Abstract
The nonclassical major histocompatibility complex (MHC) Qa-1b accommodates monomorphic leader peptides and functions as a ligand for germ line receptors CD94/NKG2, which are expressed by natural killer cells and CD8+ T cells. We here describe that the conserved peptides are replaced by a novel peptide repertoire of surprising diversity as a result of impairments in the antigen-processing pathway. This novel peptide repertoire represents immunogenic neoantigens for CD8+ T cells, as we found that these Qa-1b–restricted T cells dominantly participated in the response to tumors with processing deficiencies. A surprisingly wide spectrum of target cells, irrespective of transformation status, MHC background, or type of processing deficiency, was recognized by this T cell subset, complying with the conserved nature of Qa-1b. Target cell recognition depended on T cell receptor and Qa-1b interaction, and immunization with identified peptide epitopes demonstrated in vivo priming of CD8+ T cells. Our data reveal that Qa-1b, and most likely its human homologue human leukocyte antigen-E, is important for the defense against processing-deficient cells by displacing the monomorphic leader peptides, which relieves the inhibition through CD94/NKG2A on lymphocytes, and by presenting a novel repertoire of immunogenic peptides, which recruits a subset of cytotoxic CD8+ T cells.
Qa-1b and its human homologue human leukocyte antigen (HLA)-E are conserved MHC class I like molecules, often categorized as “nonclassical” or class Ib MHC. Similar to the classical MHC molecules, they accommodate small peptides in their binding grooves and present these on the cell surface (Jensen et al., 2004; Rodgers and Cook, 2005). Crystal structures of HLA-E show that the overall fold of the peptide–MHC complex is very similar to that of the classical MHC class I molecules (O’Callaghan et al., 1998). The unique feature of Qa-1b and HLA-E resides in the very limited number of alleles that is present in the population (Grimsley et al., 2002; Hermel et al., 2004): as few as two alleles exist in the human population, differing in just one amino acid; this difference has hardly any functional consequence (Strong et al., 2003). The monomorphic nature of Qa-1b and HLA-E is underlined by the finding that they are predominantly filled with one peptide ligand, which is derived from the signal sequence of classical MHC class I molecules, often referred to as Qa-1 determinant modifier (Qdm; Aldrich et al., 1994; DeCloux et al., 1997). These signal peptides are strikingly conserved in all tested mammalian species (Kurepa et al., 1998), pointing at an important function of this peptide–MHC combination for the immune system. A substantial body of evidence reveals that it regulates NK cells and CTLs via the heterodimeric receptors CD94/NKG2A and CD94/NKG2C (Braud et al., 1998a; Vance et al., 1999; Rodgers and Cook, 2005). These germline-encoded receptors specifically engage the Qdm peptide when bound to Qa-1b/HLA-E because residues at peptide position 5 and 8 are part of the surface interface of the CD94/NKG2 receptors (Kaiser et al., 2008; Petrie et al., 2008). Interestingly, this interaction is quite similar to the footprint of the hyper variable regions of the T cell receptor on peptide–MHC class I (Rudolph and Wilson, 2002). Absence of these Qdm side chain residues results in lack of receptor engagement (Valés-Gómez et al., 1999; Miller et al., 2003), indicating that this innate receptor is truly peptide specific. Qdm–Qa-1b complexes act as remote sensors of integrity of the MHC class I antigen-processing machinery, because defects in proteasomal cleavage, TAP-mediated peptide transport, signal peptidases or tapasin-mediated peptide loading all result in failure to process and present Qdm peptides (Braud et al., 1998b; Lemberg et al., 2001; Bland et al., 2003; Li et al., 2004). The lack of Qdm–Qa-1b complexes at the cell surface is sensed by NK cells expressing CD94/NKG2 receptors, and allows for immunosurveillance of such processing-deficient cells (Lu et al., 2006). Thus, this receptor/ligand system is a major molecular mechanism behind the “missing self” principle of NK cell reactivity (Ljunggren and Kärre, 1990).
In addition to their role in the innate immune system, Qa-1b and HLA-E serve the adaptive response as well. T cell receptor recognition of these nonclassical MHCs has been described in the context of immunity against intracellular pathogens, e.g., Listeria, Salmonella, and Mycobacterium tuberculosis (Seaman et al., 1999; Lo et al., 2000; Heinzel et al., 2002), suggesting that foreign antigens can replace the “self” Qdm peptides and be recognized by CD8+ T cells. HLA-E–restricted T cells specific for the human Qdm homologue do exist, but have only been reported in response to allogeneic MHC (García et al., 2002; Romagnani et al., 2002). Furthermore, Qa-1b–restricted CD8+ T cells have been implicated in the regulation of experimental autoimmune diseases (for review see Jiang and Chess, 2000; Lu et al., 2006). The antigenic targets of these regulatory T cells are most likely of self origin, but different from Qdm. These data reveal that Qa-1b/HLA-E–restricted T cells are present in the host, but that the specificity, diversity, and function of this T cell subset await further investigation.
Here, we describe the existence of a surprisingly broad peptide repertoire that is presented by Qa-1b on cells with impairments in the antigen-processing machinery. These peptides replace Qdm and are targeted by a unique population of CD8+ cytotoxic T cells. Normal cells with intact processing machinery were not recognized by these Qa-1b–restricted CTL, but partial defects readily resulted in the appearance of the immunogenic self peptides, which are derived from housekeeping proteins. Interestingly, we show that these Qa-1b–restricted T cells are abundantly present in the immune response to processing deficient tumors. Our data show that the nonclassical Qa-1b molecule plays a prominent role in the adaptive immune response as a restriction element for T cells and presents a much larger peptide repertoire than thus far anticipated.
RESULTS
Qa-1b–restricted T cells are frequently present in the immune response against TAP-deficient tumors
We previously described that the immune response against TAP-deficient tumors comprises Qa-1b–restricted T cells among T cells with MHC restriction to the classical class I molecules (van Hall et al., 2006). This prompted us to determine the relative contribution of this Qa-1b–restricted subset to the overall response. Mice were immunized with syngeneic TAP-deficient RMA-S cells expressing the co-stimulatory molecule CD80. Ex vivo–cultured spleens were examined for IFN-γ production against the parental RMA-S lymphoma, the β2m-negative C4.4-25 line, and a panel of single class I–expressing EC7.1 cells, which are MHC class I–loss variants of RMA-S (Howell et al., 2000; Table I). All polyclonal ex vivo cultures responded to RMA-S, but not against the β2m-negative lymphoma, indicating that they exhibited specific T cell reactivity against TAP-deficient tumors, and not merely reflected natural killer (NK) cell reactivity. Dissection of these IFN-γ responses revealed a codominance of Qa-1b–restricted activity compared with those restricted by the classical Db- and Kb-molecules, as the majority of the examined 12 independent cultures produced high IFN-γ levels when incubated with EC7.1.Qa-1b (Table I). These data suggested that the nonclassical MHC class I Qa-1b frequently acts as an antigen presentation molecule for T cells, and can even overrule the classical MHC class I molecules.
Table I.
Detection of Qa-1b–directed IFN-γ responses in ex vivo cultures
| Target cellsa | Independent T cell cultureb | |||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| RMA-S | 5,723c | 10,113 | 1,204 | 6,582 | 12,199 | 4,896 | 2,894 | 10,389 | 1,359 | 11,593 | 4,320 | 1,567 |
| C4.4-25 | 97 | 84 | 92 | 82 | 164 | 61 | 23 | 18 | 106 | 61 | 47 | 14 |
| EC7.1 | 364 | 712 | 310 | 216 | 1,311 | 799 | 273 | 81 | 485 | 564 | 418 | 38 |
| EC7.1.Kb | 5,886 | 11,270 | 538 | 7,184 | 16,031 | 2,526 | 441 | 8,309 | 405 | 11,111 | 1,552 | 837 |
| EC7.1.Db | 888 | 7,507 | 645 | 690 | 10,113 | 2,125 | 1,129 | 604 | 648 | 8,314 | 1,561 | 446 |
| EC7.1.Qa-1b | 2,053 | 5,579 | 3,330 | 6,677 | 16,265 | 3,108 | 2,999 | 272 | 1,410 | 8,827 | 2,697 | 7,039 |
| EC7.1.cont | 380 | 700 | 386 | 250 | 1,370 | 1,065 | 418 | 63 | 591 | 620 | 401 | 56 |
| Medium | 287 | 120 | 287 | 152 | 276 | 156 | 177 | 38 | 218 | 110 | 74 | 13 |
RMA-S is TAP2 deficient; C4.4-25 is β2m deficient; EC7.1 is a MHC class I–deficient variant of RMA-S. Single class I genes were reconstituted in EC7.1.
12 mice were immunized with RMA-S.B7 and spleens were stimulated twice in vitro with RMA-S.B7 before testing.
Concentration of IFN-γ released by ex vivo cultures (pg/ml).
Critical involvement of Qa-1b and the T cell receptor in target cell recognition
To characterize these Qa-1b–restricted T cells, we isolated several T cell clones from the aforementioned cultures and extensively explored the expression and function of surface receptors. All clones displayed a classical CTL phenotype and expressed CD3, CD8αβ, and TCRαβ (Fig. S1). Different rearranged TCR Vβ segments were used (unpublished data), validating the independent origin of the clones and indicating that Qa-1b recognition does not constrain the TCR repertoire. In addition to the T cell lineage receptors, the T cell clones also expressed CD94, NKG2A, NKG2C, and NKG2D, receptors that are frequently found on NK cells, which interact with nonclassical MHC class I molecules, including Qa-1b. Importantly, NK lineage markers NK1.1 and DX5, and CD16, CD32, and CD244 (2B4) were not expressed. Neither were receptors of the lectin Ly49 family, which interact with classical MHC class I (Fig. S1 and not depicted). We concluded that our Qa-1b–restricted T cells are indistinguishable at the phenotypic level from conventional CD8+ T cells restricted by classical MHC (McMahon and Raulet, 2001), but are clearly distinct from the reported Qa-1–recognizing intestinal γδ+ T cells (Davies et al., 2004) and Qa-1–restricted regulatory CD8αα+ T cell subset (Tang et al., 2006).
To carefully determine the Qa-1b restriction of our isolated T cell clones, we examined their reactivity against two cell panels, based on the TAP-negative EC7.1 lymphoma and TAP-deficient B78H1 melanoma (Fig. 1; Howell et al., 2000; Chiang et al., 2003). Both cell lines are also devoid of MHC class I proteins and single class I molecules were reconstituted by gene transfer. All isolated T cell clones exhibited cytolytic activity (Fig. 1 A) and produced IFN-γ (Fig. 1 B and Fig. S2) against the Qa-1b–expressing cells, but not against cells in which only Db or Kb were introduced. These data convincingly demonstrated the Qa-1b restriction of our T cells and excluded cross-reactivity to the classical class I molecules. Qdm-specific control CTL did not recognize the TAP-deficient tumor targets, as the presentation of Qdm in Qa-1b is TAP dependent (Aldrich et al., 1994). Exogenous loading of Qdm conferred reactivity for this CTL clone (Fig. 1, right). A direct interaction with Qa-1b was revealed by the finding that anti–Qa-1b antibody strongly blocked the recognition by our CTL (Fig. 2 A).
Figure 1.
Selective recognition of Qa-1b-expressing target cells. Isolated T cell clones were tested against panels of TAP-deficient lymphoma (EC7.1) and melanoma (B78H1) cells. Both cell lines are also deficient in MHC class I (Howell et al., 2000; Chiang et al., 2003) and were reconstituted with constructs encoding H-2Db, -Kb, or Qa-1b. Cytotoxic activity against Qa-1b–expressing EC7.1 cells (A) and IFN-γ production against Qa-1b-expressing B78H1 cells (B) by three independent T cell clones. Control Qdm-specific CTL failed to recognize the Qa-1b expressing targets because of the absence of TAP, unless pulsed with the Qdm peptide (right). Means and standard deviations of triplicate wells are shown for one out of three comparable experiments.
Figure 2.
The T cell receptor, but not NKG2A/C, mediates reactivity of the T cells. (A) Blocking antibody against Qa-1b demonstrated a direct role of this molecule for T cell recognition. Experiment was performed using EC7.1.Qa-1b target cells and repeated three times with similar outcome. (B) Blocking CD3 or CD8 with monoclonal antibodies decreased T cell reactivity against TAP-deficient EC7.1.Qa-1b target cells. Antibodies against NKG2A/C did not alter the T cell response, indicating that only the T cell receptor is critically involved in mediating reactivity. Data are representative of four experiments. (C) Exogenous peptide loading competes with endogenously presented epitopes on EC7.1.Qa-1b cells and inhibits the recognition of these target cells by Qa-1b–restricted CTL (left). Qdm (AMAPRTLLL) or Qdm L8K (AMAPRTLKL) peptides were loaded exogenously at the indicated concentrations on EC7.1.Qa-1b cells and IFN-γ release by CTL was measured. Control peptide was the Kb-binding 8-mer SIINFEKL from OVA. The Qdm L8K mutant peptide–Qa-1b complexes fail to interact with CD94/NKG2A or CD94/NKG2C (Kraft et al., 2000), indicating that the loaded peptides interfere with T cell receptor–mediated reactivity. Qdm-specific control CTL (right) was activated by the peptides. Means and standard deviations of triplicate wells are shown for one out of three comparable experiments.
Next, we examined the involvement of antigen-receptors in target cell recognition. Although CD94/NKG2A and CD94/NKG2C do engage Qa-1b (Rodgers and Cook, 2005) and were expressed by our isolated T cell clones, their involvement was not likely, because TAP-negative cells are devoid of peptide ligands for these receptors (Gays et al., 2001). Indeed, blocking with NKG2A/C-specific antibodies did not alter the response, whereas anti-CD3 and -CD8 antibodies clearly inhibited the recognition of EC7.1.Qa-1b cells (Fig. 2 B). A role for NKG2D was also not likely, as our RMA-based lymphoma panel lacks NKG2D ligands (Diefenbach et al., 2000). Together, these data indicated that the TCR governed the reactivity of our Qa-1b–restricted CTL.
Furthermore, exogenous loading with excessive amounts of competing Qdm peptides inhibited T cell recognition of EC7.1.Qa-1b cells (Fig. 2 C). Similar competition was found when the Qdm variant L8K was used (AMAPRTLKL), but not with irrelevant control peptide (Fig. 2 C, left). Qdm-specific control CTL confirmed that both peptides are efficiently loaded on the target cells (Fig. 2 C, right). The fact that the position 8 substitution in the peptide L8K prevents engagement to CD94/NKG2A receptors (Kraft et al., 2000), but retained the capacity to inhibit T cell reactivity in our assays, substantiated our findings that target cell recognition is regulated by TCR-mediated interaction with Qa-1b and not by NK receptors.
TAP- and MHC heavy chain deficiencies are targeted by Qa-1b–restricted CTL
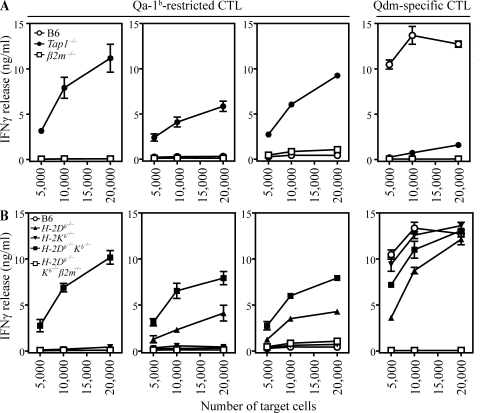
To evaluate the appearance of the peptide epitopes on target cells that express natural levels of Qa-1b, we tested B cell blasts from spleens of wild-type, TAP1-deficient, and β2m-deficient mice as targets for three independently derived CTL clones (Fig. 3 A). TAP1-deficient cells were efficiently recognized by all three clones, but wild-type and β2m-deficient cells were not. Qdm-specific control CTL selectively responded to wild-type cells, indicating that the absence of Qdm peptides caused by the TAP defect promoted the presentation of the new peptide epitopes. Qa-1b surface levels on TAP1-deficient cells were lower than on wild-type cells, but were clearly higher than the background levels on β2m-deficient cells (unpublished data).
Figure 3.
Qa-1b–restricted CTL are reactive against cells with defined processing deficiencies. (A) LPS-stimulated B cell blasts from wild-type (B6), TAP1−/−, and β2m−/− mice were used as targets for Qa-1b–restricted CTL (left) or control Qdm-specific CTL (right). (B) LPS-stimulated B cell blasts from wild-type (B6), MHC class I knockout, and MHC class I/β2m knockout mice were tested for recognition by Qa-1b–restricted CTL (left) or control Qdm-specific CTL (right). Graphs display representative experiments out of four performed. Means and standard deviations of triplicates are shown.
We also analyzed CTL reactivity against MHC class I heavy chain deficiencies with the use of B cell blasts from H-2Db and Kb knockout mice. Qdm peptides are derived from the leader sequence of H-2Db (Aldrich et al., 1994), and loss of this allele resulted in recognition by two of the three CTL clones, whereas deficiency in Kb heavy chains did not result in recognition (Fig. 3 B). These subtle differences between the CTL clones suggested that the cognate peptides are differentially influenced by loss of MHC class I heavy chains. Strikingly, all three CTL clones strongly responded to cells devoid of both alleles (H-2DbKb knockout; Fig. 3 B). Control blasts lacking β2m and H-2DbKb (triple knockout) were not recognized. These results revealed an unexpected influence of the Kb molecule on the Qa-1b–presented antigenic peptides, in that removal of Kb molecules from H-2Db–deficient cells strongly enhanced epitope display (Fig. 3 B). The underlying mechanism of this effect is currently elusive. The degree of CTL response to H-2DbKb knockout cells was comparable to TAP-deficient cells (Fig. 3 A), indicating that both defects led to optimal presentation of the new Qa-1b peptides. Interestingly, Qdm-specific CTL were able to differentiate between these two target populations, confirming previous findings that this CTL clone is also reactive to TAP-dependent peptides other than Qdm (Cotterill et al., 1997; Gays et al., 2001). Together, these results demonstrated that our Qa-1b–restricted CTL interact with peptides on activated B cells with defects in the antigen presentation route.
CTL reactivity against tumors with partial processing impairments
We then determined CTL reactivity against tumor cells with TAP dysfunction and tumor cells with undefined deficiencies in the processing machinery. Ad5-transformed mouse embryo cells (Ad5MEC) and chemically induced fibrosarcoma cells (methylcholantrene [MCA]), both derived from TAP1-knockout mice, were recognized by the Qa-1b–restricted CTL (Fig. 4 A). Correction of the TAP1 defect by gene transfer resulted in decreased recognition by our CTL and strongly increased recognition by the Qdm-specific control CTL (Fig. 4 A). Thus, the alternative Qa-1b–binding peptide antigens are expressed in a wide array of hematopoietic and nonhematopoietic tissues of normal and transformed cells, including B cells, fibroblasts, lymphoma, melanoma, and sarcoma, and emerge at the cell surface of cells with genetic disruption of the TAP peptide transporter.
Figure 4.
Partial deficiencies in the processing pathway of tumor cells induce the novel Qa-1b–presented peptide-epitopes. (A) Qa-1b–restricted CTL respond to Ad5-transformed mouse cells (Ad5MEC) and fibrosarcoma cells (MCA) that are derived from TAP1 knockout mice. Recognition was lost upon gene transfer of mouse TAP1 in these cell lines (+ TAP1). Qdm-specific control CTL displayed opposing specificity, indicating that the Qdm peptide is only presented on TAP-proficient tumor cells (right). (B) Four colon carcinoma cell lines from BALB/c (C26 and CC36) or C57BL/6 (MC38 and CMT93) background were used as targets for both CTL types. Pretreatment with IFN-γ to boost the antigen-processing and presentation machinery resulted in decreased reactivity by the Qa-1b–restricted CTL, whereas gene transfer of the viral TAP-inhibitor UL49.5 (van Hall et al., 2007) led to strongly increased recognition. Again, Qdm-specific control CTL displayed opposing specificity (right). Means and standard deviations of triplicates are shown from one representative experiment out of four.
However, human tumors mostly display partial deficiencies in their processing pathway that originate at the transcriptional level, resulting in an impairment, but not a complete shutdown, in the generation of the human counterpart of Qdm–Qa-1b complexes. To assess whether the CTL were also capable of recognizing such tumor cells, we tested four chemically induced mouse colon carcinomas, which are known to possess peptide presentation capacity (Rodolfo et al., 1994; Yang and Perry-Lalley, 2000). Qdm-specific control CTL, indeed, recognized all four cell lines, demonstrating functional processing machinery (Fig. 4 B, right). Interestingly, our Qa-1b–restricted CTL responded to all carcinoma lines, albeit to varying extents (Fig. 4 B, left). Treatment of the carcinomas with the immunostimulatory cytokine IFN-γ, which strongly improves the processing and presentation capacity of cells, resulted in decreased recognition, pointing at partial processing defects in the carcinomas that were corrected by IFN-γ. In line with this idea, gene transfer of the viral TAP inhibitor UL49.5 (van Hall et al., 2007) led to a strongly increased recognition by our CTL. As expected, the Qdm-specific control CTL clone exhibited the opposite reactivity profile (Fig. 4 B, right). Of note, the four colon carcinoma lines were derived from two mouse strains with different MHC typing, BALB/c with H-2d alleles (C26 and CC36) and C57BL/6 with H-2b alleles (MC38 and CMT93), nicely illustrating the conserved nature of Qa-1, which is the same in these strains.
We concluded that our Qa-1b–restricted CTL recognize peptide epitopes that even appear on the surface of cells with mild or partial processing impairments, implying that a mixture of Qdm and TAP-independent peptides can coexist in Qa-1b.
Determination of the Qa-1b–presented peptide repertoire
We set out to determine the nature of the TAP-independent peptides that are presented by Qa-1b via biochemical purification and tandem mass spectrometry (MS). Because of the lack of suitable Qa-1b–specific antibodies, we made use of a chimeric MHC class I molecule in which the peptide binding domains (α 1 and 2) of Qa-1b were coupled to the constant α 3 domain of Db, against which good precipitating antibodies are available. This chimeric Qa-1b/Db molecule was introduced into TAP2-negative EC7.1 cells and to compare the corresponding TAP-dependent peptide repertoire, we also created a TAP2-reconstituted variant of EC7.1.Qa-1b/Db cells. Flow cytometry data confirmed that EC7.1.Qa-1b/Db cells only expressed this chimeric molecule on the cell surface and not the endogenous Db molecule, excluding the risk that we would purify Db-binding peptides with our α 3–directed antibody (Fig. 5 A). Analysis of TAP2 expression in EC7.1.Qa-1b/Db and in the TAP-transfected variant confirmed the absence of TAP2 protein in the first and successful reconstitution in the other (Fig. 5 B). Analysis with our Qa-1b–restricted CTL and Qdm-specific CTL confirmed that our EC7.1 cell lines represented two extremes of the antigen-processing status, in that the TAP-negative line did not present Qdm and that reconstitution of TAP2 led to a disappearance of the newly defined peptides and that the chimeric molecule functionally presented Qa-1b–bound peptides (Fig. 5 C).
Figure 5.
Identification of the TAP-independent peptide repertoire of Qa-1b. (A) The chimeric Qa-1b/Db class I molecule was expressed in EC7.1 cells, which are TAP- and MHC class I–negative. TAP-positive counterparts were generated by introduction of the TAP2 gene. Surface display on EC7.1.Qa-1b/Db cells of the chimeric molecule and absence of the endogenous Db molecule was determined by flow cytometry with Qa-1b– and Dbα3-specific antibodies and Dbα2-specific antibody, respectively, as indicated in the histogram plots (clone KH95 and also H131-31; not depicted). Staining of TAP-reconstituted cells gave comparable results. Control TAP-proficient RMA cells displayed endogenous Qa-1b and Db molecules. (B) TAP expression was analyzed on RMA, RMA-S, EC7.1, and EC7.1.TAP2 cells by Western blot using the antibody TAP2.688 against mouse TAP2. Results confirmed the lack of TAP2 expression on EC7.1 and RMA-S cells. (C) The chimeric construct was recognized by CTL clones: the TAP-negative variant only by Qa-1b–restricted CTL (black bars) and the TAP-positive variant only by Qdm-specific CTL (gray bars). Means and standard deviations of triplicate wells are shown from one representative experiment out of four. (D) Peptide purification and MS analysis revealed the wide diversity of the TAP-negative peptide repertoire, whereas the TAP-positive peptide repertoire was mainly limited to the Qdm peptide. Data were collected from four independent experiments in the case of TAP-negative repertoire and two independent experiments for TAP-positive repertoire. Number of different peptides with indicated length is depicted of 84 identified peptides that are listed in Table II.
MS of peptide species from purified Qa-1b molecules demonstrated an evident difference between the two peptide repertoires. The peptide repertoire of TAP-positive cells was strikingly dominated by the Qdm peptide AMAPRTLLL (Table II). Six other peptide sequences were identified, but these represented only minor constituents (together <5% of the total ion count). In contrast, Qa-1b from TAP-negative cells was filled with a remarkable diversity of peptides from endogenous proteins (Table II). No peptide clearly stood out in terms of quantity. More than 150 tandem mass fragmentation profiles sufficiently matched with the mouse IPI protein database (http://www.ebi.ac.uk/IPI/IPImouse.html) and analysis of their corresponding synthetic peptides confirmed the amino acid sequence of 84 peptides. The other peptide masses could not be matched with a known peptide sequence, but did not correspond to Qdm or previously defined Qa-1b–binding peptides (Rodgers and Cook, 2005). The lengths of the TAP-independent peptides ranged from 8 to 18 amino acids, but the majority were 9 amino acids long (Fig. 5 D). Seven peptides were found in length variants (Table II, numbers 65 to 84), and six of these families varied at their C terminus, reminiscent of leader peptide processing (Martoglio and Dobbertein, 1998). However, only one of these families was actually encoded in the leader domain, suggesting that amino acid trimming at the C terminus might be a feature of TAP-independent peptides in general. Strikingly, no clear binding motif could be elucidated in our peptide repertoire, although previous studies with synthetic peptide libraries showed a dominant role for position 2 (M or L; Kraft et al., 2000). The C terminus of the peptides frequently contained an aliphatic amino acid (L, I, M, A, and F; Table II), as found for most mouse MHC class I alleles.
Table II.
List of Qa-1b-binding peptides isolated from TAP-positive and TAP-negative tumor cells
| Peptide number | Peptide sequence | Peptide location | Protein length | UniProtKB or SwissProt number | Protein description |
| TAP-positive | |||||
| 1 | AMAPRTLLL | 3-11 | 362 | P01899 | H-2 class I histocompatibility antigen, D-B alpha chain |
| 2 | AQAERTPEL | 691-699 | 1274 | A2RT67 | DENN domain-containing protein 3 |
| 3 | IINTHTLLL | 1302-1310 | 1657 | Q80UW7 | IQ motif containing GTPase activating protein 1 |
| 4 | PKFEVIDKPQS | 98-108 | 108 | P97450 | ATP synthase-coupling factor 6, mitochondrial |
| 5 | PTEEESPV | 486-493 | 493 | P53986 | Monocarboxylate transporter 1 |
| 6 | QAIPQGAIQ | 246-254 | 461 | Q8BG99 | Homeobox protein PKNOX2 |
| 7 | QLQPQQPLPQPQ | 125-136 | 672 | Q9WVH4 | Forkhead protein FKHR2 |
| TAP-negative | |||||
| 1 | AAIENIEHL | 1193-1201 | 1392 | Q6PB66 | Leucine-rich PPR motif-containing protein, mitochondrial |
| 2 | AALKLGQEL | 799-807 | 1271 | Q9JJ28 | Protein flightless-1 homologue |
| 3 | AAPTNANSLNSTF | 454-466 | 575 | Q8BT14 | CCR4-NOT transcription complex subunit 4 |
| 4 | AAPTSPDHSPA | 699-709 | 709 | Q66L44 | Protein Dos |
| 5 | AAVIAHDFL | 153-161 | 292 | P30282 | G1/S-specific cyclin-D3 |
| 6 | AGIENDEAF | 44-52 | 248 | B1AWD9 | Clathrin light polypeptide |
| 7 | AGPENSSKI | 383-346 | 2075 | Q80XK6 | Autophagy-related protein 2 homologue B |
| 8 | AGQFNQDYL | 45-53 | 503 | Q921F1 | Annexin A11 |
| 9 | AGVRNPQQHL | 515-524 | 636 | Q8BN32 | Pabpc1 protein |
| 10 | ASLQNFNISNL | 2103-2113 | 2128 | B2RRJ7 | Wnk1 protein |
| 11 | KSISNPPGSNL | 2115-2125 | 2128 | B2RRJ7 | Wnk1 protein |
| 12 | ASQQNSEEM | 202-210 | 210 | Q9CXE2 | B-cell CLL/lymphoma 7 protein family member A |
| 13 | ASVLNVNHI | 2195-2203 | 2603 | Q99NH0 | Ankyrin repeat domain-containing protein 17 |
| 14 | ASYRAQPSVSL | 270-280 | 573 | Q62019 | 16 kD protein |
| 15 | ATPGRLIDFL | 256-265 | 648 | Q5U222 | Ddx5 protein |
| 16 | AVSEGTKAVTKYTSAK | 111-126 | 126 | Q8CGP1 | Histone H2B type 1-K |
| 17 | FAPLPRLPTL | 17-26 | 156 | Q9CR21 | Acyl carrier protein, mitochondrial |
| 18 | FAPVNVTTEVKSVE | 280-293 | 462 | P10126 | Elongation factor 1-alpha 1 |
| 19 | FAYEGRDYI | 137-145 | 184 | Q62143 | Qa-2 cell surface antigen |
| 20 | FGPVNHEEL | 33-41 | 147 | P46414 | Cyclin-dependent kinase inhibitor 1B |
| 21 | FQIVNPHLL | 634-642 | 792 | Q6NZB3 | Ribonucleoside-diphosphate reductase |
| 22 | FQVTHTVAL | 113-121 | 361 | Q9QYA2 | Mitochondrial import receptor subunit TOM40 homologue |
| 23 | GGPINPATA | 1036-1044 | 1107 | Q80X50 | Ubiquitin-associated protein 2-like |
| 24 | GLGVLLAF | 5-12 | 113 | Q8QZT4 | Crumbs protein homologue 3 |
| 25 | HSIQNSQDM | 61-69 | 210 | Q91YN9 | BAG family molecular chaperone regulator 2 |
| 26 | IQKTPQIQVY | 21-30 | 119 | P01887 | Beta-2-microglobulin |
| 27 | KAPPPLPPLVVF | 27-38 | 499 | P16277 | Tyrosine-protein kinase BLK |
| 28 | KAPTNEFYA | 190-198 | 198 | O35988 | Syndecan-4 |
| 29 | KCSVSIQVVDVNDNYPEL | 328-345 | 794 | Q91XZ8 | Protocadherin beta 22 |
| 30 | KSAVGHEYV | 96-107 | 486 | Q922I8 | Hematopoietic cell specific Lyn substrate 1 |
| 31 | LAIRNDEEL | 93-101 | 137 | Q64426 | Histone H2A |
| 32 | LVRPGTALEL | 2294-2303 | 2883 | NP_076331 | desmoplakin |
| 33 | METLTATPQ | 976-984 | 1241 | B2RXW8 | Ppfia1 protein |
| 34 | NSIRNLDTI | 100-108 | 475 | P28658 | Ataxin-10 |
| 35 | PADIVKNLK | 12-20 | 341 | Q8VDZ8 | Calcium binding protein 39 |
| 36 | PDTGISSKA | 52-60 | 126 | Q8CGP2 | Histone H2B type 1-P |
| 37 | PEAFPALA | 385-392 | 392 | Q9CY58 | Plasminogen activator inhibitor 1 RNA-binding protein |
| 38 | PNKLVELNK | 138-146 | 943 | Q6DFV7 | Nuclear receptor coactivator 7 |
| 39 | PPSAKAAID | 97-105 | 323 | Q5SSG6 | TATA box binding protein (TBP)-associated factor |
| 40 | PPTAKAAVE | 425-433 | 661 | Q5SUS9 | Ewing sarcoma breakpoint region 1 |
| 41 | PVKAVEIEI | 155-163 | 930 | Q7TSZ1 | Xeroderma pigmentosum, complementation group C |
| 42 | RSPENPPSKEL | 171-181 | 181 | Q9CQA0 | Centromere protein M |
| 43 | RSPGNSPTPM | 186-195 | 469 | Q8CI61 | BAG family molecular chaperone regulator 4 |
| 44 | SALINLSSF | 7-15 | 161 | Q8VE65 | Transcription initiation factor TFIID subunit 12 |
| 45 | SAPENAVRM | 28-36 | 126 | O35127 | Protein C10 |
| 46 | SAPSNFEHR | 12-20 | 593 | Q8BTW9 | Serine/threonine-protein kinase PAK 4 |
| 47 | SAPTGSGKTL | 227-236 | 639 | Q6P9R1 | ATP-dependent RNA helicase DDX51 |
| 48 | SAVISLEGKPL | 156-166 | 166 | P18760 | Cofilin-1 |
| 49 | SAVSNNYIQTL | 168-178 | 911 | P30999 | Catenin delta-1 |
| 50 | SHRKFSAPR | 2-10 | 403 | Q3U9L3 | ribosomal protein L3P family member |
| 51 | SLGINPHVL | 966-974 | 1170 | B2RQL0 | Nup98 protein |
| 52 | SLGKNPTDAYL | 60-70 | 172 | Q3THE2 | Myosin regulatory light chain MRLC2 |
| 53 | SQPESKVFYL | 110-119 | 245 | P63101 | Protein kinase C inhibitor protein 1 |
| 54 | SSTTNPKLSTL | 726-736 | 959 | Q5NBZ5 | Eukaryotic translation initiation factor 4E nuclear import factor 1 |
| 55 | STIRLLTSL | 458-466 | 545 | P80318 | T-complex protein 1 subunit gamma |
| 56 | TNPESKVFYL | 155-160 | 284 | P68254 | 14-3-3 protein theta |
| 57 | TQGQNIQHL | 431-439 | 2326 | Q80YT7 | Myomegalin |
| 58 | VAVTNGPRS | 371-379 | 393 | Q9D6J1 | LAG1 longevity assurance homologue 4 |
| 59 | VPPVQVSPLIKFGRY | 2-16 | 71 | Q5EBI8 | ATP synthase, H+ transporting, mitochondrial F1F0 complex |
| 60 | VQVSNFKSGKGDSTL | 254-268 | 579 | Q80WJ7 | Metastasis adhesion protein |
| 61 | VSLLDIDHL | 473-481 | 679 | Q8BQM4 | HEAT repeat-containing protein 3 |
| 62 | VSLLNPPETL | 411-420 | 422 | Q8BRG1 | Cyclin A2, isoform CRA |
| 63 | VTLVNHGSTF | 15-24 | 640 | P59110 | SUMO-1 protease 2 |
| 64 | YGYSNRVVDLM | 316-326 | 333 | P16858 | Glyceraldehyde-3-phosphate dehydrogenase |
| 65 | AAPRSGPSV | 611-619 | 641 | Q923D5 | WW domain-binding protein 11 |
| 66 | AAPRSGPSVA | 611-620 | 641 | Q923D5 | WW domain-binding protein 11 |
| 67 | AGIENKFGL | 648-656 | 676 | Q9QXX4 | Calcium-binding mitochondrial carrier protein Aralar2 |
| 68 | AGIENKFGLYL | 648-658 | 676 | Q9QXX4 | Calcium-binding mitochondrial carrier protein Aralar2 |
| 69 | AGIENKFGLYLP | 648-659 | 676 | Q9QXX4 | Calcium-binding mitochondrial carrier protein Aralar2 |
| 70 | TKAVTKYTSSK | 117-127 | 127 | A0JNS9 | Histone H2B |
| 71 | GTKAVTKYTSSK | 116-127 | 127 | A0JNS9 | Histone H2B |
| 72 | SEGTKAVTKYTSSK | 114-127 | 127 | A0JNS9 | Histone H2B |
| 73 | VSEGTKAVTKYTSSK | 113-127 | 127 | A0JNS9 | Histone H2B |
| 74 | AVSEGTKAVTKYTSSK | 112-127 | 127 | A0JNS9 | Histone H2B |
| 75 | PEPAKSAPAPK | 2-12 | 126 | Q64478 | Histone H2B type 1-H |
| 76 | PEPAKSAPAPKKG | 2-14 | 126 | Q64478 | Histone H2B type 1-H |
| 77 | LGVKNSEPA | 59-67 | 576 | Q80UG5 | Septin-9 |
| 78 | LGVKNSEPAA | 59-68 | 576 | Q80UG5 | Septin-9 |
| 79 | SSPANISSLEFEDA | 583-596 | 773 | P51125 | Calpastatin |
| 80 | SSPANISSLEFEDAK | 583-597 | 773 | P51125 | Calpastatin |
| 81 | SSPANISSLEFEDAKLS | 583-599 | 773 | P51125 | Calpastatin |
| 82 | SSPANISSLEFEDAKLSA | 583-600 | 773 | P51125 | Calpastatin |
| 83 | KPLFEILNG | 1138-1146 | 1735 | Q9IZU3 | Endogenous mouse mammary tumor virus Mtv1 |
| 84 | KPLFEILNGD | 1138-1147 | 1735 | Q9IZU3 | Endogenous mouse mammary tumor virus Mtv1 |
We concluded that the presence of the high-affinity binding peptide Qdm in the groove of Qa-1b prevents the accommodation and display of a broad array of alternative peptides and that this new repertoire only emerges upon processing defects.
Immunogenicity of the Qa-1b-binding peptides
A direct consequence of the absence of this alternative peptide repertoire under physiological conditions is that this repertoire may comprise neoantigens for the CD8+ T cell system. To estimate the immunogenicity of the identified Qa-1b–binding peptides, we tested them in a matrix-based screening approach in which each peptide was systematically represented in two different peptide pools containing five peptides each. None of our Qa-1b–restricted T cell clones recognized peptides from the identified repertoire, so polyclonal responder T cells from mice immunized with TAP-deficient EC7.1.Qa-1b.B7 tumors were applied, assuring that the peptide-directed responses were induced by naturally presented antigens. Responses of 25 independent T cell cultures were compiled and final immunogenicity scores revealed several immunogenic peptides (Fig. 6 A). These peptide antigens had no common amino acid motif and were derived from different housekeeping proteins (see corresponding numbers in Table II), again pointing at the broad diversity of this antigen repertoire. To assess the potency of the five most immunogenic peptide sequences (number 3, 12, 17, 29, and 41) to induce CD8+ T cell responses in vivo, we immunized mice with the peptides in conjunction with imiquimod as adjuvant. After two vaccinations, clear populations of peptide-specific CD8+ T cells were detectable in the blood (Fig. 6, B–D). The peptides with the largest T cell populations also scored high in the immunogenicity screen in which tumor-stimulated T cells were applied (Fig. 6 A). Interestingly, peptide immunizations in H-2Db−/−Kb−/− mice did not lead to a population of IFN-γ–positive CD8+ T cells, indicating that the T cell repertoire in these knockout mice is tolerant for the novel Qa-1b binding peptides (Fig. S3). This is in line with the finding that H-2Db−/−Kb−/− target cells efficiently presented these peptides (Fig. 3 B), which are part of the normal Qa-1b peptide repertoire inducing central tolerance in these mice. Importantly, the vaccine-induced CD8+ T cells in wild-type mice exhibited strong cytolytic reactivity in vivo against target cells that were loaded with the relevant peptide (Fig. 6, E–G). Unloaded target cells and cells loaded with Qdm peptide were not killed in vivo. Vaccination with the Qdm peptide did not trigger any T cell immunity, illustrating that the T cell repertoire for this sequence does not exist or is functionally tolerant.
Figure 6.
Immunogenicity of Qa-1b–presented peptides in vitro and in vivo. (A) Immunogenicity of 59 peptides was determined by compiling the IFN-γ response of 25e T cell cultures, which were induced by immunization with irradiated TAP-negative EC7.1.Qa-1b.B7 cells. Responses were measured using a matrix-based approach in which each peptide was represented in two independent pools. T cell responses against each pool were scored from zero to three and compiled scores from the two pools were multiplied. (B–D) Percentage of IFN-γ–producing CD8+ T cells in the blood of mice that were naive (B) or immunized with the indicated peptide (C). Blood samples were taken and cells were stimulated overnight with medium or specific peptide, stained with antibodies, and analyzed by flow cytometry. Collected data from peptide-specific frequencies of CD8+ cells from mice are depicted in (D). Each data point represents one mouse, and data from two independent experiments is shown. (E–G) Cytotoxic reactivity in vivo in the same mice as shown in D. Specific killing was determined in naive (E) or immunized mice (F) by comparing the numbers of CFSEhigh targets, which were loaded with relevant peptides, to CFSElow targets, which were not loaded with peptide. CFSEintermediate targets were loaded with Qdm and were always comparable to targets without peptide. The means of percentage in vivo killing is depicted with standard deviations (G).
In conclusion, a broad repertoire of Qa-1b–binding peptide-epitopes of self origin emerges at the surface of cells with impairments in the antigen-processing pathway and that this novel repertoire replaces the TAP-dependent Qdm. These results underscore a role of Qa-1b in adaptive immunity and infer that T cell responses are evoked via this nonclassical MHC molecule to prevent outgrowth of processing-deficient cells in the body.
DISCUSSION
The nonclassical MHC class I molecule Qa-1b normally presents monomorphic Qdm peptides derived from the leader sequences of classical MHC. This Qdm peptide–MHC complex functions as a remote sensor of the class I processing pathway integrity through detection by CD94/NKG2A receptors on NK cells. Failure to present Qdm, caused by impairments in proteolysis, TAP, or other means, increases the susceptibility of cells to attack by these lymphocytes. We show that Qdm under such conditions is replaced by a surprisingly broad repertoire of alternative self-peptides. This novel peptide repertoire comprises neoantigens for which the T cell system is not hampered by immunological tolerance and can therefore induce a vigorous response of Qa-1b–restricted CD8+ CTL. This antigen presenting function of Qa-1b places this conserved MHC molecule at the intersection of innate and adaptive immunity. On one hand, it plays a role as a monomorphic inhibiting ligand when Qdm is presented, and, on the other hand, as a display system of an immunogenic self-peptide repertoire. Our data suggest that the immune system is equipped with multiple mechanisms to remove processing-deficient cells from the body.
The finding that Qa-1b is capable of presenting other peptides than Qdm is supported by previous work from several groups. The leader of the CMV protein UL40 was found to be presented, as well as peptides from Salmonella, Listeria, Epstein-Barr virus, and Mycobacterium tuberculosis (Rodgers and Cook, 2005). Furthermore, an increase of Hsp60 in the cellular stress response was shown to result in replacement of the human Qdm homologue with the Hsp60 signal peptide (Michaëlsson et al., 2002). The unique feature, however, of the peptide repertoire we have elucidated in our current work is the fact that these peptides are derived from endogenous housekeeping proteins and only emerge in Qa-1b on the cell surface when the processing pathway is hampered. We demonstrated the display of these peptides in cells with partial and mild deficiencies that do not completely prevent Qdm from appearance, but a complete block in the processing pathway, e.g., loss of TAP1 or TAP2, resulted in the most efficient presentation of the novel peptides, demonstrating that TAP-independent processing mechanisms are responsible for the replacement of Qdm. Surprisingly, cell surface expression of Qa-1b is less affected by TAP deficiency than classical MHC class I molecules (Kambayashi et al., 2004), and HLA-E surface expression has also been described in the absence of functional TAP proteins or class I–derived signal peptides (Furukawa et al., 1999; Palmisano et al., 2005). Qa-1b and HLA-E, indeed, seem to efficiently egress from the ER in the absence of TAP (Robinson et al., 1998; LoMonaco et al., 2008). These data implicate that our mouse studies are relevant for the human setting. Two alternative processing routes have been described by which peptides can bypass the TAP transporter: N-terminal signal peptides, which are processed by signal peptide cleaving enzymes (Martoglio and Dobbertein, 1998) and C-terminal ends of ER-resident proteins, can also be loaded without TAP function, although the responsible proteolytic enzymes have not been identified for this pathway (Snyder et al., 1998). 40% of the 84 identified peptides in our search were indeed located at the N or C terminus. Interestingly, one of the most immunogenic Qa-1b peptide epitopes FAPLPRLPTL is encoded in the N-terminal signal sequence of a mitochondrial carrier protein (UniProt Q9CR21). The fact that 60% of the peptides cannot be explained by these two processing mechanisms suggests the existence of other yet uncharacterized processing pathways for MHC class I antigen presentation. It has been proposed that autophagy might contribute to MHC class I–restricted antigen presentation (Vyas et al., 2008). Autophagy is a crucial cellular mechanism responsible for the clearance of old or damaged cellular components, including organelles and macromolecules. This pathway might lead to the generation of peptides susceptible for MHC class I loading. Previous characterizations of peptides bound to classical MHC class I from TAP-deficient cells revealed similar features: predominance of signal peptides, increased peptide lengths, and selective presentation on TAP-negative variants (Suri et al., 2006; Weinzierl et al., 2008). The Qa-1b peptide repertoires present on TAP-positive versus TAP-negative cells were extremely different (Table II), in that the high-affinity binding monomorphic Qdm strongly dominated the TAP-positive pool for >95%, whereas a broad diversity of other peptides was present in the TAP-negative pool. Previous comparisons of peptide repertoires presented by classical MHC class I molecules showed much more overlap between the TAP-positive and -negative pools (Suri et al., 2006; Weinzierl et al., 2008). This indicates that the high-affinity binding Qdm competes with the TAP-independent peptides, and thereby prevents their presentation under normal circumstances. However, even mild or partial processing deficiencies in tumor cells already led to the presentation of both types of peptides, as illustrated by the fact that Qdm-specific CTL and our Qa-1b–restricted CTL were able to recognize the four colon tumors (Fig. 4). Importantly, Qdm is not immunogenic because of the widespread presentation in the normal host, including thymus (Sullivan et al., 2002), and thereby prevent tolerance mechanisms for CTL specificities that recognize the new emerging peptide-epitopes. We indeed found that T cells specific for these novel Qa-1b–peptides are deleted in mice that do present these peptides in Qa-1b molecules during T cell development, as in H-2Db−/−Kb−/− mice (Fig. S3).
Our Qa-1b–restricted T cells exhibit a normal phenotype for CD8+ T lymphocytes (Fig. S1) and use rearranged αβTCRs, and thereby distinguish themselves from reported Qa-1–recognizing intestinal γδ+ T cells (Davies et al., 2004) and the Qa-1–restricted regulatory CD8αα+ T cell subset (Tang et al., 2006). Target cell recognition was mediated by the TCR and this receptor did not cross-react with classical class I molecules, indicating that also, at the functional level, this subset is not different from conventional CD8+ T lymphocytes. This is in line with data from mice lacking classical MHC class I molecules arguing that ∼10% of the total CD8+ T cell subset is restricted by nonclassical molecules (Pérarnau et al., 1999; Seaman et al., 1999; Ureta-Vidal et al., 1999; Vugmeyster et al., 1998). This T cell repertoire is heterogeneous, as witnessed by a diverse usage of TCRVβ segments (Pérarnau et al., 1999; Ureta-Vidal et al., 1999). Selection of Qa-1b–restricted T cells in the thymus, indeed, does occur (Sullivan et al., 2002), but it remains puzzling how a monomorphic Qdm–Qa-1b complex can mediate positive selection resulting in such a broad repertoire. Furthermore, the relationship between the suppressive Qa-1b–restricted T cells (Jiang and Chess, 2000; Lu et al., 2006) and the tumor-reactive T cells described here remains elusive.
We speculate that comparable T cell subsets exist in human beings, especially because Qa-1b and HLA-E are rather conserved in structure and function (Rodgers and Cook, 2005). TCR-mediated recognition of HLA-E has been previously described (García et al., 2002; Heinzel et al., 2002; Romagnani et al., 2002), and it is highly likely that a subset similar to our Qa-1b-CTL, which responds against tumors with processing impairments, is present in humans. HLA-E surface expression is ubiquitous in the body and even detected in the absence of the human Qdm homologue (Furukawa et al., 1999; Palmisano et al., 2005), although MHC class I leaders strongly stimulate HLA-E display (Braud et al., 1998b; Lee et al., 1998). The lack of polymorphism in the human population infers that identified neoantigens presented in HLA-E constitute universal epitopes that might be exploited for the therapy of frequently occurring tumor immune escape variants and persistent infections by viruses encoding immune evasion proteins.
MATERIALS AND METHODS
Cell lines and mice.
The origin and culturing of most cell lines used in this study was described previously (van Hall et al., 2006). C4.4-25− is a β2m-deficient variant of EL4. RMA-S.B7-1 is a CD80 transfectant of RMA-S. EC7.1 is a Kb- and Db-negative variant of RMA-S (Howell et al., 2000). B78H1 is a TAP-deficient and MHC class I–deficient melanoma (Chiang et al., 2003). Tap1−/− and wild-type mouse embryo fibroblasts were immortalized by the adenovirus type 5 E1 gene (Ad5MEC, clone XC3). Tap1−/− fibrosarcoma was induced with MCA (clone MCB6TAP). Gene transfer of mouse TAP1, mouse TAP2, H-2Db, H-2Kb, and Qa-1b (T23 gene) was performed with retroviruses based on the MuLV vector LZRS. Generation of CTL clones was performed according to previous description (van Hall et al., 2006). In brief, syngeneic C57BL/6 mice were immunized with irradiated RMA-S.B7-1 cells and in vitro stimulated weekly with a mixture of RMA-S.B7-1 and EC7.1.Qa-1b.B7-1 cells, naive splenocytes, and IL-2. Qa-1b–restricted CTL clone B12i that is reactive with the TAP-dependent AMAPRTLLL peptide was isolated from a B6.Tla mouse (Cotterill et al., 1997).
All mice were purchased from Iffa-Credo and housed in the animal facility of the Leiden University Medical Center under specific pathogen–free conditions and used between 8 and 12 wk of age. TAP1-knockout mice were purchased from Jackson ImmunoResearch Laboratories and MHC class I knockout animals were obtained from F. Lemonnier (Institut Pasteur, Paris, France; Pérarnau et al., 1999). Experiments were performed in accordance with Dutch national legislation and institutional guidelines and were approved by the animal ethical committee of the Leiden University Medical Center.
CTL assays and flow cytometry.
CTL activity was measured by chromium (51Cr) release assay or IFN-γ ELISA as previously described (van Hall et al., 2000). Data shown represent mean values obtained from triplicate test-wells, and the error bars represent standard deviation of these values. For antibody blocking, CTL were pretreated with 20 µg/ml antibodies, washed, and added as responders to target cells for IFN-γ release. The following antibodies were used in functional blocking assays: hamster anti-CD3 (Fab2 fragments of clone 145-2C11), control hamster anti-TNP, rat anti-CD4 (clone GK1.4), rat anti-CD8 (clone 2.43) anti-NKG2A/C (clone 20d5), and anti-Qa-1b (clone 6A8.6F10.1A6). For flow cytometry analysis, the following monoclonal antibodies were purchased from BD: Ly49AB6 (clone A1), Ly49C/I (clone 5E6), Ly49G (clone LGL-1), Qa-1b (clone 6A8.6F10.1A6), NKG2AB6 (clone 16a11), CD94 (clone 18d3), CD3 (clone 145-2C11), CD8α (clone Ly2), CD8β (clone Ly-3.2.), CD4 (clone GK1.4), CD16/CD32 (clone 2.4G2), NK1.1 (clone PK136), CD49b (clone DX5), and CD244 (clone 2B4). From eBioscience we purchased NKG2D (clone CX5).
Peptide elution, HPLC, and MS.
Peptides were eluted out of purified chimeric Qa-1b/Db molecules from EC7.1 cells. The genomic H-2Db construct in which the α 1 and α 2 domains were exchanged with those of Qa-1b was provided by P.J. Dyson (Imperial College, Hammersmith Hospital, London, UK; Cotterill et al., 1997). This genomic construct was expressed in HeLa cells, and cDNA was then cloned into the retroviral construct LZRS. EC7.1 cells transduced with this construct were twice sorted by FACS. TAP-positive counterparts were generated by retroviral gene transfer of the mouse TAP2 gene. Immunoprecipitation of Qa-1b/Db molecules was performed with protein A beads covalently coupled with anti-Db mAb 28–14-8S from two and four independent lysates of 40.109 TAP-positive and TAP-negative EC7.1 cells, respectively, as previously described (van Hall et al., 2000). The peptide pools were prefractionated on a C18 RP-HPLC system (200 µm × 15 cm; Reprosil-C18-AQ 3 µm; Dr. Maisch GmbH). Fractions were reduced to near dryness, diluted in 95/3/0.1 vol/vol/vol water/acetonitril/formic acid and subsequently analyzed by tandem MS. Peptides were analyzed by nanoflow liquid chromatography using an Agilent 1100 HPLC system (Agilent Technologies), as previously described (van Hall et al., 2006), and coupled on line to a 7-tesla LTQ-FT mass spectrometer (Thermo Electron). The end of the nanocolumn was drawn to a tip (internal dimaeter [ID] ∼5 µm), from which the eluent was sprayed into the mass spectrometer. Peptides were trapped at 5 µl/min on a 1-cm column (100-µm ID; ReproSil-Pur C18-AQ, 3 µm) and eluted to a 15 cm column (50-µm ID; ReproSil-Pur C18-AQ, 3 µm) at 150 nl/min in a 60-min gradient from 0 to 50% acetonitrile in 0.1% formic acid.
The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition. Full scan MS spectra were acquired in the FT-ICR with a resolution of 25,000 at a target value of 3,000,000. The two most intense ions were then isolated for accurate mass measurements by a selected ion monitoring scan in FT-ICR with a resolution of 50,000 at a target accumulation value of 50,000. The selected ions were then fragmented in the linear ion trap using collision-induced dissociation at a target value of 10,000. In a post-analysis process, raw data were converted to peak lists using Bioworks Browser software, Version 3.1. For peptide/protein identification, MS/MS data were submitted to the mouse IPI database using Mascot Version 2.1 (Matrix Science) with the following settings: 5 ppm and 0.8-D deviation for precursor and fragment masses, respectively; no enzyme was specified. All peptides with a mascot ion score >30 were manually inspected. This resulted in an initial set of 150 peptides. These peptides were synthesized, and their MS/MS spectra compared with the MS/MS spectra of the Qa-1b–eluted peptides. This resulted in 84 confirmed peptide identifications.
Immunogenicity screen of Qa-1b presented peptides.
C57BL/6 mice were immunized twice with irradiated EC7.1.B7-1 cells with stable expression of Qa-1b. Spleens of immunized mice were cultured with the same cells for 1-2 wk, and reactivity of the cultures was tested against the EC7.1 cell panel to confirm Qa-1b restriction of the T cells. Reactivity against the identified peptides was assessed using a matrix-based system with pools of five different peptides and in which each peptide was represented in two independent pools. The peptide pools were loaded on spleen cells and IFN-γ production by T cells was measured with sandwich ELISA after 18 h. Reactivity against each pool was arbitrarily scored from zero to three, according to the mean background response. 25 independent cultures were assayed in this way, and the scores of each pool were compiled. The final immunogenicity score for each peptide was then obtained by multiplying the scores of two pools containing that particular peptide.
Peptide immunizations and in vivo cytotoxicity assay.
For peptide immunizations, C57BL/6 and H-2Db−/−Kb−/− mice were injected s.c. at the flank with 50 µg peptide mixed with 50 µg helper peptide from MuLV (H19; van Hall et al., 2006) in PBS on day 0 and 7. Immediately after injection, 60 mg Aldara cream (3M Pharmaceutical) containing 5% imiquimod was applied to the skin at the injection site. Mice received i.p. injections of 500,000 IU recombinant human IL-2 (Novartis) on the day of the second vaccination and on the day thereafter. T cell frequencies were determined from blood lymphocytes after 4 d and from spleens after 7 d with intracellular cytokine staining, as previously described (Bijker et al., 2007). In short, cells were cultured overnight with medium or 5 µg/ml of peptide and stained for CD8 and intracellular IFN-γ. Killing capacity of the peptide-induced T cells was determined in immunized animals using differentially CFSE-labeled and peptide-pulsed splenocytes, as described (Bijker et al., 2007). In brief, spleens from CD90.1 congenic C57BL/6 mice were passed through nylon wool and labeled with 5 µM CFSE and pulsed with indicated peptides, labeled with 0.5 µM CFSE and pulsed with control Qdm peptide, or labeled with 0.05 µM CFSE without peptide. These three target cell populations were washed, mixed, and 107 cells were injected i.v. per recipient mouse. The spleens of recipient mice were harvested after 2 d, stained with CD90.1-specific antibodies, and analyzed with flow cytometry. Percentage killing was calculated as the ratio between the numbers of peptide pulsed targets and control pulsed targets.
Online supplemental material.
Fig. S1 shows flow cytometry analysis of one representative Qa-1b–restricted CTL clone for several T cell lineage and NK cell surface markers. Fig. S2 depicts the T cell reactivity against a panel of EC7.1 cells (TAP- and MHC class I-negative; Howell et al., 2000) that were reconstituted with single constructs encoding H-2Db, -Kb, or -Qa-1b. In Fig. S3, the immunogenicity of Qa-1b–presented peptides in H-2Db−/−Kb−/− knockout mice is shown from blood lymphocytes of immunized mice. Lymphocytes incubated with PMA/ionomycin mitogens served as positive control. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20091429/DC1.
Acknowledgments
The authors would like to thank Dr. F. Lemonnier for providing MHC class I knockout mice, Margit Lampen for assistance with experiments, and Drs. Van Bergen, Koning, Toes, and Melief for critical reading of the manuscript.
Financial support was received from the Macropa foundation, the AICR (09-776), and the Dutch Cancer Society (UL 2007-3897).
Authors declare no financial conflict of interest.
Footnotes
Abbreviations used:
- HLA
- human leukocyte antigen
- MCA
- methylcholantrene
- MS
- mass spectrometry
- Qdm
- Qa-1 determinant modifier
References
- Aldrich C.J., DeCloux A., Woods A.S., Cotter R.J., Soloski M.J., Forman J. 1994. Identification of a Tap-dependent leader peptide recognized by alloreactive T cells specific for a class Ib antigen. Cell. 79:649–658 doi: 10.1016/0092-8674(94)90550-9 [DOI] [PubMed] [Google Scholar]
- Bijker M.S., van den Eeden S.J., Franken K.L., Melief C.J., Offringa R., van der Burg S.H. 2007. CD8+ CTL priming by exact peptide epitopes in incomplete Freund’s adjuvant induces a vanishing CTL response, whereas long peptides induce sustained CTL reactivity. J. Immunol. 179:5033–5040 [DOI] [PubMed] [Google Scholar]
- Bland F.A., Lemberg M.K., McMichael A.J., Martoglio B., Braud V.M. 2003. Requirement of the proteasome for the trimming of signal peptide-derived epitopes presented by the nonclassical major histocompatibility complex class I molecule HLA-E. J. Biol. Chem. 278:33747–33752 doi: 10.1074/jbc.M305593200 [DOI] [PubMed] [Google Scholar]
- Braud V.M., Allan D.S., O’Callaghan C.A., Söderström K., D’Andrea A., Ogg G.S., Lazetic S., Young N.T., Bell J.I., Phillips J.H., et al. 1998a. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 391:795–799 doi: 10.1038/35869 [DOI] [PubMed] [Google Scholar]
- Braud V.M., Allan D.S.J., Wilson D., McMichael A.J. 1998b. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr. Biol. 8:1–10 doi: 10.1016/S0960-9822(98)70014-4 [DOI] [PubMed] [Google Scholar]
- Chiang E.Y., Henson M., Stroynowski I. 2003. Correction of defects responsible for impaired Qa-2 class Ib MHC expression on melanoma cells protects mice from tumor growth. J. Immunol. 170:4515–4523 [DOI] [PubMed] [Google Scholar]
- Cotterill L.A., Stauss H.J., Millrain M.M., Pappin D.J., Rahman D., Canas B., Chandler P., Stackpoole A., Simpson E., Robinson P.J., Dyson P.J. 1997. Qa-1 interaction and T cell recognition of the Qa-1 determinant modifier peptide. Eur. J. Immunol. 27:2123–2132 doi: 10.1002/eji.1830270902 [DOI] [PubMed] [Google Scholar]
- Davies A., Lopez-Briones S., Ong H., O’Neil-Marshall C., Lemonnier F.A., Nagaraju K., Metcalf E.S., Soloski M.J. 2004. Infection-induced expansion of a MHC Class Ib-dependent intestinal intraepithelial gammadelta T cell subset. J. Immunol. 172:6828–6837 [DOI] [PubMed] [Google Scholar]
- DeCloux A., Woods A.S., Cotter R.J., Soloski M.J., Forman J. 1997. Dominance of a single peptide bound to the class I(B) molecule, Qa-1b. J. Immunol. 158:2183–2191 [PubMed] [Google Scholar]
- Diefenbach A., Jamieson A.M., Liu S.D., Shastri N., Raulet D.H. 2000. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat. Immunol. 1:119–126 doi: 10.1038/77793 [DOI] [PubMed] [Google Scholar]
- Furukawa H., Yabe T., Akaza T., Tadokoro K., Tohma S., Inoue T., Tokunaga K., Yamamoto K., Geraghty D.E., Juji T. 1999. Cell surface expression of HLA-E molecules on PBMC from a TAP1-deficient patient. Tissue Antigens. 53:292–295 doi: 10.1034/j.1399-0039.1999.530310.x [DOI] [PubMed] [Google Scholar]
- García P., Llano M., de Heredia A.B., Willberg C.B., Caparrós E., Aparicio P., Braud V.M., López-Botet M. 2002. Human T cell receptor-mediated recognition of HLA-E. Eur. J. Immunol. 32:936–944 doi: [DOI] [PubMed] [Google Scholar]
- Gays F., Fraser K.P., Toomey J.A., Diamond A.G., Millrain M.M., Dyson P.J., Brooks C.G. 2001. Functional analysis of the molecular factors controlling Qa1-mediated protection of target cells from NK lysis. J. Immunol. 166:1601–1610 [DOI] [PubMed] [Google Scholar]
- Grimsley C., Kawasaki A., Gassner C., Sageshima N., Nose Y., Hatake K., Geraghty D.E., Ishitani A. 2002. Definitive high resolution typing of HLA-E allelic polymorphisms: identifying potential errors in existing allele data. Tissue Antigens. 60:206–212 doi: 10.1034/j.1399-0039.2002.600302.x [DOI] [PubMed] [Google Scholar]
- Heinzel A.S., Grotzke J.E., Lines R.A., Lewinsohn D.A., McNabb A.L., Streblow D.N., Braud V.M., Grieser H.J., Belisle J.T., Lewinsohn D.M. 2002. HLA-E–dependent presentation of Mtb-derived antigen to human CD8+ T cells. J. Exp. Med. 196:1473–1481 doi: 10.1084/jem.20020609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermel E., Hart A.J., Gunduz I., Acton H., Kim C., Wurth M., Uddin S., Smith C., Fischer Lindahl K., Aldrich C.J. 2004. Polymorphism and conservation of the genes encoding Qa1 molecules. Immunogenetics. 56:639–649 doi: 10.1007/s00251-004-0722-x [DOI] [PubMed] [Google Scholar]
- Howell D., Levitt J.M., Foster P.A., Guenther M.M., Shawar S.M., Rich R.R., Rodgers J.R. 2000. Heterogeneity of RMA-S cell line: derivatives of RMA-S cells lacking H2-Kb and H2-Db expression. Immunogenetics. 52:150–154 doi: 10.1007/s002510000254 [DOI] [PubMed] [Google Scholar]
- Jensen P.E., Sullivan B.A., Reed-Loisel L.M., Weber D.A. 2004. Qa-1, a nonclassical class I histocompatibility molecule with roles in innate and adaptive immunity. Immunol. Res. 29:81–92 doi: 10.1385/IR:29:1-3:081 [DOI] [PubMed] [Google Scholar]
- Jiang H., Chess L. 2000. The specific regulation of immune responses by CD8+ T cells restricted by the MHC class Ib molecule, Qa-1. Annu. Rev. Immunol. 18:185–216 doi: 10.1146/annurev.immunol.18.1.185 [DOI] [PubMed] [Google Scholar]
- Kaiser B.K., Pizarro J.C., Kerns J., Strong R.K. 2008. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc. Natl. Acad. Sci. USA. 105:6696–6701 doi: 10.1073/pnas.0802736105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambayashi T., Kraft-Leavy J.R., Dauner J.G., Sullivan B.A., Laur O., Jensen P.E. 2004. The nonclassical MHC class I molecule Qa-1 forms unstable peptide complexes. J. Immunol. 172:1661–1669 [DOI] [PubMed] [Google Scholar]
- Kraft J.R., Vance R.E., Pohl J., Martin A.M., Raulet D.H., Jensen P.E. 2000. Analysis of Qa-1(b) peptide binding specificity and the capacity of CD94/NKG2A to discriminate between Qa-1-peptide complexes. J. Exp. Med. 192:613–624 doi: 10.1084/jem.192.5.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa Z., Hasemann C.A., Forman J. 1998. Qa-1b binds conserved class I leader peptides derived from several mammalian species. J. Exp. Med. 188:973–978 doi: 10.1084/jem.188.5.973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Goodlett D.R., Ishitani A., Marquardt H., Geraghty D.E. 1998. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 160:4951–4960 [PubMed] [Google Scholar]
- Lemberg M.K., Bland F.A., Weihofen A., Braud V.M., Martoglio B. 2001. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J. Immunol. 167:6441–6446 [DOI] [PubMed] [Google Scholar]
- Li L., Sullivan B.A., Aldrich C.J., Soloski M.J., Forman J., Grandea A.G., III, Jensen P.E., Van Kaer L. 2004. Differential requirement for tapasin in the presentation of leader- and insulin-derived peptide antigens to Qa-1b-restricted CTLs. J. Immunol. 173:3707–3715 [DOI] [PubMed] [Google Scholar]
- Ljunggren H.G., Kärre K. 1990. In search of the ‘missing self ’: MHC molecules and NK cell recognition. Immunol. Today. 11:237–244 doi: 10.1016/0167-5699(90)90097-S [DOI] [PubMed] [Google Scholar]
- Lo W.-F., Woods A.S., DeCloux A., Cotter R.J., Metcalf E.S., Soloski M.J. 2000. Molecular mimicry mediated by MHC class Ib molecules after infection with gram-negative pathogens. Nat. Med. 6:215–218 doi: 10.1038/72329 [DOI] [PubMed] [Google Scholar]
- Lo Monaco E., Sibilio L., Melucci E., Tremante E., Suchànek M., Horejsi V., Martayan A., Giacomini P. 2008. HLA-E: strong association with β2-microglobulin and surface expression in the absence of HLA class I signal sequence-derived peptides. J. Immunol. 181:5442–5450 [DOI] [PubMed] [Google Scholar]
- Lu L., Werneck M.B.F., Cantor H. 2006. The immunoregulatory effects of Qa-1. Immunol. Rev. 212:51–59 doi: 10.1111/j.0105-2896.2006.00418.x [DOI] [PubMed] [Google Scholar]
- Martoglio B., Dobbertein B. 1998. Signal sequences: more than just greasy peptides. Trends Cell Biol. 8:410–415 [DOI] [PubMed] [Google Scholar]
- McMahon C.W., Raulet D.H. 2001. Expression and function of NK cell receptors in CD8+ T cells. Curr. Opin. Immunol. 13:465–470 doi: 10.1016/S0952-7915(00)00242-9 [DOI] [PubMed] [Google Scholar]
- Michaëlsson J., Teixeira de Matos C., Achour A., Lanier L.L., Kärre K., Söderström K. 2002. A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition. J. Exp. Med. 196:1403–1414 doi: 10.1084/jem.20020797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.D., Weber D.A., Ibegbu C., Pohl J., Altman J.D., Jensen P.E. 2003. Analysis of HLA-E peptide-binding specificity and contact residues in bound peptide required for recognition by CD94/NKG2. J. Immunol. 171:1369–1375 [DOI] [PubMed] [Google Scholar]
- O’Callaghan C.A., Tormo J., Willcox B.E., Braud V.M., Jakobsen B.K., Stuart D.I., McMichael A.J., Bell J.I., Jones E.Y. 1998. Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E. Mol. Cell. 1:531–541 doi: 10.1016/S1097-2765(00)80053-2 [DOI] [PubMed] [Google Scholar]
- Palmisano G.L., Contardi E., Morabito A., Gargaglione V., Ferrara G.B., Pistillo M.P. 2005. HLA-E surface expression is independent of the availability of HLA class I signal sequence-derived peptides in human tumor cell lines. Hum. Immunol. 66:1–12 doi: 10.1016/j.humimm.2004.10.006 [DOI] [PubMed] [Google Scholar]
- Pérarnau B., Saron M.F., San Martin B.R., Bervas N., Ong H., Soloski M.J., Smith A.G., Ure J.M., Gairin J.E., Lemonnier F.A. 1999. Single H2Kb, H2Db and double H2KbDb knockout mice: peripheral CD8+ T cell repertoire and anti-lymphocytic choriomeningitis virus cytolytic responses. Eur. J. Immunol. 29:1243–1252 doi: [DOI] [PubMed] [Google Scholar]
- Petrie E.J., Clements C.S., Lin J., Sullivan L.C., Johnson D., Huyton T., Heroux A., Hoare H.L., Beddoe T., Reid H.H., et al. 2008. CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. J. Exp. Med. 205:725–735 doi: 10.1084/jem.20072525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson P.J., Travers P.J., Stackpoole A., Flaherty L., Djaballah H. 1998. Maturation of Qa-1b class I molecules requires β 2-microglobulin but is TAP independent. J. Immunol. 160:3217–3224 [PubMed] [Google Scholar]
- Rodgers J.R., Cook R.G. 2005. MHC class Ib molecules bridge innate and acquired immunity. Nat. Rev. Immunol. 5:459–471 doi: 10.1038/nri1635 [DOI] [PubMed] [Google Scholar]
- Rodolfo M., Castelli C., Bassi C., Accornero P., Sensi M., Parmiani G. 1994. Cytotoxic T lymphocytes recognize tumor antigens of a murine colonic carcinoma by using different T-cell receptors. Int. J. Cancer. 57:440–447 doi: 10.1002/ijc.2910570324 [DOI] [PubMed] [Google Scholar]
- Romagnani C., Pietra G., Falco M., Millo E., Mazzarino P., Biassoni R., Moretta A., Moretta L., Mingari M.C. 2002. Identification of HLA-E-specific alloreactive T lymphocytes: a cell subset that undergoes preferential expansion in mixed lymphocyte culture and displays a broad cytolytic activity against allogeneic cells. Proc. Natl. Acad. Sci. USA. 99:11328–11333 doi: 10.1073/pnas.172369799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph M.G., Wilson I.A. 2002. The specificity of TCR/pMHC interaction. Curr. Opin. Immunol. 14:52–65 doi: 10.1016/S0952-7915(01)00298-9 [DOI] [PubMed] [Google Scholar]
- Seaman M.S., Pérarnau B., Lindahl K.F., Lemonnier F.A., Forman J. 1999. Response to Listeria monocytogenes in mice lacking MHC class Ia molecules. J. Immunol. 162:5429–5436 [PubMed] [Google Scholar]
- Snyder H.L., Bacík I., Yewdell J.W., Behrens T.W., Bennink J.R. 1998. Promiscuous liberation of MHC-class I-binding peptides from the C termini of membrane and soluble proteins in the secretory pathway. Eur. J. Immunol. 28:1339–1346 doi: [DOI] [PubMed] [Google Scholar]
- Strong R.K., Holmes M.A., Li P., Braun L., Lee N., Geraghty D.E. 2003. HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J. Biol. Chem. 278:5082–5090 doi: 10.1074/jbc.M208268200 [DOI] [PubMed] [Google Scholar]
- Sullivan B.A., Kraj P., Weber D.A., Ignatowicz L., Jensen P.E. 2002. Positive selection of a Qa-1-restricted T cell receptor with specificity for insulin. Immunity. 17:95–105 doi: 10.1016/S1074-7613(02)00343-6 [DOI] [PubMed] [Google Scholar]
- Suri A., Walters J.J., Levisetti M.G., Gross M.L., Unanue E.R. 2006. Identification of naturally processed peptides bound to the class I MHC molecule H-2Kd of normal and TAP-deficient cells. Eur. J. Immunol. 36:544–557 doi: 10.1002/eji.200526235 [DOI] [PubMed] [Google Scholar]
- Tang X., Maricic I., Purohit N., Bakamjian B., Reed-Loisel L.M., Beeston T., Jensen P., Kumar V. 2006. Regulation of immunity by a novel population of Qa-1-restricted CD8alphaalpha+TCRalphabeta+ T cells. J. Immunol. 177:7645–7655 [DOI] [PubMed] [Google Scholar]
- Ureta-Vidal A., Firat H., Pérarnau B., Lemonnier F.A. 1999. Phenotypical and functional characterization of the CD8+ T cell repertoire of HLA-A2.1 transgenic, H-2KbnullDbnull double knockout mice. J. Immunol. 163:2555–2560 [PubMed] [Google Scholar]
- Valés-Gómez M., Reyburn H.T., Erskine R.A., López-Botet M., Strominger J.L. 1999. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 18:4250–4260 doi: 10.1093/emboj/18.15.4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hall T., van Bergen J., van Veelen P.A., Kraakman M., Heukamp L.C., Koning F., Melief C.J.M., Ossendorp F., Offringa R. 2000. Identification of a novel tumor-specific CTL epitope presented by RMA, EL-4, and MBL-2 lymphomas reveals their common origin. J. Immunol. 165:869–877 [DOI] [PubMed] [Google Scholar]
- van Hall T., Wolpert E.Z., van Veelen P., Laban S., van der Veer M., Roseboom M., Bres S., Grufman P., de Ru A., Meiring H., et al. 2006. Selective cytotoxic T-lymphocyte targeting of tumor immune escape variants. Nat. Med. 12:417–424 doi: 10.1038/nm1381 [DOI] [PubMed] [Google Scholar]
- van Hall T., Laban S., Koppers-Lalic D., Koch J., Precup C., Asmawidjaja P., Offringa R., Wiertz E.J. 2007. The varicellovirus-encoded TAP inhibitor UL49.5 regulates the presentation of CTL epitopes by Qa-1b1. J. Immunol. 178:657–662 [DOI] [PubMed] [Google Scholar]
- Vance R.E., Jamieson A.M., Raulet D.H. 1999. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J. Exp. Med. 190:1801–1812 doi: 10.1084/jem.190.12.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vugmeyster Y., Glas R., Pérarnau B., Lemonnier F.A., Eisen H., Ploegh H. 1998. Major histocompatibility complex (MHC) class I KbDb -/- deficient mice possess functional CD8+ T cells and natural killer cells. Proc. Natl. Acad. Sci. USA. 95:12492–12497 doi: 10.1073/pnas.95.21.12492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas J.M., Van der Veen A.G., Ploegh H.L. 2008. The known unknowns of antigen processing and presentation. Nat. Rev. Immunol. 8:607–618 doi: 10.1038/nri2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzierl A.O., Rudolf D., Hillen N., Tenzer S., Van Endert P., Schild H., Rammensee H.G., Stevanovic S. 2008. Features of TAP-independent MHC class I ligands revealed by quantitative mass spectrometry. Eur. J. Immunol. 38:1–8 doi: 10.1002/eji.200838136 [DOI] [PubMed] [Google Scholar]
- Yang J.C., Perry-Lalley D. 2000. The envelope protein of an endogenous murine retrovirus is a tumor-associated T-cell antigen for multiple murine tumors. J. Immunother. 23:177–183 doi: 10.1097/00002371-200003000-00001 [DOI] [PubMed] [Google Scholar]