Abstract
High-affinity antibodies are generated by somatic hypermutation with nucleotide substitutions introduced into the IgV in a semirandom fashion, but with intrinsic mutational hotspots strategically located to optimize antibody affinity maturation. The process is dependent on activation-induced deaminase (AID), an enzyme that can deaminate deoxycytidine in DNA in vitro, where its activity is sensitive to the identity of the 5′-flanking nucleotide. As a critical test of whether such DNA deamination activity underpins antibody diversification and to gain insight into the extent to which the antibody mutation spectrum is dependent on the intrinsic substrate specificity of AID, we investigated whether it is possible to change the IgV mutation spectrum by altering AID’s active site such that it prefers a pyrimidine (rather than a purine) flanking the targeted deoxycytidine. Consistent with the DNA deamination mechanism, B cells expressing the modified AID proteins yield altered IgV mutation spectra (exhibiting a purine→pyrimidine shift in flanking nucleotide preference) and altered hotspots. However, AID-catalyzed deamination of IgV targets in vitro does not yield the same degree of hotspot dominance to that observed in vivo, indicating the importance of features beyond AID’s active site and DNA local sequence environment in determining in vivo hotspot dominance.
After initial encounter of B cells with antigen, their IgV genes are subjected to somatic hypermutation (SHM), a process that underpins antibody affinity maturation. The isotype of the antibody produced can also be altered from IgM to IgG, IgA, or IgE by class-switch recombination (CSR). Both SHM and CSR are dependent on the protein activation-induced deaminase (AID). Although homology of AID to the RNA-editing enzyme APOBEC1 led to the early suggestion that AID also functioned by editing RNA (Muramatsu et al., 2000), much evidence indicates that AID functions by directly deaminating deoxycytidine residues within the immunoglobulin locus to yield deoxyuridine (Alt and Honjo, 2007; Di Noia and Neuberger, 2007). Nevertheless, although the supporting evidence is extensive, the DNA deamination mechanism has not gained universal acceptance. There has been discussion as to whether the uracil-excision activity of uracil-DNA glycosylase is essential for its function in antibody diversification (Begum et al., 2004, 2007, 2009; Stivers, 2004; Nagaoka et al., 2005; Di Noia et al., 2007) and, more recently, the dependence of antibody diversification on the DNA deaminase activity of AID has also been called into question (Shivarov et al., 2008).
It has long been known that somatic mutations are not randomly distributed along the IgV gene. Although most nucleotide positions in the IgV can be targeted during SHM, some positions are intrinsically more mutable than others, with the SHM process exhibiting a clear preference for certain major hotspots (Berek and Milstein, 1987; Sharpe et al., 1991; Rogozin and Kolchanov, 1992; Betz et al., 1993b). Thus, for example, a cytosine (C) residue in the IgV is especially likely to be mutated during SHM if it forms part of a WRCY consensus (where W = adenosine/thymine, R = purine, and Y = pyrimidine; Rogozin and Kolchanov, 1992; Betz et al., 1993a; Dörner et al., 1998; Milstein et al., 1998; Oprea and Kepler, 1999). Indeed, most of the dominant mutational hotspots within IgV genes conform to the WRCY consensus, although not all WRCY consensuses form major hotspots. The location of these hotspots has been proposed to have been selected during evolution because either they are located at positions where amino acid substitutions are likely to be particularly effective in allowing affinity maturation or they are at codons where single-nucleotide substitutions are likely to yield a range of potentially useful amino acid substitutions and a reduced likelihood of generating stop codons (Chang and Casali, 1994; Wagner et al., 1995; Jolly et al., 1996; Kepler, 1997). The CDR1 in both VH and VL is a preferred target of somatic hypermutation in vivo and is typically rich in AGY serine codons, many of which conform well (on the opposite strand) to the WRC consensus for AID targets (Betz et al., 1993b; Cowell et al., 1999).
Extensive biochemical studies have revealed that when acting on a DNA target in vitro recombinant AID also targets C residues for deamination in a context-dependent manner with WRC being a favored consensus in single-stranded DNA substrates (Pham et al., 2003; Bransteitter et al., 2004; Yu et al., 2004; Larijani et al., 2005). Thus, there appears to be a broad correlation between the in vitro target preferences of AID and the pattern of antibody hotspots in vivo. However, comparison of mutation spectra (albeit on different DNA target sequences in vivo and in vitro) suggests that the two are not identical. Thus, for example, mutational targeting in IgV genes appears to show sensitivity to the nucleotide flanking the 3′-side of the targeted C, whereas such sensitivity is not apparent from in vitro deamination studies (Pham et al., 2003; Beale et al., 2004; Rogozin and Diaz, 2004).
Here, to gain insight into the contribution of the target preference of AID’s catalytic site to the overall spectrum of antibody hypermutation and as a critical test of the DNA deamination mechanism of antibody gene diversification, we have generated mutant AIDs with altered sequence preferences and observed how this affects the spectrum of antibody hypermutation.
RESULTS
Active-site mutants of AID
The human APOBEC3 proteins, like AID, are able to deaminate C in DNA, but whereas AID prefers to target C residues flanked by a 5′-flanking purine, the APOBEC3s largely prefer a 5′-pyrimidine flank with individual APOBEC3s differing with regard to the details of this 5′-flanking nucleotide preference. Comparison of human APOBEC3 gene sequences previously led us to speculate that a stretch of ∼8 amino acids located some 60 residues from the C-terminal end of the protein domain played an important role in determining this flanking nucleotide preference; we supported this speculation by showing that the target specificity of APOBEC3F could indeed be altered by specific amino acid changes in this region (Langlois et al., 2005). Subsequently, in light of the crystal structure of APOBEC2 (Prochnow et al., 2007) together with that of the TadA tRNA-adenosine deaminase in complex with an oligonucleotide substrate (Losey et al., 2006), we suggested this amino acid stretch in both AID and APOBEC3s likely formed a contact with the DNA substrate (Conticello et al., 2007). Thus, to generate AID variants with altered target specificity, we focused on this stretch of amino acids and replaced AID residues 115–123 with the corresponding portion of APOBEC3s (Fig. 1 A). Kohli et al. (2009) have recently adopted a similar approach to modify AID’s substrate preference.
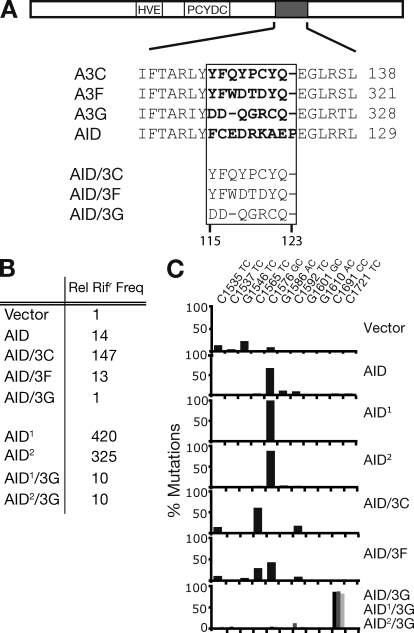
Figure 1.
Changing the target specificity of AID. (A) Depiction of AID cDNA showing the Zn coordination domain (HVE and PCYDC), and the region containing a putative substrate contact loop (gray), with alignment to equivalent regions of APOBEC3C, 3F, and 3G shown below. Residues predicted to be in the substrate contact loop are highlighted in bold. In the AID chimeras, residues 115–123 of AID were replaced by equivalent residues in APOBEC3C/F/G as indicated. (B) Bacterial mutator activity of variant AIDs was determined by the mean frequency from 12 independent cultures with which they yielded colonies resistant to rifampicin (Rifr), expressed relative to that given by the vector-only control. AID1 and AID2 are previously described upmutants of wild-type AID (AID1: K10E/T82I/E156G; AID2: K34E/E156G/R157T; Wang et al., 2009). (C) Target specificity of the various AID-derived deaminases as judged by the distribution of rpoB mutations in rifr resistant colonies. Transition mutations at any one of eleven C:G pairs within rpoB can give rise to Rifr. Mutations at a specific C:G pair are expressed as a percentage of the total number of Rifr colonies scored for each deaminase. Results with AID/3G are shaded black, AID1/3G in dark gray, and AID2/3G in light gray (all largely target C1691).
AID mutants in which residues 115–123 had been replaced by equivalent regions from APOBEC3C, APOBEC3F, and APOBEC3G were cloned into a bacterial expression plasmid, and their mutator activity assayed by monitoring the frequency with which they yielded colonies resistant to rifampicin after transformation into E. coli. Although the AID/3C and AID/3F proteins retained good mutator activity, AID/3G gave rifampicin-resistant colonies at a frequency indistinguishable over background (Fig. 1 B). Rifampicin resistance is conferred by one of a limited number of mutations in rpoB with the nature of the mutations obtained giving insight into the target specificity of the deaminase (Harris et al., 2002). Wild-type AID prefers to deaminate the C residues at rpoB position 1576 (C1576), which has a 5′-flanking purine (G) residue. In contrast, the AID variants in which residues 115–123 had been replaced by corresponding regions from APOBEC3C/F/G showed a preference (as do the APOBEC3s themselves) for pyrimidines at the −1 position. Thus, AID/3C and AID/3F show a shift in the spectrum of rpoB mutations to favor targets with a 5′-T (C1535, C1565, and C1592), whereas the AID/3G transformants almost solely targeted C1691, which has a 5′-C (Fig. 1 C).
Although the mutator activity of AID/3G is sufficient to yield a shift in the distribution of rpoB mutations observed in rifampicin-resistant E. coli, its mutator activity is nevertheless considerably lower than that of the wild-type enzyme because it does not yield a total frequency of mutation to rifampicin-resistance that is above background (Fig. 1). This led us to anticipate that it might be difficult to obtain a good database of IgV mutations when this AID mutant was expressed in B cells. To avoid this problem, we generated two AID/3G upmutants (which we designate AID1/3G and AID2/3G) in which three additional amino acid substitutions (AID1: K10E, T82I, and E156G; AID2: K34E, E156G, and R157T) were introduced into these proteins, substitutions that we have previously shown to increase AID’s specific activity without detectably affecting its target preference (Wang et al., 2009). Both these AID/3G upmutants appear to retain the parental AID/3G protein’s preference for a 5′-flanking C residue as judged by the rpoB mutation spectrum (Fig. 1 C).
We also prepared AID variants (designated AID*, AID*/3F, AID1*/3G, etc.) in which the C-terminal portion of AID (which includes its nuclear export sequence) has been deleted because previous work with wild-type AID (Barreto et al., 2003) has indicated that such deletions could also yield significantly increased mutation frequencies in B cell transfectants. The C-terminal truncation does not yield a detectable effect on AID mutational target site preference in bacterial mutation assays (Fig. S1).
Modified AIDs exhibit altered mutation spectra in vitro
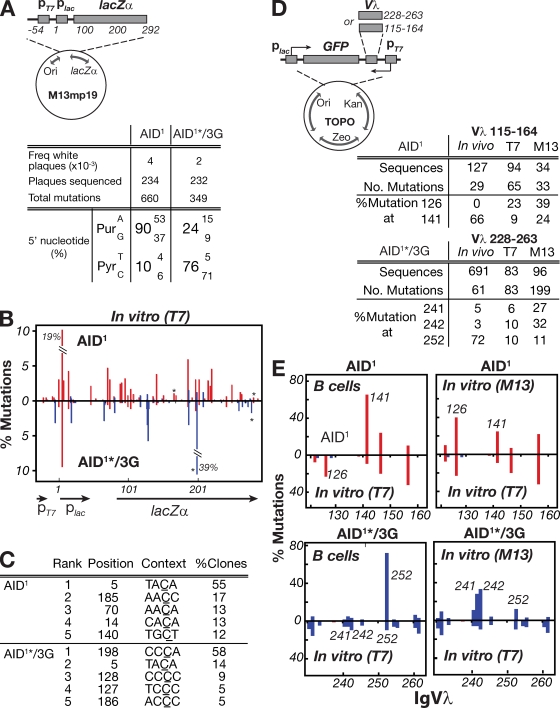
To analyze the biochemical target specificity of the mutant AIDs in greater detail, the various AID enzymes were partially purified from E. coli extracts as recombinant GST-fusion proteins (Fig. S1 A) and used to deaminate single-stranded lacZ target DNA in the context of the M13 gapped duplex assay (Bebenek and Kunkel, 1995; Pham et al., 2003). In this assay, recombinant GST-AID is incubated with gapped duplex M13lacZ DNA (Fig. 2 A), which is then transformed into E. coli; white (as opposed to blue) plaques reveal mutated clones.
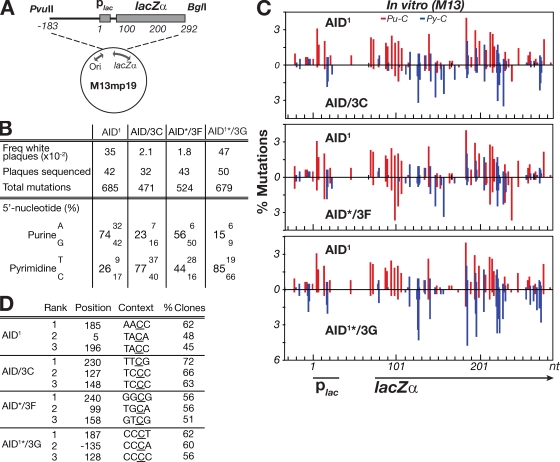
Figure 2.
Mutation spectrum of modified AIDs assayed in vitro on a gapped duplex lacZ target. (A) Depiction of the M13mp19lacZ gapped duplex substrate DNA. (B) 5′-flanking nucleotide preferences of the C mutations produced by the variant AID deaminases. The spectra shown for the AID/3G chimera is from an AID1*/3G derivative in which the asterisk denotes that the protein has been truncated at amino acid position 190 of AID (removing the nuclear export sequence) and is shown to allow comparison with the same AID variant analyzed in transfected DT40 B cells (see Fig. 3). The C-terminal truncations do not detectably alter the patterns of in vitro mutational targeting. (C) Distribution of mutations over a 310-nt stretch of the single-stranded lacZ target. The numbers of independent mutations at each nucleotide position are expressed as a percentage of the total mutation database (as analyzed over the entire 475-nt single-stranded target). Nucleotide position 1 is defined as the start of the lac promoter. Mutations at C residues flanked by a 5′-purine (Pu-C) are shown in red, those flanked by a 5′-pyrimidine (Py-C) in blue. (D) Identity of the three most frequently targeted residues by each deaminase, with targeting expressed as the percentage of clones analyzed in which the relevant cytosine (underlined) was mutated.
Analysis of 30–50 mutated M13lacZ clones in each experiment yielded databases of 471–685 mutations, all of which were transitions at C:G pairs (Fig. 2 B). In the case of AID1, 74% of the C mutations were at sites flanked by a 5′-purine. In contrast, the AID mutants carrying transplanted segments from APOBEC3 proteins showed a shift toward a preference for a flanking pyrimidine, especially marked in the case of the AID/3C and AID/3G proteins (85 and 77% pyrimidine, respectively; Fig. 2 B). This change in flanking nucleotide preference is accompanied by a change in the distribution of mutations along lacZ (Fig. 2, C and D). Given that for most of the AID variants, the mutated sequences carried an average of 10–16 transition mutations over the 475-nt stretch of single-stranded substrate analyzed (Fig. S1 B), the mutations observed will largely reflect the intrinsic preference of the mutational process without extensive skewing by virtue of the selection for lacZ inactivation.
Modified AIDs give altered IgV mutation spectra in B cells
To ascertain whether changing the catalytic specificity of AID results in an alteration in the distribution of nucleotide substitutions introduced during SHM in B cells, we expressed the mutant AIDs in an AID-deficient, ψV-deleted chicken DT40 B cell line. In this cell-line, the mutations are largely restricted to nucleotide substitutions at C:G pairs, with little contribution from polymerase η–triggered hypermutation (Sale et al., 2001; Di Noia and Neuberger, 2002; Arakawa et al., 2004), meaning that mutations at C:G can largely be ascribed to the direct effects of AID rather than possibly being a consequence of a second phase of mutation creation. The frequency of SHM at the IgV can be inferred from the frequency of generation of sIgM-loss variants (Buerstedde et al., 1990; Sale et al., 2001). As judged by this assay, both AID/3C and AID/3F are proficient in SHM. Indeed, AID/3C is even more potent than the wild-type enzyme, especially when account is taken of the lower abundance of the AID/3C polypeptide in the B cell extracts (Fig. 3 A). The low abundance of AID/3C is evident in multiple independent transfectants (unpublished data) and parallels observations in previous work (Wang et al., 2009) that AID mutants displaying increased specific activity are usually expressed at lower abundance in B cell transfectants. The reason for this low expression is unclear but it might reflect cytotoxicity of excessive DNA deaminase activity.
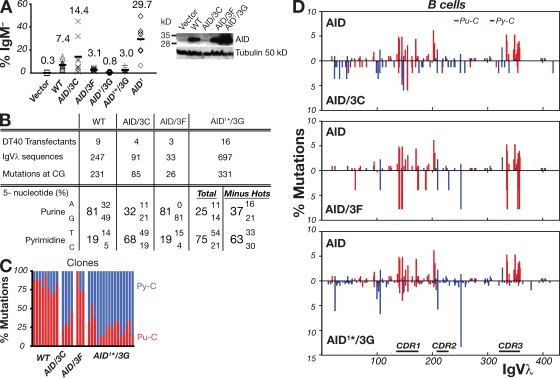
Figure 3.
Modified AIDs give altered IgVλ hypermutation spectra in B cells. (A) Hypermutation of IgVλ was assayed by monitoring surface IgM-loss in AID−/− ψV−/− sIgM+ DT40 cells that had been stably transfected with constructs coexpressing the indicated AID mutants together with GFP. For each construct, the percentage of surface IgM-loss variants in 8–12 independent clonal transfectants were determined 3 wk after subculturing. On the right, Western blots representative of multiple clones show AID abundance in the DT40 cell extracts, with tubulin as loading control. (B) 5′-flanking nucleotide preferences of the IgVλ C mutations produced by the variant AID deaminases in the DT40 clonal transfectants. The compilations are based on mutations detected in unsorted DT40 cells analyzed 8 wk after transfection, except for AID/3C, where sequences from both unsorted and sorted sIgM− populations contributed to the mutation database. In the case of AID1*/3G, the nucleotide preferences are given based on an analysis of all the mutations in the dataset, as well as from an analysis in which the four major hotspots were removed from the calculations. (C) Percentage of mutated C residues flanked by 5′-purine (red) or 5′-pyrimidine (blue) in IgVλ sequences analyzed from individual expanded DT40 clonal transfectants represented by each bar. (D) Distribution of IgVλ mutations in the DT40 transfectants, in each comparing the spectrum achieved with a modified AID (below the line) to that achieved with wild-type AID (above the line). Mutations (which were >95% at C:G pairs) were computed as being caused by C deamination with those Cs flanked by a 5′-purine (Pu-C) indicated in red and those by a 5′-pyrimidine (Py-C) in blue. Further details on the mutations obtained with these deaminases, as well as with AID1 are shown in Fig. S2 and Fig. S3.
In contrast to the AID/3C and AID/3F mutants, the AID1/3G mutant gave only a very low frequency of sIgM-loss variants. However, as anticipated, this frequency was considerably enhanced by deleting the AID C-terminal portion (Fig. 3 A).
To characterize the IgV gene hypermutation spectrum in the DT40 B cell transfectants expressing the various modified AID proteins, the IgVλ segment from multiple independent transfectants for each expression construct was PCR amplified and sequenced after 8 wk of clonal expansion. The results reveal that the modifications of the AID active site result in a substantial alteration to the IgVλ mutation spectrum. Thus, AID/3C and AID1*/3G largely target C residues with a 5′-flanking pyrimidine residue (68 and 75%, respectively) in contrast to the wild-type enzyme in which only 19% of the mutations are targeted to C residues with a 5′-flanking pyrimidine (Fig. 3 B). This striking change in mutation spectrum is evident both in the composite datasets as well as in each of those from independent clones (Fig. 3 C). In contrast, AID/3F maintains the preference of the parental enzyme for a flanking purine residue, but (as found in the in vitro assay on the gapped duplex lacZ substrate; Fig. 2 B) there is a shift toward a preference for a flanking guanine rather than adenine (Fig. 3 B).
The change in mutational targeting as judged by the nature of the 5′-flanking nucleotide broadly correlates with an altered mutational spectrum as judged by the distribution of nucleotide substitutions along the IgVλ segment (Fig. 3 D). Thus, for example, the IgVλ mutation hotspots are found at distinct locations when comparing wild-type AID with AID1*/3G. With wild-type AID (as well as in the AID1 upmutant; Fig. 3 D, Fig. S2, and Fig. S3), clusters of hotspots are evident within CDR1, toward the 5′-side of CDR2 and also within CDR3, with these hotspots mostly conforming to a WRC consensus as observed previously (Sale et al., 2001; Arakawa et al., 2004; Saribasak et al., 2006; Wang et al., 2009). In contrast, the IgVλ mutations obtained using AID1*/3G show reduced clustering in CDR1 and CDR3, with a focusing on hotspots with a 5′-pyrimidine flank and which are located in regions (FR1 and FR3) that are relatively spared by the wild-type enzyme (Fig. 3 D and Fig. S3). Thus, changing the active site of AID modifies the mutation spectrum that is obtained both by DNA deamination in vitro and by antibody hypermutation in B cell transfectants.
Different hotspots dominate the B cell and in vitro mutation spectra
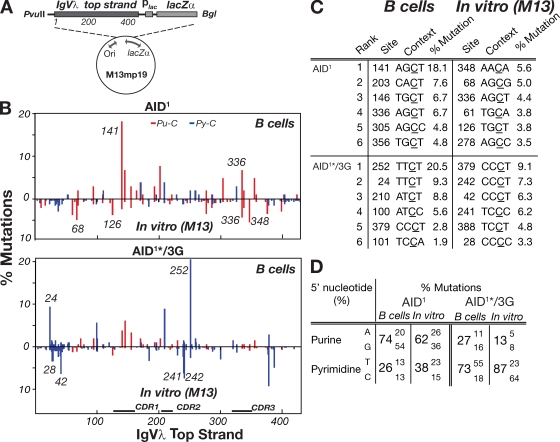
Although the active site modifications in AID/3C and AID1*/3G have led to a shift from a preference for a flanking 5′-purine to a flanking 5′-pyrimidine in both in vivo (DT40 IgVλ) and in vitro (gapped-duplex lacZ) mutation assays, the nature of the shift in the two assays is not equivalent. Thus, for AID/3G, although T is the flanking pyrimidine of choice in the DT40 IgVλ mutation spectrum, a flanking C is preferred in the in vitro assay (Fig. 2 B and Fig. 3 B). This discrepancy is substantially caused by the skewing effect of a few major hotspots in the DT40 IgVλ spectrum (Fig. 3 B), suggesting that some aspect of hypermutation in B cells might result in the creation of dominant hotspots which are not recapitulated in the in vitro gapped duplex assay.
To confirm this, we performed the gapped duplex mutation assay on an IgVλ (rather than lacZ) target sequence, comparing the resulting in vitro mutation spectrum to that observed on the equivalent (nontranscribed) IgVλ DNA strand in DT40 B cells (Fig. 4, A–D). Significant differences in mutational targeting are indeed observed; for example, compare the relative targeting in vivo and in vitro at Vλ positions 126/141 or 336/348 with wild-type AID, or compare targeting at 24/28/42 as well as at 241/242/252 with AID1*/3G (Fig. 4 B). These differences are similarly evident if the highly mutated sequences are excluded from the database used to deduce the patterns of in vitro mutational targeting.
Figure 4.
Different IgVλ hotspots dominate the mutation spectra in B cells and in the gapped duplex assay. (A) The M13mp19-IgVλ-lacZ gapped duplex substrate DNA is depicted with more detailed information provided in Fig. S5 A. Nucleotide position 1 is equivalent to the first nucleotide of the in vivo IgVλ analyzed in Fig. 3. (B) The graphs compare the distribution of IgVλ mutations obtained with AID1 or AID1*/3G in the gapped duplex assay (shown below the line) with the distribution of IgVλ top strand C mutations obtained with the same deaminases in DT40 B cells (shown above the line). The distributions of IgVλ top strand C mutations in DT40 cells for AID1*/3G and AID1 derive from the same mutation databases used in Fig. 3 and Fig. S2, respectively, although those figures portray all C mutations (i.e., whichever DNA strand they have occurred upon). The positions of some individual hotspots are indicated in italics. Red, 5′-purine flank (Pu-C); blue, 5′-pyrimidine flank (Py-C). (C) The location and local context of the most frequently mutated C residues along the IgVλ top strand in the gapped duplex and DT40 B cell mutation assays are compared. Mutation load at each nucleotide position is represented as a percentage of the total mutations in each dataset. (D) The percentages of total C mutations in the gapped duplex assays and DT40 B cells (top strand) at C residues with each of the four possible 5′-flanking bases are compared for AID1 and AID1*/3G.
To find out whether the differences in targeting could reflect that mutation in vivo likely occurs while transcribing double-stranded DNA, whereas the gapped duplex assay uses a single-stranded DNA target, we investigated mutational targeting using an assay described by Bransteitter et al. (2004) in which recombinant AID is incubated with double-stranded DNA at the same time as the target gene (lacZ) within the substrate is being transcribed from a linked T7 polymerase promoter. In such an assay, AID1*/3G clearly differs from wild-type AID, still preferring a 5′-pyrimidine, especially 5′-C, rather than the 5′-T that is observed in DT40 B cells (Fig. 5, A–C). To assess mutational targeting within an IgVλ substrate in an in vitro transcription-coupled assay, we modified the T7-linked assay to create a substrate in which unselected mutations in short segments of IgVλ can be scored in clones that have suffered mutational inactivation of a closely linked GFP reporter gene. However, in such assays we found that, as in the gapped duplex assay, we still did not recapture the relative dominance of major hotspots at IgVλ positions 141 (with wild-type AID) or 252 (with AID1*/3G) that was observed during hypermutation in DT40 cells (Fig. 5, D and E). In fact, the mutational targeting in the transcription-linked assay looks rather more similar to that obtained in the gapped duplex assay than to the pattern of mutational targeting observed in DT40 B cells. Thus, neither in vitro assay fully recapitulates the pattern of IgV hotspot dominance observed in B cells.
Figure 5.
Mutation spectra from in vitro transcription-coupled mutation assays. (A) The T7-lacZ transcription-coupled substrate in which a T7 promoter is inserted upstream of the lac promoter in M13mp19 (Fig. S5 B). Nucleotide position 1 is defined as the start of the lac promoter. The adjacent table shows the 5′-flanking nucleotide preferences of the C mutations produced by GST-AID1 or GST-AID1*/3G in lac target DNA during in vitro transcription by phage T7 RNA polymerase. (B) Mutation distribution along the T7 transcription-coupled lacZ target DNA with mutation at each nucleotide position expressed as the percentage of total mutations. Two heavily mutated positions are off-scale: their percentage mutations are indicated. Because mutation analysis was restricted to Lac− plaques, this selection results in a skewing in favor of lac-inactivating mutations, although most mutated templates carried 1–3 mutations in the target region. Positions at which C deamination yields a stop codon are indicated by an asterisk. (C) The location and local context of the five most frequently mutated C residues along the transcribed lac target. All the mutated residues shown are located on the top (nontranscribed) strand. (D) Transcription-linked mutation of a T7-linked GFP-Vλ target. The substrate DNA is a derivative of plasmid pCR-Blunt II-TOPO in which a region of IgVλ (residues 115–164 or 228–263) has been inserted between the T7 promoter and a GFP reporter with T7-catalyzed transcription of the IgVλ fragment being in the same sense as IgVλ transcription in B cells (Fig. S5 C). For each construct (pCR-GFP-Vλ115−164-T7 and pCR-GFP-Vλ228−263-T7), the tables compare the percentage of total mutations within the target Vλ region that occur at selected individual positions with the equivalent percentage mutation values for the same positions over the same target regions in the M13 or B cell mutation assays. (E) Comparison of the mutation distributions obtained in the T7-coupled mutation assay over IgVλ residues 115–164 (for AID1) or 228–263 (for AID1*/3G) to the distributions obtained in DT40 B cells (left) or in the gapped duplex assay (right). In these comparisons, analysis is restricted to nontranscribed strand mutations. Positions of individual hotspots are indicated in italics. Red, 5′-purine flank; blue, 5′-pyrimidine flank.
AIDs with altered target preference permit class switching
The preference of wild-type AID for a WRC (and especially AGCT) target sequence accords well with the sequence of the short consensus repeat in the Sμ switch region (GAGCT). We were therefore interested to know whether the AID mutants that preferred a 5′-flanking pyrimidine would nevertheless retain the ability to trigger switching. Both AID/3C and AID/3F were able to potentiate switching after retroviral transduction into LPS-activated AID-deficient B cells, although this occurred at a somewhat reduced efficiency (Fig. S4). The AID1/3G variant showed greatly reduced switching activity, but we cannot distinguish the extent to which this is caused by its altered target site preference as opposed to its reduced deaminase activity.
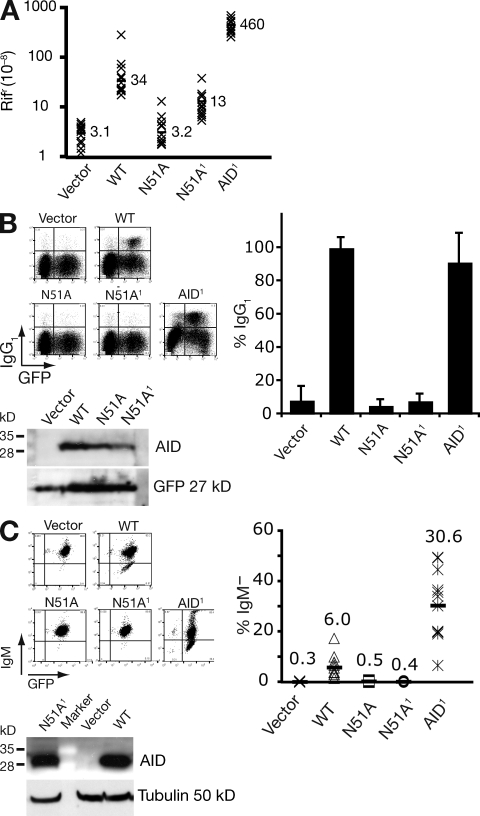
N51A mutation in AID reduces both DNA deamination and class switching
The finding that modifying AID’s active site such that it prefers a 5′-flanking pyrimidine leads to corresponding change in the IgV gene mutation spectrum in B cells lends further strong support to the DNA deamination model of antibody diversification. It is, however, difficult to reconcile with the claim that a single-point mutation (N51A) in AID can abolish its DNA deaminase activity while still allowing AID to retain 50% of its activity in class-switch recombination (Shivarov et al., 2008). We therefore decided to reexamine the effect of the N51A mutation on AID activity. In the context of human AID, the N51A mutation reduces the DNA deaminase activity of AID down to background as judged by the bacterial mutation assay (Fig. 6 A). However, we suspect that this mutation does not totally destroy AID activity. Mutator activity was still detected after introduction of the N51A substitution into our AID1 upmutant (which is substantially more active than wild-type AID as judged in the rifampicin resistance assay; Fig. 6 A). Nevertheless, neither AID[N51A] nor AID1[N51A] exhibited any detectable antibody diversification activity in either switching or somatic hypermutation assays (Fig. 6, B and C). Thus, the N51A mutation substantially reduces AID’s DNA deaminase activity (though probably not wholly destroying it) with the same mutant displaying a lack of detectable activity in antibody diversification assays. We therefore see no evidence in these experiments of CSR being triggered by an AID mutant lacking DNA deaminase activity.
Figure 6.
AID N51A mutants retain DNA deaminase activity. (A) Bacterial mutator activity of AID variants monitored by frequency of Rifr colonies in overnight cultures with mean frequencies (x10−8) from 12 independent cultures indicated. The N51A mutant of AID1 is abbreviated N51A1. (B) Switching to IgG1 in LPS+IL-4 cultures of AID-deficient B cells that have been transduced with retroviruses encoding the various AIDs as indicated together with a linked IRES-GFP. Representative flow cytometry plots are presented along with histograms showing the results of four experiments (mean and SD, with 2–3 mice per construct per experiment). AID abundance in the B cell extracts 3 d after retroviral transduction was monitored by Western blotting; the blot was reprobed with anti-GFP antibodies as a control. (C) The frequency of surface IgM-loss variants in transfectants of AID−/− ψV−/− sIgM+ DT40 that coexpress the indicated AID mutant and GFP is presented both with a histogram showing the average percentage sIgM-loss variants in 12 independent clonal transfectants as well as by representative flow cytometry plots. Representative Western blots show AID abundance in the DT40 cell extracts with tubulin as loading control.
DISCUSSION
Altering the active site of AID such that it preferentially deaminates deoxycytidine residues in DNA that are flanked by a 5′-pyrimidine (rather than purine) results in an analogous change in the IgVλ gene hypermutation spectrum in B cell transfectants. This provides strong support to the idea that the action of AID in antibody diversification in vivo involves its direct interaction with the DNA substrate.
The fact that by changing AID residues 115–123 it is possible to change the enzyme’s preference for 5′-flanking nucleotide is in agreement with our earlier observations that target site preference changes (both in vitro and in vivo) also result from modifications in the equivalent region of APOBEC3 deaminases (Langlois et al., 2005). Furthermore, the altered in vitro deamination specificity of our AID/3F and AID/3G enzymes coincides with the altered deamination preferences as analyzed by in vitro deamination assays of very similar AID variants that were recently described by Kohli et al. (2009). However, although the AID variants made by us and by Kohli et al. are very similar, they are not identical, which presumably accounts for apparent minor differences in their catalytic activity/specificity.
Recent work in which it has been proposed that substitution of Asn51 by alanine in mouse AID results in loss of DNA deaminase activity, but retention of immunoglobulin class-switching (Shivarov et al., 2008), is, however, clearly at odds with the DNA deamination model of antibody diversification. We revisited the effect of the N51A substitution, but in the context of human AID, where we have previously generated upmutants that allow for more sensitive detection of DNA deaminase activity. In this context, we find that the N51A substitution causes a very substantial reduction (though not total ablation) of DNA deaminase activity and also results in a very substantial reduction in its ability to trigger class-switching. We therefore do not consider that the previous results of Shivarov et al. (2008) indicate that AID’s function in antibody diversification is dissociated from its DNA deaminase activity, but instead believe that these authors’ results reflect differences in the dynamic ranges of the assays used for investigating these processes.
Although the active site of AID clearly plays a major role in determining the identity of the deamination targets acted upon, the hierarchy of target preferences of AID as judged by its in vitro action on single-stranded or transcribing double-stranded substrates does not recapture the detailed pattern of AID-dependent hypermutation seen in B cells. Mutations are more evenly distributed among the various target C residues in the IgV in the in vitro assays as compared with IgV hypermutation in B cells, where dominant hotspots can more readily be discerned (Fig. 4). Thus, the preferential focusing of SHM on the IgV CDR1 seen in previous studies (Betz et al., 1993a; Tomlinson et al., 1996) probably does not merely derive from the fact that CDR1 contains a cluster of AID consensus target sequences. Indeed, even with the AID/3G mutant that shows no affinity for WRC or CDR1 targets in in vitro DNA deamination assays, circumstances in B cells nevertheless appear to encourage a small amount of mutation accumulation in CDR1 (Fig. 3 D and Fig. 4 B).
What could be responsible for these differences in the mutational targeting at the IgV observed when comparing the B cell hypermutation and in vitro deamination assays? Although there could be a contribution from differential DNA repair, this is unlikely to be the sole explanation because the mutation spectrum in the in vitro assays is examined after transformation into E. coli cells deficient in the uracil-DNA glycosylase repair enzyme; previous work has also revealed that dominant IgV gene mutational hotspots observed during hypermutation in normal B cells are still evident in repair-deficient B cells (Di Noia and Neuberger, 2002; Rada et al., 2004; Saribasak et al., 2006; Shen et al., 2006). A distinct possibility is that part of the difference might be caused by some posttranslational modification of AID that occurs in B cells, but is not present on the recombinant AID used in our in vitro assays. Indeed, the detailed target preference of AID has been shown to be affected (at least in vitro) by single amino acid substitutions at serine 38 (Pham et al., 2008), a residue known to undergo phosphorylation in vivo (Basu et al., 2005). We suspect, however, that an even greater responsibility is likely to lie in a difference of the nature of the DNA targets analyzed in vitro and in vivo. The former consists of single- or transcribing double-stranded naked DNA; the latter is likely to be in transcriptionally active chromatin.
Although evolutionary selection could well have favored the emergence of hotspots at selected positions within the IgV to assist antibody maturation, such selection would presumably not contribute in noncoding flanking sequences. Consistent with this, a recent analysis by MacCarthy et al. (2009) of mutation accumulation in the JH 3′-flank did not reveal the same degree of discrepancy between in vivo and in vitro mutation spectra that we observe here within the body of IgVλ. What then could be the molecular mechanisms contributing to hotspot dominance within the IgV region in vivo? Ronai et al. (2007) have detected regions within the IgV in vivo that exhibit increased local single-strandedness, and therefore presumably have increased susceptibility to AID-catalyzed deamination; others have also predicted the formation of stable secondary structures correlating with the intrinsic mutability of nucleotides at the CDRs during high levels of transcription (Wright et al., 2008a, 2008b).
Nevertheless, we suspect that hotspot dominance might not simply be a function of local sequence but might also depend on position within the transcription unit: hotspot dominance is still evident in heterologous sequences that have been introduced into the Ig locus in place of the V segment (Yélamos et al., 1995). One possibility is that hotspot dominance in vivo is affected by nucleosome phasing at the 5′-end of the transcription unit. Indeed, recent work by Shen et al. (2009) provides compelling evidence that transcription plays a major role in allowing AID to act on chromatinized substrates. It will be interesting in future work to extend on the approach pioneered by Shen et al. (2009) to ask whether it is possible to reproduce the in vivo pattern of IgV mutational hotspots in vitro using chromatinized targets with differently phased or modified nucleosomes or undergoing transcription by different transcription complexes; such studies could give valuable insights into the nature of the DNA substrate during antibody hypermutation in vivo.
Understanding the targeting of AID may not only give insight into the mechanism of creation of mutational hotspots in immunoglobulin genes, whose location appears to have been selected by coevolution of AID and the IgV gene target. One might also gain wider insights into the targeting of mutation during evolution. Thus, although many distinct mutagenic processes contribute to organismal evolution, AID-catalyzed DNA deamination provides a uniquely tractable system for experimental investigation. For example, the conclusions reached regarding the differential mutability of serine AGY and TCN codons in IgV gene SHM (Wagner et al., 1995) appear to also apply to their differential mutability of these two categories of serine codon during the genome evolution of organisms (Collins and Jukes, 1994). This is consistent with the suggestion of shared features between mutational targeting in SHM and in meiotic mutation (Oprea et al., 2001). Thus, studies of the effects of nucleosomes and nucleosome phasing, histone modifications, DNA-binding proteins, transcription, etc., on AID-dependent mutational hotspots could well give insight into wider aspects of mutational targeting and genome evolution. The information could also be relevant to the study of off-target AID hotspots, for example, in protooncogenes with potential relevance to the creation of breakpoints for chromosomal translocations (Tsai et al., 2008).
The ability to alter the pattern of IgV gene mutational hotspots by altering the active-site of AID should also allow the creation of engineered mouse strains that display an altered distribution of mutational hotspots within their IgV genes. It will be interesting to discover the effects of such an altered distribution on the efficiency of the maturation of high-affinity antibodies.
MATERIALS AND METHODS
Rifampicin resistance assay.
E. coli strain KL16 (Hfr [PO-45] relA1 spoT1 thi-1) transformed with pTrc99/AID plasmids was grown overnight to saturation in LB medium supplemented with ampicillin (100 µg ml−1) and isopropyl β-D-1-thiogalactopyranoside (IPTG; 1 mM), and plated on LB low-salt agar containing ampicillin (100 µg ml−1) and rifampicin (50 µg ml−1). Mutation frequencies were measured by determining the median number of colony-forming cells that survived selection per 107 viable cells plated with each median determined from 12 independent cultures. The identity of mutations was determined by sequencing the relevant section of rpoB (typically from 25 to 200 individual colonies) after PCR amplification using oligonucleotides 5′-TTGGCGAAATGGCGGAAAACC-3′ and 5′-CACCGACGGATACCACCTGCTG-3′).
Assaying deaminase target specificity in vitro.
For assay on a gapped duplex substrate, a 475-nt single-stranded gap in M13mp19 DNA was generated by annealing replicative form M13mp19 DNA that had been digested with PvuII and BglI to single-stranded M13mp19 DNA extracted from phage as previously described (Bebenek and Kunkel, 1995). Deamination of this gapped substrate (0.5 µg) by GST-AID fusion protein (200 ng; semipurified as previously described; Wang et al., 2009) was monitored at 37°C in 10 µl reaction buffer (8 mM-Tris, pH 8.0, 8 mM-KCl, 10 mM-NaCl, 2.5 mM-EDTA, and 0.2 mM-dithiothreitol) containing 0.5 µg µl−1 RNase A essentially as described by Pham et al. (2003). Deamination reactions were terminated by diluting samples to 100 µl with water and, after phenol extraction, the DNA was transfected into E. coli strain BD1528 F– Δ(codB-lacI)3 ung-1 hsdR514) and plated together with α-complementation E. coli strain TG1 (supE thi-1 Δ(lac-proAB) hsdΔ5 [F′traD36 proAB+ lacIq ZΔM15]), Xgal (0.8 mg ml−1), and IPTG (80 µg ml−1) onto minimal glucose agar. DNA from white or light blue plaques was sequenced using primers 5′-GCAAACCAGCGTGGACCGCTTGC-3′ (forward) and 5′-GGACGACGACCGTATCGGCCTCAG-3′ (reverse). Mutations were analyzed using Sequencher software (Gene Codes Corporation).
For the T7 transcription-coupled assays, semipurified GST-AID (200 ng) was incubated with 1 µg DNA substrate (either M13mp19, in which a 27-nt fragment containing the T7 promoter had been inserted upstream of lacZα [Fig. S5 B], or plasmid pCR-Blunt II-Topo (Invitrogen), in which a GFP gene and either 49- or 35-nt fragments [positions 115–164 or 228–263] of chicken IgVλ were placed between the lac and T7 promoters [Fig. S5C]) at 37°C in 10 µl reaction buffer (40 mM-Tris, 6 mM-MgCl2, 10 mM-dithiothreitol, and 2 mM-spermidine, pH 7.9) containing RNase A (0.5 µg µl−1), 0.6 mM rNTP mix and 2 U T7 RNA polymerase (NEB). For the M13mp19-T7-lacZ transcription substrate, the extraction and visualization of mutated DNA products were as described for the gapped duplex assay. For the pCR-Topo-T7-Vλ-GFP reporter substrates, the extracted DNA products were transformed into BD1528, plated onto TYE plates supplemented with 50 µg ml−1 kanamycin, and incubated at 37°C until colonies turned green. DNA from colorless colonies was sequenced as described for the gapped duplex assay.
Assaying antibody diversification.
AID-induced SHM was monitored by measuring the frequency of sIgM-loss variants in AID−/− ψV−/− sIgM+ DT40 cells transfected with AID-encoding vectors based on pExpressPuro2 (Arakawa et al., 2004) in which AID and GFP transcription units are coexpressed from a vector including a puromycin-resistance cassette (gift from J.-M. Buerstedde, Institute of Molecular Radiobiology, Munich, Germany). For each construct, the percentage of sIgM– cells was monitored in 8–12 independent transfectants that had been expanded under selection (0.5 µg ml−1 puromycin) for 3 wk before flow cytometry. After 8 wk of culturing, mutations in the IgVλ region were characterized by sequencing genomic DNA that was PCR-amplified from either 100,000 unsorted or (GFP+, sIgM–)-sorted cell equivalents (forward 5′-CAGGAGCTCGCGGGGCCGTCACTGATTGCCG-3′; reverse 5′-GCGCAAGCTTCCCCAGCCTGCCGCCAAGTCCAAG-3′). To minimize double counting of mutations from dynastically related sequences, identical mutations in sequences from a single B cell culture that shared more than one common mutation were computed only once.
To assay class-switching, surface IgG1 expression was analyzed by flow cytometry in B cells that had been purified from AID−/− mice and cultured in the presence of LPS+IL-4 (48 h) after a 24-h infection with AID-encoding retroviruses, as previously described (Geisberger et al., 2009). The abundance of AID in extracts prepared by heating 106 cells in 50 µl of reducing SDS-sample buffer was monitored following SDS-PAGE by Western blot analysis using rabbit anti-AID antiserum (Abcam); GFP was detected using HRP conjugated goat anti-GFP antiserum (Abcam); tubulin was detected using mouse antitubulin antibody (Abcam).
Online supplemental material.
Fig. S1 shows an analysis of the semipurified GST-AIDs on SDS/PAGE, provides information on the mutation loads produced by them in the lacZ gapped-duplex assay, and compares the spectrum of AID1/3G and AID1*/3G to show that the C-terminal truncation does not affect the pattern of in vitro mutational targeting. Fig. S2 shows a comparison of the IgVλ mutation spectrum in DT40 transfectants expressing wild-type AID or AID1. Fig. S3 provides a compilation of the IgVλ mutations identified in DT40 cells expressing the various modified AID proteins. Fig. S4 presents the results of class-switching assays performed with modified AID proteins. Fig. S5 provides the DNA sequences of the various substrates used in the in vitro DNA deamination assays. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092238/DC1.
Acknowledgments
We thank Gareth Williams (Cambridge, UK) for helpful suggestions.
This work was supported by the James Baird and Frank Elmore funds and the Medical Research Council Laboratory of Molecular Biology block grant.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AID
- activation-induced deaminase
- CSR
- class-switch recombination
- SHM
- somatic hypermutation
References
- Alt F.W., Honjo T. 2007. Advances in Immunology Vol. 94: AID for Immunoglobulin Diversity; Elsevier, Amsterdam [Google Scholar]
- Arakawa H., Saribasak H., Buerstedde J.M. 2004. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2:E179 10.1371/journal.pbio.0020179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto V., Reina-San-Martin B., Ramiro A.R., McBride K.M., Nussenzweig M.C. 2003. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell. 12:501–508 10.1016/S1097-2765(03)00309-5 [DOI] [PubMed] [Google Scholar]
- Basu U., Chaudhuri J., Alpert C., Dutt S., Ranganath S., Li G., Schrum J.P., Manis J.P., Alt F.W. 2005. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 438:508–511 10.1038/nature04255 [DOI] [PubMed] [Google Scholar]
- Beale R.C., Petersen-Mahrt S.K., Watt I.N., Harris R.S., Rada C., Neuberger M.S. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585–596 10.1016/j.jmb.2004.01.046 [DOI] [PubMed] [Google Scholar]
- Bebenek K., Kunkel T.A. 1995. Analyzing fidelity of DNA polymerases. Methods Enzymol. 262:217–232 10.1016/0076-6879(95)62020-6 [DOI] [PubMed] [Google Scholar]
- Begum N.A., Kinoshita K., Kakazu N., Muramatsu M., Nagaoka H., Shinkura R., Biniszkiewicz D., Boyer L.A., Jaenisch R., Honjo T. 2004. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science. 305:1160–1163 10.1126/science.1098444 [DOI] [PubMed] [Google Scholar]
- Begum N.A., Izumi N., Nishikori M., Nagaoka H., Shinkura R., Honjo T. 2007. Requirement of non-canonical activity of uracil DNA glycosylase for class switch recombination. J. Biol. Chem. 282:731–742 10.1074/jbc.M607439200 [DOI] [PubMed] [Google Scholar]
- Begum N.A., Stanlie A., Doi T., Sasaki Y., Jin H.W., Kim Y.S., Nagaoka H., Honjo T. 2009. Further evidence for involvement of a noncanonical function of uracil DNA glycosylase in class switch recombination. Proc. Natl. Acad. Sci. USA. 106:2752–2757 10.1073/pnas.0813252106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berek C., Milstein C. 1987. Mutation drift and repertoire shift in the maturation of the immune response. Immunol. Rev. 96:23–41 10.1111/j.1600-065X.1987.tb00507.x [DOI] [PubMed] [Google Scholar]
- Betz A.G., Neuberger M.S., Milstein C. 1993a. Discriminating intrinsic and antigen-selected mutational hotspots in immunoglobulin V genes. Immunol. Today. 14:405–411 10.1016/0167-5699(93)90144-A [DOI] [PubMed] [Google Scholar]
- Betz A.G., Rada C., Pannell R., Milstein C., Neuberger M.S. 1993b. Passenger transgenes reveal intrinsic specificity of the antibody hypermutation mechanism: clustering, polarity, and specific hot spots. Proc. Natl. Acad. Sci. USA. 90:2385–2388 10.1073/pnas.90.6.2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R., Pham P., Calabrese P., Goodman M.F. 2004. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J. Biol. Chem. 279:51612–51621 10.1074/jbc.M408135200 [DOI] [PubMed] [Google Scholar]
- Buerstedde J.M., Reynaud C.A., Humphries E.H., Olson W., Ewert D.L., Weill J.C. 1990. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 9:921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B., Casali P. 1994. The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol. Today. 15:367–373 10.1016/0167-5699(94)90175-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins D.W., Jukes T.H. 1994. Rates of transition and transversion in coding sequences since the human-rodent divergence. Genomics. 20:386–396 10.1006/geno.1994.1192 [DOI] [PubMed] [Google Scholar]
- Conticello S.G., Langlois M.A., Neuberger M.S. 2007. Insights into DNA deaminases. Nat. Struct. Mol. Biol. 14:7–9 10.1038/nsmb0107-7 [DOI] [PubMed] [Google Scholar]
- Cowell L.G., Kim H.J., Humaljoki T., Berek C., Kepler T.B. 1999. Enhanced evolvability in immunoglobulin V genes under somatic hypermutation. J. Mol. Evol. 49:23–26 10.1007/PL00006530 [DOI] [PubMed] [Google Scholar]
- Di Noia J., Neuberger M.S. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 419:43–48 10.1038/nature00981 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Neuberger M.S. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22 10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Williams G.T., Chan D.T., Buerstedde J.M., Baldwin G.S., Neuberger M.S. 2007. Dependence of antibody gene diversification on uracil excision. J. Exp. Med. 204:3209–3219 10.1084/jem.20071768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörner T., Foster S.J., Farner N.L., Lipsky P.E. 1998. Somatic hypermutation of human immunoglobulin heavy chain genes: targeting of RGYW motifs on both DNA strands. Eur. J. Immunol. 28:3384–3396 [DOI] [PubMed] [Google Scholar]
- Geisberger R., Rada C., Neuberger M.S. 2009. The stability of AID and its function in class-switching are critically sensitive to the identity of its nuclear-export sequence. Proc. Natl. Acad. Sci. USA. 106:6736–6741 10.1073/pnas.0810808106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S., Petersen-Mahrt S.K., Neuberger M.S. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell. 10:1247–1253 10.1016/S1097-2765(02)00742-6 [DOI] [PubMed] [Google Scholar]
- Jolly C.J., Wagner S.D., Rada C., Klix N., Milstein C., Neuberger M.S. 1996. The targeting of somatic hypermutation. Semin. Immunol. 8:159–168 10.1006/smim.1996.0020 [DOI] [PubMed] [Google Scholar]
- Kepler T.B. 1997. Codon bias and plasticity in immunoglobulins. Mol. Biol. Evol. 14:637–643 [DOI] [PubMed] [Google Scholar]
- Kohli R.M., Abrams S.R., Gajula K.S., Maul R.W., Gearhart P.J., Stivers J.T. 2009. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J. Biol. Chem. 284:22898–22904 10.1074/jbc.M109.025536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois M.A., Beale R.C., Conticello S.G., Neuberger M.S. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 33:1913–1923 10.1093/nar/gki343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larijani M., Frieder D., Basit W., Martin A. 2005. The mutation spectrum of purified AID is similar to the mutability index in Ramos cells and in ung(-/-)msh2(-/-) mice. Immunogenetics. 56:840–845 10.1007/s00251-004-0748-0 [DOI] [PubMed] [Google Scholar]
- Losey H.C., Ruthenburg A.J., Verdine G.L. 2006. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13:153–159 10.1038/nsmb1047 [DOI] [PubMed] [Google Scholar]
- MacCarthy T., Kalis S.L., Roa S., Pham P., Goodman M.F., Scharff M.D., Bergman A. 2009. V-region mutation in vitro, in vivo, and in silico reveal the importance of the enzymatic properties of AID and the sequence environment. Proc. Natl. Acad. Sci. USA. 106:8629–8634 10.1073/pnas.0903803106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein C., Neuberger M.S., Staden R. 1998. Both DNA strands of antibody genes are hypermutation targets. Proc. Natl. Acad. Sci. USA. 95:8791–8794 10.1073/pnas.95.15.8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Nagaoka H., Ito S., Muramatsu M., Nakata M., Honjo T. 2005. DNA cleavage in immunoglobulin somatic hypermutation depends on de novo protein synthesis but not on uracil DNA glycosylase. Proc. Natl. Acad. Sci. USA. 102:2022–2027 10.1073/pnas.0409491102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea M., Kepler T.B. 1999. Genetic plasticity of V genes under somatic hypermutation: statistical analyses using a new resampling-based methodology. Genome Res. 9:1294–1304 10.1101/gr.9.12.1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea M., Cowell L.G., Kepler T.B. 2001. The targeting of somatic hypermutation closely resembles that of meiotic mutation. J. Immunol. 166:892–899 [DOI] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Petruska J., Goodman M.F. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 424:103–107 10.1038/nature01760 [DOI] [PubMed] [Google Scholar]
- Pham P., Smolka M.B., Calabrese P., Landolph A., Zhang K., Zhou H., Goodman M.F. 2008. Impact of phosphorylation and phosphorylation-null mutants on the activity and deamination specificity of activation-induced cytidine deaminase. J. Biol. Chem. 283:17428–17439 10.1074/jbc.M802121200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnow C., Bransteitter R., Klein M.G., Goodman M.F., Chen X.S. 2007. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 445:447–451 10.1038/nature05492 [DOI] [PubMed] [Google Scholar]
- Rada C., Di Noia J.M., Neuberger M.S. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171 10.1016/j.molcel.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Diaz M. 2004. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 172:3382–3384 [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Kolchanov N.A. 1992. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta. 1171:11–18 [DOI] [PubMed] [Google Scholar]
- Ronai D., Iglesias-Ussel M.D., Fan M., Li Z., Martin A., Scharff M.D. 2007. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J. Exp. Med. 204:181–190 10.1084/jem.20062032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale J.E., Calandrini D.M., Takata M., Takeda S., Neuberger M.S. 2001. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 412:921–926 10.1038/35091100 [DOI] [PubMed] [Google Scholar]
- Saribasak H., Saribasak N.N., Ipek F.M., Ellwart J.W., Arakawa H., Buerstedde J.M. 2006. Uracil DNA glycosylase disruption blocks Ig gene conversion and induces transition mutations. J. Immunol. 176:365–371 [DOI] [PubMed] [Google Scholar]
- Sharpe M.J., Milstein C., Jarvis J.M., Neuberger M.S. 1991. Somatic hypermutation of immunoglobulin kappa may depend on sequences 3′ of C kappa and occurs on passenger transgenes. EMBO J. 10:2139–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H.M., Tanaka A., Bozek G., Nicolae D., Storb U. 2006. Somatic hypermutation and class switch recombination in Msh6(-/-)Ung(-/-) double-knockout mice. J. Immunol. 177:5386–5392 [DOI] [PubMed] [Google Scholar]
- Shen H.M., Poirier M.G., Allen M.J., North J., Lal R., Widom J., Storb U. 2009. The activation-induced cytidine deaminase (AID) efficiently targets DNA in nucleosomes but only during transcription. J. Exp. Med. 206:1057–1071 10.1084/jem.20082678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivarov V., Shinkura R., Honjo T. 2008. Dissociation of in vitro DNA deamination activity and physiological functions of AID mutants. Proc. Natl. Acad. Sci. USA. 105:15866–15871 10.1073/pnas.0806641105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stivers J.T. 2004. Comment on “Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch.” Science. 306:2042 10.1126/science.1104396 [DOI] [PubMed] [Google Scholar]
- Tomlinson I.M., Walter G., Jones P.T., Dear P.H., Sonnhammer E.L., Winter G. 1996. The imprint of somatic hypermutation on the repertoire of human germline V genes. J. Mol. Biol. 256:813–817 10.1006/jmbi.1996.0127 [DOI] [PubMed] [Google Scholar]
- Tsai A.G., Lu H., Raghavan S.C., Muschen M., Hsieh C.L., Lieber M.R. 2008. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 135:1130–1142 10.1016/j.cell.2008.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S.D., Milstein C., Neuberger M.S. 1995. Codon bias targets mutation. Nature. 376:732 10.1038/376732a0 [DOI] [PubMed] [Google Scholar]
- Wang M., Yang Z., Rada C., Neuberger M.S. 2009. AID upmutants isolated using a high-throughput screen highlight the immunity/cancer balance limiting DNA deaminase activity. Nat. Struct. Mol. Biol. 16:769–776 10.1038/nsmb.1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright B.E., Schmidt K.H., Davis N., Hunt A.T., Minnick M.F. 2008a. II. Correlations between secondary structure stability and mutation frequency during somatic hypermutation. Mol. Immunol. 45:3600–3608 10.1016/j.molimm.2008.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright B.E., Schmidt K.H., Minnick M.F., Davis N. 2008b. I. VH gene transcription creates stabilized secondary structures for coordinated mutagenesis during somatic hypermutation. Mol. Immunol. 45:3589–3599 10.1016/j.molimm.2008.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yélamos J., Klix N., Goyenechea B., Lozano F., Chui Y.L., González Fernández A., Pannell R., Neuberger M.S., Milstein C. 1995. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature. 376:225–229 10.1038/376225a0 [DOI] [PubMed] [Google Scholar]
- Yu K., Huang F.T., Lieber M.R. 2004. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 279:6496–6500 10.1074/jbc.M311616200 [DOI] [PubMed] [Google Scholar]