Abstract
Cdx2 is an intestine-specific transcription factor known to regulate proliferation and differentiation. We have reported previously that Cdx2 limits the proliferation of human colon cancer cells by inhibiting the transcriptional activity of the β-catenin–T-cell factor (TCF) bipartite complex. Herein we further elucidate this mechanism. Studies with a classic Cdx2 target gene and a canonical Wnt/β-catenin/TCF reporter suggest that Cdx2 regulates these promoters by distinctly different processes. Specifically, inhibition of β-catenin/TCF activity by Cdx2 does not require Cdx2 transcriptional activity. Instead, Cdx2 binds β-catenin and disrupts its interaction with the DNA-binding TCF factors, thereby silencing β-catenin/TCF target gene expression. Using Cdx2 mutants, we map the Cdx2 domains required for the inhibition of β-catenin/TCF activity. We identify a subdomain in the N-terminus that is highly conserved and when mutated significantly reduces Cdx2 inhibition of β-catenin/TCF transcriptional activity. Mutation of this subdomain also abrogates Cdx2’s anti-proliferative effects in colon cancer cells. In summary, we conclude that Cdx2 binds β-catenin and disrupts the β-catenin–TCF complex. Considering the pivotal role of β-catenin/TCF activity in driving proliferation of normal intestinal epithelial and colon cancer cells, our findings suggest a novel mechanism for Cdx2-mediated regulation of Wnt/β-catenin signaling and cell proliferation.
Introduction
Despite an intense research effort, colon cancer remains a major cause of cancer morbidity and mortality worldwide. Much has been learned about the sequence by which most colorectal cancers arise from normal epithelium. This progression has been termed the adenoma–carcinoma sequence (1). Progression along this pathway to cancer occurs through the accumulation of multiple somatic mutations, ultimately leading to malignant transformation and the formation of an invasive colon cancer.
One of the more important pathways mutated in the progression to colorectal cancer is the Wnt/β-catenin/T-cell factor (TCF) pathway. When the Wnt receptor is activated, the β-catenin protein is stabilized and translocates to the nucleus where it partners with a TCF/lymphocyte enhancer factor (LEF) family member to bind DNA and activate target gene expression. Mutations in this pathway lead to the stabilization of intracellular β-catenin and unrestricted activation of target genes (2,3). Aberrant activity from the Wnt/β-catenin/TCF pathway is nearly always present in human colon cancers. However, signaling from this pathway is also required to maintain the proliferative compartment of normal intestinal crypts (4). It remains an open question as to how this critical pathway is constrained in the normal intestine to prevent neoplasia.
As with the process of colon carcinogenesis, the molecular mechanisms governing development and differentiation of the intestinal epithelium are becoming understood. One important factor promoting differentiation is the homeodomain protein Cdx2 (5). Cdx2 is a transcription factor required for the intestine-specific expression of a number of genes (6–10). Cdx2 is required for normal intestinal cell development (11) and also promotes the morphologic maturation of intestinal epithelial cells (6,12,13). Observations from a number of laboratories suggest that Cdx2 expression can regulate epithelial cell proliferation and in certain contexts can limit tumor development (14–16). However, the mechanism governing this effect is unknown.
Studies from our laboratory have investigated this question. In previous studies, we determined that colon cancer cell proliferation was reduced and β-catenin/TCF transcriptional activity was inhibited by the expression of Cdx2 (17,18) or the closely related homologue Cdx1 (17–20). Cdx1 or Cdx2 expression resulted in diminished expression of the Wnt target genes LEF-1, Cyclin D1 and c-Myc, as well as a reduction of cell proliferation. This finding has subsequently been confirmed by others in endometrial carcinoma cells (21). In both studies, Cdx expression was associated with diminished β-catenin responsiveness of Wnt targeted promoters like cyclin D1, Cdx1 and p14ARF (17,21). However, in neither study was the mechanism whereby Cdx2 inhibits Wnt/β-catenin/TCF transcriptional activity elucidated. In the present study, we extend these observations by demonstrating that Cdx2 inhibition of β-catenin/TCF transcriptional activity does not require Cdx2 transactivator function. Rather, Cdx2 directly binds β-catenin and disrupts the β-catenin–TCF protein complex. This is a novel and unexpected function for Cdx2, suggesting that Cdx2’s contributions to intestinal cell differentiation may include other functional protein–protein interactions beyond those required for its well-studied role as an intestine-specific transcription factor.
Materials and methods
Cell culture and transfections
DLD1, Caco2 and 293T cells were obtained from the American Type Culture Collection or the Cell Center (University of Pennsylvania). Cells were all maintained as recommended by the American Type Culture Collection.
The 293T cells were transfected using FuGENE 6 (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Gemany). Cells were grown to 50–70% confluence and were transfected with 100 ng of pRC expression vectors, 100 ng of TOPFLASH and 200 ng of pCineo-S33Y-β-catenin (both kindly provided by Dr Ken Kinzler, The Johns Hopkins University), 1 ng of pRL-CMV Renilla control reporter (Promega, Madison, WI) and 700 μg of pCR2.1 (Stratagene, La Jolla, Calif) as carrier. Expression plasmids for Flag-Cdx2 and Flag-Cdx1 were described previously (10,22). For studies examining sucrase isomaltase (SI) promoter activation (6,23), 100 ng of the SI-luc reporter was transfected in place of TOPFLASH. Media were changed at 24 h, and at 48 h, the cells were harvested. Luciferase assays were performed using the dual luciferase assay kit (Promega). Luciferase activity was normalized to both total protein concentration and the transfection control Renilla luciferase levels. Cellular DNA content was quantified as described (20). DLD1 cells were transfected by electroporation as described previously (17,20). Statistical testing was by analysis of variance and Tukey rank mean testing for all assays.
The DNA content studies by flow cytometry were performed exactly as described (17).
Immunoblot and immunoprecipitation analyses
The 293T cells were transfected as before (19,20), and at 48 h, whole-cell protein lysates were prepared and the products analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotting. Cytoplasmic and nuclear fractions were prepared as described previously (24). The antibodies for Cdx1 (CPSP) and Cdx2 (CNL) have been described previously (24,25). The β-catenin antibody was mouse monoclonal (Transduction Laboratories, BD Biosciences, San Diego, CA). For western blot loading control, we used the actin:A-4700 antibody (Sigma-Aldrich, St. Louis, MO).
For the β-catenin/TCF co-precipitation study, 293T cells were transfected as before, using a c-Myc-tagged TCF4 along with pCineo-S33Y-β-catenin (both kindly provided by Ken Kinzler, Johns Hopkins) along with our Cdx2 expression vectors. At 48 h post-transfection, cells were harvested and lysed in mammalian protein extract reagent (M-PER; Pierce Protein Research Products, Thermo Scientific, Rockford, IL). Immunoprecipitation was performed by using the ProFound c-Myc-Tag IP/CoIP kit according to the manufacturer's instructions. The precipitated products were analyzed by western blotting by using an anti-β-catenin polyclonal antibody (sc-7199; Santa Cruz Biotechnology, Santa Cruz, CA). For the β-catenin-Cdx2/1 co-immunoprecipitation, two approaches were used. One was to use Flag-tagged Cdx proteins. The 293T cells were transfected as before with Flag-Cdx and pCineo-S33Y-β-catenin. At 48 h, cells were harvested and lysed in lysis buffer containing 50 mM Tris–HCl, pH 7.4, 150 mM NaCL, 1 mM ethylenediaminetetraacetic acid and 1% Triton X-100. Immunoprecipitation was performed by using FLAG-tagged protein immunoprecipitation kit (Sigma). For the Caco2 cell study, the cells were cultured to total confluence for 7 days, then harvested and lysed in lysis buffer containing 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40 and 0.1% sodium deoxycholate. Immunoprecipitation was performed by using monoclonal Cdx2 (MU392A-UC; BioGenex Laboratories, San Ramon, CA) and Protein G-Agarose (Roche). The precipitated products were analyzed by western blotting by using an anti-β-catenin polyclonal antibody (sc-7199; Santa Cruz Biotechnology). CatCLEF and VP16-LEF were transfected as described. CatCLEF was kindly provided by Brigid Hogan, Duke University (26,27). VP16-LEF was generously provided by Peter Klein, University of Pennsylvania (17).
Serial and site-directed mutagenesis
To map the region in Cdx2 protein responsible for the inhibitory effect on the β-catenin/TCF transcriptional activity, we initially used polymerase chain reaction-truncated mutants that were subsequently subcloned into the pRC-CMV vector. Primers utilized for this are listed in supplementary Table S1 (Supplementary Table S1 is available at Carcinogenesis Online) or published elsewhere (24). After determining that the protein region between amino acids 15 and 50 was required for the inhibitory effect, we used the QuikChange II site-directed mutagenesis kit (Stratagene) to generate five mutants within this region in which seven sequential amino acids were mutated to an alanine or glycine. All mutants were fully sequenced to confirm accuracy of the mutagenesis. The sequence of the primers used for the mutagenesis is listed in Table S1.
Glutathione-S transferase pull-down
For in vitro binding assay, 35S-labeled Cdx1, Cdx2 and TCF4 proteins were produced in vitro by using the TNT T7 Quick-coupled transcription/translation system (Promega). Bacterial expression constructs pGEX-KT/CAG456-β-catenin (kindly provided by Dr David L. Rimm, Yale University) directed the synthesis of Glutathione-S transferase (GST)–β-catenin fusion protein. Equal amounts of GST–β-catenin fusion protein or GST alone, as a negative control, was incubated with in vitro synthesized 35S-labeled Cdx1, Cdx2 or Tcf4 in HND buffer (10 mg/ml Bovine Serum Albumin, 20 mM (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), 50 mM NaCl, 0.1% Nonidet P-40, 5 mM Dithiothreitol) on a rotator for 1 h at 4°C. After washing the beads four times in 500 μl of MTPBS buffer (phosphate buffered saline + 0.1% Nonidet P-40), bound 35S-labeled proteins were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and visualized by autoradiography.
Results
Cdx2 inhibition of β-catenin/TCF is sensitive to protein levels in human colon cancer cells
We have demonstrated previously that expression of the homeodomain transcription factor Cdx2, or the closely related homologue Cdx1, could inhibit Wnt/β-catenin/TCF signaling in many cell types including human colon cancer cells (HCT116, DLD1, HT-29 and Colo 205), immortalized and transformed 293T cells and in Xenopus embryos (12,17,19,20). Others have reported a similar effect of Cdx2 in human endometrial cancers (21). In our last published study, we noted unexpectedly that Cdx1 was a more potent inhibitor of β-catenin/TCF transcriptional activity than its homologue Cdx2 in colon cancer cells (17). This was unexpected because Cdx2 is typically a better transcriptional activator of intestine-specific genes (data not shown). In the same study, we also noted that in non-colon cancer models (Xenopus embryos and 293T cells), Cdx2 was a better inhibitor of β-catenin/TCF transcriptional activity. To understand these contrasting observations better, we directly compared Cdx1 and Cdx2’s ability to activate SI promoter activity (a canonical Cdx-responsive reporter) (6,14,23) with their ability to inhibit the β-catenin/TCF reporter TOPFLASH (28) in both DLD1 cells (a human colon cancer cell line) and 293T cells (a non-colon cancer cell line).
We observed that Cdx2 remained a potent activator of the SI promoter in DLD1 cells, nearly 10-fold greater than Cdx1 (Figure 1A). This is typical for most colon cancer cells and may be due to Cdx2’s ability to synergize with co-factors GATA4 and HNF-1α (23,29). However, despite this significant activity, Cdx2 was less effective than Cdx1 at inhibiting TOPFLASH in those same cells. In the 293T cells, a different pattern emerged. Both Cdx1 and Cdx2 are potent activators of the SI reporter construct (Figure 1A). Moreover, both are potent inhibitors of TOPFLASH reporter activity (Figure 1A). However, Cdx2 is a much better inhibitor of TOPFLASH than Cdx1 (99.5% reduction versus 92% inhibition, respectively). This confirms our earlier findings that in non-colon cancer cells, Cdx2 is a more potent inhibitor of Wnt/β-catenin/TCF than Cdx1 (17).
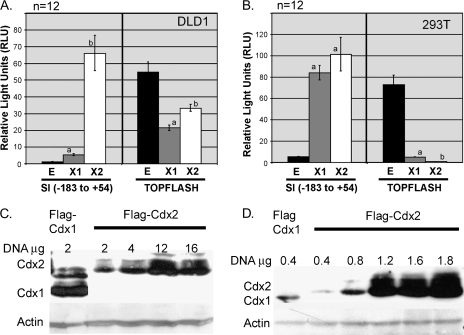
Fig. 1.
Comparison of TOPFLASH inhibition with SI promoter activation in DLD1 and 293T cells. (A and B) Luciferase activity was determined after Flag-Cdx1 (X1; gray bar) or Flag-Cdx2 (X2; white bar) expression vectors or an empty vector control (E; black bar), were co-transfected with TOPFLASH and SI reporters; 2 μg or 400 ng of the expression vectors were transfected into DLD1 or 293T cells (400 ng of expression vectors). a, Significantly differs from empty control and Cdx1, P < 0.01. b, Significantly differs from empty control, P < 0.05. (C and D) Protein levels of Flag-Cdx1 and Flag-Cdx2 after transfection of DLD1 and 293T cells.
The vectors we used to express Cdx1 and Cdx2 included a Flag-tag at the Cdx N-terminus. We next compared Cdx1 and Cdx2 protein levels using a Flag antibody. Transfecting equal amounts of Cdx1 and Cdx2 expression vector DNA did not yield equal amounts of Cdx protein. Greater protein levels of Cdx1 resulted from the otherwise equivalent transfection in both cell lines (Figure 1B). In 293T cells, Cdx1 and Cdx2 protein levels could be matched by doubling the amount of the Cdx2 expression vector transfected. However, in DLD1 cells, increasing the amount of Cdx2 expression vector by as much as 8-fold did not yield significantly greater Cdx2 levels. We suspect that this limitation of Cdx2 protein levels in colon cancer cells may explain why Cdx2 is a less effective inhibitor of TOPFLASH than Cdx1. We conclude that Cdx2-mediated inhibition of Wnt/β-catenin signaling is mechanistically distinct from its activation of transcriptional targets. Moreover, this mechanism is very sensitive to Cdx2 protein levels, much more so than the classic transcriptional activator function.
The classical transcriptional activation domain is not required for Cdx2-mediated inhibition of Wnt/β-catenin transcriptional activity
Since it appeared that SI induction and TOPFLASH inhibition by Cdx2 were mechanistically different, we next tested several Cdx2 mutants for their ability to activate the SI reporter or inhibit TOPFLASH. The response of the SI reporter to the Cdx2 mutants was as reported previously (24), while the response of TOPFLASH was revealing. Cdx2 is a known phosphoprotein, and phosphorylation at Ser60 has been associated with reduced SI promoter activation (24). While mutation of this site as well as several other serine phosphorylation sites (Cdx2S60 and Cdx2S56,57,58,60) did appear to weakly increase SI promoter luciferase activity, the difference was not statistically significant. Also, these mutations had no significant effect upon TOPFLASH reporter inhibition (Figure 2A). Deletion of the Cdx2 C-terminus (Cdx2CD) reduced SI reporter activity by ∼50%. This was unexpected based on previous studies in NIH3T3 cells that found no effect from the C-terminus truncation on SI reporter activation (24). Studies following up on this modest reduction are planned. However, this truncation had no effect on TOPFLASH inhibition (Figure 2A). In contrast, deletion of Cdx2’s N-terminus (Cdx2ND), containing the transcription activation domain, completely abrogated both SI activation and TOPFLASH inhibition. Surprisingly, though, when we deleted just the canonical transcription activation domain between amino acids 55 and 136 (Cdx2Δ55–136) (24) while we completely abrogate Cdx2 transcriptional activity and SI promoter induction, this truncation has no effect on TOPFLASH inhibition. The Cdx2Δ55–136 mutant remained equal to the wild-type Cdx2 protein with regard to TOPFLASH inhibition (Figure 2A).
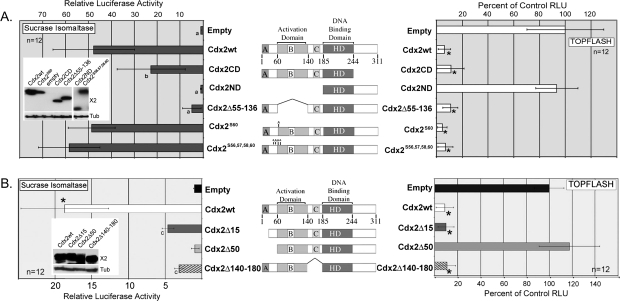
Fig. 2.
The inhibition of TOPFLASH by Cdx2 requires distinct domains in the N-terminus. The Cdx2-responsive reporter SI-Luc or the β-catenin/TCF reporter TOPFLASH were transfected into 293T cells to determine the effect of Cdx2 mutations upon their transcriptional activity. (A) Luciferase activity after co-transfection with Cdx2 wild-type and truncation mutants. (B) Additional Cdx2 truncation mutants were generated and tested for their ability to activate SI-Luc or inhibit TOPFLASH. In Cdx2 diagram, HD: homeodomain; A, B,C: conserved domains. Insets: Western blots showing equal protein levels of new mutants and wild-type Cdx2 after transfection. Blots were probed for Cdx2 with tubulin as a loading control. Cdx2ND and Cdx2S56,57,58,60 were probed using a polyclonal antibody against the Cdx2 C-terminus. a, significantly differs from Cdx2 wild-type, P < 0.001; b, significantly differs from Cdx2 wild-type and Empty vector, P < 0.05; c, Differs from Empty vector and Cdx2Δ50, P < 0.01. *Significantly differs from empty vector control, P < 0.001.
To better define the region of Cdx2 required for β-catenin/TCF inhibition, we generated additional truncation mutants by polymerase chain reaction mutagenesis. We separately deleted the first 15 amino acids (Cdx2Δ15), 50 amino acids (Cdx2Δ50) and the region between amino acids 140 and 180 (Cdx2Δ140–180; Figure 2B). All three mutants were Flag tagged and yielded proteins detectable by western blot. Moreover, all three were fully localized to the nucleus post-transfection (data not shown). Surprisingly, all three Cdx2 mutants had significantly diminished capacity to activate the SI reporter despite the fact that these deletions fell outside the transcription activation domain (24). In contrast, TOPFLASH inhibition was largely maintained by the mutant constructs. Both the Cdx2Δ15 and Cdx2Δ140–180 mutants fully inhibited the TOPFLASH reporter (Figure 2B). However, the Cdx2Δ50 mutant lacked any ability to limit TOPFLASH and was no different from the empty vector construct. Taken as a whole, these studies suggest that a subdomain located between amino acids 15 and 50 in Cdx2 is required for β-catenin/TCF inhibition.
Cdx2 expression disrupts the β-catenin–TCF protein complex
A number of mechanisms have been reported that limit Wnt signaling. These mechanisms either block signaling by the receptor (30,31), regulate β-catenin protein levels (32), inhibit its translocation to the nucleus (33,34), disrupt the β-catenin–TCF protein complex (35,36), or block β-catenin/TCF binding to DNA (37,38). In our prior studies, we had demonstrated that Cdx expression did not significantly alter β-catenin protein levels or its ability to localize to the nucleus (17). We therefore considered other mechanisms.
To determine whether Cdx2 was a competitive inhibitor of β-catenin/TCF by either binding TCF-responsive elements or blocking (through other means) TCF-binding DNA targets, we co-transfected two TCF–LEF fusion proteins along with Cdx2 into 293T cells. VP16-LEF is a fusion of the viral VP16 transactivation domain with the DNA-binding domain from LEF-1 (17). CatCLEF fuses the β-catenin transactivation domain to the LEF-1 DNA-binding domain (26,27). Cdx2 co-expression could not diminish TOPFLASH activation by either of these two chimeric proteins (Figure 3A). Thus, Cdx2 cannot block TCF/LEF binding to DNA, suggesting that this is not the mechanism by which Cdx2 inhibits Wnt/β-catenin. Cdx2’s failure to inhibit either chimera suggested that Cdx2 functions at an earlier step, possibly by inhibiting β-catenin–TCF complex formation.
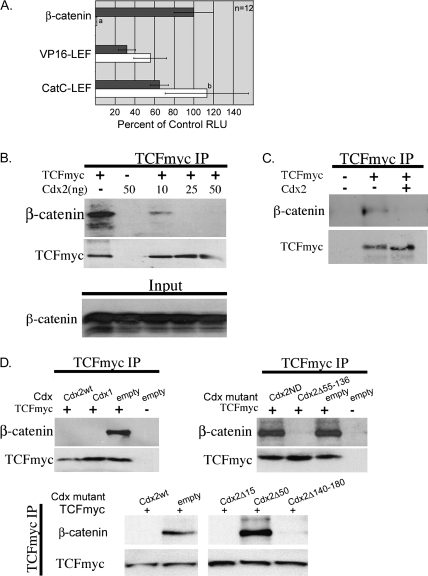
Fig. 3.
The inhibition of TOPFLASH by Cdx2 correlates with disruption of the β-catenin–TCF complex. (A) Constitutively activated chimeric LEF/TCF proteins VP16-Lef1 (17) and CatCLef (26) or the stabilized β-catenin mutant (S33Y) were transfected into 293T cells along with TOPFLASH and an expression vector for Cdx2 (white bar) or an empty control vector (gray bar). Luciferase activity was determined as before; a, differs from all other transfections, P < 0.001; b, differs from CatCLef and empty vector, P < 0.05; (B) A myc-tagged TCF4 and the stabilized β-catenin mutant were co-transfected into 293T cells along with an empty vector control or increasing amounts of a Cdx2 expression vector. Then myc-TCF4 was immunoprecipitated, and the products were analyzed for the presence of β-catenin. (C) Similar study except it was carried out in DLD1 colon cancer cells without the addition of the β-catenin mutant S33Y. (D) Wild-type Cdx1 and Cdx2 truncation mutants are tested for their ability to disrupt the β-catenin–TCF co-immunoprecipitation complex in 293T cells; 50 ng of each Cdx expression vector was transfected for this study.
We tested for this using a myc-tagged TCF4 in 293T cells. After co-transfection of myc-TCF4 along with the S33Y-β-catenin mutant that is stabilized from GSK-3β phosphorylation and proteosome degradation, we could immunoprecipitate the myc-tagged protein and identify robust β-catenin co-precipitation (Figure 3B). Co-transfection of as little as 10 ng of a Cdx2 expression vector nearly completely blocked this interaction and increasing it to 25 ng completely abrogated the complex formation. This was not due to effects on β-catenin or myc-TFC protein levels, as evidenced by controls. Similar results were obtained in DLD1 cells (Figure 3C), suggesting that this disruption is a general phenomenon. Moreover, when we tested the Cdx2 truncation mutants in this assay, we noted that the ability to disrupt the β-catenin-TCF complex correlated with the inhibition of TOPFLASH activity (Figure 3D). Wild-type Cdx1 and Cdx2 proteins, as well as truncation mutants Cdx2Δ15, Cdx2Δ55–136 and Cdx2Δ140–180 completely blocked β-catenin co-precipitation with myc-TCF, whereas the Cdx2ND and Cdx2Δ50 mutants did not. In summary, Cdx2 expression blocks Wnt signaling by disruption of the β-catenin–TCF complex.
The homeodomain transcription factor Cdx2 binds β-catenin
These findings together suggest that Cdx2 might bind β-catenin or TCF4 to inhibit complex formation and transcriptional activity. To investigate for this, we transfected Flag-Cdx2 along with S33Y-β-catenin into 293T cells and immunoprecipitated either with anti-Flag or anti-β-catenin. We found that β-catenin binds with Cdx2, as both proteins are precipitated when either is pulled down (Figure 4A). However, Cdx2 did not co-precipitate with the myc-tagged TCF4 vector (data not shown). Similarly, using endogenously expressed proteins, immunoprecipitation of CDX2 from 7-day post-confluent Caco2 cells also pulled down β-catenin (Figure 4B). Thus, the interaction between Cdx and β-catenin is not due to transfection-mediated protein over-expression and can be established using endogenous CDX2 and β-catenin proteins.
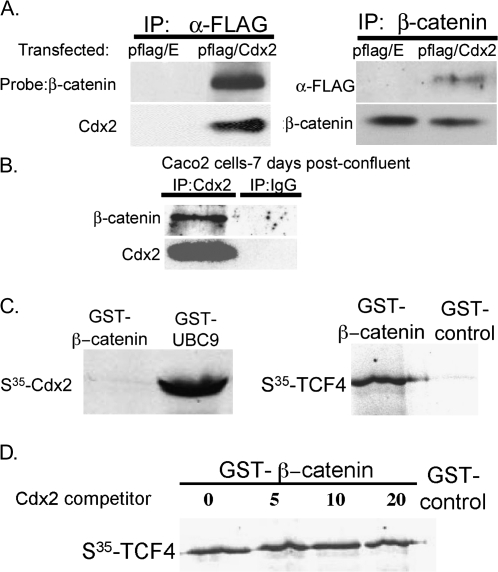
Fig. 4.
Cdx2 protein binds β-catenin in vivo but not in vitro. (A) A Flag-tagged Cdx2 or the empty Flag vector were co-transfected along with β-catenin (S33Y) into 293T cells, and FLAG or β-catenin immunoprecipitations were performed. (B) β-catenin co-immunoprecipitates with Cdx2 in human Caco2 cells. (C) 35S-labeled Cdx2 was incubated with a full-length GST–β-catenin, or with the control Cdx2-binding GST–UBC9 protein, and then GST pull-down was performed. As a control, the GST–β-catenin construct was incubated with an 35S-labeled TCF4 protein. (D) Increasing amounts of an in vitro translated Cdx2 protein were pre-incubated with GST–β-catenin before the addition of 35S-TCF4.
To determine if Cdx2 directly binds β-catenin, we performed GST pull-down studies. GST–β-catenin could not pull-down 35S-labeled Cdx2 protein, although an interaction with a control 35S-labeled TCF4 was established (Figure 4C). The 35S-labeled Cdx2 did interact with a positive control GST–UBC9, confirming that the in vitro translated Cdx2 protein and assay conditions were functioning. Moreover, the in vitro synthesized Cdx2 could not disrupt the GST–β-catenin/35S-TCF4 interaction, as pre-incubation of GST–β-catenin with increasing amounts of in vitro synthesized Cdx2 had no impact on the 35S-TCF4 pull-down (Figure 4D). Together with the cell culture studies, these observations suggest that Cdx2 requires another factor or protein modification in order to bind β-catenin.
Cdx2 transcriptional activity and TOPFLASH suppression requires a highly conserved subdomain located in the N-terminus
Using truncation mutants, we grossly mapped a subdomain required for the inhibition of β-catenin/TCF to between amino acids 15 and 50 (Figure 2). To better define this region, we generated a series of five site-directed mutants. Seven amino acids in each mutant were mutated to alanine or glycine (Figure 5A). All mutants were carefully sequenced, and production of nuclear-localizing proteins of appropriate sizes was confirmed by western blot and immunofluorescence (data not shown). We then utilized these mutants in transient transfection studies with TOPFLASH and SI reporters in 293T cells, as was done before. Mutant 1 behaved much as the wild-type Cdx2 protein (Figure 5B). In contrast, mutants 2, 3, 4 and 5 were all significantly different from Cdx2 wild-type and empty expression vector controls (P < 0.05). Mutant 4, in particular (amino acids 37–43), had very significant findings. Mut4 was distinctly different from all other constructs used in this study, including the other mutants (P < 0.05). Mut4 transactivation of the SI reporter was only 20% of the wild-type protein (Figure 5B). Moreover, Mut4 inhibition of TOPFLASH was diminished by one-third (Figure 5B). These findings indicate that a subdomain critical to Cdx2 transcriptional activity and TOPFLASH suppression is located between amino acids 23 and 50, with the most critical region between amino acids 37 and 43.
Fig. 5.
A subdomain located in the N-terminus is required for Cdx2 transcriptional activity and TOPFLASH inhibition. (A) N-terminal amino acid sequences of five Cdx2 mutants generated for this study. Mutant sequences in bold and underlined. (B) Cdx2 mutants were tested for their ability to transactivate an SI reporter or block β-catenin/TCF-mediated TOPFLASH activation as before. S33Y-β-catenin was co-transfected into 293T cells along with wild-type Cdx2 (white bar), mutant Cdx2s (gray bars), or the empty vector control (black bar); a, significantly different from wild-type Cdx2 and empty vector controls, P < 0.005; b, differs from all other expression vectors, P < 0.001; c, significantly differs from wild-type Cdx2 and empty vector controls, P < 0.05. (C) Alignment of murine Cdx2 with other Cdx sequences using MacVector (Oxford Molecular). Genebank accession numbers indicated. Conserved subdomain indicated by box.
To determine whether this region of Cdx2 is conserved in any fashion, we aligned Cdx2 amino acid sequences using MacVector software (Oxford Molecular Group, Campbell, CA). We noted that the Cdx2 sequences across highly diverged species remained remarkably conserved (Figure 5C). This high-degree of conservation has been noted by others (5). However, the amino acid sequence between amino acids 25 and 45 is nearly perfectly conserved. More remarkably, when we aligned the murine Cdx2 sequence with Cdx1 and Cdx4 sequences, we found a region between amino acids 33 and 43 that remained highly conserved between these more distant Caudal homologues and paralogues (Figure 5C). In light of the fact that Cdx1 is also an effective inhibitor of Wnt/β-catenin/TCF4 signaling, we suspect that the conserved domain shared by these factors plays a critical role in that process.
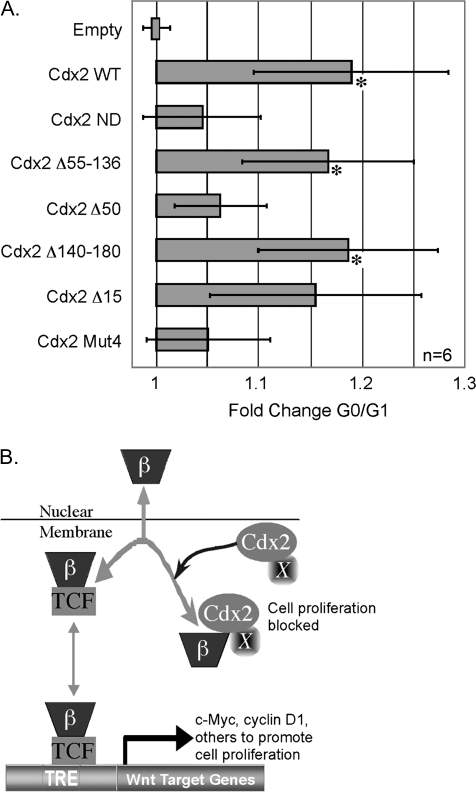
Lastly, to establish a functional role for this domain in the inhibition of cell proliferation, we transfected the truncation mutants along with Cdx2Mut4 into HCT 116 cells. Previously, we had shown that transfection of Cdx1 or Cdx2 into these cells inhibited TOPFLASH reporter activity and induced a G0/G1 accumulation of cells, consistent with a growth arrest (17). As we had reported previously, the wild-type Cdx2 protein induces an 18% increase in G0/G1 cells (Figure 6A). The Cdx2 mutants Cdx2Δ15, Cdx2Δ55–136 and Cdx2Δ140–180, which efficiently inhibited TOPFLASH and disrupted the β-catenin–TCF complex, induced a similar level of G0/G1 accumulation of cells. In contrast, neither Cdx2ND, Cdx2Δ50 nor Cdx2Mut4 were significantly different from each other or from the empty vector control (Figure 6A). This further substantiates that Cdx2 inhibition of TOPFLASH correlates with the inhibition of cell proliferation and that a subdomain critical for this effect is located in the protein's N-terminus and centered between amino acids 36 and 43. We conclude that a subdomain important both for normal Cdx2 transcriptional activity and the inhibition of β-catenin/TCF4 transcriptional activity and cell proliferation is centered between amino acids 33 and 43.
Fig. 6.
Inhibition of cancer cell proliferation by Cdx2 requires a conserved subdomain in the N-terminus. (A) HCT116 cells were transfected with a GFP expression vector as well as wild-type Cdx2, the Cdx2 truncation mutants, with Cdx2Mut4, or the empty vector as control. At 48 h, the cells were stained with propidium iodide and DNA content quantitated in the GFP+ cells. *Significantly differs from empty vector, Cdx2ND, and Cdx2Δ50, P < 0.05 by. (B) Model of Cdx2 inhibition of Wnt/β-catenin transcriptional activity. β-catenin translocates to the nucleus where it partners with a TCF family member, bind DNA and activates target genes. In the presence of Cdx2 expression and an as yet unidentified factor or post-translational modification (X), β-catenin associates with Cdx2. This interaction prevents β-catenin/TCF transcriptional activity.
Discussion
One important question that has not been well studied to date is how developmental mechanisms that promote intestinal cell differentiation can regulate the Wnt/β-catenin/TCF pathway. The Caudal related homeodomain protein Cdx2 is an intestine-specific transcription factor that directs the expression of the intestinal cell phenotype by enhancing the expression of numerous intestine-specific genes and promoting the emergence of a mature, columnar cell morphology (5,10–14,18). We and others have demonstrated previously that Cdx1 or Cdx2 expression could reduce colon cancer cell proliferation by inhibiting β-catenin/TCF transcriptional activity (17,19–21). This was associated with the diminished expression of the Wnt target genes LEF-1, Cyclin D1 and c-Myc, as well as a reduction of β-catenin responsiveness of Wnt targeted promoters like cyclin D1, Cdx1 and p14ARF (17,21). The present study advanced this work by identifying a mechanism for this effect, namely that these important developmental regulators may antagonize Wnt/β-catenin/TCF transcriptional activity by directly binding β-catenin and disrupting the β-catenin–TCF protein complex. This is a very novel finding and suggests that Cdx2 may influence development, gene expression patterns and cell differentiation by means other than as a transcription activator.
Cdx2-mediated inhibition of β-catenin/TCF is molecularly distinct from its role as a transcriptional activator
Several lines of data strongly suggest that the mechanisms whereby Cdx2 induces SI promoter activity and inhibits β-catenin/TCF are quite different. Previous studies by us in colon cancer cell lines as well as non-colorectal cancer systems suggested that there was a disconnect between the two processes. In the present study, using Flag-tagged Cdx1 and Cdx2 proteins, we determined that the inhibition of TOPFLASH was very dependent upon protein levels, whereas SI activation was not. Studies with Cdx2 truncation mutants firmly established that these were distinctly different processes since Cdx2 transcriptional activity was not required for TOPFLASH inhibition or the reductions in cell proliferation, but is necessary for SI induction. Together, these findings establish that Cdx2 inhibition of β-catenin/TCF and transactivation of target genes like SI utilizes distinctly different mechanisms. These are important differences that colon cancer cells appear able to exploit, specifically suppressing TOPFLASH inhibition while maintaining SI activation. This undoubtedly has important implications for colon carcinogenesis. These observations also have clear mechanistic implications we explored in the subsequent studies described above.
A model for Cdx2 inhibition of Wnt/β-catenin/TCF signaling
Our findings have significantly extended our previously published work and the work of others which first identified the inhibitory effect of Cdx1 and Cdx2 upon Wnt/β-catenin/TCF signaling (17,19,20). Our data argue that the inhibitory effect is mediated by the Cdx2 protein interacting with β-catenin, leading to disruption of the β-catenin–TCF complex (Figure 6B). However, since we can only demonstrate this interaction in vivo, we believe that another protein factor or some post-translational modification of Cdx2 or β-catenin is required to promote this interaction. We are presently pursuing studies to identify this unknown factor. Moreover, it is interesting to note that while classic Cdx2 transcriptional activity is not required for the inhibition of β-catenin/TCF, every Cdx2 mutation that disrupts TOPFLASH inhibition also significantly degrades Cdx2 transcriptional activity. Therefore, our work yields novel insights not only into the regulation of Wnt/β-catenin/TCF activity by Cdx2, but the regulation of classic Cdx2 transcriptional activity as well.
Although Wnt signaling is known to inhibit the actions and expression of homeodomain transcription factors (39,40), to our knowledge, this is the first example of a homeobox protein inhibiting β-catenin–TCF by a protein interaction. These studies also shed important new light on a paradox regarding the role of Cdx2 in human colorectal cancer. Historically, Cdx2 was thought to be a ‘tumor suppressor’ due to its ability to reduce proliferation of some colon cancer cells and its reported loss in human colon polyps and cancers (14,41,42). Most importantly, transgenic mice heterozygous for a deletion of Cdx2 formed many more polyps than non-transgenic controls when crossed with the tumorigenic Apc+/Δ716 mice or when treated with the carcinogen azoxymethane (15,16). However, several recent studies have begun to cast doubt on Cdx2 as a classic tumor suppressor, with at least two studies, suggesting that Cdx2 may possess tumorigenic properties (43–45). In these studies, it is suggested that Cdx2 expression enhances cell survival, possibly by inducing the expression of the anti-apoptotic Bcl-2 protein. If, indeed, Cdx2 possesses both tumor-promoting and inhibitory properties, our studies suggest that colon cancer cells may be able to separate them, retaining the former whereas disabling the latter.
The antitumor property studied here, the anti-proliferative effect of Cdx2, is dependent upon a protein–protein interaction between Cdx2 and β-catenin that disrupts the pro-proliferative β-catenin–TCF complex. This is probably a stochiometric relationship and, therefore, processes that increase β-catenin levels or diminish Cdx2 can promote transformation. Thus, we see β-catenin levels increased and Cdx1 protein expression silenced in the majority of human colon cancers (46,47). Moreover, the Cdx2 protein appears to be less stable in colon cancer cells due to ubiquination and proteosome degradation (48,49). The ultimate result is diminished capacity for Cdx-mediated inhibition of β-catenin/TCF while retaining significant Cdx2 transcriptional activity. This may then serve to promote carcinogenesis by preserving the expression of anti-apoptotic factors like Bcl-2 (45,50).
In summary, we establish a novel mechanism by which the intestine-specific transcription factor Cdx2 inhibits intestinal cell proliferation and β-catenin/TCF function. An evolutionarily conserved subdomain localized to the N-terminus of Cdx2 mediates its interaction with β-catenin. This interaction is capable of disrupting β-catenin binding to TCF, leading to loss of the proliferative Wnt/β-catenin/TCF stimulus. We conclude that Cdx2 inhibition of Wnt/β-catenin/TCF signaling and proliferation in colon cancer and intestinal epithelial cells is dependent upon Cdx2 binding β-catenin and disrupting its interaction with the DNA-binding TCF factor family.
Supplementary material
Supplementary Table S1 can be found at http://carcin.oxfordjournals.org/.
Funding
The National Institute of Diabetes and Digestive and Kidney Diseases (DK068366 to J. L.); National Cancer Institute Program (Project P01 DE12467); the Morphology, Cell Culture and Molecular Biology Core Facilities of the Center for Molecular Studies in Digestive and Liver Disease at the University of Pennsylvania (P30-DK50306).
Supplementary Material
Acknowledgments
We would like to thank Oren Mushin for his technical assistance.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- GST
Glutathione-S transferase
- LEF
lymphocyte enhancer factor
- SI
sucrase isomaltase
- TCF
T-cell factor
References
- 1.Lynch JP, et al. The genetic pathogenesis of colorectal cancer. Hematol. Oncol. Clin. North Am. 2002;16:1–36. doi: 10.1016/s0889-8588(02)00029-1. [DOI] [PubMed] [Google Scholar]
- 2.Schneikert J, et al. The canonical Wnt signalling pathway and its APC partner in colon cancer development. Gut. 2007;56:417–425. doi: 10.1136/gut.2006.093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Korinek V, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 5.Guo RJ, et al. The role of Cdx proteins in intestinal development and cancer. Cancer Biol. Ther. 2004;3:593–601. doi: 10.4161/cbt.3.7.913. [DOI] [PubMed] [Google Scholar]
- 6.Suh E, et al. A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol. Cell. Biol. 1994;14:7340–7351. doi: 10.1128/mcb.14.11.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang DT, et al. Expression of the gut-enriched Kruppel-like factor (Kruppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene. 2001;20:4884–4890. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinoi T, et al. CDX2 regulates liver intestine-cadherin expression in normal and malignant colon epithelium and intestinal metaplasia. Gastroenterology. 2002;123:1565–1577. doi: 10.1053/gast.2002.36598. [DOI] [PubMed] [Google Scholar]
- 9.Hecht A, et al. Regulation of sucrase and lactase in developing rats: role of nuclear factors that bind to two gene regulatory elements. Gastroenterology. 1997;112:803–812. doi: 10.1053/gast.1997.v112.pm9041242. [DOI] [PubMed] [Google Scholar]
- 10.Funakoshi S, et al. Repression of the Desmocollin 2 gene in colorectal cancer cells is relieved by the homeodomain transcription factors Cdx1 and Cdx2. Mol. Cancer Res. 2008;6:1478–1490. doi: 10.1158/1541-7786.MCR-07-2161. [DOI] [PubMed] [Google Scholar]
- 11.Gao N, et al. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller MS, et al. Cdx1 or Cdx2 expression activates E-Cadherin-mediated cell-cell adhesion and compaction in human COLO 205 cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287:G104–G14. doi: 10.1152/ajpgi.00484.2003. [DOI] [PubMed] [Google Scholar]
- 13.Soubeyran P, et al. Cdx1 promotes differentiation in a rat intestinal epithelial cell line. Gastroenterology. 1999;117:1326–1338. doi: 10.1016/s0016-5085(99)70283-0. [DOI] [PubMed] [Google Scholar]
- 14.Suh E, et al. An intestine-specific homeobox gene regulates proliferation and differentiation. Mol. Cell. Biol. 1996;16:619–625. doi: 10.1128/mcb.16.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoki K, et al. Colonic polyposis caused by mTOR-mediated chromosomal instability in Apc+/Delta716 Cdx2+/− compound mutant mice. Nat. Genet. 2003;35:323–330. doi: 10.1038/ng1265. [DOI] [PubMed] [Google Scholar]
- 16.Bonhomme C, et al. The Cdx2 homeobox gene has a tumour suppressor function in the distal colon in addition to a homeotic role during gut development. Gut. 2003;52:1465–1471. doi: 10.1136/gut.52.10.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo RJ, et al. Cdx1 inhibits human colon cancer cell proliferation by reducing β-catenin/TCF transcriptional activity. J. Biol. Chem. 2004;279:36865–36875. doi: 10.1074/jbc.M405213200. [DOI] [PubMed] [Google Scholar]
- 18.Ezaki T, et al. The homeodomain transcription factors Cdx1 and Cdx2 induce E-cadherin adhesion activity by reducing beta- and p120-catenin tyrosine phosphorylation. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293:G54–G65. doi: 10.1152/ajpgi.00533.2006. [DOI] [PubMed] [Google Scholar]
- 19.Lynch J, et al. The caudal-related homeodomain protein Cdx1 inhibits proliferation of intestinal epithelial cells by down-regulation of D-type cyclins. J. Biol. Chem. 2000;275:4499–4506. doi: 10.1074/jbc.275.6.4499. [DOI] [PubMed] [Google Scholar]
- 20.Lynch J, et al. Cdx1 inhibits the proliferation of human colon cancer cells by reducing cyclin D1 gene expression. Oncogene. 2003;22:6395–6407. doi: 10.1038/sj.onc.1206770. [DOI] [PubMed] [Google Scholar]
- 21.Saegusa M, et al. A functional role of Cdx2 in beta-catenin signaling during transdifferentiation in endometrial carcinomas. Carcinogenesis. 2007;28:1885–1892. doi: 10.1093/carcin/bgm105. [DOI] [PubMed] [Google Scholar]
- 22.Crissey MA, et al. The homeodomain transcription factor Cdx1 does not behave as an oncogene in normal mouse intestine. Neoplasia. 2008;10:8–19. doi: 10.1593/neo.07703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boudreau F, et al. Hepatocyte nuclear factor-1 alpha, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J. Biol. Chem. 2002;277:31909–31917. doi: 10.1074/jbc.M204622200. [DOI] [PubMed] [Google Scholar]
- 24.Rings EH, et al. Phosphorylation of the serine 60 residue within the cdx2 activation domain mediates its transactivation capacity. Gastroenterology. 2001;121:1437–1450. doi: 10.1053/gast.2001.29618. [DOI] [PubMed] [Google Scholar]
- 25.Silberg DG, et al. CDX1 protein expression in normal, metaplastic, and neoplastic human alimentary tract epithelium. Gastroenterology. 1997;113:478–486. doi: 10.1053/gast.1997.v113.pm9247467. [DOI] [PubMed] [Google Scholar]
- 26.Okubo T, et al. Hyperactive Wnt signaling changes the developmental potential of embryonic lung endoderm. J. Biol. 2004;3:11. doi: 10.1186/jbiol3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galceran J, et al. Rescue of a Wnt mutation by an activated form of LEF-1: regulation of maintenance but not initiation of Brachyury expression. Proc. Natl. Acad. Sci. USA. 2001;98:8668–8673. doi: 10.1073/pnas.151258098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korinek V, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 29.Boudreau F, et al. Sucrase-isomaltase gene transcription requires the hepatocyte nuclear factor-1 (HNF-1) regulatory element and is regulated by the ratio of HNF-1 alpha to HNF-1 beta. J. Biol. Chem. 2001;276:32122–32128. doi: 10.1074/jbc.M102002200. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki H, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 31.Kuhnert F, et al. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl. Acad. Sci. USA. 2004;101:266–271. doi: 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stone CD, et al. Gut-enriched Kruppel-like factor regulates colonic cell growth through APC/beta-catenin pathway. FEBS Lett. 2002;530:147–152. doi: 10.1016/s0014-5793(02)03449-x. [DOI] [PubMed] [Google Scholar]
- 33.Gottardi CJ, et al. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J. Cell. Biol. 2001;153:1049–1060. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orsulic S, et al. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J. Cell Sci. 1999;112:1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- 35.Tago K, et al. Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev. 2000;14:1741–1749. [PMC free article] [PubMed] [Google Scholar]
- 36.Zorn AM, et al. Regulation of Wnt signaling by Sox proteins: xSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol. Cell. 1999;4:487–498. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]
- 37.Sampson EM, et al. Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. EMBO J. 2001;20:4500–4511. doi: 10.1093/emboj/20.16.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van de Wetering M, et al. Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol. Cell Biol. 1996;16:745–752. doi: 10.1128/mcb.16.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLin VA, et al. Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development. 2007;134:2207–2217. doi: 10.1242/dev.001230. [DOI] [PubMed] [Google Scholar]
- 40.Kahler RA, et al. Lymphoid enhancer factor-1 and beta-catenin inhibit Runx2-dependent transcriptional activation of the osteocalcin promoter. J. Biol. Chem. 2003;278:11937–11944. doi: 10.1074/jbc.M211443200. [DOI] [PubMed] [Google Scholar]
- 41.Ee HC, et al. Cdx-2 homeodomain protein expression in human and rat colorectal adenoma and carcinoma. Am. J. Pathol. 1995;147:586–592. [PMC free article] [PubMed] [Google Scholar]
- 42.Mallo GV, et al. Expression of the Cdx1 and Cdx2 homeotic genes leads to reduced malignancy in colon cancer-derived cells. J. Biol. Chem. 1998;273:14030–14036. doi: 10.1074/jbc.273.22.14030. [DOI] [PubMed] [Google Scholar]
- 43.Witek ME, et al. The putative tumor suppressor Cdx2 is overexpressed by human colorectal adenocarcinomas. Clin. Cancer Res. 2005;11:8549–8556. doi: 10.1158/1078-0432.CCR-05-1624. [DOI] [PubMed] [Google Scholar]
- 44.Rawat VP, et al. Ectopic expression of the homeobox gene Cdx2 is the transforming event in a mouse model of t(12;13)(p13;q12) acute myeloid leukemia. Proc. Natl. Acad. Sci. USA. 2004;101:817–822. doi: 10.1073/pnas.0305555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dang LH, et al. CDX2 has tumorigenic potential in the human colon cancer cell lines LOVO and SW48. Oncogene. 2006;25:2264–2272. doi: 10.1038/sj.onc.1209247. [DOI] [PubMed] [Google Scholar]
- 46.Wong NA, et al. Loss of CDX1 expression in colorectal carcinoma: promoter methylation, mutation, and loss of heterozygosity analyses of 37 cell lines. Proc. Natl. Acad. Sci. USA. 2004;101:574–579. doi: 10.1073/pnas.0307190101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suh ER, et al. DNA methylation down-regulates CDX1 gene expression in colorectal cancer cell lines. J. Biol. Chem. 2002;227:35795–35800. doi: 10.1074/jbc.M205567200. [DOI] [PubMed] [Google Scholar]
- 48.Boulanger J, et al. Cdk2-dependent phosphorylation of homeobox transcription factor CDX2 regulates its nuclear translocation and proteasome-mediated degradation in human intestinal epithelial cells. J. Biol. Chem. 2005;280:18095–18107. doi: 10.1074/jbc.M502184200. [DOI] [PubMed] [Google Scholar]
- 49.Gross I, et al. Phosphorylation of the homeotic tumor suppressor Cdx2 mediates its ubiquitin-dependent proteasome degradation. Oncogene. 2005;24:7955–7963. doi: 10.1038/sj.onc.1208945. [DOI] [PubMed] [Google Scholar]
- 50.Arcinas M, et al. Molecular mechanisms of transcriptional control of bcl-2 and c-myc in follicular and transformed lymphoma. Cancer Res. 2001;61:5202–5206. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.