Soluble ERAD substrates require the Hrd1 E3 ligase for degradation compared with membrane-anchored peptides that use GP78.
Abstract
Sophisticated quality control mechanisms prolong retention of protein-folding intermediates in the endoplasmic reticulum (ER) until maturation while sorting out terminally misfolded polypeptides for ER-associated degradation (ERAD). The presence of structural lesions in the luminal, transmembrane, or cytosolic domains determines the classification of misfolded polypeptides as ERAD-L, -M, or -C substrates and results in selection of distinct degradation pathways. In this study, we show that disposal of soluble (nontransmembrane) polypeptides with luminal lesions (ERAD-LS substrates) is strictly dependent on the E3 ubiquitin ligase HRD1, the associated cargo receptor SEL1L, and two interchangeable ERAD lectins, OS-9 and XTP3-B. These ERAD factors become dispensable for degradation of the same polypeptides when membrane tethered (ERAD-LM substrates). Our data reveal that, in contrast to budding yeast, tethering of mammalian ERAD-L substrates to the membrane changes selection of the degradation pathway.
Introduction
Accumulation of misfolded proteins hampers the function of the ER and elicits a variety of stress responses that might eventually result in cell death. The capacity to rapidly remove folding-defective polypeptides from the ER lumen is therefore crucial to maintain cell homeostasis (Malhotra and Kaufman, 2007; Ron and Walter, 2007; Yoshida, 2007). Polymerogenic, misfolded proteins such as ATZ, a folding-defective, aggregation-prone mutant of the secretory protein α1-antitrypsin, and serpins are removed from the ER upon activation of multiple proteasome-dependent and proteasome-independent disposal pathways including autophagy (Perlmutter, 2006; Shen et al., 2006; Granell and Baldini, 2008; Kroeger et al., 2009; Rutledge et al., 2009). Normally, however, misfolded proteins produced in the ER are extracted from folding machineries and are dislocated across the ER membrane to be degraded by cytosolic 26S proteasomes in a series of tightly regulated events collectively defined as ER-associated degradation (ERAD; McCracken and Brodsky, 1996). Most of the polypeptides entering the ER lumen are modified with preassembled glucose3-mannose9-N-acetylglucosamine2-oligosaccharides, which are covalently attached to Asn side chains in Asn-Xxx-Ser/Thr sequons emerging in the ER lumen. Stepwise processing of protein-bound oligosaccharides eventually determines the fate of the covalently linked polypeptide chains (for review see Molinari, 2007). For instance, degradation of terminally misfolded glycoproteins from the ER requires extensive demannosylation of the protein-bound oligosaccharides, which precedes polypeptide dislocation across the ER membrane and degradation (Hebert and Molinari, 2007; Nakatsukasa and Brodsky, 2008; Lederkremer, 2009). Current models claim that dislocation across the ER membrane follows specific pathways regulated by luminal, transmembrane, and cytosolic complexes built around membrane-embedded E3 ubiquitin ligases (Kostova et al., 2007; Nakatsukasa and Brodsky, 2008; Ravid and Hochstrasser, 2008; Hirsch et al., 2009). In Saccharomyces cerevisiae, disposal of transmembrane proteins with cytosolic defects (ERAD-C substrates) requires intervention of the DOA10 pathway. Membrane-anchored polypeptides with transmembrane lesions (ERAD-M substrates) as well as membrane-tethered and soluble polypeptides with lesions in the ER lumen (ERAD-L substrates) are cleared from the ER through the HRD1 pathway (Taxis et al., 2003; Huyer et al., 2004; Vashist and Ng, 2004; Carvalho et al., 2006; Denic et al., 2006; Gauss et al., 2006; Willer et al., 2008).
The mammalian system is more complex. The mammalian ER membrane contains several E3 ubiquitin ligases. Few of them (e.g., RNF5/RMA1, TEB4, TRC8, and RFP2) are still poorly characterized and/or have a restricted number of substrates (Younger et al., 2006; Lerner et al., 2007; Morito et al., 2008; Wang et al., 2008; Hirsch et al., 2009; Stagg et al., 2009). Others, such as synoviolin/HRD1 and GP78, are better characterized and regulate, in a concerted action with several interacting partners, disposal of numerous folding-defective polypeptides used as model ERAD substrates (Kostova et al., 2007; Nakatsukasa and Brodsky, 2008; Ravid and Hochstrasser, 2008; Hirsch et al., 2009).
Individual substrates follow preferential routes for dislocation across the mammalian ER membrane. Consistently, inactivation of the GP78 pathway is sufficient to substantially delay disposal of conventional ERAD substrates such as TCRα, CD3-δ, and CFTRΔF508 (Fang et al., 2001; Zhong et al., 2004; Ballar et al., 2006; Chen et al., 2006; Vij et al., 2006; Tsai et al., 2007; Morito et al., 2008), but it does not affect disposal of other classical ERAD substrates such as null Hong Kong (NHK) or Ri332. However, inactivation of HRD1 or of components of the multimeric complex built around this E3 ligase has no consequence on disposal of GP78 clients (Nadav et al., 2003; Song et al., 2005; Cao et al., 2007; Yang et al., 2007; Morito et al., 2008; Riemer et al., 2009) but specifically interferes with disposal of folding-defective polypeptides such as NHK, Ri332, Igµs, IgκLC, IgγHC, and IgλLC (Liang et al., 2003; Mueller et al., 2006; Okuda-Shimizu and Hendershot, 2007; Yang et al., 2007; Bernasconi et al., 2008; Brodsky and Fisher, 2008; Cattaneo et al., 2008; Christianson et al., 2008; Hosokawa et al., 2008; Alcock and Swanton, 2009; and references therein). Because the reasons behind this substrate specificity are unknown, it is impossible to predict which pathway will be used by a given folding-defective polypeptide.
ERAD substrates recruitment to HRD1 occurs either directly or indirectly through transient associations of the HRD1 cofactor SEL1L with luminal acceptors such as OS-9, XTP3-B, EDEM1, BiP, or GRP94 (Mueller et al., 2006, 2008; Christianson et al., 2008; Hosokawa et al., 2008; Cormier et al., 2009). Few of these components do not participate in the GP78 or in other dislocation complexes (Hosokawa et al., 2008; Morito et al., 2008; Alcock and Swanton, 2009). Therefore, certainly the different composition of the complexes built around membrane-embedded E3 ubiquitin ligases determines substrate selection, but which substrate feature is relevant to determine whether a misfolded polypeptide will preferentially use one or the other ERAD pathway is unknown.
To better understand this, we compared the requirements for efficient disposal of two canonical, N-glycosylated, membrane-anchored ERAD substrates, BACE476 (Molinari et al., 2002) and CD3-δ (Yang et al., 1998), with the requirements for efficient disposal of their variants lacking the transmembrane anchor (BACE476Δ and CD3-δΔ; Fig. 1). For all of these proteins, extensive demannosylation is required for ERAD. Our data reveal that only degradation of the soluble (nontransmembrane) variants of BACE476 and CD3-δ strictly depend on several participants of the HRD1 pathway regulating ERAD, namely the E3 ubiquitin ligase HRD1, the HRD1-associated cargo receptor SEL1L, and the ERAD lectins OS-9 and XTP3-B. Disposal of the membrane-tethered variants of the same folding-defective polypeptides remained unperturbed upon inactivation of HRD1, SEL1L, and, significantly, OS-9 and XTP3-B. Thus, in contrast to yeast (Quan et al., 2008; Clerc et al., 2009), substrate demannosylation in the mammalian ER is not (only) required to generate a signal for disposal decoded by the ERAD lectins of the OS-9 family. In fact, at least when the folding-defective glycopolypeptide is tethered at the ER membrane, intervention of OS-9 and XTP3-B becomes dispensable for efficient disposal. Moreover, and again in contrast to yeast (Taxis et al.,2003; Willer et al., 2008), the presence or the absence of a membrane anchor alters selection of the disposal pathway used by ERAD-L substrates in mammalian cells. This was confirmed by the finding that the crucial dependency on components of the HRD1 pathway for degradation of NHK, another classical ERAD-L substrate (Liu et al., 1997), was substantially relieved when the protein was anchored at the ER membrane. Our data lead us to group polypeptides with structural lesions in the ER lumen in two subclasses, namely ERAD-LS substrates (for soluble ERAD-L substrates whose disposal is strictly dependent on the HRD1 pathway) and ERAD-LM substrates (for membrane-tethered ERAD-L substrates for which alternative ERAD pathways can be activated to ensure efficient disposal). Our data further highlight the more significant complexity and the somewhat different mechanisms regulating protein quality control in the mammalian versus the budding yeast ER.
Figure 1.
Schematic representation of the seven canonical ERAD substrates used in this study. BACE476, CD3-δ, NHKBACE, and NHKCD3δ are type I membrane proteins (ERAD-LM substrates); BACE476Δ, CD3-δΔ, and NHK are the corresponding soluble ERAD-LS substrates.
Results
Consequences of HRD1 deletion on disposal of membrane-anchored and soluble BACE476
Availability of mouse embryonic fibroblasts (MEFs) lacking HRD1 (Yagishita et al., 2005) prompted us to assess the involvement of this membrane-embedded E3 ubiquitin ligase in disposal of folding-defective polypeptides from the ER lumen. Hrd1−/− cells are more sensitive to ER stress-induced apoptosis, but they are not under ER stress (Fig. S2 C; Yagishita et al., 2005).
Tissue-specific versions of the human β-secretase (BACE501; Vanoni et al., 2008) are among the best-characterized ERAD substrates. For example, the type I membrane glycoprotein BACE476 and its soluble variant BACE476Δ originate from alternative splicing of the BACE transcripts, resulting in a 25-residue deletion in the protein's luminal ectodomain. This deletion prevents attainment of the native structure when the polypeptide is ectopically expressed in cultured mammalian cells (Molinari et al., 2002). BACE variants are defined as canonical ERAD substrates because their degradation requires extensive demannosylation of the protein-bound oligosaccharides (Fig. S1; Molinari et al., 2002), is accelerated in cells expressing high levels of EDEM1 and EDEM2 (Molinari et al., 2003; Olivari et al., 2005, 2006), is delayed in cells with low content of EDEM proteins (Molinari et al., 2003), and is performed in the cytosol upon P97-facilitated extraction by 26S proteasomes (Fig. S1; Molinari et al., 2002; Wang et al., 2006).
BACE476 (Fig. 2 A) and BACE476Δ (Fig. 2 B) were individually expressed in wild-type MEFs (wt; lanes 1–3) and MEFs lacking HRD1 (Hrd1−/−; lanes 4–6). 17 h after transfection with appropriate expression plasmids, cells were pulsed for 10 min with [35S]methionine and cysteine and were subsequently chased in normal cell culture media for the times shown in Fig. 2 (A and B). At the end of each chase time, cells were detergent solubilized, and the residual amount of radiolabeled ERAD substrate was immunoisolated from postnuclear supernatants (PNSs) with specific antibodies (Fig. S2 A), separated on SDS polyacrylamide gels, and quantified.
Figure 2.
Involvement of HRD1 in disposal of membrane-tethered and soluble BACE476. (A) Radiolabeled BACE476 was immunoisolated after the indicated chase times from wild-type MEFs (wt; lanes 1–3) or MEFs lacking HRD1 (Hrd1−/−; lanes 4–6). Relevant bands were quantified and plotted. (B) Same as described in A for BACE476Δ. (C) Radiolabeled BACE476Δ was immunoisolated after the indicated chase times from cells lacking HRD1 transfected with an empty plasmid (mock; lanes 1–3), a plasmid for expression of wild-type HRD1 (lanes 4–6), or a plasmid for expression of inactive HRD1 (HRD1*; lanes 7–9). Relevant bands were quantified and plotted. Error bars represent standard deviation (n = 2). Molecular mass markers are shown on the left for all gels (given in kilodaltons).
The kinetics of BACE476 disposal was essentially the same in cells with (Fig. 2 A, lanes 1–3) and without HRD1 (Fig. 2 A, lanes 4–6). In both cell lines, only ∼30% of the initial amount of radiolabeled polypeptide was immunoisolated from PNS after a 6-h chase (Fig. 2 A, quantification). However, HRD1 deletion substantially delayed disposal of the same protein lacking a transmembrane anchor. In fact, only ∼20% of the initial amount of BACE476Δ was immunoisolated from lysates of wt cells after a 4-h chase (Fig. 2 B, lane 3 and quantification), whereas cells lacking HRD1 still contained ∼80% of the initial amount of labeled protein (Fig. 2 B, lane 6 and quantification). Therefore, HRD1 deletion selectively impaired disposal of the soluble variant of BACE476.
The labeled ERAD candidates had faster mobility at the end (Fig. 2, A and B, lanes 3 and 6) than at the beginning of the chase (Fig. 2, A and B, lanes 1 and 4). This is caused by the progressive demannosylation of ER-retained, misfolded polypeptides that reduces their apparent molecular weight. Extensive demannosylation interrupts futile folding attempts in the calnexin chaperone system and promotes substrate deviation in the ERAD pathway (for review see Molinari, 2007). Consistently, the progressive increase in BACE electrophoretic mobility and BACE disposal were inhibited by kifunensine, a specific inhibitor of α1,2-mannosidases (Fig. S1, A and B, lane 3). Deletion of HRD1 substantially delayed BACE476Δ disposal without affecting the enhancement of BACE mobility, thus the extensive demannosylation that precedes BACE ERAD. This shows that, in the series of luminal events leading to BACE destruction, i.e., (a) folding attempts phase in the calnexin cycle (Molinari et al., 2002), (b) extraction from the folding machinery facilitated by substrate demannosylation (Molinari et al., 2002, 2003), (c) delivery at the ER membrane, (d) and dislocation across the ER membrane, the deletion of HRD1 only interferes with late events, as expected for a component of the dislocation machinery.
HRD1 activity is required for efficient disposal of BACE476Δ
To avoid spurious phenotypes possibly caused by uncharacterized differences between wild-type and knockout cell lines, we confirmed that HRD1 is required for efficient disposal of BACE476Δ by reproducing the experiments shown in Fig. 2 (A and B) in a single cell line. To this end, we compared degradation of BACE476Δ in Hrd1 knockout cells mock transfected (Fig. 2 C, lanes 1–3) and in the same cell line expressing an active (HRD1; Fig. 2 C, lanes 4–6; and Fig. S2 B) or inactive form (the C307S mutant HRD1*; Fig. 2 C, lanes 7–9; and Fig. S2 B; Amano et al., 2003) of HRD1. In all experiments, BACE476Δ was efficiently retained in the ER and degraded as confirmed by the lack of BACE476Δ secretion (Fig. S2 D). BACE476Δ had t1/2 > 240 min in cells lacking HRD1 (for determination of protein t1/2 see Materials and methods; Fig. 2, B [lanes 4–6] and C [lanes 1–3]). Back transfection of the active form of HRD1 accelerated disposal of BACE476Δ to a rate (t1/2 = 150 min; Fig. 2 C, lanes 4–6) approaching the disposal rate measured in wild-type cells (Fig. 2 B, lanes 1–3 and quantifications). In cells expressing the inactive form of HRD1, disposal of BACE476Δ (t1/2 > 240 min; Fig. 2 C, lanes 7–9) was as slow as in cells lacking HRD1 (Fig. 2, B [lanes 4–6] and C [lanes 1–3]). We concluded that an active HRD1 pathway is required for efficient degradation of BACE476Δ from the mammalian ER. Experiments performed to assess consequences of ectopic expression of active or inactive HRD1 on disposal of BACE476 in cells lacking this E3 ubiquitin ligase confirmed dispensability of the HRD1 activity when the same folding-defective ERAD substrate was tethered at the ER membrane (Fig. S3 A).
Consequences of HRD1 deletion on disposal of membrane-anchored and soluble CD3-δ
CD3-δ is a type I membrane glycoprotein, which is typically part of the T cell receptor–CD3 complex involved in T cell development and signal transduction. When expressed individually, the CD3-δ orphan subunit cannot attain its native structure and is degraded from the ER with mechanisms that have been thoroughly characterized. In particular, CD3-δ degradation requires demannosylation and is operated by cytosolic proteasomes (Yang et al., 1998). CD3-δ is a client of the E3 ubiquitin ligase GP78 (Fang et al., 2001; Zhong et al., 2004; Ballar et al., 2006; Chen et al., 2006; Tsai et al., 2007), and it has been reported that RFP2, another E3 ligase, contributes to its disposal (Lerner et al., 2007). Down-regulation of HRD1 does not affect CD3-δ degradation (Yang et al., 2007), although it has recently been reported that down-regulation of HRD1 might result in increased intracellular levels of GP78 (Shmueli et al., 2009; Ballar et al., 2010). Therefore, it was not unexpected that CD3-δ clearance from the ER lumen occurred with similar kinetics in wild-type cells (Fig. 3 A, lanes 1–3) and in cells lacking HRD1 (Fig. 3 A, lanes 4–6).
Figure 3.
Involvement of HRD1 in disposal of membrane-tethered and soluble CD3-δ. (A) Radiolabeled CD3-δ was immunoisolated after the indicated chase times from wild-type MEFs (wt; lanes 1–3) or from MEFs lacking HRD1 (Hrd1−/−; lanes 4–6). Relevant bands were quantified and plotted. (B) Same as described in A for CD3-δΔ. (C) Radiolabeled CD3-δΔ was immunoisolated after the indicated chase times from cells lacking HRD1 transfected with an empty plasmid (mock; lanes 1–3), a plasmid for expression of wild-type HRD1 (lanes 4–6), or a plasmid for expression of inactive HRD1 (HRD1*; lanes 7–9). Relevant bands were quantified and plotted. Molecular mass markers are shown on the left for all gels (given in kilodaltons).
The anchorless variant of CD3-δ was also efficiently retained in the ER as confirmed by lack of secretion in the extracellular media (Fig. S2 E). As for the membrane-tethered version of the protein, CD3-δΔ degradation required demannosylation and was operated by cytosolic proteasomes. Significantly, however, deletion of the CD3-δ membrane anchor converted this GP78/RFP2 client protein to an HRD1 client. In fact, degradation of the anchorless CD3-δΔ was defective in cells lacking HRD1 (Fig. 3 B, lanes 4–6 and quantifications). HRD1 deletion inhibited CD3-δΔ disposal without affecting the extensive substrate demannosylation that signals prolonged ER retention of this folding-defective polypeptide.
As in the case of BACE476Δ, to avoid spurious phenotypes possibly caused by uncharacterized differences between wild-type and knockout cell lines, we confirmed involvement of HRD1 by showing that CD3-δΔ disposal was defective in cells lacking HRD1 or expressing the inactive form of HRD1 (t1/2 > 120 min; Fig. 3 C, lanes 1–3 and 7–9, respectively) but progressed with wild-type kinetics (t1/2 < 60 min; Fig. 3 C, lanes 4–6) upon back transfection of active HRD1 in the knockout cells. The finding that an active HRD1 pathway is required for efficient degradation of CD3-δΔ but not for disposal of the same protein when associated with the ER membrane was confirmed by assessing the dispensability of HRD1 for CD3-δ disposal (Fig. S3 B).
HRD1 and GP78 requirements for disposal of membrane-tethered and soluble BACE476
So far, we showed that deletion of HRD1 significantly interferes with disposal of the anchorless variants of BACE476 and CD3-δ (ERAD-LS substrates), leaving unaffected disposal of polypeptides with the same luminal domain, but tethered at the ER membrane (ERAD-LM substrates). This may indicate that HRD1 is not involved in disposal of membrane-anchored ERAD-L substrates or that upon HRD1 deletion, surrogate pathways regulated by other membrane-embedded E3 ligases are engaged to ensure disposal.
To verify this, we next assessed the consequences of the individual and combined down-regulation of HRD1 and GP78 in clearance of membrane-tethered and soluble BACE476 from the ER. Down-regulation of HRD1 obtained by specific RNAi (Fig. 4 A) slightly delayed disposal of BACE476 (Fig. 4 B, lanes 4–6 and quantification of siHRD1). Down-regulation of GP78 (Fig. 4 A) had no effect (Fig. 4 B, lanes 7–9 and quantification of siGP78). However, the combined inactivation of both HRD1 and GP78 (Fig. 4 A) substantially delayed BACE476 disposal (Fig. 4 B, lanes 10–12 and quantifications siHRD1 + siGP78). The delay of BACE476 disposal obtained upon combined inactivation of the HRD1 and of the GP78 pathways was comparable with the delay obtained upon cell exposure to classical ERAD inhibitors such as kifunensine or PS341 (Fig. S1 A). Collectively, these data show that GP78 can replace the inactive HRD1 and that, vice versa, HRD1 can replace inactive GP78 to ensure efficient disposal of BACE476.
Figure 4.
Consequences of HRD1 and GP78 down-regulation on degradation of membrane-tethered and soluble variants of BACE476 and CD3-δ. (A) The efficiency of siRNA-based HRD1 and GP78 down-regulations were checked by immunoblotting. Tubulin is a loading control. (B) Radiolabeled BACE476 was immunoisolated after the indicated chase times from cells expressing a scrambled siRNA (siSCR; lanes 1–3), an siRNA targeting HRD1 (siHRD1; lanes 4–6), GP78 (siGP78; lanes 7–9), or both HRD1 and GP78 (siHRD1/siGP78; lanes 10–12). Relevant bands were quantified and plotted. (C–E) Same as described in B for BACE476Δ, CD3-δ, and CD3-δΔ, respectively. Molecular mass markers are shown on the left for all gels (given in kilodaltons).
On the contrary, HRD1 down-regulation was sufficient to delay BACE476Δ disposal to a similar extent as α1,2-mannosidases or proteasome inactivation (Fig. 4 C, lanes 4–6 and quantifications; and Fig. S1 B). Down-regulation of GP78 had no effect on BACE476Δ degradation (Fig. 4 C, lanes 7–9), and combined inactivation of the two E3 ubiquitin ligases (Fig. 4 C, lanes 10–12) had the same inhibitory effect as the individual down-regulation of HRD1 (Fig. 4 C, quantification). This is in agreement with a model claiming that the presence or absence of a membrane anchor determines requirements for disposal of ERAD-L substrates. More precisely, deletion of the membrane anchor converted BACE476 from an HRD1/GP78 client into an obligate client of the HRD1 pathway.
HRD1 and GP78 requirements for efficient disposal of membrane-tethered and soluble CD3-δ
Next, we assessed the contribution of HRD1 and GP78 in clearance from the ER of CD3-δ and CD3-δΔ. Down-regulation of HRD1 did not affect degradation of CD3-δ (Fig. 4 D, lanes 4–6 and quantifications). Down-regulation of GP78 did delay CD3-δ disposal (Fig. 4 D, lanes 7–9), and the additional inactivation of HRD1 did not further protect this folding-defective polypeptide from ERAD (Fig. 4 D, lanes 10–12 and quantifications). These data are consistent with findings reporting that CD3-δ is a client of the GP78 machinery (Fang et al., 2001; Zhong et al., 2004; Ballar et al., 2006; Chen et al., 2006; Tsai et al., 2007) and with data showing that CD3-δ disposal remains unaffected upon inactivation of the HRD1 pathway (Fig. 3 A; Yang et al., 2007). The incomplete inhibition of CD3-δ disposal upon inactivation of GP78 (Fig. 4 D) is consistent with data showing that at least another E3 ligase, RFP2 (Lerner et al., 2007), contributes to CD3-δ clearance from the ER lumen.
In agreement with our model predicting that changes in substrate tethering to the membrane determine selection of the ERAD pathway regulating clearance of ERAD-L substrates from the ER, HRD1 down-regulation was sufficient to substantially delay CD3-δΔ disposal (Fig. 4 E, lanes 4–6 and quantifications), whereas down-regulation of GP78 had no effect (Fig. 4 E, lanes 7–9). Combined inactivation of the two ligases (Fig. 4 E, lanes 10–12) had the same inhibitory effect on CD3-δΔ disposal as the individual down-regulation of HRD1. Thus, deletion of the membrane anchor converted CD3-δ from a GP78/RFP2 client into an obligate client of the HRD1 pathway.
SEL1L is required for efficient disposal of ERAD-LS substrates
Our data show a stringent requirement of HRD1 for degradation of the ERAD-LS substrates BACE476Δ and CD3-δΔ. The same polypeptides might exploit the HRD1 and/or the GP78 and/or other E3 ligases for efficient disposal when anchored at the ER membrane. The complex built around HRD1 contains several proteins that are excluded from the GP78 complex. Among them, SEL1L plays a crucial role in protein disposal by recruiting ERAD substrates either directly or indirectly through association with the ERAD lectins OS-9 and XTP3-B (Mueller et al., 2006, 2008; Christianson et al., 2008; Hosokawa et al., 2008) through association with the member of the glycosyl hydrolase 47 family EDEM1 (Cormier et al., 2009) or through association with luminal chaperones such as BiP or GRP94.
Our finding that only ERAD-LS substrates were strictly dependent on active HRD1 prompted us to verify the involvement of SEL1L in clearance of ERAD-LS versus ERAD-LM substrates. To this end, kinetics of protein disposal was compared in mock-treated cells (Fig. 5, siSCR) and in cells with reduced expression of SEL1L (Fig. 5, siSEL1L). Down-regulation of SEL1L (Fig. 5 A) only marginally slowed disposal of the ERAD-LM substrate BACE476 (Fig. 5 B, lanes 4–6 and quantifications) but substantially delayed degradation of the ERAD-LS substrate BACE476Δ (Fig. 5 C, lanes 4–6 and quantifications). Similarly, down-regulation of SEL1L had no effect on degradation of the ERAD-LM substrate CD3-δ (Fig. 5 D, lanes 4–6 and quantifications) but fully protected the anchorless variant of the same folding-defective polypeptide from degradation (Fig. 5 E, lanes 4–6 and quantifications). Collectively, these findings showed that down-regulation of SEL1L recapitulates the phenotype obtained upon deletion (Figs. 2 and 3) or down-regulation of HRD1 (Fig. 4). Inactivation of HRD1 or SEL1L had little or no effect on disposal of ERAD-LM substrates but dramatically interfered with clearance from the ER lumen of the same polypeptides when detached from the ER membrane (ERAD-LS substrates).
Figure 5.
Disposal of soluble misfolded polypeptides relies on SEL1L and both OS-9 + XTP3-B. (A) The efficiency of siRNA-based SEL1L, OS-9, and XTP3-B down-regulations were checked by immunoblotting. Tubulin is a loading control. (B) Radiolabeled BACE476 was immunoisolated after the indicated chase times from cells expressing a scrambled siRNA (siSCR; lanes 1–3), an siRNA targeting SEL1L (siSEL1L; lanes 4–6), OS-9 (siOS-9; lanes 7–9), XTP3-B (siXTP3-B; lanes 10–12), or both XTP3-B and OS-9 (siXTP3-B/siOS-9; lanes 13–15). Relevant bands were quantified and plotted. (C–E) Same as described in B for BACE476Δ, CD3-δ, and CD3-δΔ, respectively. Molecular mass markers are shown on the left for all gels (given in kilodaltons).
OS-9 and XTP3-B are interchangeable ERAD shuttles required for efficient disposal of ERAD-LS but not ERAD-LM substrates
The mammalian splice variants of OS-9 and XTP3-B constitute a group of proteins that have lectin-like domains with homology to the mannose-6-phosphate receptor family (Munro, 2001). So far, OS-9 or XTP3-B intervention in ERAD has only been shown for the model ERAD substrates NHK or Ri332 (Bernasconi et al., 2008; Christianson et al., 2008; Hosokawa et al., 2008; Mueller et al., 2008; Alcock and Swanton, 2009; Hosokawa et al., 2009). The individual down-regulation of these ERAD lectins has generally no or mild phenotypes, and attempts to establish the involvement of OS-9 or XTP3-B variants in disposal of other glycosylated folding-defective polypeptides have been unsuccessful, thus questioning their relevance as general regulators of protein disposal from the mammalian ER. Our data confirm that individual down-regulation of OS-9 (Fig. 5, A, B, and E, lanes 7–9 and quantifications) or XTP3-B (Fig. 5, A, B, and E, lanes 10–12 and quantifications) did not significantly delay disposal of BACE476, BACE476Δ, CD3-δ, and CD3-δΔ. However, combined down-regulation of OS-9 and XTP3-B specifically retarded disposal of the HRD1 client BACE476Δ (Fig. 5 C, lanes 13–15 and quantifications) to a similar extent as down-regulation of SEL1L (lanes 4–6) and HRD1 (Fig. 4 C, lanes 4–6). Combined down-regulation of OS-9 and XTP3-B also partially protected CD3-δΔ from disposal (Fig. 5 E, lanes 13–15). Thus, OS-9 and XTP3-B have redundant activities in the ER lumen so that their combined inactivation might be required to substantially delay disposal. NHK is an exception to this rule because individual down-regulation of OS-9 is sufficient to significantly delay ERAD (Bernasconi et al., 2008; Christianson et al., 2008). Moreover, our data show that OS-9 and XTP3-B intervention is only required for disposal of ERAD-LS substrates that must be shuttled from the ER lumen to the ER membrane-embedded HRD1 complex for efficient clearance from the ER. For this reason, we define OS-9 and XTP3-B as ERAD shuttles. Disposal of the same glycopolypeptides tethered to the ER membrane (ERAD-LM substrates) progressed unperturbed even upon substantial reduction of the cellular content of OS-9 and XTP3-B. This shows that the two ERAD shuttles do not intervene in disposal of membrane-tethered N-glycosylated polypeptides with luminal defects or that they can efficiently be replaced by a surrogate factor upon their combined inactivation (see Discussion).
Down-regulation of SEL1L and HRD1 allows identification of OS-9.1 as delivery factor for BACE476Δ
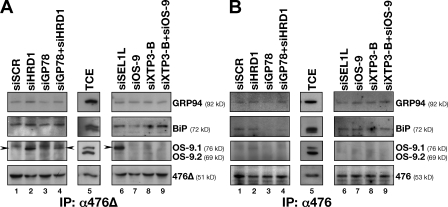
In an attempt to identify ERAD factors interacting with ERAD-LS versus ERAD-LM substrates, BACE476Δ was tagged with an EFRH tetrapeptide (which is recognized by β1, a monoclonal antibody described in Paganetti et al., 2005), whereas BACE476 was HA tagged. The two proteins were ectopically coexpressed in mock-treated cells (siSCR; Fig. 6, A and B, lane 1; Fig. S4) or in cells with reduced levels of HRD1 (Fig. 6, A and B, lane 2), GP78 (Fig. 6, A and B, lane 3), HRD1 and GP78 (Fig. 6, A and B, lane 4), SEL1L (Fig. 6, A and B, lane 6), OS-9 (Fig. 6, A and B, lane 7), XTP3-B (Fig. 6, A and B, lane 8), and OS-9 and XTP3-B (Fig. 6, A and B, lane 9). Cells were lyzed under conditions that preserve many substrate–chaperone interactions (Molinari et al., 2002). BACE476Δ-β1 (Fig. 6 A) and BACE476-HA (Fig. 6 B) were individually immunoisolated with the appropriate anti-tag antibody together with their interacting partners. The amount of proteins separated in SDS-PAGE and blotted on a PVDF membrane was normalized to ensure equal loading of BACE in each lane (Fig. 6, A and B, bottom). The PVDF membrane was probed with antibodies to GRP94, BiP, and OS-9 to determine whether these ER chaperones, claimed to be involved in ERAD, were found in complexes sufficiently stable to survive the cell lysis and immunoprecipitation protocols. The case for GRP94 is unclear, as variations in the amount of this chaperone coprecipitated with BACE476Δ-β1 (Fig. 6 A) or BACE476-HA (Fig. 6 B) were too small to be considered significant. Variations were also small for BiP. However, a stabilization of BACE476Δ–BiP complexes was observed upon inactivation of HRD1 (Fig. 6 A, lane 2), HRD1 + GP78 (Fig. 6 A, lane 4), SEL1L (Fig. 6 A, lane 6), and upon combined inactivation of OS-9 + XTP3-B (Fig. 6 A, lane 9). These variations were reproducible even when BACE476Δ-β1 was individually expressed in cells subjected to transient interferences (unpublished data). Stabilization of BACE476Δ–BiP complexes upon disassembly of the HRD1 dislocon shows that BiP contributes to delivery of terminally misfolded polypeptides to this machinery.
Figure 6.
Trapping of BACE476Δ by OS-9.1 upon inactivation of the HRD1 pathway. (A) BACE476Δ was immunoisolated from detergent extracts of cells incubated with a scrambled siRNA, and siRNA targeting HRD1, GP78, GP78, and HRD1 (lanes 1–4) and SEL1L, OS-9, XTP3-B, and XTP3-B + OS-9 (lanes 6–9, respectively). Proteins were separated in SDS polyacrylamide gels and transferred on PVDF. The membranes were blotted with antibodies recognizing endogenous GRP94, BiP, OS-9.1, and OS-9.2 and BACE476Δ as a loading control. (B) Same as described in A for BACE476. Arrowheads indicate the coprecipitated OS-9.1. TCE, total cell extract; IP, immunoprecipitation.
Our analysis also revealed a significant stabilization of BACE476Δ–OS-9.1 complexes in cells subjected to HRD1 (Fig. 6 A, lane 2), HRD1 + GP78 (less evident; Fig. 6 A, lane 4), or SEL1L down-regulations (Fig. 6 A, lane 6). Under the same conditions, and actually in the same cells because BACE476Δ-β1 and BACE476-HA were coexpressed, we were unable to detect stabilization of complexes between BACE476 and the ER chaperones tested (Fig. 6 B).
The stabilization of BACE476Δ–OS-9.1 complexes (and possibly of BACE476Δ–XTP3-B complexes [an antibody to detect endogenous XTP3-B is unfortunately not available]) when SEL1L or HRD1 is not available for substrate delivery is consistent with a sequential involvement of these components during channeling of BACE476Δ into the cytosol for disposal.
Disposal of the classical ERAD-LS substrate NHK and of its two ERAD-LM variants, NHKBACE and NHKCD3δ
So far, we showed that deletion of the membrane anchor of ERAD-L substrates confers a strong dependency on the HRD1 machinery independent of the mechanisms regulating disposal of the membrane-tethered version of the proteins under investigation. To challenge our model, we decided to determine requirements for disposal of a soluble ERAD substrate and to verify how requirements would change upon addition of a transmembrane anchor. To this end, we selected a classical ERAD-L substrate, the NHK variant of the secretory protein α1-antitrypsin (Liu et al., 1997). As previously shown in other cell lines (Bernasconi et al., 2008), we confirm that a fraction of NHK escapes ER retention and is secreted extracellularly (∼15% of the labeled protein in HeLa cells; Fig. S2 F). The expectations were that disposal of this ERAD-LS protein should substantially be delayed upon inactivation of the E3 ligase HRD1 and that the strict dependency on HRD1 should be relieved when the folding-defective polypeptide is converted in a membrane-tethered ERAD-LM protein. Reduction of the intralumenal level of HRD1 (Fig. 7 A, lanes 4–6) substantially inhibited NHK degradation without affecting secretion of the protein (Fig. S2 F). The combined inactivation of the GP78 pathway did not further protect the protein from disposal (Fig. 7 A, lanes 10–12). The intervention of HRD1 was not surprising because both SEL1L (Christianson et al., 2008; unpublished data) and OS-9 (Bernasconi et al., 2008; Christianson et al., 2008) have been shown to participate in NHK disposal. Thus, NHK behaves as a bonafide ERAD-LS protein as defined above, which shows strict dependency on the HRD1 pathway for disposal.
Figure 7.
Involvement of HRD1 and GP78 in disposal of soluble and membrane-tethered NHK variants. (A) Radiolabeled NHK was immunoisolated after the indicated chase times from cells expressing a scrambled siRNA (siSCR; lanes 1–3), an siRNA targeting HRD1 (siHRD1; lanes 4–6), GP78 (siGP78; lanes 7–9), or both HRD1 and GP78 (siHRD1/siGP78; lanes 10–12). Relevant bands were quantified and plotted. (B and C) Same as described in A for NHKBACE and NHKCD3δ, respectively. Molecular mass markers are shown on the left for all gels (given in kilodaltons).
Tethering of NHK at the ER membrane was obtained by appending the membrane anchor of BACE501 (NHKBACE; Fig. 1) or the membrane anchor of CD3-δ (NHKCD3δ; Fig. 1) at the C terminus of NHK (see Materials and methods). The membrane anchor of BACE501 does not contain misfolded or retention signals. Consistently, BACE501 is efficiently transported at the cell surface when the protein is ectopically expressed in cultured cells (Vanoni et al., 2008). However, it is not entirely clear whether the transmembrane domain of CD3-δ is free of misfolding signal. Anchoring of NHKBACE and of NHKCD3δ at the ER membrane was confirmed by subcellular fractionation as described previously (Fig. S5, A and B, respectively; Olivari et al., 2005). Both proteins were degraded from cells with slightly faster kinetics compared with their soluble counterpart (Fig. 7, A [NHK], B [NHKBACE], and C [NHKCD3δ]; compare lanes 1–3). Degradation was efficiently inhibited upon inactivation of ER mannosidases and cytosolic proteasomes (unpublished data). In agreement with our model, conversion of NHK in an ERAD-LM substrate substantially relieved HRD1 dependency. In fact, disposal of both NHKBACE (Fig. 7 B) and NHKCD3δ (Fig. 7 C) was unaffected upon reduction of the intraluminal level of HRD1. Disposal of the membrane-tethered versions of NHK was only partially delayed upon inactivation of both the HRD1 and GP78 pathways (Fig. 7, B and C, lanes 10–12), hinting at the possible intervention of alternative ERAD pathways in disposal of NHK when anchored at the ER membrane.
Discussion
The protein folding, quality control, and disposal machineries operating in the mammalian ER lumen determine the fate of thousands of gene products, each one characterized by unique structural, biophysical, and biochemical properties. The covalent addition of preassembled glucose3-mannose9-N-acetylglucosamine2-oligosaccharides on nascent polypeptide chains (Fig. 8 A) and their processing generate unique signals decoded by ER-resident lectins, glucosyl transferases, and glycosyl hydrolases that determine prolongation or interruption of folding and onset of disposal (Aebi et al., 2009; for review see Molinari, 2007). Current models claim that a series of α1,2-mannosidases (MnsIp and HtmIp in S. cerevisiae; Quan et al., 2008; Clerc et al., 2009), ERManI, EDEM proteins, and/or Golgi mannosidases in mammalian cells (Liu et al., 1997; Hirao et al., 2006; Olivari et al., 2006; Hosokawa et al., 2007; for review see Aebi et al., 2009) slowly remove terminal α1,2-bonded mannose residues (Fig. 8 A, dark green) from oligosaccharides displayed on terminally misfolded polypeptides. Demannosylated oligosaccharides recruit ERAD lectins such as Yos9p in S. cerevisiae and OS-9 and XTP3-B splice variants in mammals (Bernasconi et al., 2008; Christianson et al., 2008; Hosokawa et al., 2008, 2009; Mueller et al., 2008; Quan et al., 2008; Tamura et al., 2008; Wang and Ng, 2008; Alcock and Swanton, 2009; Clerc et al., 2009). ERAD lectins and luminal chaperones such as BiP, GRP94, and protein disulfide isomerase deliver terminally misfolded polypeptides to dislocons at the ER membrane. Dislocons contain a membrane-embedded E3 ubiquitin ligase and several adaptor molecules with substrate-recognition/modifying domains facing the luminal and/or cytosolic side of the ER membrane. Altogether, these multimeric protein complexes regulate the export of terminally misfolded polypeptides across the ER membrane and their polyubiquitylation that facilitates proteasomal intervention (Kostova et al., 2007; Nakatsukasa and Brodsky, 2008; Ravid and Hochstrasser, 2008; Hirsch et al., 2009).
Figure 8.
Structure and composition of N-linked glycans. (A) The Asn-linked core oligosaccharide is composed of two N-acetylglucosamines (squares), nine mannoses (circles; the cleavable α1,2-bonded mannoses are shown in dark green), and three glucoses (triangles). The linkages are indicated, and letters a–n are assigned. a–c define the three oligosaccharide branches. (B) Aberrant oligosaccharide transferred to nascent polypeptide chains in B3F7 cells (Moremen and Molinari, 2006).
In S. cerevisiae, the primary role of demannosylation of folding-defective polypeptides is the exposure of the α1,6-bonded mannose residue j (Fig. 8 A) that recruits the ERAD lectin Yos9p (Quan et al., 2008; Clerc et al., 2009). Our data show that in mammalian cells, even though demannosylation is required for disposal of N-glycosylated ERAD-LS (for soluble; detached from the ER membrane) and ERAD-LM (for membrane tethered) substrates, OS-9 and XTP3-B are only required for disposal of ERAD-LS substrates. We have defined OS-9 and XTP3-B as ERAD shuttles because they associate with soluble, misfolded polypeptides in the ER lumen and deliver them at dislocons embedded in the ER membrane. Our data show that OS-9 and XTP3-B are interchangeable in this function, thus explaining the mild, if any phenotype caused by their individual inactivation reported so far in the literature.
At least for ERAD-LM substrates, the primary role of demannosylation might not be the recruitment of OS-9 and XTP3-B because the two ERAD shuttles are dispensable for disposal of N-glycosylated polypeptides tethered at the ER membrane. One could envision that another lectin decodes the ERAD signal generated upon demannosylation of ERAD-LM substrates when OS-9 and XTP3-B are not available. If this would hold true, this yet to be characterized factor cannot act as surrogate ERAD lectin for soluble, misfolded glycopolypeptides.
Alternatively, the primary role of demannosylation of ERAD-LM substrates could be to elicit extraction of folding-defective polypeptides from the calnexin chaperone system, which is a rate-limiting step in disposal of N-glycosylated proteins (for review see Molinari, 2007). The mammalian ER contains the quality control enzyme UDP-glucose–glycoprotein glucosyl transferase (UGT1; Parodi, 2000). The activity of UGT1 preserves the glucose residue l (Fig. 8 A), which is required for the association of nonnative polypeptides with the lectin chaperones calnexin and calreticulin (Hammond et al., 1994). Retention of nonnative polypeptides in the calnexin chaperone system protects them from degradation (Caramelo and Parodi, 2008; Aebi et al., 2009). Therefore, removal of mannose g, which is the only hexose that can be reglucosylated by the UGT1, causes the irreversible extraction of terminally misfolded polypeptides from the folding machinery and is crucial to initiate a series of events eventually leading to polypeptide disposal. Consistently, pharmacologic inactivation of α1,2-mannosidases (Molinari et al., 2002) or reduction of their intraluminal level (Molinari et al., 2003) substantially delays release of terminally misfolded polypeptides from calnexin. As a further indication that removal of mannose g is relevant for mammalian ERAD, recent data have shown that substrate demannosylation is required in mutant cell lines characterized by addition on nascent chains of aberrant oligosaccharides in which mannose g is the only removable α1,2-bonded mannose (Fig. 8 B; Ermonval et al., 2001; Moremen and Molinari, 2006; Olivari et al., 2006). In S. cerevisiae, a functional orthologue for UGT1 and the possibility for nonnative polypeptides to be retained in the calnexin cycle are missing. Removal of mannose g as a signal required for polypeptide disposal would therefore make little sense, whereas removal of mannose residues i and k is crucial because it allows an active segregation of misfolded polypeptides upon Yos9p association (Aebi et al., 2009).
The mammalian ER membrane hosts several E3 ubiquitin ligases that might potentially contribute to dislocation/disposal of misfolded proteins from the ER (Kostova et al., 2007; Nakatsukasa and Brodsky, 2008; Ravid and Hochstrasser, 2008; Hirsch et al., 2009). Individual or combined inactivation of two of them, namely HRD1 and GP78, was sufficient to significantly delay disposal of classical ERAD-L substrates such as BACE, CD3-δ, and NHK variants. For folding-defective polypeptides detached from the ER membrane (ERAD-LS substrates), efficient disposal required HRD1, SEL1L, and OS-9 or XTP3-B. The stringent requirement for components of the HRD1 pathway was relieved when the very same luminal domains were tethered to the ER membrane. Thus, ERAD-LM substrates do use, or can eventually engage, alternative ERAD pathways when the HRD1 pathway has been shut down. Such complexity is absent or has not been reported so far in the case of S. cerevisiae in which soluble and membrane-tethered ERAD-L substrates are degraded by a unique pathway built around the E3 ubiquitin ligase Hrd1p (Taxis et al., 2003; Vashist and Ng, 2004; Carvalho et al., 2006; Willer et al., 2008).
It can be envisioned that access to membrane-embedded dislocons might be facilitated by lateral diffusion in the lipid bilayer when folding-defective polypeptides are tethered to the ER membrane. However, misfolded proteins floating in the ER lumen might be more dependent on luminal ERAD shuttles (e.g., OS-9 and XTP3-B) that direct them from the ER lumen to dislocons embedded in the ER membrane. HRD1 accessory factors such as OS-9, XTP3-B, and their membrane receptor SEL1L are excluded from complexes containing GP78 or other E3 ubiquitin ligases (Hosokawa et al., 2008; Morito et al., 2008; Alcock and Swanton, 2009). Thus, the HRD1 complex seems better equipped to recruit soluble folding-defective polypeptides at the ER membrane.
Notably, misfolded proteins lacking a transmembrane anchor might fall outside the ERAD-LS group and show little or no dependency on conventional ERAD shuttles that would channel them to the HRD1 complex. For example, ATZ and serpins do not belong to the ERAD-LS group because they form polymeric deposits that cells remove from the ER by activating multiple proteasome-dependent and proteasome-independent disposal pathways (see Introduction; Perlmutter, 2006; Shen et al., 2006; Granell and Baldini, 2008; Kroeger et al., 2009; Rutledge et al., 2009). Another example is ApoB, a protein that lacks a transmembrane anchor but is actually membrane associated in the lipid ligand–deficient state that triggers its disposal (Brodsky and Fisher, 2008), and as such, may engage alternative, ERAD shuttle–independent disposal pathways.
It remains to be established what determines the ERAD pathway selection for ERAD-LM substrates. For example, why membrane-anchored canonical ERAD substrates such as CD3-δ (Figs. 3–5) require GP78 and/or RFP2 for efficient disposal (Fang et al., 2001; Zhong et al., 2004; Ballar et al., 2006; Chen et al., 2006; Lerner et al., 2007; Tsai et al., 2007), whereas others such as BACE476 (Figs. 2, 4, and 5), NHKBACE, and NHKCD3δ (Fig. 7) can use the HRD1, the GP78, and possibly other pathways. Certainly, our observations are consistent with the emerging picture that protein quality control in the mammalian ER conserves regulatory factors and basic mechanisms operating in the ER of the budding yeast but is characterized by additional levels of complexity. This is probably required to ensure efficiency of protein biogenesis and maintenance of homeostasis in the context of multicellular systems.
Materials and methods
Expression plasmids, antibodies, and inhibitors
The pCIneo plasmids encoding for CD3-δ and XTP3-B were provided by S. Fang (University of Maryland Biotechnology Institute, Medical Biotechnology Center, Baltimore, MD) and C. Niehrs (German Cancer Research Center, Heidelberg, Germany), respectively. Plasmids for expression of BACE, NHK, and HRD1 variants were described previously (Molinari et al., 2003; Yagishita et al., 2005; Bernasconi et al., 2008). CD3-δΔ was prepared by inserting a stop codon (primer sequence: 5′-GACTCGGGCACCATGGAGTTCCGACACTGAGCTGGTGTCATCTTC-3′) before the membrane anchor of CD3-δ using the site-directed mutagenesis kit (Agilent Technologies). The membrane-tethered variants of NHK (NHKBACE and NHKCD3δ) were generated by appending at the protein C terminus the amino acids PQTDESTLMTIAYVMAAICALFMLPLCLMVCQWRCLRCLRQQHDDFADDISLLK (which correspond to residues 448–501 of BACE501) or VELDSGTMA GVIFIDLIATLLLALGVYCFAGHETGRPSGAAEVQALLKNEQLYQPLRDREDTQYSRLGGNWPRNKKS (which correspond to residues 96–173 of CD3-δ). Transmembrane segments are italicized. DNA preparations were obtained using commercially available purification kits (Sigma-Aldrich). The nucleotide sequences of all plasmids were verified on both strands. Antibodies to BACE, NHK, and OS-9 were described previously (Bernasconi et al., 2008). Antibody to tubulin was obtained from ABM. Antibodies to GP78 and SEL1L were provided by Y. Ye (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD) and H. Ploegh (Whitehead Institute for Biomedical Research, Cambridge, MA). Antibodies to BiP, EDEM1, and GRP94 were obtained from Santa Cruz Biotechnology, Inc., Sigma-Aldrich, and Thermo Fisher Scientific, respectively. The proteasome inhibitor PS341 was provided by Millenium Pharmaceuticals, Inc. and was used at a concentration of 9 µM. Kifunensine was obtained from TRC, Inc. and was used at a concentration of 100 µM.
Cell Lines, transient transfections, RNAis, metabolic labeling, immunoprecipitations, immunoblots, and analysis of data
Hrd1−/− and wild-type MEF cells were grown in DME supplemented with 10% FBS. Cells at 80–90% confluence in a 6-cm tissue culture plate were transfected with the expression plasmid of interest (4 µg for single transfections and 6 µg total DNA for double transfections) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Experiments were normally performed 17 h after transfection. Expression of all proteins has been confirmed by immunoprecipitation (Fig. S2). For siRNA-based interference, HeLa cells were grown in MEM Alpha supplemented with 10% FBS. Cells at 50% confluence in a 3.5-cm tissue culture plate were transfected with 50 pmol/dish siRNA duplex (Applied Biosystems) using Lipofectamine 2000 according to the manufacturer's instructions. 4 h after transfection, the medium was replaced with MEM Alpha supplemented with 1% nonessential amino acids (Invitrogen). 30 h after siRNA transfection, cells were transfected with the expression plasmids of interest. Experiments were performed 48 h after RNAi transfection. siRNA targeting sequences used are as follows: HRD1, 5′-GGUGAUGGGCAAGGUGUUC-3′; GP78, 5′-GACGGAUUCAAGUACCUUU-3′; SEL1L, 5′-GGCUAUACUGUGGCUAGAA-3′; OS-9.1 and OS-9.2, 5′-CAUCAUCCAGGAGACAGAG-3′; XTP3-B1 and XTP3-B2, 5′-AGCAGUUGUUCCUACAGAA-3′. When detection of endogenous target proteins was impossible, the efficiency of siRNA was confirmed by the significant reduction in expression of ectopically expressed targets (i.e., HRD1, GP78, and XTP3-B).
18 h after transfection, cells were starved for 20 min in Met/Cys-free medium, pulsed for 10 min with 50 µCi [S35]Met/Cys, and chased for the indicated times with MEM Alpha supplemented with 5 mM cold Met/Cys. PNS was prepared by solubilization of cells in 400 µl/3.5-cm dish (or 800 µl/6-cm dish) ice-cold 2% CHAPS (Anatrace) in Hepes-buffered saline (HBS), pH 6.8, containing 20 mM N-ethylmaleimide and protease inhibitors. CHAPS-insoluble material was separated by centrifugation at 10,000 g for 10 min. Immunoprecipitations were performed by adding protein A beads (1:10 wt/vol swollen in HBS; Sigma-Aldrich) with the selected antibody and incubated for 2 h at 4°C.
Immunoprecipitates were extensively washed (three times for 10 min) with 0.5% CHAPS in HBS, resuspended in sample buffer, boiled for 5 min, and finally separated in SDS-PAGE. Gels were exposed to films (BioMax; Kodak) and scanned with a scanner (Agfa). Relevant bands were quantified by ImageQuant software (Molecular Dynamics). Because of the number of Petri dishes that can be handled in parallel, to analyze all conditions in a single transfection round, we had to perform three-point kinetics. Protein half-lives were confirmed by separate, independent experiments using additional chase times. Immunoblots were performed using the SNAP i.d. protein detection system (Millipore). All primary antibodies were used at 1:200–1:333 dilutions. Secondary antibodies were HRP conjugated and used at 1:10,000 dilutions. The ECL Plus detection system was obtained from GE Healthcare.
Subcellular fractionation and separation of membrane versus luminal content
Metabolically labeled cells expressing NHKBACE and NHKCD3δ were extensively washed with isotonic buffer and resuspended in 800 µl of homogenization buffer (10 mM triethanolamine, 10 mM acetic acid, 250 mM sucrose, 20 mM NEM, 1 mM EDTA, and a cocktail of protease inhibitors, pH 7.4; Olivari et al., 2005). Cells were broken with 10 passages through a 25G1 needle. After nuclear supernatants were subjected to ultracentrifugation (45 min at 200,000 g in TLA 120.2), the endomembrane-containing pellet was extensively washed, resuspended in 500 µl 100 mM Na2CO3, and incubated for 25 min on ice for carbonate extraction. After an additional ultracentrifugation step (45 min for 200,000 g), the supernatant was harvested. The endomembrane fraction was washed once with 100 mM Na2CO3 (35 min at 200,000 g) and resuspended in 800 µl of lysis buffer. Insoluble material was removed after 25 min on ice by 10 min centrifugation at 200,000 g. The ER luminal content and the endomembrane fraction were subjected to immunoprecipitation against calnexin, calreticulin, and NHKBACE or NHKCD3δ.
Semiquantitative RT-PCRs
MEF cells were mock treated or incubated for 12 h with 2.5 µg/ml tunicamycin. Cells were lyzed in TRIzol reagent (Invitrogen), and RNA was isolated according to the instructions of the manufacturer. 2 µg RNA was used for cDNA synthesis using reverse transcription (SuperScriptII; Invitrogen) and oligo(dT) (Invitrogen). RT-PCR was performed using TaqDNA polymerase (Invitrogen) with transcript-specific primers: unspliced + spliced Xbp1 (forward), 5′-AAACAGAGTAGCAGCTCAGACTGC and (reverse) 5′-TGGCTGGATGAAAGCAGGTT-3′; BiP (forward), 5′-GAGTTCTTCAATGGCAAGGA-3′ and (reverse) 5′-CCAGTCAGATCAAATGTACCC-3′; β-actin (forward), 5′-CTTTCTGGGTATGGAATCCT-3′ and (reverse) 5′-GGAGCAATGATCTTGATCTT-3′.
Online supplemental material
Fig. S1 shows that α1,2-mannosidases and proteasomes are required for disposal of both ERAD-LM and ERAD-LS substrates. Fig. S2 shows control of model ERAD substrate expression, unfolded protein response activity in Hrd1−/− cells, and analysis of secretion of soluble ERAD substrates. Fig. S3 illustrates dispensability of HRD1 for disposal of membrane-anchored BACE476 and CD3-δ. Fig. S4 shows how various siRNAs affect BiP level. Fig. S5 shows that NHKBACE and NHKCD3δ are membrane-tethered versions of NHK. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200910042/DC1.
Acknowledgments
We thank S. Fang, C. Niehrs, H. Ploegh, Y. Ye, and A.M. Weissman (Center for Cancer Research, National Cancer Institute, Frederick, MD) for sharing reagents and advices. We also give a special thank to D. Presotto, C. Caprara, L. Brambilla, T. Pertel, P. Paganetti, G. Noseda, and S. Monti.
M. Molinari is supported by grants from The Foundation for Research on Neurodegenerative Diseases, Onelife Advisors SA, The Swiss National Center of Competence in Research on Neural Plasticity and Repair, The Swiss National Science Foundation, The Synapsis Foundation, and The Bangerter-Rhyner Foundation.
Footnotes
Abbreviations used in this paper:
- ERAD
- ER-associated degradation
- HBS
- Hepes-buffered saline
- MEF
- mouse embryonic fibroblast
- NHK
- null Hong Kong
- PNS
- postnuclear supernatant
References
- Aebi M., Bernasconi R., Clerc S., Molinari M. 2009. N-glycan structures: recognition and processing in the ER. Trends Biochem. Sci. 10.1016/j.tibs.2009.10.001 [DOI] [PubMed] [Google Scholar]
- Alcock F., Swanton E. 2009. Mammalian OS-9 is upregulated in response to endoplasmic reticulum stress and facilitates ubiquitination of misfolded glycoproteins. J. Mol. Biol. 385:1032–1042 10.1016/j.jmb.2008.11.045 [DOI] [PubMed] [Google Scholar]
- Amano T., Yamasaki S., Yagishita N., Tsuchimochi K., Shin H., Kawahara K., Aratani S., Fujita H., Zhang L., Ikeda R., et al. 2003. Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel pathogenic factor for arthropathy. Genes Dev. 17:2436–2449 10.1101/gad.1096603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballar P., Shen Y., Yang H., Fang S. 2006. The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation. J. Biol. Chem. 281:35359–35368 10.1074/jbc.M603355200 [DOI] [PubMed] [Google Scholar]
- Ballar P., Ors A.U., Yang H., Fang S. 2010. Differential regulation of CFTRDeltaF508 degradation by ubiquitin ligases gp78 and Hrd1. Int. J. Biochem. Cell Biol. 42:167–173 10.1016/j.biocel.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Bernasconi R., Pertel T., Luban J., Molinari M. 2008. A dual task for the Xbp1-responsive OS-9 variants in the mammalian endoplasmic reticulum: inhibiting secretion of misfolded protein conformers and enhancing their disposal. J. Biol. Chem. 283:16446–16454 10.1074/jbc.M802272200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J.L., Fisher E.A. 2008. The many intersecting pathways underlying apolipoprotein B secretion and degradation. Trends Endocrinol. Metab. 19:254–259 10.1016/j.tem.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Wang J., Qi W., Miao H.H., Wang J., Ge L., DeBose-Boyd R.A., Tang J.J., Li B.L., Song B.L. 2007. Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell Metab. 6:115–128 10.1016/j.cmet.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Caramelo J.J., Parodi A.J. 2008. Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 283:10221–10225 10.1074/jbc.R700048200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T.A. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 126:361–373 10.1016/j.cell.2006.05.043 [DOI] [PubMed] [Google Scholar]
- Cattaneo M., Otsu M., Fagioli C., Martino S., Lotti L.V., Sitia R., Biunno I. 2008. SEL1L and HRD1 are involved in the degradation of unassembled secretory Ig-mu chains. J. Cell. Physiol. 215:794–802 10.1002/jcp.21364 [DOI] [PubMed] [Google Scholar]
- Chen B., Mariano J., Tsai Y.C., Chan A.H., Cohen M., Weissman A.M. 2006. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl. Acad. Sci. USA. 103:341–346 10.1073/pnas.0506618103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J.C., Shaler T.A., Tyler R.E., Kopito R.R. 2008. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10:272–282 10.1038/ncb1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S., Hirsch C., Oggier D.M., Deprez P., Jakob C., Sommer T., Aebi M. 2009. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 184:159–172 10.1083/jcb.200809198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier J.H., Tamura T., Sunryd J.C., Hebert D.N. 2009. EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Mol. Cell. 34:627–633 10.1016/j.molcel.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V., Quan E.M., Weissman J.S. 2006. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 126:349–359 10.1016/j.cell.2006.05.045 [DOI] [PubMed] [Google Scholar]
- Ermonval M., Kitzmüller C., Mir A.M., Cacan R., Ivessa N.E. 2001. N-glycan structure of a short-lived variant of ribophorin I expressed in the MadIA214 glycosylation-defective cell line reveals the role of a mannosidase that is not ER mannosidase I in the process of glycoprotein degradation. Glycobiology. 11:565–576 10.1093/glycob/11.7.565 [DOI] [PubMed] [Google Scholar]
- Fang S., Ferrone M., Yang C., Jensen J.P., Tiwari S., Weissman A.M. 2001. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 98:14422–14427 10.1073/pnas.251401598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R., Sommer T., Jarosch E. 2006. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 25:1827–1835 10.1038/sj.emboj.7601088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granell S., Baldini G. 2008. Inclusion bodies and autophagosomes: are ER-derived protective organelles different than classical autophagosomes? Autophagy. 4:375–377 [DOI] [PubMed] [Google Scholar]
- Hammond C., Braakman I., Helenius A. 1994. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. USA. 91:913–917 10.1073/pnas.91.3.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert D.N., Molinari M. 2007. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 87:1377–1408 10.1152/physrev.00050.2006 [DOI] [PubMed] [Google Scholar]
- Hirao K., Natsuka Y., Tamura T., Wada I., Morito D., Natsuka S., Romero P., Sleno B., Tremblay L.O., Herscovics A., et al. 2006. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J. Biol. Chem. 281:9650–9658 10.1074/jbc.M512191200 [DOI] [PubMed] [Google Scholar]
- Hirsch C., Gauss R., Horn S.C., Neuber O., Sommer T. 2009. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 458:453–460 10.1038/nature07962 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., You Z., Tremblay L.O., Nagata K., Herscovics A. 2007. Stimulation of ERAD of misfolded null Hong Kong alpha1-antitrypsin by Golgi alpha1,2-mannosidases. Biochem. Biophys. Res. Commun. 362:626–632 10.1016/j.bbrc.2007.08.057 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. 2008. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 283:20914–20924 10.1074/jbc.M709336200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Kamiya Y., Kamiya D., Kato K., Nagata K. 2009. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J. Biol. Chem. 284:17061–17068 10.1074/jbc.M809725200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer G., Piluek W.F., Fansler Z., Kreft S.G., Hochstrasser M., Brodsky J.L., Michaelis S. 2004. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 279:38369–38378 10.1074/jbc.M402468200 [DOI] [PubMed] [Google Scholar]
- Kostova Z., Tsai Y.C., Weissman A.M. 2007. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin. Cell Dev. Biol. 18:770–779 10.1016/j.semcdb.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger H., Miranda E., MacLeod I., Pérez J., Crowther D.C., Marciniak S.J., Lomas D.A. 2009. Endoplasmic reticulum-associated degradation (ERAD) and autophagy cooperate to degrade polymerogenic mutant serpins. J. Biol. Chem. 284:22793–22802 10.1074/jbc.M109.027102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer G.Z. 2009. Glycoprotein folding, quality control and ER-associated degradation. Curr. Opin. Struct. Biol. 19:515–523 10.1016/j.sbi.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Lerner M., Corcoran M., Cepeda D., Nielsen M.L., Zubarev R., Pontén F., Uhlén M., Hober S., Grandér D., Sangfelt O. 2007. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol. Biol. Cell. 18:1670–1682 10.1091/mbc.E06-03-0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J.S., Kim T., Fang S., Yamaguchi J., Weissman A.M., Fisher E.A., Ginsberg H.N. 2003. Overexpression of the tumor autocrine motility factor receptor Gp78, a ubiquitin protein ligase, results in increased ubiquitinylation and decreased secretion of apolipoprotein B100 in HepG2 cells. J. Biol. Chem. 278:23984–23988 10.1074/jbc.M302683200 [DOI] [PubMed] [Google Scholar]
- Liu Y., Choudhury P., Cabral C.M., Sifers R.N. 1997. Intracellular disposal of incompletely folded human alpha1-antitrypsin involves release from calnexin and post-translational trimming of asparagine-linked oligosaccharides. J. Biol. Chem. 272:7946–7951 10.1074/jbc.272.12.7946 [DOI] [PubMed] [Google Scholar]
- Malhotra J.D., Kaufman R.J. 2007. The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 18:716–731 10.1016/j.semcdb.2007.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken A.A., Brodsky J.L. 1996. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 132:291–298 10.1083/jcb.132.3.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M. 2007. N-glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 3:313–320 10.1038/nchembio880 [DOI] [PubMed] [Google Scholar]
- Molinari M., Galli C., Piccaluga V., Pieren M., Paganetti P. 2002. Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J. Cell Biol. 158:247–257 10.1083/jcb.200204122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M., Calanca V., Galli C., Lucca P., Paganetti P. 2003. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 299:1397–1400 10.1126/science.1079474 [DOI] [PubMed] [Google Scholar]
- Moremen K.W., Molinari M. 2006. N-linked glycan recognition and processing: the molecular basis of endoplasmic reticulum quality control. Curr. Opin. Struct. Biol. 16:592–599 10.1016/j.sbi.2006.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D.M., Tanaka K., Iwai K., Nagata K. 2008. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRDeltaF508. Mol. Biol. Cell. 19:1328–1336 10.1091/mbc.E07-06-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B., Lilley B.N., Ploegh H.L. 2006. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J. Cell Biol. 175:261–270 10.1083/jcb.200605196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B., Klemm E.J., Spooner E., Claessen J.H., Ploegh H.L. 2008. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. USA. 105:12325–12330 10.1073/pnas.0805371105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. 2001. The MRH domain suggests a shared ancestry for the mannose 6-phosphate receptors and other N-glycan-recognising proteins. Curr. Biol. 11:R499–R501 10.1016/S0960-9822(01)00302-5 [DOI] [PubMed] [Google Scholar]
- Nadav E., Shmueli A., Barr H., Gonen H., Ciechanover A., Reiss Y. 2003. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem. Biophys. Res. Commun. 303:91–97 10.1016/S0006-291X(03)00279-1 [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K., Brodsky J.L. 2008. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 9:861–870 10.1111/j.1600-0854.2008.00729.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda-Shimizu Y., Hendershot L.M. 2007. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol. Cell. 28:544–554 10.1016/j.molcel.2007.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari S., Galli C., Alanen H., Ruddock L., Molinari M. 2005. A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J. Biol. Chem. 280:2424–2428 10.1074/jbc.C400534200 [DOI] [PubMed] [Google Scholar]
- Olivari S., Cali T., Salo K.E., Paganetti P., Ruddock L.W., Molinari M. 2006. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun. 349:1278–1284 10.1016/j.bbrc.2006.08.186 [DOI] [PubMed] [Google Scholar]
- Paganetti P., Calanca V., Galli C., Stefani M., Molinari M. 2005. β-site specific intrabodies to decrease and prevent generation of Alzheimer's Abeta peptide. J. Cell Biol. 168:863–868 10.1083/jcb.200410047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi A.J. 2000. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem. J. 348:1–13 10.1042/0264-6021:3480001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter D.H. 2006. The role of autophagy in alpha-1-antitrypsin deficiency: a specific cellular response in genetic diseases associated with aggregation-prone proteins. Autophagy. 2:258–263 [DOI] [PubMed] [Google Scholar]
- Quan E.M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., Weissman J.S. 2008. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell. 32:870–877 10.1016/j.molcel.2008.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T., Hochstrasser M. 2008. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9:679–690 10.1038/nrm2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer J., Appenzeller-Herzog C., Johansson L., Bodenmiller B., Hartmann-Petersen R., Ellgaard L. 2009. A luminal flavoprotein in endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. USA. 106:14831–14836 10.1073/pnas.0900742106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8:519–529 10.1038/nrm2199 [DOI] [PubMed] [Google Scholar]
- Rutledge A.C., Qiu W., Zhang R., Kohen-Avramoglu R., Nemat-Gorgani N., Adeli K. 2009. Mechanisms targeting apolipoprotein B100 to proteasomal degradation: evidence that degradation is initiated by BiP binding at the N terminus and the formation of a p97 complex at the C terminus. Arterioscler. Thromb. Vasc. Biol. 29:579–585 10.1161/ATVBAHA.108.181859 [DOI] [PubMed] [Google Scholar]
- Shen Y., Ballar P., Fang S. 2006. Ubiquitin ligase gp78 increases solubility and facilitates degradation of the Z variant of alpha-1-antitrypsin. Biochem. Biophys. Res. Commun. 349:1285–1293 10.1016/j.bbrc.2006.08.173 [DOI] [PubMed] [Google Scholar]
- Shmueli A., Tsai Y.C., Yang M., Braun M.A., Weissman A.M. 2009. Targeting of gp78 for ubiquitin-mediated proteasomal degradation by Hrd1: cross-talk between E3s in the endoplasmic reticulum. Biochem. Biophys. Res. Commun. 390:758–762 10.1016/j.bbrc.2009.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B.L., Sever N., DeBose-Boyd R.A. 2005. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell. 19:829–840 10.1016/j.molcel.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Stagg H.R., Thomas M., van den Boomen D., Wiertz E.J., Drabkin H.A., Gemmill R.M., Lehner P.J. 2009. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J. Cell Biol. 186:685–692 10.1083/jcb.200906110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Cormier J.H., Hebert D.N. 2008. Sweet bays of ERAD. Trends Biochem. Sci. 33:298–300 10.1016/j.tibs.2008.04.013 [DOI] [PubMed] [Google Scholar]
- Taxis C., Hitt R., Park S.H., Deak P.M., Kostova Z., Wolf D.H. 2003. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 278:35903–35913 10.1074/jbc.M301080200 [DOI] [PubMed] [Google Scholar]
- Tsai Y.C., Mendoza A., Mariano J.M., Zhou M., Kostova Z., Chen B., Veenstra T., Hewitt S.M., Helman L.J., Khanna C., Weissman A.M. 2007. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat. Med. 13:1504–1509 10.1038/nm1686 [DOI] [PubMed] [Google Scholar]
- Vanoni O., Paganetti P., Molinari M. 2008. Consequences of individual N-glycan deletions and of proteasomal inhibition on secretion of active BACE. Mol. Biol. Cell. 19:4086–4098 10.1091/mbc.E08-05-0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Ng D.T. 2004. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165:41–52 10.1083/jcb.200309132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij N., Fang S., Zeitlin P.L. 2006. Selective inhibition of endoplasmic reticulum-associated degradation rescues DeltaF508-cystic fibrosis transmembrane regulator and suppresses interleukin-8 levels: therapeutic implications. J. Biol. Chem. 281:17369–17378 10.1074/jbc.M600509200 [DOI] [PubMed] [Google Scholar]
- Wang S., Ng D.T. 2008. Lectins sweet-talk proteins into ERAD. Nat. Cell Biol. 10:251–253 10.1038/ncb0308-251 [DOI] [PubMed] [Google Scholar]
- Wang Q., Li L., Ye Y. 2006. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J. Cell Biol. 174:963–971 10.1083/jcb.200605100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Dong H., Soroka C.J., Wei N., Boyer J.L., Hochstrasser M. 2008. Degradation of the bile salt export pump at endoplasmic reticulum in progressive familial intrahepatic cholestasis type II. Hepatology. 48:1558–1569 10.1002/hep.22499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer M., Forte G.M., Stirling C.J. 2008. Sec61p is required for ERAD-L: genetic dissection of the translocation and ERAD-L functions of Sec61P using novel derivatives of CPY. J. Biol. Chem. 283:33883–33888 10.1074/jbc.M803054200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagishita N., Ohneda K., Amano T., Yamasaki S., Sugiura A., Tsuchimochi K., Shin H., Kawahara K., Ohneda O., Ohta T., et al. 2005. Essential role of synoviolin in embryogenesis. J. Biol. Chem. 280:7909–7916 10.1074/jbc.M410863200 [DOI] [PubMed] [Google Scholar]
- Yang M., Omura S., Bonifacino J.S., Weissman A.M. 1998. Novel aspects of degradation of T cell receptor subunits from the endoplasmic reticulum (ER) in T cells: importance of oligosaccharide processing, ubiquitination, and proteasome-dependent removal from ER membranes. J. Exp. Med. 187:835–846 10.1084/jem.187.6.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Zhong X., Ballar P., Luo S., Shen Y., Rubinsztein D.C., Monteiro M.J., Fang S. 2007. Ubiquitin ligase Hrd1 enhances the degradation and suppresses the toxicity of polyglutamine-expanded huntingtin. Exp. Cell Res. 313:538–550 10.1016/j.yexcr.2006.10.031 [DOI] [PubMed] [Google Scholar]
- Yoshida H. 2007. ER stress and diseases. FEBS J. 274:630–658 10.1111/j.1742-4658.2007.05639.x [DOI] [PubMed] [Google Scholar]
- Younger J.M., Chen L., Ren H.Y., Rosser M.F., Turnbull E.L., Fan C.Y., Patterson C., Cyr D.M. 2006. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 126:571–582 10.1016/j.cell.2006.06.041 [DOI] [PubMed] [Google Scholar]
- Zhong X., Shen Y., Ballar P., Apostolou A., Agami R., Fang S. 2004. AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. J. Biol. Chem. 279:45676–45684 10.1074/jbc.M409034200 [DOI] [PubMed] [Google Scholar]