Abstract
Cancer is a genetic disease in which the interplay between alterations in protein-coding genes and non-coding RNAs (ncRNAs) plays a fundamental role. In recent years, the full coding component of the human genome was sequenced in various cancers, whereas such attempts related to ncRNAs are still fragmentary. We screened genomic DNAs for sequence variations in 148 microRNAs (miRNAs) and ultraconserved regions (UCRs) loci in patients with chronic lymphocytic leukemia (CLL) or colorectal cancer (CRC) by Sanger technique and further tried to elucidate the functional consequences of some of these variations. We found sequence variations in miRNAs in both sporadic and familial CLL cases, mutations of UCRs in CLLs and CRCs and, in certain instances, detected functional effects of these variations. Furthermore, by integrating our data with previously published data on miRNA sequence variations, we have created a catalog of DNA sequence variations in miRNAs/ultraconserved genes in human cancers. These findings argue that ncRNAs are targeted by both germ line and somatic mutations as well as by single-nucleotide polymorphisms with functional significance for human tumorigenesis. Sequence variations in ncRNA loci are frequent and some have functional and biological significance. Such information can be exploited to further investigate on a genome-wide scale the frequency of genetic variations in ncRNAs and their functional meaning, as well as for the development of new diagnostic and prognostic markers for leukemias and carcinomas.
Introduction
Since the completion of the human genome sequencing, researchers have focused considerable effort on identifying cancer-related somatic mutations in protein-coding genes, such as mutational analysis of the serine/threonine kinome (1) or the tyrosine phosphatome in colorectal cancers (CRCs) (2) as well as the genomic landscapes of human breast cancers (BCs), CRCs, glioblastomas and pancreatic cancers (3–6). These findings offer a considerable amount of information important for the functional understanding of proteome alterations in human cancers. In recent years, the addition of non-coding RNAs (ncRNAs) to the cancer genetics puzzle has provided stimulus for further studies. The main investigations have focused on variations in the expression of microRNA (miRNA) genes, the most extensively studied category of ncRNAs (7,8). In fact, since their initial identification in B cell chronic lymphocytic leukemia (CLL) cases (9), the most common form of adult leukemia, researchers have detected miRNAs alterations in all types of investigated human tumors (10–12).
MiRNAs consist of small sequences of 19–25 nucleotide (nt) non-coding genes and are found in the genomes of animals and plants (8). MiRNAs are cleaved from 60 to 110 nt hairpin precursors (pre-miRNAs) that are produced from large transcripts (pri-miRNAs) that can be thousands of bases long (7). MiRNAs regulate gene expression by reducing the amount of transcribed messenger RNA and/or translated protein and a lot of efforts are aimed to identify the spectrum of miRNA targets important in cancer (12,13). The frequency and significance of genome sequence variations [including mutations and single-nucleotide polymorphisms (SNPs)] in ncRNAs, particularly miRNAs, is unknown at present time. We previously identified germ line and somatic mutations in miRNA genes in ∼10% of patients with CLL, including a germ line mutation in the primary transcript of the tumor suppressor miR-16-1, which is located 7 nt downstream from the pre-miRNA [that we named (C-to-T) + 7 bp in 3′] (14). This mutation reduced expression of miR-16 and the clustered miR-15a in leukemia cells, and of note, one of two patients with CLL harboring it had a family history of cancer: the proband had CLL and BC, whereas her mother died with CLL and her sister died with BC (14). In addition, another group analyzed the sequence of the mouse miR-16-1 locus located near D14Mit160 marker and demonstrated a point mutation in New Zeland Black mice that are naturally developing at late age a B cell malignancy similar to human CLL (15). This point mutation was not present in any of the other mouse strains they analyzed, including the nearest neighbor strain, New Zeland White. Because the affected nucleotide is located in the 3′-flanking region of miR-16-1 6 nt after the precursor, the investigators examined the expression of miR-16 in various tissues in New Zeland Black mice using reverse transcription–polymerase chain reaction (PCR) and northern blot analysis and found decreased expression of it in lymphoid tissue. The delivery of exogenous miR-16 to an New Zeland Black malignant B-1 cell line resulted in cell cycle alterations and increased apoptosis (15). Taken together, these two studies—one of human CLL and the other of a mouse model of human CLL—confirm that miR-16 is the first miRNA proven to be involved in cancer predisposition.
Recently, we found a new category of ncRNAs abnormally expressed in human CLL and CRC cells, which we named ultraconserved genes (UCGs) (16). These genes are transcripts of ultraconserved regions (UCRs), which are completely conserved in the human, mouse and rat genomes (17). Because of a high degree of conservation, the UCRs may be important to the ontogeny and phylogeny of mammals. In recent years, authors have extensively reported genome-wide identification of mutations in the coding component of the human genome, whereas the knowledge regarding sequence variations in ncRNAs is still fragmentary. Herein, we describe the first step toward cataloging DNA variations in miRNAs and UCRs in human cancers by analyzing the genomic sequence of 120 miRNAs expressed in CLL cells and 28 UCRs in both CLL and CRC samples.
Materials and methods
Patient samples and cell lines
Thirty-nine peripheral blood samples from patients with CLL and three samples of mononuclear cells obtained from normal individuals were collected at the CLL Research Consortium Institutions after they gave their written informed consent to participate. Briefly, after the samples were obtained, mononuclear cells were isolated by using Ficoll–Hypaque gradient centrifugation (Amersham Pharmacia Biotech, GE Healthcare, Pittsburgh, PA) and then processed for RNA and DNA extraction according to standard protocols (18). Samples were also obtained from 35 patients with CRC and 175 cancer-free control individuals as described previously (14). The tumor tissues from CRC patients were macroscopically dissected for enrichment in epithelial component and further checked by microscopy and selected for the study only if contained at least 60–70% malignant cells. All the subjects were white Caucasians as indicated by medical records for the patients with CLL and CRC and interviews for the controls. The clinical characteristics of analyzed cancers are presented in Table I. MEG-01 chronic leukemia cells, HEK-293 fetal kidney cells and HeLa cervical cancer cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained according to instructions.
Table I.
Clinical data of cancer patients included in the present study
| CLL | |
| No. of patients | 39 |
| Gender | |
| Male | 20 |
| Women | 19 |
| Age | |
| >60 | 22 |
| <60 | 17 |
| ZAP70 expression (%) | |
| >20 | 19 |
| <20 | 20 |
| IgG status | |
| MUT | 18 |
| Unmutated | 21 |
| Family history of malignancy (including CLL) | |
| Positive | 13 |
| Negative or unknown | 26 |
| CRC | |
| No. of patients | 35 |
| Gender | |
| Male | 19 |
| Women | 16 |
| Age | |
| >50 | 30 |
| <50 | 5 |
| Location | |
| Right colon | 14 |
| Left colon | 21 |
| Histotype | |
| Adenocarcinoma | 29 |
| Non-adenocarcimona (mucinous or medulary carcinoma) | 6 |
| Stage | |
| I | 3 |
| II | 21 |
| III | 10 |
| IV | 1 |
Detection of miRNA mutations
The genomic region corresponding to each pre-miRNA and at least 100 bp on the 5′ and 3′ ends of the region in the CLL cells obtained from the study patients and in normal mononuclear cells obtained from three control subjects were amplified and sequenced using the Applied Biosystems Model 377 DNA sequencing system. The Sequencher 4.1 software program (Gene Codes Corporation, Ann Arbor, MI) was used to analyze the sequencing data. When a change in the normal genomic sequence was found, it was confirmed using a second PCR followed by sequencing using both amplification primers and consequently, a panel of DNAs from blood samples obtained from 98 control subjects were screened to identify if the specific change is a polymorphism. For a small number of variations that were further selected for functional investigation, we increased the number of controls up to 220. If two attempts to amplify and/or sequence a specific PCR product failed then we discarded that sample/sequence from the final counts.
Detection of UCR mutations
A sequence variation screening in general population was performed using a panel of DNAs from blood samples obtained from 175 normal individuals consisting of the 98 controls described above and 77 other control subjects, all of whom were described previously (14). We used a larger panel of controls than in the miRNA study, as sequence variations in UCRs have yet to be thoroughly investigated. Twenty-eight UCRs randomly selected from a total of 482 as reported (17) were sequenced. For the screening of cancer patients, the genomic regions corresponding to the same selected UCRs from the 39 patients with CLL and 35 patients with CRC, three samples of mononuclear cells obtained from normal individuals and five paired normal colonic mucosa samples including at least 40 bp on the 5′ and 3′ ends of the regions were integrally amplified. Direct sequencing was performed using the Applied Biosystems Model 377 DNA sequencing system. When a sequence variation was found, it was confirmed using a second PCR followed by sequencing using both amplification primers. When available, corresponding normal genomic DNA from the individuals with UCR sequence alterations was analyzed.
In vitro studies of the effects of miRNA mutants
MiR-185 and miR-206 expression vectors containing an ∼800 bp genomic sequence, one wild-type (WT) sequence and one mutated (MUT) sequence were constructed in a sense orientation using the mammalian expression vector pSR-GFP-Neo (Oligoengine, Seattle, WA). Sequenced constructs of miR-185 and miR-206 were transfected into MEG-01 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. MEG-01 cells were also transfected with miR-15/16 cluster construct inserted into the pcDNA 3.1 vector or pSR-GFP-Neo using Lipofectamine 2000 according to the manufacturer's protocol. Northern blotting and quantitative reverse transcription-PCR were performed to assess the expression of specific miRNAs as described previously (14,16).
In silico studies of putative interactions between UCGs and miRNAs
We investigated the functional implications only for the UCGs, defined as transcribed UCRs for which we reported DNA sequence changes. Two independent algorithms were used: initially, the pattern of complementarity and the interaction energy were evaluated using the RNA22 batch script (19) applied to the UCR alleles known to be transcribed in at least one type of normal tissue or one type of human cancer according to our previous study (16). This was done independently for each of the alleles identified in patients with cancer and in the general population according to the complementarity with the active sequence of the miRNAs reported in miRBase (version 11.0; Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK) (20). Additionally, the sequence variations found in UCGs were analyzed independently for their ability to affect the binding of putative miRNA interactions using the Miranda prediction program (21). First, for each UCR mutation that we found, two sequences centered on the MUT base and flanked by 25 nt on both sides were retrieved. Second, the UCR sequences (MUT and WT) paired with all of the miRNAs reported in miRBase were scanned, which identified the putative miRNA interactors. Third, the Miranda output was filtered, selecting only the miRNAs that displayed a difference in interactions score and interaction energy between the two paired sequences. The following Miranda setting parameters were used: interaction energy, −16; score, 80; gap-open penalty, −2 and gap-extend penalty, −8. We used as cutoff value 10% variation as by our unpublished data related to effects of SNP in target sites for miRNAs, this is the lower value that still gives experimental effects on luciferase assays for miRNA-target interaction.
UCR cloning
Uc.276 was amplified by PCR (Platinum Taq High Fidelity; Invitrogen) from HCT116 genomic DNA (5′ primer: CGGCTGGATGCACATTATCTT and 3′ primer: TCATCTTAACACATTTCCAGCCC). The amplified product was XbaI cloned into the 3′-untranslated regions of the pGL3-control vector (Promega, Madison, WI), after an intermediate cloning step in TOPO TA vector 2.1. Plasmid DNA was purified using the high-speed maxi purification kit (Qiagen, Valencia, CA), and all of the inserts were checked by enzymatic digestion and sequencing for cloning directionality and sequence exactness, respectively. The point mutation of uc.276 was generated with the Quickchange Mutagenesis Kit (Stratagene, La Jolla, CA).
Luciferase assay
For functional reporter silencing assay, HeLa cells were reverse transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions in 96-well plates (50 × 105 cells per well). In each well, 0.1 μg of pGL3 (luciferase) construct together with 0.02 μg of pRLTK (renilla) vector (Promega) were cotransfected with 50 nM of pre-miRNA or negative (scrambled) control (Applied Biosystem/Ambion Austin, TX). Thirty-six hours after transfection, cells were lysed in 20 μl of passive lysis buffer according to the dual luciferase reporter assay protocol (Promega) and processed for luciferase activity; measurements were taken with Veritas luminometer (Turner Biosytems, Sunnyvale, CA). The relative reporter activity was obtained by normalization to the renilla activity and compared with scrambled. Luciferase assays were performed at least four times in replicates of eight.
Statistical analyses
Results were expressed as mean ± SD. Student's two-sided t-test was used to compare values of test and control samples. P < 0.05 indicated significant difference.
The catalog of DNA sequence variations in ncRNAs
It was implemented by searching the PubMed database (http://www.pubmed.com) for publications using the key words ‘miRNA’, ‘mutation’ and ‘SNP’. Information on ncRNA sequence variations was extracted from the resulting full text articles and supplementary data. NcRNA and SNP identifiers, SNP database National Center for Biotechnology Information accession numbers, cancer type and the reference were included.
Results
Sequence variations in miRNAs are present in both sporadic and familial CLL cases
Based on microarray expression data we obtained for mononuclear cells, normal B lymphocytes and normal CD5-positive B cells (14,22,23), we found that 120 miRNAs were expressed at values >1.5 times higher than the background signal array intensity and we selected all for the mutation screening. Data on 42 miRNAs that we previously analyzed in a larger series of patients with CLL, including all cases in the present study (Table I) were reported in (14). Thus, in the present study, we analyzed for sequence abnormalities in CLL cells using direct sequencing the remaining set of 78 miRNAs and we were able to successfully amplify by PCR products for 72 miRNAs. We sequenced a total of 1 337 276 bp in the blood samples obtained from patients with CLL and 586 471 bp in the blood samples obtained from normal controls. We identified variations from the normal sequences in 24 (27% of 72) miRNAs that were not reported previously in (24) and (14) and some of the miRNA has more than one changes: 18 were located downstream from the pre-miRNA sequence (up to ∼200 bp), 16 were located upstream of the pre-miRNA sequence (up to ∼200 bp) and two were located in pre-miRNA sequence (supplementary Table 1 is available at Carcinogenesis Online). As we found 21 of these variations in the National Center for Biotechnology Information SNP database (http://www.ncbi.nlm.nih.gov/SNP/) and they therefore represented polymorphisms, we decided to perform sequencing of a larger set of control DNA samples for seven miRNAs with no previously reported sequence variations (supplementary Table 1 is available at Carcinogenesis Online). We identified three new sequence variations in the transcript for miR24-1 (a G-to-A substitution located 152 bp down the pre-miRNA and a C-to-T substitution located 40 bp down the pre-miRNA) and miR-185 (a C-to-T substitution located 73 bp up the pre-miRNA), which we found in patients with CLL but not in 98 normal controls and can therefore be considered new mutations or very rare polymorphisms (Table II and supplementary Table 1 is available at Carcinogenesis Online). All 39 of the CLL patients harbored SNPs or mutations in the miRNA genomic regions, with 1–26 sequence variations per patient and the number of variations was higher in Zeta-chain-associated protein kinase 70-positive patients (169 in 19 Zeta-chain-associated protein kinase 70-positive and 148 in 20 Zeta-chain-associated protein kinase 70-negative, 8.7 versus 7.4 per patient, respectively, P not significant).
Table II.
Sequence variations in the human miRNAome in CLL cases with family history of cancers including CLL
| MiRNA | Chromosome location | Variation | CLL | Controls | MiRNA array expressiona | Clinical history |
| miR-16-1 | 13q14.2 | Germ line pri-miRNA (C-to-T) + 7 bp in 3′ | 2/75 | 0/160 | Reduced to15% and 40% of normal, respectively | One patient: previous BC; mother died of CLL; sister died of BC |
| miR-24-1 | 9q22.32 | pri-miRNA (C-to-T) + 40 bp in 3′ | 2/19 | 0/222 | NA | Familial CLL |
| miR-27b | 9q22.32 | Germ line pri-miRNA (G-to-A) + 50 bp in 3′ | 1/75 | 0/160 | Normal | Mother, throat and lung cancer at 58; father, lung cancer at 57 |
| miR-29b-2 | 1q32.2 | pri-miRNA (G-to-T) + 212 in 3′ | 1/75 | 0/160 | Reduced to 75% | Sister, BC at 88 (still living); brother, ‘some type of blood cancer’ at 70 |
| miR-29b-2 | 1q32.2 | pri-miRNAs ins (+A) + 107 in 3′ | 3/75 | 0/160 | Reduced to 80% | For two patients: family history of unspecified cancer |
| miR-29c | 1q32.2 | pri-miRNA (G-to-A) 31 in 5′ | 2/75 | 1/160 | NA | For one patient: paternal grandmother, CLL; sister, BC |
| miR-95 | 4p16.1 | pri-miRNA (A-to-G) 40 in 3′ | 10/29 | 2/93 | Upregulated 0–30% | Father died of CLL at 79 |
| miR-95 | 4p16.1 | pri-miR (A-to-C) 100 in 3′ | 16/27 | 46/89 | Upregulated 0–30% | Father died of CLL at 79 |
| miR-96 | 7q32.2 | pri-miRNA 36 (G-to-A) | 1/35 | 2/91 | Normal | Father diagnosed with melanoma at 60 |
| miR-122a | 18q21.31 | pri-miRNA 53 (C-to-T) | 1/75 | 2/160 | Reduced to 33% | Paternal uncle, colon cancer |
| miR-187 | 18q12.2 | pri-miRNA 34 (G-to-A) | 1/75 | 1/160 | NA | Grandfather polycythemia vera; father history of cancer but not lymphoma |
| miR-206 | 6p12.2 | pri-miRNA 33 (G-to-T) | 3/103 | 3/243 | Reduced to 25% | Prostate cancer; mother, esophageal cancer; brother, prostate cancer; sister, BC |
| miR-220a | Xq25 | pri-miRNA 33 (G-to-A) | 1/28 | 1/83 | Downregulated 20–50% | Mother died of CLL; patient has BC, survivor after 5 years treated with tamoxifen; sister died of BC |
We had clinical data for the majority of the CLL patients and we found that 13 of the patients harboring sequence variations in miRNAs also had a familial history of various types of cancer, including CLL (Tables I and II). Because we previously proposed that cancer predisposition can be linked to alterations in miRNAs (13,25), we decided to further investigate the 13 probands (one per family) from familial CLL kindreds (defined as at least two first-grade relatives with CLL). We obtained DNA from buccal swabs and investigated for the presence of germ line variations in the same set of miRNAs. Interestingly, although the number of familial CLL individuals was small, we found three DNA sequence variations (Table I and supplementary Table 1 is available at Carcinogenesis Online), one of which was present in miR-24-1 (a C-to-T substitution located 40 bp after the pre-miRNA) and was also found in the other CLL affected member from the same family but not in the only non-affected member available for the study. This shows a linkage of the miRNA alteration with the malignant phenotype in this particular family with CLL. None of the 175 normal controls had this variation, and we did not find it in the SNP database (http://www.ncbi.nlm.nih.gov/SNP/). Of the other two DNA sequence variations, we did not identify the 152 bp G-to-A substitution down the pre-miR-24-1 in a second affected member, whereas we found the 17 bp C-to-G substitution down the pre-miR-146a in normal controls (supplementary Table 1 is available at Carcinogenesis Online). Therefore, the C-to-T substitution located 40 bp down the pre-miR-24-1 germ line mutation is the second example of a miRNA mutation involved in familial CLL in addition to the initial one identified in miR-16-1 (14).
UCRs are mutated in human cancers
Because we previously found variations in UCG expression in patients with CLL or CRC (16), we decided to investigate whether the genomic sequences in UCRs harbor mutations or polymorphisms in CLL and CRC cells. Before screening patients with cancer, we focused on answering the question whether UCR variations occur in the general population. It has been reported that UCRs exhibit almost no natural variation in the general population, as only 6 of 106 767 examined ultraconserved bases were validated SNPs (17). Because it was recently reported on the creation of a haplotype map of the human genome with a more detailed description of the frequency of SNPs (26), we decided to sequence a set of randomly selected 28 UCRs in a panel of 95 normal white Caucasian individuals. We sequenced 9634 bp of human ultraconserved genomic sequences for a total of 892 525 kb sequence and found six SNPs or one per 1572 ultraconserved nucleotides. This is about six times lower than the frequency of SNPs identified across 10 regions on the human genome haplotype map, which was one SNP per 279 bp (Table III). These data confirm that, although less conserved than initially suggested, the UCRs are sites of very low natural variation in the general population.
Table III.
UCR sequence variations in the general population and patients with CLL or CRCa
| UCR name | Type | Chromosomal location | Neighboring coding genes | Population (n = 175) | Cancer Patients (n = 74) | Expressed in normal tissues | |
| Upstream | Downstream | ||||||
| UC.021 | N/I | 1p33 | SPATA6 | FLJ11588 | NORM | 190 (T-to-C) (CRC) | Yes |
| UC.025 | N/I | 1p32.3 | DMRTA2 | LOC51336 | NORM | NORM | No |
| UC.033 | E | 1p21.3 | BC030757 | DPYD | NORM | NORM | Yes |
| UC.072 | N/I | 2q22.3 | AK126774 | ZFHX1B | 380 (G-to-A) | 380 (G-to-A) (CLL) | No |
| UC.099 | N | 2q31.1 | DLX1 | DLX2 | NORM | NORM | Yes |
| UC.123 | N | 3q22.3 | MGC34923 | SOX14 | NORM | NORM | Yes |
| UC.129 | E | 3q25.2 | AY358260 | P2RY1 | NORM | NORM | Yes |
| UC.159 | N | 5q14.1 | AK128395 | AP3B1 | 103 (DEL TT) × 2 | 103 (DEL TT) (4× CLL and CRC) | Yes |
| UC.185 | E | 5q35.3 | COL23A1 | ZNF354A | NORM | NORM | Yes |
| UC.189 | E | 6p21.31 | STK38 | CDKN1A | 83 (G-to-A) | NORM | Yes |
| UC.190 | N/I | 6p21.1 | AJ225109 | BC009628 | NORM | NORM | Yes |
| UC.206 | N | 7p15.3 | SP8 | SP4 | NORM | 324 (G-to-A) (CLL) | Yes |
| UC.243 | P | 8q21.13 | AF130052 | PXMP3 | NORM | 146 (G-to-A) (CLL) | No |
| UC.269 | N/I | 9q33.3 | AK123000 | LHX2 | 16 (T-to-C) 4× | NORM | Yes |
| UC.276 | P | 9q33.3 | LOC51145 | C9orf28 | 335 (G-to-A) | 90 (A-to-G) (CRC) | Yes |
| UC.279 | N/I | 9q33.3 | LOC51145 | C9orf28 | NORM | NORM | Yes |
| UC.300 | N/I | 10q24.31 | HIF1AN | C10orf6 | NORM | NORM | Yes |
| UC.328 | P | 11p13 | ELP4 | RCN1 | NORM | 179 (G-to-T) (2× CRC) | No |
| UC.341 | E | 12q13.13 | HOXC11 | HOXC9 | 291 (C-to-A) | NORM | Yes |
| UC.351 | N | 13q21.33 | DACH | FLJ22624 | NORM | NORM | No |
| UC.419 | E | 17q23.2 | AK126318 | Dlc2 | NORM | NORM | Yes |
| UC.461 | N/I | Xp22.11 | PCYT1B | ARX | NORM | NORM | Yes |
| UC.463 | N/I | Xp22.11 | PCYT1B | ARX | NORM | NORM | Yes |
| UC.464 | N/I | Xp22.11 | PCYT1B | ARX | NORM | NORM | Yes |
| UC.465 | N/I | Xp22.11 | PCYT1B | ARX | NORM | NORM | Yes |
| UC.469 | P | Xp22.11 | POLA | ARX | NORM | NORM | Yes |
| UC.479 | E | Xq25 | GLUD2 | THOC2 | NORM | NORM | Yes |
| UC.483 | P | 3p24.3 | AK125129 | SATB1 | NORM | 166 (A-to-T) (CRC) | Yes |
N, non-exonic(intronic or intergenic); P, possible exonic; E, exonic location; NORM, normal sequence.
The position of sequence variation refers to the UCR in genomic sense orientation, the expression column refers to microarray data, and the number of individuals with a specific alteration of UCR was reported when >1.
Consequently, we genotyped 39 peripheral blood samples obtained from the patients with CLL and tumor tissue samples obtained from 35 patients with CRC. We used genomic sequences on the same 28 UCRs used for the general population study. We sequenced a total of 703 502 kb of tumoral genomic DNA. We found sequence abnormalities in 11 UCRs, six of them specifically found only in the 74 cancer patients (9.5%)—two with CLL and four with CRC—and none identified in a larger set of 175 normal controls (Table III). All four CRC alterations were found in adenocarcinomas stage II and III patients but due to the limited number of analyzed cases in the present study, we cannot draw any conclusion about correlation with stage. In addition, we found the same UCR variations in the germ line DNA of the same three patients for whom blood DNA was available, suggesting that they were germ line alterations or very rare polymorphisms (frequency <1%) (Table III). Because we found none of these six UCR sequence variations in the large cohort of controls that we analyzed, the mutation hypothesis seems more probable.
We found also rare polymorphisms in UCRs. For example, in uc.159(N), we found a deletion of 2 nt at position 103, 103 (del TT), approximately three times more often in patients with cancer than in the normal controls [4/72 (6%) and 2/95 (2%), respectively; P = 0.253]. We identified also sequence variations in UCRs in normal population that were not identified in cancer patients (Table III). Globally, we identified one UCR mutation at every 90 kb of sequenced tumoral DNA, which is roughly 10 times more frequent than previous estimates of non-functional alterations in the cancer genome (∼1 per Mb) (27). For the investigated 28 UCRs, we found about three times more frequent somatic variations in cancer patients versus normal controls [12/2016 of analyzed cancer sequences versus 9/5460 of analyzed normal sequences contained variations, respectively (P < 0.05, Fisher's exact test)]. Therefore, UCR mutations occur more often in patients with CLL or CRC than in the general population, and some of them may be germ line mutations.
Functional consequences of variations in ncRNAs
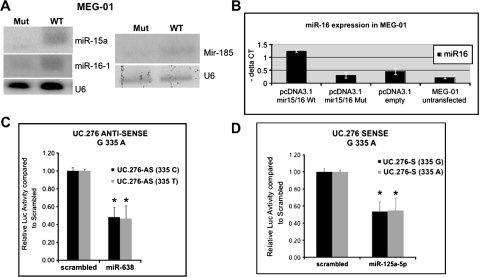
To determine whether the sequence variations in ncRNAs have a functional effect, first, we transfected MEG-01 cells with both WT and MUT alleles cloned in expression vectors (see Material and Methods) for miR-16-1, miR-185 and miR-206, the first two having sequence variations only in cancer patients but not in controls. We used miR-206 also because the identified genomic variation represents a previously unreported SNP (supplementary Table 1 is available at Carcinogenesis Online). MEG-01 is a good model for CLL, as these cells lack expression of miR-16 on chromosome 13 because of genomic rearrangements of this locus on both alleles, reproducing the homozygous 13q del identified in ∼10% of patients with CLL (28). In silico studies of the effects on the folding energy of the hairpin structure of the two types of alleles found differences in the folding energy between WT and MUT for miR-16-1—a decrease from −31 to −37 kcal/mol (−19%)—and few or no significant differences for miR-206 (a decrease from −43 to −45 kcal/mol) and miR-185 (no difference in the −29 kcal/mol free energy). The quantitative reverse transcription–PCR and northern blot analysis showed a reduction in the expression of the MUT alleles of miR-16-1, miR-15a, and to a lesser extent, miR-185 with respect to both WT and empty vector-transfected cells (Figure 1). We identified no consistent or reproducible expression effects of miR-206 using northern blotting. Notably, for miR-16-1, we observed the same reduction on the expression of mature miR-16-1 and miR-15a in a non-cancerous cell model, HEK-293 fetal kidney cells (14). We found that the MUT position of miR-16 (C-to-T substitution 7 nt after the pre-miRNA) is highly conserved during evolution (http://genome.ucsc.edu/), whereas this is not the case for miR-185 (C-to-T substitution 73 nt down the pre-miRNA) or miR-206 (G-to-T substitution 49 nt before the pre-miRNA). Consistent with this high conservation, we found that the MUT transcript of miR-15a/16 cluster has a significant lower effect on reducing the levels of anti-apoptotic protein BCL2 and consequently to activate apoptosis in respect with the WT gene (28).
Fig. 1.
Functional consequences of sequence variation in miRNAs and UCGs. (A) Reduction in active molecule expression for miR-15a/16-1 cluster and for miR-185 by northern blottings. (B) Expression reduction for the (C-to-T) +7 bp in 3′ of miR-15a/16-1 cluster by quantitative RT–PCR in MEG-01 leukemia cells in respect with WT cluster and empty vector; standard deviations of three measurements are presented. (C–D) Interaction between miRNAs and UCG by in vitro luciferase assay for the two allelic variants of the uc.276; the asterisk indicates P < 0.05.
Second, we examined the possible effects of sequence variations in UCGs. Because we recently identified a new type of gene regulation by miRNAs, specifically, the direct interaction between miRNAs and other ncRNAs, the UCGs (16), we analyzed these variations in the genomic sequence of UCGs to determine whether they influence the putative interactions with miRNA. Using in silico miRNA-target predictions (see Materials and Methods), we found that the sequence comprising the uc.276 G-to-A substitution at position 335 found in the general population when transcribed in the sense orientation is predicted to interact with miR-125a, whereas when it is transcribed in the anti-sense orientation it is able to bind to miR-638. Also in uc.276, the A-to-G substitution at position 90, which is found in patients with CRC only, disturbs the putative interaction with miR-214, which is overexpressed in solid tumors (10) and decreases apoptosis in HeLa cells (29) and with the newly cloned miR-887. Therefore, we cloned two alleles of uc.276 corresponding to variations at position 335 in luciferase vectors both in the sense and anti-sense orientation and focused on investigating the interaction with miR-125a and miR-638, respectively. We observed that these miRNAs significantly interact with uc.276 (P always <0.0001) and repress luciferase activity (Figure 1) but the differences between the two alleles, although consistent between at least six independent triplicate experiments, were small (<5%) and not significant (Figure 1C and 1D). Therefore, further detailed functional studies are needed to understand the real influence of sequence variations on UCG expression.
A catalog of DNA sequence variations in ncRNAs in human cancers
The results described above suggest that genomic variations in ncRNAs are not simple bystanders during tumorigenesis but may be selected for during malignant transformation and may have functional effects. Therefore, we collected published information on mutations and/or SNPs in miRNAs and UCRs that can serve as a research tool for scientists studying ncRNAs. We identified reported sequence variations with proven functional effects for nine miRNA genes. A detailed description of the sequence variations in 86 miRNAs and UCGs is included in Table IV and supplementary Table 2 (available at Carcinogenesis Online).
Table IV.
A catalog of sequence variations in miRNAs and UCGs—DNA sequence variations with functional effects in ncRNAs
| miRNA | Variation | Location | Type of cancer | Functional consequences | Reference |
| let-7e | (G-to-A) + 19 nt | 3′ of miRNA (pri-miRNA) | Human cancers | Decreased expression of mature miRNA | (30) |
| miR-15a/16-1 cluster | Human germ line (C-to-T) +7 nt; NZB specific (A-to-T) +6 nt | 3′ of miRNA (pri-miRNA) | Sporadic and familial CLL; B-lymphoproliferative disease in mice | Decreased expression of mature miRNA and failure to decrease BCL2 protein levels; decreased levels of miR-16 expression in lymhoproliferative tissues | (14,15) |
| miR-17 | C/T | Pri-miRNA | BC | Conformational changes in the predicted secondary structures with consequently alteration of the mature miR-17 | (31) |
| miR-30c-1 | G/A | Pre-miRNA | BC | Conformational changes in the predicted secondary structures with consequently alteration of the mature miR-30c-1 | (31) |
| miR-125a | SNP | Mature miR position 8 | Not reported | Alteration of pri-miRNA processing | (32) |
| miR-146 | rs2910164 G/C | Precursor | Papillary thyroid carcinoma predisposition; hepatocellular carcinoma; familial BC/ovarian cancer | Decreased expression of mature miRNA; G-allelic miR-146a precursor increased production of mature miR-146a compared with C-allelic one | (33–35) |
| miR-146a* | rs2910164 G/C | Precursor | Thyroid cancer | Epistasis through the production of additional mature miRs: miR-146a*G and miR-146a*C | (36) |
| mir-196a | rs11614913 CC | Mature miR position 12 | Non-small cell lung cancer | Decreased expression of mature miRNA | (37) |
| miR-196a2 | rs11614913 CC; rs11614913 C/T | Pre-miRNA | Lung cancer; esophageal cancer | Might affect mature miR-196a expression and target mRNA-binding activity; affect esophageal cancer risk | (38,39) |
NZB, New Zeland Black.
Discussion
The functions of ncRNAs, defined as RNAs that does not encode for proteins, are still not well known. Authors recently reported the role of RNA molecules in controlling expression of various genes involved in physiology and development (40,41). MiRNAs make up one of the most abundant classes of regulatory genes play a role in developmental timing, hematopoiesis, cell death, cell proliferation, stress response and stem cell division (7) and are abnormally expressed in different types of cancer (12,13). In the present study, we report three new findings. First, we identified new sequence variations in miRNAs in patients with CLL and CRC. The collection of the data presented herein and in our previous mutation study (14) represents the first step in developing a catalog of mutations in miRNAs in human leukemias and carcinomas. The fact that globally sequence variations (either SNPs or mutations) in the investigated ncRNAs are significantly more frequent in cancer samples than in normal controls means that such variations are selected during the tumorigenesis and play a role in the establishment of the malignant phenotype. Further supporting this view, we identified five miRNAs (miR-16-1, miR-24-1, miR-27b, miR-29b-2 and miR-29c) harboring mutations in CLL patients with familial history but not in normal controls, including some miRNAs that are frequently deleted in CLLs (such as miR-16-1) or in other types of leukemias (such as miR-29b-2 and miR-29c) (Table II). These data support the hypothesis that cancer predisposition involves abnormalities in miRNAs and therefore, the present study offers for the near future the scientific ground for the development of new diagnostic markers. Of note, none of the identified sequence variations were located in the active sequence of a miRNA, in agreement with ours and other previously published studies in CLL (14), human cancers (32) and ovarian cancers (34).
Second, we identified new sequence variations in UCRs including non-coding UCGs in patients with CLL and CRC. We recently identified that a large number of genomic UCRs encode for a particular set of ncRNAs whose expression is altered in human cancers. Genome-wide profiling revealed that these UCRs have distinct signatures in human leukemias and carcinomas (16). UCRs are frequently located at fragile sites and genomic regions involved in cancers. We identified UCRs whose expression may be regulated by miRNAs abnormally expressed in human CLL cases, and we proved that inhibition of an overexpressed UCR induces apoptosis in CRC cells (16). These findings argue that ncRNAs are involved in tumorigenesis to a greater extent than previously thought. SNPs are the most abundant DNA variations in the human genome and contribute to human phenotypic differences. In the present study, we identified for the first time mutations in both expressed and unexpressed UCRs, meaning that somatic and germ line mutations occur in these evolutionary conserved regions in CLL and CRC cases. Furthermore, polymorphisms in UCRs may also alter various biological processes by influencing the processing and/or target selection of miRNAs. A recent study linked SNPs in UCRs with predisposition to familial BC risk (42). Two of six SNPs initially reported in (17) showed an association with familial BC risk. Yang et al. (42) also reported that whereas rs9572903 had only a borderline significant association with BC risk, the frequency of the rare allele of rs2056116 was higher in patients with BC than in controls, indicating an increased familial BC risk. Interestingly, both of these SNPs are located in UCRs that were not expressed in any of the 19 normal tissue samples or 177 cancer samples that we analyzed using both microarray and quantitative reverse transcription–PCR as previously reported (16). Interestingly, Catucci et al. (43) did not support the association of these two SNPs and BC risk in a population of 737 Italian female familial BC cases negative for mutations in BRCA1/2 genes compared with 1245 Italian female blood donors. The conflicting results could be explained by differences in the allele frequencies between the German and the Italian control populations (0.39 versus 0.34, respectively, P = 0.00007) and by statistical fluctuations. Large multiple-center studies are warranted to evaluate the effect of UCRs/UCGs SNPs on cancer risk.
Finally, identification of the functional implications is central to prove the importance of these sequence variations in ncRNAs. In the present study, we further tested in vitro that miRNAs could directly interact with UCGs (such as miR-125a-5p with uc.276-S and miR-638 with uc.276-AS). The two alleles have only slightly different interaction energy and not significant different levels of reduction of luciferase activity. As UCGs could be regulators of protein-coding gene expression as miRNAs are, small differences in interaction with miRNAs could consistently indirectly affect the protein-coding gene regulation by the slightly different amount of UCG alleles, but this hypotheses should be confirmed by further studies. Whereas some of the identified mutations and SNPs in miRNAs do not affect gene expression (24,44), we identified a functional germ line variation in miR-16-1 in patients with CLL (including one from a kindred of familial CLL and BC) that was proven to be functional and was also identified in a mouse strain in which a CLL-like disease naturally develops (15). Other examples support a functional role of miRNA sequence variations: an SNP located in the 3′ region of miR-125 in normal individuals blocks processing of primary miRNA to the precursor gene, reducing the effect of the target inhibition by this miRNA (32). More recently, it was reported that a G-to-A mutation 19 nt downstream from the miRNA let-7e, a member of the largest family of tumor suppressor miRNAs (45), led to a significant reduction in its expression in vivo (37). Furthermore, researchers found new SNPs and novel mutations distributed in the regions of primary miRNAs, precursor miRNAs and even mature miRNAs that are present in specific human cancers (30), common genetic variants in pre-miRNAs that are associated with increased risk of BC in Chinese women (46) and a common SNP (rs 2910164) in pre-miR-146a that decreases mature miRNA expression and predisposes individuals to papillary thyroid carcinoma (33) (Table IV). The SNP rs 2910164 was found also by us in a CLL sample (supplementary Table 1 is available at Carcinogenesis Online). Although such variations are frequently located outside the precursor or active sequence of miRNAs, they are included in the primary transcripts of the miRNAs as we showed previously (see ref. 14), and therefore, may affect the various steps in miRNA maturation, not only the target selection of miRNAs.
In conclusion, sequence variations in miRNAs and UCGs occur frequently in patients with CLL or CRC and may have functional consequences and target significant genes for human tumorigenesis (47–51). It is only a matter of time until the catalog of DNA sequence variations in ncRNAs that we describe herein will contain an overwhelming amount of information.
Supplementary material
Supplementary Tables 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
The University of Texas M. D. Anderson Research Trust; University of Texas System Regents Research Scholar; Ladjevardian Regents Research Scholar Fund; Institutional Research Grant; Cancer Center Support Grant (New Faculty Award) to G.A.C.; National Cancer Institute to C.M.C.
Supplementary Material
Acknowledgments
We thank Dr Muller Fabbri for the critical reading of the manuscript and useful comments. We thank Don Norwood, Department of Scientific Publications, The University of Texas M. D. Anderson Cancer Center, for expert editorial assistance.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BC
breast cancer
- CRC
colorectal cancer
- CLL
chronic lymphocytic leukemia
- miRNA
microRNA
- MUT
mutated
- ncRNA
non-coding RNA
- nt
nucleotide
- PCR
polymerase chain reaction
- SNP
single-nucleotide polymorphism
- UCG
ultraconserved gene
- UCR
ultraconserved region
- WT
wild-type
References
- 1.Parsons DW, et al. Colorectal cancer: mutations in a signaling pathway. Nature. 2005;436:792–796. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 3.Wood LD, et al. The Genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 4.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, et al. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, et al. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 15.Raveche ES, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calin GA, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 17.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 18.Lagos-Quintana M, et al. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miranda KC, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkm952. (Database issue), D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu CG, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl Acad. Sci. USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwai N, et al. Polymorphisms in human pre-miRNAs. Biochem. Biophys. Res. Commun. 2005;331:1439–1444. doi: 10.1016/j.bbrc.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 25.Calin GA, et al. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 26.Altshuler D, et al. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bardelli A, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 28.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng AM, et al. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu M, et al. Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs. Carcinogenesis. 2008;29:1710–1716. doi: 10.1093/carcin/bgn073. [DOI] [PubMed] [Google Scholar]
- 31.Shen J, et al. Novel genetic variants in microRNA genes and familial breast cancer. Int. J. Cancer. 2009;124:1178–1182. doi: 10.1002/ijc.24008. [DOI] [PubMed] [Google Scholar]
- 32.Duan R, et al. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 33.Jazdzewski K, et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl Acad. Sci. USA. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu T, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–2131. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 35.Shen J, et al. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 36.Jazdzewski K, et al. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc. Natl Acad. Sci. USA. 2009;106:1502–1505. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Z, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J. Clin. Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye Y, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev. Res. (Phila. Pa) 2008;1:460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian T, et al. Functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol. Biomarkers Prev. 2009;18:1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 40.Sevignani C, et al. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm. Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stefani G, et al. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 42.Yang R, et al. SNPs in ultraconserved elements and familial breast cancer risk. Carcinogenesis. 2008;29:351–355. doi: 10.1093/carcin/bgm290. [DOI] [PubMed] [Google Scholar]
- 43.Catucci I, et al. SNPs in ultraconserved elements and familial breast cancer risk. Carcinogenesis. 2009;30:544–545. doi: 10.1093/carcin/bgn289. [DOI] [PubMed] [Google Scholar]
- 44.Diederichs S, et al. Sequence variations of microRNAs in human cancer: alterations in predicted secondary structure do not affect processing. Cancer Res. 2006;66:6097–6104. doi: 10.1158/0008-5472.CAN-06-0537. [DOI] [PubMed] [Google Scholar]
- 45.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 46.Hu Z, et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 47.Fabbri M, et al. MicroRNAs and genomic variations: from Proteus tricks to Prometheus gift. Carcinogenesis. 2009;30:912–917. doi: 10.1093/carcin/bgp063. [DOI] [PubMed] [Google Scholar]
- 48.Saunders MA, et al. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl Acad. Sci. USA. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glinsky GV. An SNP-guided microRNA map of fifteen common human disorders identifies a consensus disease phenocode aiming at principal components of the nuclear import pathway. Cell Cycle. 2008;7:2570–2583. doi: 10.4161/cc.7.16.6524. [DOI] [PubMed] [Google Scholar]
- 50.Landi D, et al. A catalog of polymorphisms falling in microRNA-binding regions of cancer genes. DNA Cell Biol. 2007;27:35–43. doi: 10.1089/dna.2007.0650. [DOI] [PubMed] [Google Scholar]
- 51.Yang H, et al. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–2537. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.