic dynein1 has previously been suggested as an MT minus end–directed molecular motor for both autophagosomes and LEs/lysosomes (Johansson et al., 2007; Kimura et al., 2008). RILP and ORP1L bound to Rab7 are responsible for the recruitment of the dynactin–dynein complex to the surface of LEs/lysosomes (Johansson et al., 2007), whereas little is known about the adaptor complex that links autophagosomes to microtubular molecular motors. Competition between FYCO1–kinesin and RILP–dynactin–dynein1 complexes for Rab7 binding and recruitment to LEs/lysosomes is a plausible mechanism for reciprocal regulation of the number of molecular motors with opposite direction specificity on the surface of these vesicles.
Based on our experimental data, we suggest the following model of FYCO1 function. At sufficient nutrient supply, FYCO1 preferentially resides on the membranes of perinuclear ER in a conformation that prevents its binding to kinesins. Upon amino acid starvation, it binds to the MT plus end–directed motors and redistributes preautophagosomal membranous compartments to the sites of autophagosome formation throughout the cytosol. After the formation of autophagosomes, FYCO1 competes with the dynein recruitment complex for binding to Rab7 and membrane recruitment and provides regulated bidirectional transport of autophagosomes along the MT track.
We identified a hypothetical FYCO1 orthologue in the genome of the morphologically simplest multicellular metazoan Trichoplax adhaerens (NCBI accession no. XP_002109478.1) but failed to find orthologues in the single-cell choanoflagellate Monosiga brevicollis or yeasts. Interestingly, the dependence of autophagy on the MT cytoskeleton seems also to be restricted to metazoans. Although an intact MT network is required for starvation-induced autophagy in mammalian cells (Fass et al., 2006; Köchl et al., 2006), no such dependence was observed in Saccharomyces cerevisiae (Kirisako et al., 1999). Multiple studies suggest that the actin cytoskeleton rather than MTs is important for several types of autophagy in yeasts (Reggiori et al., 2005; Monastyrska et al., 2008).
FYCO1, similar to EEA1 and RUFY proteins, has two predicted Rab-binding sites flanking the CC region, potentially enabling binding to Rab proteins located on different vesicles tethering these together. Indeed, overexpressed FYCO1 is localized onto the surface of LEs/lysosomes of up to 2 µm in diameter in HeLa cells and up to 15 µm in HEK293 cells, suggesting that it promotes the fusion and enlargement of these compartments. The nature of the compartments that undergo fusion is presently unknown. Despite tight clustering of FYCO1-decorated vesicles, we observed very few homotypic fusions between these structures, suggesting that FYCO1 does not facilitate the homotypic fusion of autophagosomes/LEs/lysosomes. In conclusion, in this study, we identified FYCO1, a novel protein harboring a long CC, binding to PI3P via its FYVE domain and acting as an LC3 and Rab7 effector protein with MT plus end–directed transport and possible membrane-tethering functions.
Materials and methods
Antibodies and reagents
The following antibodies were used: anti-p62 C-terminal guinea pig polyclonal antibody (Progen Biotechnik GmbH); anti-EEA1 mouse monoclonal antibodies (BD); anti-GST rabbit polyclonal and anti–c-myc mouse monoclonal antibodies (Santa Cruz Biotechnology, Inc.); anti-LC3 rabbit polyclonal antibody (Cell Signaling Technology); anti-GFP rabbit polyclonal antibody (Abcam); anti-MBP mouse monoclonal antibody (Sigma-Aldrich); anti-FYCO1 mouse polyclonal antibody (Abnova); anti-Atg16L rabbit polyclonal antibody (Cosmo Bio Co., Ltd.); anti-LAMP1 rabbit polyclonal antibody (a gift from H. Stenmark, Norwegian Radium Hospital, Oslo, Norway); and HRP-conjugated anti–mouse and anti–rabbit polyclonal antibody (BD). The following fluorescent secondary antibodies were used: Alexa Fluor 488–conjugated goat anti–mouse and goat anti–rabbit IgG, Alexa Fluor 555–conjugated goat anti–mouse and goat anti–guinea pig IgG, Alexa Fluor 633–conjugated goat anti–guinea pig IgG, and Alexa Fluor 680–conjugated goat anti–mouse IgG (Invitrogen); and IRDye800-conjugated goat anti–rabbit antibody (Rockland Immunochemicals, Inc.). LysoTracker red and Alexa Fluor 647–dextran (10,000 D) were obtained from Invitrogen. BafA1, cytochalasin B, and vinblastine were purchased from Sigma-Aldrich. Colcemid and latrunculin B were obtained from EMD. DRAQ5 was purchased from Biostatus Limited. l-[35S]methionine was obtained from PerkinElmer. PIP Strips were purchased from Echelon Biosciences, Inc.
Cell culture and transfection
HeLa cells were grown in Eagle’s minimum essential medium supplemented with 10% fetal calf serum, nonessential amino acids, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen). HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and the aforementioned antibiotics. Subconfluent HeLa and HEK293 cells were transfected using Metafectene Pro (Biontex Laboratories GmbH). Stable mCherry-FYCO1–transfected HeLa cells were selected using G418, and mCherry-positive cells were sorted on FACSAria (BD). Stable 3xFlag-FYCO1– and GFP-FYCO1–transfected HEK293 cells were generated using the FlpIn recombination system (Invitrogen) according to the manufacturer’s protocol. Two different siRNAs used in this study (Applied Biosystems Silencer Predesigned siRNA with the sequence 5′-GGGCCGGCAAGAACCUAUUTT-3′ and Thermo Fisher Scientific ON-TARGET plus SMARTpool FYCO1) were transfected using Lipofectamine RNAiMAX at a final concentration of 20 nM twice with 24-h intervals.
Plasmid constructs
Plasmids used in this study are listed in Tables S1 and S2 and the supplemental text. All constructs were made either by conventional cloning or by Gateway recombination cloning. Point mutants were made using the QuikChange site-directed mutagenesis kit (Agilent Technologies). Oligonucleotides for mutagenesis, PCR, and DNA sequencing reactions were obtained from Operon. All plasmid constructs were verified by sequencing (BigDye; Applied Biosystems) and/or restriction digestion. The schematic structure of some of the deletion mutants of FYCO1 used in this study is depicted in Fig. S1.
Confocal microscopy analyses
Cells grown on 8-well coverglass slides (Nunc) were directly examined under the microscope or fixed for 10 min in 4% paraformaldehyde, washed with PBS, permeabilized with 40 µg/ml digitonin in PBS for 5 min at room temperature, and blocked with 3% serum in PBS for 30 min. Subsequently, cells were incubated at room temperature with primary and secondary antibodies for 30 min each (Lamark et al., 2003). Live cells were placed in Hanks’ medium with or without amino acids and serum at 37°C and imaged for up to 1 h. Images were collected using a microscope (Axiovert 200; Carl Zeiss, Inc.) with a 40× 1.2 W C-Apochromat objective equipped with an LSM510-META confocal module using the LSM 5 software version 3.2 (Carl Zeiss Inc.) or a confocal microscope (TCS SP5; Leica) with a 60× 1.2 W objective equipped with incubation chamber with CO2 and temperature control. Images were processed using Canvas version 9 (ACD Systems).
Immunoprecipitations and immunoblots
For in vivo coimmunoprecipitation experiments, cells were lysed 24 h after transfection in HA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1 mM EGTA, 1% Triton X-100) with phosphatase inhibitor mixture set II (EMD) and Complete Mini EDTA-free protease inhibitor mixture (Roche). Cell lysates were incubated with 1 µl of anti-GFP antibodies (ab290; Abcam) overnight at 4°C followed by a 1-h incubation with 15 µl of protein A–Sepharose and washed six times with 1 ml of HA buffer. Immunoprecipitated proteins were eluted by boiling for 5 min in 20 µl of 2× SDS-PAGE loading buffer. For immunoprecipitation of 3xFlag-FYCO1, cell lysates (lysis buffer: 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, and 1% Triton X-100) were incubated with 15 µl of anti–Flag M2 agarose (Sigma-Aldrich) for 1 h at 4°C and washed six times with 1 ml of lysis buffer, and bound proteins were eluted with 0.2 mg/ml of Flag peptide (Sigma-Aldrich). Coimmunoprecipitated proteins were resolved by SDS-PAGE, transferred to nitrocellulose membrane (GE Healthcare), and detected with antigen-specific primary antibodies followed by HRP-conjugated secondary antibodies. For in vitro coimmunoprecipitation experiments, 0.5 µg of expression vectors for GFP- and myc-tagged proteins were in vitro cotranscribed/cotranslated in a total volume of 25 µl using the TNT T7 coupled reticulocyte lysate system (Promega) according to the manufacturer’s protocol. 20 µl of in vitro translated 35S-labeled proteins were diluted in 200 µl of ice-cold NETN-E buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 6 mM EDTA, 6 mM EGTA, 0.5% Nonidet P-40, and 1 mM dithiothreitol supplemented with Complete Mini EDTA-free protease inhibitor cocktail) and incubated for 1 h at 4°C with 1 µl anti-GFP (Abcam) or anti-myc (Santa Cruz Biotechnology, Inc.) antibodies, followed by incubation with 15 µl of protein A–Sepharose and washing six times with 1 ml of NETN-E buffer. Bound proteins were eluted by boiling for 5 min in 20 µl of 2× Laemmli buffer and resolved by SDS-PAGE. Protein gels were vacuum dried, and 35S-labeled proteins were detected on a bioimaging analyzer (BAS-5000; Fujifilm).
Protein–lipid overlay assay
PIP Strips were blocked in 1% nonfat dry milk solution for 1 h, incubated with a 1-µg/ml solution of the indicated MBP fusion protein for 1 h at room temperature, and immunostained with anti-MBP antibody.
GST pull-down assay and GST-LC3B affinity purification
All GST-tagged proteins were expressed in Escherichia coli BL21(DE3)pLysS and purified on glutathione–Sepharose 4 Fast Flow beads (GE Healthcare). MBP fusion proteins were expressed in E. coli DH5-α and purified on amylose resin (New England Biolabs, Inc.). 35S-labeled GFP- or myc-tagged proteins were cotranscribed/translated in vitro using the TNT T7 coupled reticulocyte lysate system. For GST pull-downs with MBP-tagged FYCO1863–1,478, 1–2 µg GST-LC3B was incubated with 1–2 µg MBP-FYCO1863–1,478 in 800 µl of NETN-E buffer for 1 h at 4°C and then washed five times with 1 ml of NETN-E buffer. For GST pull-downs with 35S-labeled proteins, in vitro translation reaction products from 0.5 µg of plasmid were incubated with 1–2 µg GST or GST-tagged proteins in 300 µl of NETN-E buffer for 1 h at 4°C, washed six times with 1 ml of NETN-E buffer, boiled with 2× SDS gel loading buffer, and subjected to SDS-PAGE. For GST pull-downs with 35S-labeled proteins, gels were stained with Coomassie blue and vacuum dried. 35S-labeled proteins were detected on a BAS-5000 bioimaging analyzer. For GST pull-downs with MBP-tagged protein, SDS-PAGE–resolved proteins were transferred to nitrocellulose membrane and detected by immunoblotting with the indicated antibodies. For affinity purification of proteins interacting with GST-LC3B, 5 µg of GST-LC3B bound to glutathione–Sepharose beads was incubated with HeLa cell lysate from ∼108 cells in HA-lysis buffer for 3 h at 4°C. Bound proteins were isolated by centrifugation of Sepharose beads at 13,000 rpm for 30 s, washed six times with 1 ml of HA buffer, eluted by boiling with 2× SDS gel loading buffer, and subjected to SDS-PAGE. Resolved proteins were detected by staining of gels with Imperial protein stain (Thermo Fisher Scientific) and mass spectrometry or by immunoblotting with anti-FYCO1 antibody (Abnova).
Mass spectrometry
Gel bands were excised and subjected to in-gel reduction, alkylation, and tryptic digestion using 2–10 ng/µl trypsin (V511A; Promega; Shevchenko et al., 1996). Peptide mixtures containing 0.1% formic acid were loaded onto a nanoACQUITY UltraPerformance LC (Waters), containing a 3-µm Symmetry C18 Trap column (180 µm × 22 mm; Waters) in front of a 3-µm Atlantis C18 analytical column (100 µm × 100 mm; Waters). Peptides were separated with a gradient of 5–95% acetonitrile, 0.1% formic acid, with a flow of 0.4 µl/min eluted to a Q-TOF Ultima Global mass spectrometer (Micromass/Waters) and subjected to data-dependent tandem mass spectrometry analysis. Peak lists were generated by the ProteinLynx Global server software (version 2.1; Waters). The resulting pkl files were searched against the Swiss-Prot 51.6 protein sequence databases using an in-house Mascot server (Matrix Sciences). Peptide mass tolerances used in the search were 50 ppm, and fragment mass tolerance was 0.1 D.
Electron microscopy
HeLa cells were fixed in a Petri dish with 4% formaldehyde/0.1% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, for 2 h at room temperature. After washing thrice in phosphate buffer, the cells were scraped off the dish in 1% gelatin/PBS, pelleted, and embedded in 10% gelatin/PBS (Peters et al., 1991). After infusion with 2.3 M sucrose for 1 h, small blocks were cut and mounted on specimen holders and frozen in liquid nitrogen. 70–100-nm cryosections were cut at −110°C (Leica UTC with FCS), collected with a 1:1 mixture of 2% methyl cellulose/2.3 M sucrose, transferred to formvar/carbon-coated grids, and labeled with primary antibodies followed by protein A–gold conjugates essentially as described by Slot et al. (1991). After embedding in 2% methyl cellulose/0.4% uranyl acetate, we observed sections at 60–80 kV in an electron microscope (JEM 1230; JEOL). Images were recorded with a digital camera (Morda; SIS) and further processed with Photoshop software (Adobe).
Online supplemental material
Fig. S1 lists the deletion mutants of FYCO1. Fig. S2 shows data and a model of how LC3 binding may regulate the membrane recruitment of FYCO1. Fig. S3 shows the alignment of 2xFYVE constructs and the CC and FYVE domains of FYCO1. Fig. S4 shows the subcellular distribution of the point mutants of 2xFYVEFYCO1 or the wild-type 2xFYVEFYCO1 after wortmannin treatment and colocalization of LC3B and FYCO11,156–1,233+1,156–1,478. Fig. S5 shows that FYCO1 preferentially interacts with the GTP-locked mutant of Rab7 and that the Rab7 recruitment property of the CC can be transferred to 2xFYVEHrs. Video 1 shows the movement of mCherry-FYCO1–positive vesicles along the MTs. Table S1 lists the plasmids used in this study. Table S2 lists descriptions of some of the cDNA constructs made by traditional subcloning or site-directed mutagenesis in this study. The supplemental text lists other cDNA constructs made by traditional subcloning or site-directed mutagenesis and Gateway LR reactions in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200907015/DC1.
Supplementary Material
Acknowledgments
We are grateful to Vesa M. Olkkonen and Harald Stenmark for generous gifts of pEGFP-ORP1L and pcDNA4HisMax-RILP and of anti-LAMP1 antibody and pEGFP-2xFYVEHrs, respectively. We thank the BioImaging and Proteomics Functional Genomics (FUGE) core facilities at the Institute of Medical Biology for use of instrumentation and expert assistance.
This work was supported by grants from the FUGE program of the Norwegian Research Council, the Norwegian Cancer Society, and the Blix Foundation to T. Johansen. S. Pankiv is a fellow of the Norwegian Cancer Society.
Footnotes
Abbreviations used in this paper:
- BafA1
- bafilomycin A1
- CC
- coiled-coil
- LE
- late endosome
- LIR
- LC3-interacting region
- MBP
- maltose-binding protein
- MIL
- membrane insertion loop
- MT
- microtubule
- PI3P
- phosphatidylinositol-3-phosphate
- RILP
- Rab-interacting lysosomal protein
References
- Blommaart E.F., Krause U., Schellens J.P., Vreeling-Sindelárová H., Meijer A.J.. 1997. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 243:240–246. 10.1111/j.1432-1033.1997.0240a.x [DOI] [PubMed] [Google Scholar]
- Burguete A.S., Fenn T.D., Brunger A.T., Pfeffer S.R.. 2008. Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell. 132:286–298. 10.1016/j.cell.2007.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I., de Gunzburg J., Goud B., Mornon J.P.. 2001. RUN domains: a new family of domains involved in Ras-like GTPase signaling. Trends Biochem. Sci. 26:79–83. 10.1016/S0968-0004(00)01730-8 [DOI] [PubMed] [Google Scholar]
- Cook J.L., Re R.N., deHaro D.L., Abadie J.M., Peters M., Alam J.. 2008. The trafficking protein GABARAP binds to and enhances plasma membrane expression and function of the angiotensin II type 1 receptor. Circ. Res. 102:1539–1547. 10.1161/CIRCRESAHA.108.176594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormont M., Mari M., Galmiche A., Hofman P., Le Marchand-Brustel Y.. 2001. A FYVE-finger-containing protein, Rabip4, is a Rab4 effector involved in early endosomal traffic. Proc. Natl. Acad. Sci. USA. 98:1637–1642. 10.1073/pnas.031586998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas J.J., Merithew E., Sudharshan E., Rajamani D., Hayes S., Lawe D., Corvera S., Lambright D.G.. 2001. Multivalent endosome targeting by homodimeric EEA1. Mol. Cell. 8:947–958. 10.1016/S1097-2765(01)00385-9 [DOI] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D., Furlán M., Colombo M.I.. 2008. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 9:230–250. [DOI] [PubMed] [Google Scholar]
- Fass E., Shvets E., Degani I., Hirschberg K., Elazar Z.. 2006. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J. Biol. Chem. 281:36303–36316. 10.1074/jbc.M607031200 [DOI] [PubMed] [Google Scholar]
- Gaullier J.M., Simonsen A., D’Arrigo A., Bremnes B., Stenmark H., Aasland R.. 1998. FYVE fingers bind PtdIns(3)P. Nature. 394:432–433. 10.1038/28767 [DOI] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., Stenmark H.. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588. 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M.G., Munafó D.B., Berón W., Colombo M.I.. 2004. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 117:2687–2697. 10.1242/jcs.01114 [DOI] [PubMed] [Google Scholar]
- Hayakawa A., Hayes S.J., Lawe D.C., Sudharshan E., Tuft R., Fogarty K., Lambright D., Corvera S.. 2004. Structural basis for endosomal targeting by FYVE domains. J. Biol. Chem. 279:5958–5966. 10.1074/jbc.M310503200 [DOI] [PubMed] [Google Scholar]
- Hirota Y., Tanaka Y.. 2009. A small GTPase, human Rab32, is required for the formation of autophagic vacuoles under basal conditions. Cell. Mol. Life Sci. 66:2913–2932. 10.1007/s00018-009-0080-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Fujita N., Kanno E., Yamamoto A., Yoshimori T., Fukuda M.. 2008. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol. Biol. Cell. 19:2916–2925. 10.1091/mbc.E07-12-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., Eskelinen E.L.. 2004. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 117:4837–4848. 10.1242/jcs.01370 [DOI] [PubMed] [Google Scholar]
- Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V.M., Neefjes J.. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7–RILP–p150Glued, ORP1L, and the receptor βlll spectrin. J. Cell Biol. 176:459–471. 10.1083/jcb.200606077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T.. 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117:2805–2812. 10.1242/jcs.01131 [DOI] [PubMed] [Google Scholar]
- Kim J., Huang W.P., Stromhaug P.E., Klionsky D.J.. 2002. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277:763–773. 10.1074/jbc.M109134200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T.. 2008. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct. Funct. 33:109–122. 10.1247/csf.08005 [DOI] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y.. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435–446. 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y.. 2000. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151:263–276. 10.1083/jcb.151.2.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33:505–516. 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Kiss H., Yang Y., Kiss C., Andersson K., Klein G., Imreh S., Dumanski J.P.. 2002. The transcriptional map of the common eliminated region 1 (C3CER1) in 3p21.3. Eur. J. Hum. Genet. 10:52–61. 10.1038/sj.ejhg.5200758 [DOI] [PubMed] [Google Scholar]
- Köchl R., Hu X.W., Chan E.Y., Tooze S.A.. 2006. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 7:129–145. 10.1111/j.1600-0854.2005.00368.x [DOI] [PubMed] [Google Scholar]
- Kukimoto-Niino M., Takagi T., Akasaka R., Murayama K., Uchikubo-Kamo T., Terada T., Inoue M., Watanabe S., Tanaka A., Hayashizaki Y., et al. 2006. Crystal structure of the RUN domain of the RAP2-interacting protein x. J. Biol. Chem. 281:31843–31853. 10.1074/jbc.M604960200 [DOI] [PubMed] [Google Scholar]
- Kutateladze T.G. 2006. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim. Biophys. Acta. 1761:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze T.G., Capelluto D.G., Ferguson C.G., Cheever M.L., Kutateladze A.G., Prestwich G.D., Overduin M.. 2004. Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 279:3050–3057. 10.1074/jbc.M309007200 [DOI] [PubMed] [Google Scholar]
- Lamark T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G., Johansen T.. 2003. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278:34568–34581. 10.1074/jbc.M303221200 [DOI] [PubMed] [Google Scholar]
- Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J.U.. 2008. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10:776–787. 10.1038/ncb1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J.. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Mansuy V., Boireau W., Fraichard A., Schlick J.L., Jouvenot M., Delage-Mourroux R.. 2004. GEC1, a protein related to GABARAP, interacts with tubulin and GABA(A) receptor. Biochem. Biophys. Res. Commun. 325:639–648. 10.1016/j.bbrc.2004.10.072 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., Gwadz M., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210. 10.1093/nar/gkn845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873. 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Mohrlüder J., Hoffmann Y., Stangler T., Hänel K., Willbold D.. 2007a. Identification of clathrin heavy chain as a direct interaction partner for the gamma-aminobutyric acid type A receptor associated protein. Biochemistry. 46:14537–14543. 10.1021/bi7018145 [DOI] [PubMed] [Google Scholar]
- Mohrlüder J., Stangler T., Hoffmann Y., Wiesehan K., Mataruga A., Willbold D.. 2007b. Identification of calreticulin as a ligand of GABARAP by phage display screening of a peptide library. FEBS J. 274:5543–5555. 10.1111/j.1742-4658.2007.06073.x [DOI] [PubMed] [Google Scholar]
- Monastyrska I., He C., Geng J., Hoppe A.D., Li Z., Klionsky D.J.. 2008. Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell. 19:1962–1975. 10.1091/mbc.E07-09-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafó D.B., Colombo M.I.. 2002. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 3:472–482. 10.1034/j.1600-0854.2002.30704.x [DOI] [PubMed] [Google Scholar]
- Nowak J., Archange C., Tardivel-Lacombe J., Pontarotti P., Pébusque M.J., Vaccaro M.I., Velasco G., Dagorn J.C., Iovanna J.L.. 2009. The TP53INP2 protein is required for autophagy in mammalian cells. Mol. Biol. Cell. 20:870–881. 10.1091/mbc.E08-07-0671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki N., Yan J., Yuasa S., Ueno T., Kominami E., Masuho Y., Koga H., Muramatsu M.. 2000. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res. Mol. Brain Res. 85:1–12. 10.1016/S0169-328X(00)00218-7 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Paz Y., Elazar Z., Fass D.. 2000. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J. Biol. Chem. 275:25445–25450. 10.1074/jbc.C000307200 [DOI] [PubMed] [Google Scholar]
- Peters P.J., Neefjes J.J., Oorschot V., Ploegh H.L., Geuze H.J.. 1991. Segregation of MHC class II molecules from MHC class I molecules in the Golgi complex for transport to lysosomal compartments. Nature. 349:669–676. 10.1038/349669a0 [DOI] [PubMed] [Google Scholar]
- Ravikumar B., Imarisio S., Sarkar S., O’Kane C.J., Rubinsztein D.C.. 2008. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J. Cell Sci. 121:1649–1660. 10.1242/jcs.025726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recacha R., Boulet A., Jollivet F., Monier S., Houdusse A., Goud B., Khan A.R.. 2009. Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain. Structure. 17:21–30. 10.1016/j.str.2008.10.014 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Shintani T., Klionsky D.J.. 2005. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 16:5843–5856. 10.1091/mbc.E05-07-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A., Schraegle S.J., Stahlberg E.A., Meier I.. 2005. Coiled-coil protein composition of 22 proteomes—differences and common themes in subcellular infrastructure and traffic control. BMC Evol. Biol. 5:66. 10.1186/1471-2148-5-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagiv Y., Legesse-Miller A., Porat A., Elazar Z.. 2000. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 19:1494–1504. 10.1093/emboj/19.7.1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarten M., Mohrlüder J., Ma P., Stoldt M., Thielmann Y., Stangler T., Hersch N., Hoffmann B., Merkel R., Willbold D.. 2009. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 5:690–698. 10.4161/auto.5.5.8494 [DOI] [PubMed] [Google Scholar]
- Seglen P.O., Gordon P.B.. 1982. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA. 79:1889–1892. 10.1073/pnas.79.6.1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M.. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858. 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- Simonsen A., Lippé R., Christoforidis S., Gaullier J.M., Brech A., Callaghan J., Toh B.H., Murphy C., Zerial M., Stenmark H.. 1998. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 394:494–498. 10.1038/28879 [DOI] [PubMed] [Google Scholar]
- Slot J.W., Geuze H.J., Gigengack S., Lienhard G.E., James D.E.. 1991. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 113:123–135. 10.1083/jcb.113.1.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Xu W., Zhang A., Huang G., Liang X., Virbasius J.V., Czech M.P., Zhou G.W.. 2001. Phox homology domains specifically bind phosphatidylinositol phosphates. Biochemistry. 40:8940–8944. 10.1021/bi0155100 [DOI] [PubMed] [Google Scholar]
- Sugawara K., Suzuki N.N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F.. 2004. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 9:611–618. 10.1111/j.1356-9597.2004.00750.x [DOI] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., Wolf D.H.. 1994. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349:275–280. 10.1016/0014-5793(94)00672-5 [DOI] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y.. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169–174. 10.1016/0014-5793(93)80398-E [DOI] [PubMed] [Google Scholar]
- Wang H., Bedford F.K., Brandon N.J., Moss S.J., Olsen R.W.. 1999. GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature. 397:69–72. 10.1038/16264 [DOI] [PubMed] [Google Scholar]
- Webb J.L., Ravikumar B., Rubinsztein D.C.. 2004. Microtubule disruption inhibits autophagosome-lysosome fusion: implications for studying the roles of aggresomes in polyglutamine diseases. Int. J. Biochem. Cell Biol. 36:2541–2550. 10.1016/j.biocel.2004.02.003 [DOI] [PubMed] [Google Scholar]
- Wu M., Wang T., Loh E., Hong W., Song H.. 2005. Structural basis for recruitment of RILP by small GTPase Rab7. EMBO J. 24:1491–1501. 10.1038/sj.emboj.7600643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J.. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102–1109. 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- Xie Z., Nair U., Klionsky D.J.. 2008. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 19:3290–3298. 10.1091/mbc.E07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yan L., Zhou Z., Yang P., Tian E., Zhang K., Zhao Y., Li Z., Song B., Han J., et al. 2009. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 136:308–321. 10.1016/j.cell.2008.12.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.