Abstract
Nuclear receptor coactivator [peroxisome proliferator-activated receptor-binding protein (PBP)/mediator subunit 1 (MED1)] is a critical component of the mediator transcription complex. Disruption of this gene in the mouse results in embryonic lethality. Using the PBP/MED1 liver conditional null (PBP/MED1ΔLiv) mice, we reported that PBP/MED1 is essential for liver regeneration and the peroxisome proliferator-activated receptor α ligand Wy-14,643-induced receptor-mediated hepatocarcinogenesis. We now examined the role of PBP/MED1 in genotoxic chemical carcinogen diethylnitrosamine (DEN)-induced and phenobarbital-promoted hepatocarcinogenesis. The carcinogenic process was initiated by a single intraperitoneal injection of DEN at 14 days of age and initiated cells were promoted with phenobarbital (PB) (0.05%) in drinking water. PBP/MED1ΔLiv mice, killed at 1, 4 and 12 weeks, revealed a striking proliferative response of few residual PBP/MED1-positive hepatocytes that escaped Cre-mediated deletion of PBP/MED1 gene. No proliferative expansion of PBP/MED1 null hepatocytes was noted in the PBP/MED1ΔLiv mouse livers. Multiple hepatocellular carcinomas (HCCs) developed in the DEN-initiated PBP/MED1fl/fl and PBP/MED1ΔLiv mice, 1 year after the PB promotion. Of interest is that all HCC developing in PBP/MED1ΔLiv mice were PBP/MED1 positive. None of the tumors was PBP/MED1 negative implying that hepatocytes deficient in PBP/MED1 are not susceptible to neoplastic conversion. HCC that developed in PBP/MED1ΔLiv mouse livers were transplantable in athymic nude mice and these maintained PBP/MED1fl/fl genotype. PBP/MED1fl/fl HCC cell line derived from these tumors expressed PBP/MED1 and deletion of PBP/MED1fl/fl allele by adeno-Cre injection into tumors caused necrosis of tumor cells. These results indicate that PBP/MED1 is essential for the development of HCC in the mouse.
Introduction
Nuclear receptors and many other regulated transcription factors control a surprisingly diverse array of genes capable of influencing many biological processes (1,2). Transcriptional activation of genes is a complex process, which involves the participation of many transcription coactivators (3–5). These coactivators bind directly to transcription factors and are involved in the remodeling of chromatin structures and/or in bridging transcription factors with RNA polymerase II and the general basal transcription machinery (4–7). Many of these transcription coactivators along with other coregulatory molecules bind singly or as preformed multisubunit protein complexes to an activated transcription factor to enhance target gene transcription (3–6). Since the cloning of the first transcription coactivator steroid receptor coactivator (7) and corepressors nuclear receptor corepressor (8) and SMRT (9), over 300 coactivators/coregulators have been identified with new members still being added to this spectrum (4). This diversity raises issues about the versatility and complexity of the coregulatory molecules in orchestrating transcription and the need to delineate specific functional roles of individual coregulators in embryogenesis, development, differentiation and oncogenesis, as well as energy and xenobiotic metabolism (4,10). Limited studies involving gene knockout mouse models show that some of the coactivators/coregulators are essential for embryonic growth and survival, whereas others are not critical for mouse embryogenesis (reviewed in ref. 10). For example, the disruption of a coactivator gene such as peroxisome proliferated-activated receptor (PPAR)-binding protein, which is also known as thyroid hormone receptor-associated protein (TRAP) 220, vitamin D3 receptor-interacting protein (DRIP) 205 or mediator subunit 1 (MED1), referred to here as peroxisome proliferator-activated receptor-binding protein (PBP)/MED1, results in embryonic lethality in the mouse around gestational day 11.5 (11–15). Likewise, germ-line deletion of coactivator peroxisome proliferator-activated receptor-interacting protein (PRIP) (ASC-2/RAP250/TRBP/NRC/NCoA6) gene and PRIP-interacting protein with methyltransferase domain (PIMT/NCoA6IP) gene in the mouse also results in embryonic lethality (16–19). On the other hand, null mutation of coactivators steroid receptor coactivator (20,21), SRC-2 (22), SRC-3 (23,24), PPAR gamma coactivator 1 alpha (25) or coactivator associated arginine methyl transferase 1 (26) results in viable phenotype with or without overt functional changes.
To analyze the roles of PBP/MED1 and PRIP in adult tissues, we generated mice for conditional gene disruption using the Cre-loxP strategy (27,28). Using the albumin enhancer and promoter-driven Cre recombinase transgene [Alb-Cre] (29) to mediate hepatocyte-specific deletion of the PBP/MED1fl/fl allele, we demonstrated that PBP/MED1 is essential for PPARα-regulated gene expression in liver (27). Likewise, hepatocyte-specific deletion of the PBP/MED1fl/fl allele resulted in near abrogation of the induction of constitutive androstane receptor (CAR)-regulated genes in liver (30). In contrast, deletion of PRIPfl/fl allele in liver did not interfere with PPARα and CAR signaling (28). The dependence of PPARα and CAR-regulated gene transcription on coactivator PBP/MED1, but not on PRIP, attests to the existence of coactivator selectivity in nuclear receptor function at least in the liver (10,31). Furthermore, PBP/MED1 null hepatocytes in PBP/MED1 liver conditional null (PBP/MED1ΔLiv) mice did not develop hepatocellular carcinoma (HCC) when chronically exposed to a PPARα ligand, indicating that PBP/MED1 is essential for this receptor-mediated liver tumorigenesis (32). PPARα ligands, such as ciprofibrate and Wy-14,643, are non-genotoxic hepatocarcinogens (33) that differ from classical DNA-damaging mutagenic chemical carcinogens (34). Typically, mutagenic chemicals and or their electrophilic metabolites interact with and damage DNA inducing mutations and initiating the neoplastic change (34). Therefore, it would be of importance to ascertain whether the PBP/MED1 null hepatocytes function as targets for a potent genotoxic carcinogen such as diethylnitrosamine (DEN) (35–38). Here, we used a DEN/phenobarbital (PB) liver tumor-induction protocol to demonstrate that PBP/MED1-deficient mouse hepatocytes fail to develop liver tumors in PBP/MED1ΔLiv mice. Initiation by DEN, followed by PB promotion in PBP/MED1ΔLiv mice, resulted in a failure of PBP/MED1 null hepatocytes to undergo proliferation. However, an occasional PBP/MED1 expressing hepatocyte with PBP/MED1fl/fl genotype that escaped Alb-Cre-mediated deletion exhibited enormous proliferative potential and such cells gave rise to liver tumors following DEN initiation and PB promotion. PB exerts liver tumor promoting effects by activating the nuclear receptor CAR (39) and since PBP/MED1 deficiency abrogates CAR function (30), it would appear that disruption of PBP/MED1 gene would have an impact on liver tumor development. Furthermore, we established a transplantable HCC with PBP/MED1fl/fl genotype in athymic nude mice and show that intratumoral injection of Adeno-Cre-mediated deletion of PBP/MED1 allele in tumor cells caused necrosis. These studies suggest PBP/MED1 is essential for development of HCC and required for the proliferative expansion of liver cancers.
Materials and methods
Generation of PBP/MED1 conditional null mutation in liver (PBP/MED1ΔLiv) and treatment with PB and DEN
The homozygous mutant mice lacking the PBP/MED1 in hepatocytes (PBP/MED1ΔLiv) were generated and bred in our laboratory as described elsewhere (27). Mice carrying PBP/MED1fl/fl (fl, for flanked by LoxP) allele were bred with a mouse containing albumin-Cre (AlbCre) transgene to generate PBP/MED1ΔLiv mice (29). All the mice used for experiments were maintained in the C57BL/6 background. Mice were housed, under 12 h light–12 h dark cycle, in a pathogen-free animal facility accredited by the Association for the Advancement and Accreditation of Laboratory Animal Care International. They were maintained on standard rodent chow (Teklad #7904; Harlan-Teklad, Indianapolis, IN) and water ad libitum. All the experiments involving PBP/MED1 liver nulls and wild-type mice were carried out using littermates. All animal procedures used in this study were reviewed and preapproved by the Northwestern University Institutional Animal Care and Use Committee. At 14 days postnatal age, the entire mouse litter received a single intraperitoneal injection of the genotoxic tumor initiator DEN (5 μg/g body weight; catalog no. N0756, Sigma, St Louis, MO; dissolved in 0.9% saline) which induces hepatocyte DNA damage through DNA adduct formation (37). Three days following injection of the tumor initiator DEN, the mice were allowed free access to drinking water containing tumor promoter PB (0.05%) until they were killed at specified intervals. Groups of six to seven mice, containing both males and females, were used for the experiments. Polymerase chain reaction (PCR) genotyping of mice was performed using the following primers flanking loxP site 1 in the floxed PBP/MED1 allele; LoxP+: 5′-TCCATCTGACCTGCTGGATGATAA-3’ and LoxP−: 5′- GGGTGTGACCCCATAATT-3′. Cre-specific primers used included Cre-F: 5′-AGGTGTAGAGAAGGCACTCAGC-3’ and Cre-R: 5′-CTAATCGCCATCTTCCAGCAGG-3′. PCR genotyping of tumor and non-tumor areas in liver were performed using the following primers flanking exons 8–10 in the floxed PBP/MED1 allele: PBPNA: 5′- GGTTATACATACCTTTCTGCTGT-3′ and PBPNS: 5′- CCGTACAGTATCAGTCACCAT-3′. These primers were also used for the genotyping of HCC cell lines derived from these tumors. Microdissection was performed to excise 1 × 1 mm tumor and non-tumorous tissue for DNA extraction. Groups of PBP/MED1fl/fl and PBP/MED1ΔLiv mice were killed at 1, 4, 12 or 52 weeks after DEN injection. At necropsy, terminal body weights, liver weights and liver morphology were evaluated. Livers were examined for the presence of liver tumors and sections of liver, liver tumors and selected samples of lung were fixed in 10% formalin or 4% paraformaldehyde and processed for light microscopic evaluation by three authors (K.M., M.S.R. and J.K.R.) with liver pathology expertise.
Bromodeoxyuridine labeling, immunohistochemical staining and terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling apoptosis assay
To assess cell proliferation, bromodexoyuridine (BrdUrd) (0.5 mg/ml) was administered in drinking water for 3 days and given a single intraperitoneal dose of BrdUrd (100 mg/kg body wt) 2 h before killing (32). Liver and lung slices were fixed in 10% formalin or 4% paraformaldehyde, processed for embedding in paraffin, sectioned and stained with either hematoxylin and eosin or processed for the immunohistochemical localization of PBP/MED1 and BrdUrd or proliferating cell nuclear antigen. The antibodies used were anti-PBP/MED1 (catalog number sc-5334, Santa Cruz Biotechnology, Santa Cruz, CA), anti-BrdUrd (BD Biosciences, San Jose, CA) and anti-PCNA (catalog number sc-25280, Santa Cruz Biotechnology). To measure apoptosis in mouse liver isolated from either untreated or DEN/PB-treated PBP/MED1fl/fl and PBP/MED1ΔLiv mice, terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling staining was performed on liver sections by using the in situ cell death detection kit from Roche Applied Science (Indianapolis, IN) according to the manufacturer's recommendations. Histological analysis and image processing were carried out using a Leica DMRE microscope equipped with Spot digital camera. The volume occupied by large PBP/MED1 positive hepatocytes in PBP/MED1ΔLiv mice was estimated by using Scion Image software (http://scion-image.software.informer.com/).
Tumor implantation in nude mice
Male athymic (nu/nu) mice, 5–6 weeks of age (Jackson Laboratory, Bar Harbor, ME), were maintained in a pathogen-free environment, according to an institutionally approved animal protocol, and given food and water ad libitum. After 1 week's acclimation, mice were anesthetized with nembutal prior to tumor implantation. Liver tumors from PBP/MED1ΔLiv mice, harvested at the end of 1 year on DEN initiation and PB promotion, were chopped into 1–2 mm cubes and one piece of tumor tissue was surgically implanted subcutaneously on the back of nude mouse. Transplanted tumors were processed for histopathological evaluation and for PBP/MED1 immunohistochemical staining. To assess the role of PBP/MED1 in tumor cell survival, we injected Adeno (Ad)-Cre-GFP viral particles (Vector Biolabs, Philadelphia, PA) into the tumor transplanted in nude mice and the tumor tissue examined for histological alterations.
Generation of PBP/MED1fl/fl HCC cell lines (MED1fl/flHCC cell line)
To derive a HCC cell line from the PBP/MED1 expressing HCC serially transplanted in athymic nude mice, PBP/MED1fl/fl tumor tissue was dispersed with collagenase or trypsin digestion into single cells or small cell clumps and these were cultured in fresh Dulbecco's modified Eagle’s medium (DMEM) containing 10% fetal bovine serum. Two cell lines are established and designated as PBP/MED1fl/flHCC1 and PBP/MED1fl/flHCC2 cell line.
Soft agar colony formation assay
Tumor cells (300) were suspended in a 1 ml mixture containing 0.35 (wt/vol) agar DMEM supplemented with 10% fetal bovine serum. This mixture was layered on a 1 ml base of 0.7% (wt/vol) agar in DMEM with 10% fetal bovine serum in a six-well plate and maintained in DMEM for 2 weeks. Colonies were grown for 8 days and live pictures were taken on phase contrast microscope. To check the affect of PBP/MED1 deletion in tumor cells, PBP/MED1fl/flHCC cells were infected with 8 × 108 virus particle [Ad/Cre-GFP or Ad/enhanced green fluorescent protein (EGFP)] and 300 cells were inoculated for 2 weeks and pictures were taken. Ad/Cre-GFP was purchased from Vector Biolabs (catalog no. 1700) while Ad/EGFP was generated as follows: coding sequence of EGFP was cloned into NotI and HindIII sites of pShuttle-CMV expression vector (Quantum Biotechnologies, Quebec, Canada). The shuttle vector and AdEASY vector (Quantum Biotechnologies) were linearized using PmeI and cotransformed into Escherichia coli strain BJ5183 for the homologous recombination. Recombinant plasmid Ad/EGFP was selected and Ad/EGFP virus was generated in HEK293A cells as per manufacturer protocol.
Quantitative PCR analysis
Expression level of PBP/MED1 messenger RNA (mRNA) was analyzed by real-time quantitative PCR (qPCR). Total RNA was extracted from transplanted tumor tissues obtained from nude mice and liver samples from PBP/MED1fl/fl and PBP/MED1ΔLiv mice using TRIzol reagent (Invitrogen, Carlsbad, CA). Complementary DNA was prepared with 2 μg of total RNA using SuperScript™ III First-Strand Synthesis System for RT–PCR (Invitrogen) and qPCR procedure was performed with SYBR Green Supermix (Applied Biosystems, Foster City, CA) and the oligonucleotide for PBP/MED1 (5′-GAGACTCCGCCCACTTACCG-3′ and 5′-GGACACTTCAAACTGGAGG-3′), insulin-like growth factor binding protein 1 (5′-TGCACCCGCCACGAGCAC-3′ and 5′-GGGCCATCAGGTGGAAGCTGTC-3′), cyclin D (5′- GTGGTGGCTGCGATGCAAGG-3′ and 5′-TGTTCCTGGCAGGCACGGAG-3′) and cyclin B1 (5′-CCATGGCGCTCAGGGTCACTAG-3′ and 5′-CCGGGCTTGGAAGCAGCAG-3′). Each sample was normalized on the basis of its endogenous 18S ribosomal content and performed in triplicate.
Results
PBP/MED1 expressing hepatocytes exhibit growth advantage in PBP/MED1ΔLiv mice during PB promotion
Previously, we demonstrated that chronic exposure to Wy-14,643, a non-genotoxic hepatocarcinogen and a potent peroxisome proliferator, results in a rapid and sustained proliferation of PBP/MED1fl/fl hepatocytes that escaped Cre-mediated gene deletion in PBP/MED1ΔLiv mouse livers (27,32). PBP/MED1-positive hepatocytes are easily discernible in Wy-14,643 treated PBP/MED1ΔLiv mouse livers because of their large size, reflecting peroxisome proliferation occurring in response to PPARα activation (27). All liver tumors that developed in PBP/MED1 null livers expressed PBP/MED1 and no tumor appeared to be derived from PBP/MED1-deficient hepatocytes (32). This led us to conclude that absence of PBP/MED1 abrogates the response to PPARα ligand signaling required for liver tumorigenesis with Wy-14,643 (32).
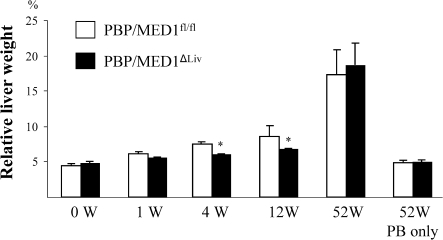
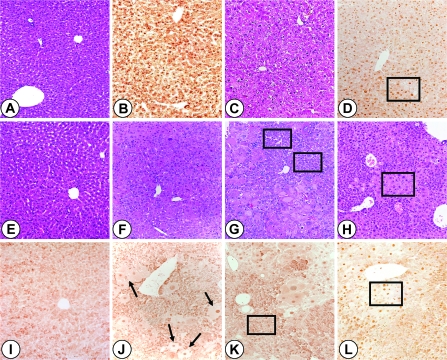
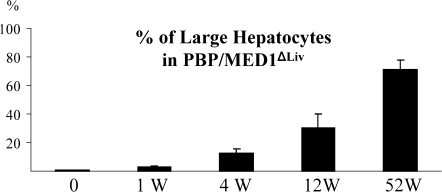
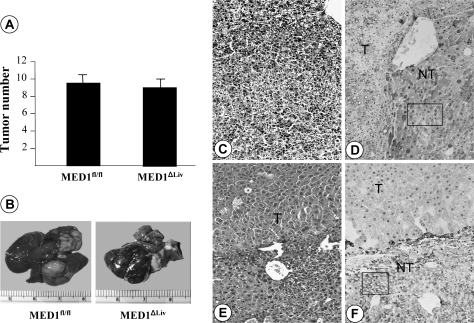
In the present study, it was decided to ascertain whether PBP/MED1 null hepatocytes function as targets for tumor development following DEN-initiation and PB promotion (35,37). DEN, in contrast to PPARα ligand Wy-14,643, is a genotoxic chemical carcinogen and the DEN-initiated hepatocytes are readily promotable when exposed to liver tumor promoter PB, an activator of nuclear receptor CAR (38). In the present study, following DEN initiation, PBP/MED1fl/fl and PBP/MED1ΔLiv mice were given PB in drinking water and mice were killed at selected intervals to examine changes in the liver. Liver weight:body weight ratio was slightly lower in PBP/MED1ΔLiv mice at 4 and 12 weeks on PB promotion and by 52 weeks, the ratios were similar and the increase in ratio at this interval is due to liver tumor burden (Figure 1). In mice given PB for 52 weeks without DEN initiation, the liver weight:body weight ratio remained lower compared with DEN-initiated animals (Figure 1). It should be noted that 1 year of PB treatment had similar effect of liver weight:body weight ratio in normal and PBP/MED1 liver null mice and this is attributed to PB accelerated expansion of the residual PBP/MED1 expressing hepatocytes in PBP/MED1ΔLiv mice. In PBP/MED1fl/fl mice, the histological appearance of liver was unremarkable at 0, 1 and 12 weeks of PB promotion following DEN initiation (Figure 2A and C). All hepatocytes revealed PBP/MED1 nuclear staining (Figure 2B and D). In PBP/MED1ΔLiv mice, nuclear staining of PBP/MED1 was not present in hepatocytes at 0 day (Figure 2E and I), except for positive staining in a rare hepatocyte that apparently escaped Cre-mediated gene deletion. By 1 week of PB promotion, large hepatocytes begin to emerge in DEN-initiated PBP/MED1ΔLiv mouse livers (Figure 2F and J) and they revealed PBP/MED1 nuclear staining implying PBPMED1fl/fl genotype of these cells, in the background of PBP/MED1-deficient hepatocytes (Figure 2J). The PBP/MED1 expressing large hepatocytes in PBP/MED1ΔLiv livers increased in number and began to dominate the liver phenotype in that they formed large expanding colonies between 12 and 52 weeks of PB promotion (Figure 2G, H, K and L). The volume occupied by large PBP/MED1-positive hepatocytes in PBP/MED1ΔLiv mice was estimated by using Scion Image software (Figure 3). The data on PBP/MED1 expressing large hepatocytes obtained at 52 weeks (Figure 3) represents only the proportion of these cells in non-neoplastic areas (see Figure 2H; Figure 4H). These results indicate that few PBP/MED1 expressing hepatocytes present in PBP/MED1ΔLiv mouse livers exhibit profound growth advantage in a milieu where the majority of hepatocytes do not express this coactivator.
Fig. 1.
Liver weight changes. Liver weight:body weight ratio at 0, 1, 4, 12 and 52 weeks in PBP/MED1fl/fl and PBP/MED1ΔLiv mice (*P < 0.05) initiated with DEN and promoted with PB. The large increase in liver weight at 52 weeks in DEN-initiated mice was due to the presence of liver tumors.
Fig. 2.
Sequential histological changes in liver of PBP/MED1fl/fl and PBP/MED1ΔLiv mice. PBP/MED1fl/fl (A–D) and PBP/MED1ΔLiv (E–L) mice initiated with DEN were promoted with PB in drinking water and killed after 0 (A, B, E and I), 1 (F and J), 12 (C, D, G and K) and 52 (H and L) weeks. Liver sections were stained with hematoxylin and eosin (A, C and E–H) or processed for immunohistochemical localization of PBP/MED1 (B, D and I–L). In PBP/MED1fl/fl livers, all hepatocyte nuclei are positive for PBP/MED1 (B and D; see boxed in D). In PBP/MED1ΔLiv mouse livers, large PBP/MED1-positive hepatocytes proliferate and expand in number (see arrows in J and boxed area in H and L). These are distinct from PBP/MED1-deficient small atrophic, relatively basophilic, PBP/MED1-deficient hepatocytes (boxed areas in G and K).
Fig. 3.
Increase in the volume of PBP/MED1-positive large hepatocytes in DEN-initiated and PB promoted PBP/MED1ΔLiv mouse livers as analyzed by Scion Image. The data obtained at 52 weeks represents only non-tumor areas.
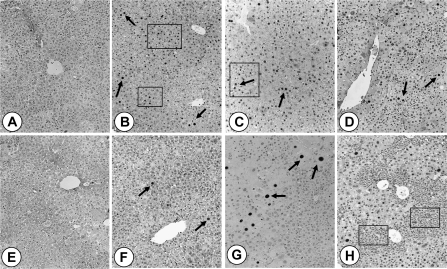
Fig. 4.
Hepatocellular proliferation in PBP/MED1fl/fl and PBP/MED1ΔLiv mouse livers following DEN initiation and PB promotion. Representative illustrations from PBP/MED1fl/fl (A–D) and PBP/MED1ΔLiv (E–H) mouse livers. Hepatocellular proliferation was assessed by BrdUrd labeling at 0 (A and E), 1 (B and F), 12 (C and G) and 52 (D and H) weeks. BrdUrd incorporation was evaluated by immunohistochemistry. Many nuclei are labeled in PBP/MED1fl/fl mouse livers (B–D) at 1 (see arrows and boxed area in B), 12 (see arrows and boxed areas in C) and 52 (arrows in D) weeks when compared with PBP/MED1ΔLiv mouse livers (F–H see arrows). In PBP/MED1ΔLiv livers, BrdUrd labeling was observed only in PBP/MED-positive large hepatocytes (see arrows in F–H). At 52 weeks, the non-tumor areas in PBP/MED1ΔLiv livers expanding populations of large hepatocytes are seen and PBP/MED1-negative hepatocytes are smaller (see boxed in H).
Hepatocellular proliferation was evaluated by BrdUrd incorporation and immunohistochemical staining (Figure 4). Significant increase in BrdUrd nuclear staining was observed at 1 week of PB treatment in PBP/MED1fl/fl livers (Figure 4B) as compared with PBP/MED1ΔLiv mouse livers (Figure 4F), which was similar to that seen in 0 day controls (Figure 4A and E). In 12 week PB promoted livers, the cell proliferation in PBP/MED1fl/fl livers was still higher than in liver conditional nulls (Figure 4C). In PBP/MED1ΔLiv mouse livers, BrdUrd nuclear staining was mostly confined to large PBP/MED1 expressing hepatocytes (Figure 4G) in comparison with random labeling of hepatocytes throughout the liver lobule in floxed controls (Figure 4C). At 52 weeks of PB promotion, hepatocellular proliferation in non-tumorous areas in PBP/MED1ΔLiv mouse livers was more than in non-tumorous areas in PBP/MED1fl/fl livers and this increased BrdUrd nuclear staining liver conditional nulls was due to proliferation of large PBP/MED1-positive hepatocytes and not in smaller hepatocyte population (Figure 4D and H). We carried out terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling staining of liver sections because hematoxylin and eosin stained sections of PBP/MED1ΔLiv mice showed apoptotic alterations involving non-large hepatocyte areas representing PBP/MED1 null cells (not illustrated). It appeared that apoptosis contributed to the progressive reduction in PBP/MED1-deficient hepatocytes and this served as a stimulus for PBP-positive cells to proliferate. On the other hand, proliferation of large PBP/MED1 expressing hepatocytes may exert compressive pressure on smaller PBP/MED1-deleted hepatocytes forcing them to undergo cell death.
PBP/MED1 is necessary for DEN-induced liver carcinogenesis
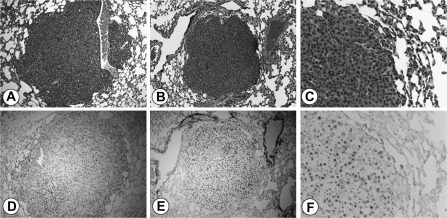
At 52 weeks, the livers of both PBP/MED1fl/fl and PBP/MED1ΔLiv mice that were initiated with DEN and promoted with PB revealed multiple grossly visible tumors that were randomly distributed among all liver lobes. Altered hepatic foci and hepatic adenomas were difficult to delineate due to proliferative expansion of hepatocytes, particularly in PBP/MED1ΔLiv mice. Accordingly, grossly and histologically distinct HCC were evaluated and the HCC load appeared similar in both groups (9.63 ± 2.69 in PBP/MED1fl/fl versus 9.13 ± 2.25 in PBP/MED1ΔLiv mice) (Figure 5A and B). Histologically, in both PBP/MED1fl/fl and PBP/MED1ΔLiv mice, a majority of liver tumors was well to moderately differentiated HCC generally with trabecular pattern (Figure 5C and E). We examined ∼30 tumors from each group for the expression of PBP/MED1 (Figure 5D and F). Interestingly, all liver tumors developed in PBP/MED1ΔLiv mice were PBP/MED1 positive, whereas the surrounding non-tumor portions of liver in these mice did not express this coactivator. Genotyping of microdissected non-tumorous areas confirmed PBP/MED1 gene deletion whereas tumors in these mice had intact gene. Large hepatocytes scattered in predominantly smaller PBP/MED1 gene deleted hepatocytes in non-tumorous areas show PBP/MED1-positive nuclear staining (Figures 2L, 4H and 5E and F). None of the tumors that developed in PBP/MED1ΔLiv mice was PBP/MED1 negative, implying that PBP/MED1 null hepatocytes are resistant to malignant change. As expected, all tumors in the PBP/MED1fl/fl mouse liver expressed this coactivator and all hepatocytes in non-tumor areas were also PBP/MED1 positive (Figure 5D). We found occasional metastasis in lungs of both PBP/MED1fl/fl and PBP/MED1ΔLiv mice bearing HCC (Figure 6A–C). Immunohistochemical staining revealed the expression of PBP/MED1 in these metastatic tumor cells in the lung (Figure 6D–F).
Fig. 5.
DEN induced liver tumors in PBP/MED1fl/fl and PBP/MED1ΔLiv mice. (A) Liver tumor incidence. Data are mean ± SD. (B) Representative gross photograph of liver from both groups. (C–F) Histological appearance (hematoxylin and eosin staining; C and E) and immunohistochemical staining for PBP/MED1 (D and F). Liver tumors (T) developing in both PBP/MED1fl/fl and PBP/MED1ΔLiv mice reveal PBP/MED1 nuclear staining. Non-tumor (NT) areas of PBP/MED1fl/fl (see boxed in D) but not PBP/MED1ΔLiv mouse livers (see boxed in F) showed PBP/MED1 nuclear staining of hepatocytes. PBP/MED1 is labeled as MED1 in panels (A and B).
Fig. 6.
HCC metastasized to lungs. Hematoxylin and eosin staining of lung metastases from PBP/MED1 fl/fl (A) and PBP/MED1ΔLiv (B and C) mice. PBP/MED1 immunostaining of metastatic tumors in the lung of PBP/MED1fl/fl (D) and PBP/MED1ΔLiv (E and F) mice.
HCC implanted from PBP/MED1ΔLiv mouse into nude mouse reveal PBP/MED1-positive phenotype
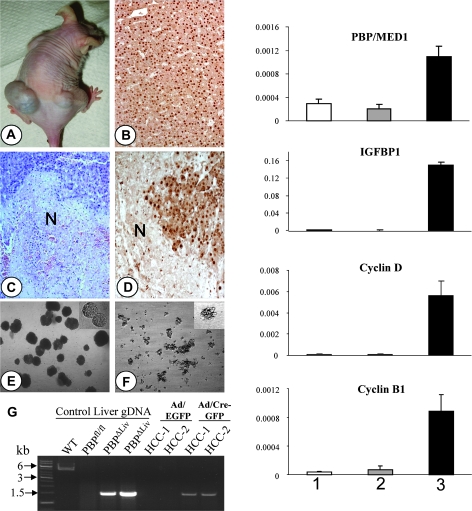
We transplanted small fragments of liver tumors obtained from PBP/MED1ΔLiv mice following DEN initiation and PB promotion into the anterior or posterior flanks of athymic nude mice (Figure 7A and B). By 4–5 weeks after initial transplantation, tumors appeared in the sites of implantation and these grew to 10–40 mm3 by 50 days. These tumors are being maintained by serial transplantation in nude mice and the sixth passage is now in progress without apparent changes in the histological pattern of tumors. Transplanted tumors revealed typical HCC appearance akin to that of HCC in DEN initiated and PB promoted PBP/MED1ΔLiv mice (Figure 7B). These transplanted HCC and PBP/MED1fl/flHCC1 and PBP/MED1fl/flHCC2 cell lines derived from these tumors expressed PBP/MED1 prominently in the nucleus of tumor cells (Figure 7C). We confirmed PBP/MED1 expression in transplanted HCC by qPCR (Figure 7). PBP/MED1 mRNA level in tumors was higher than that in the livers of both PBP/MED1fl/fl and PBP/MED1ΔLiv mice (Figure 7). Furthermore, we analyzed the expression of insulin-like growth factor binding protein 1, cyclin D and cyclin B1 by qPCR (Figure 7). mRNA content of these genes was significantly higher than that seen in PBP/MED1fl/fl and PBP/MED1ΔLiv mouse livers which is typical characteristic of tumor cells.
Fig. 7.
Liver tumor from PBP/MED1ΔLiv mouse transplanted in nude mouse. (A) Third generation transplants (in both flanks) are shown. (B) Typical histological pattern of transplanted tumor expressing PBP/MED1 as assessed by immunohistochemical staining. Panels (C) (hematoxylin and eosin stained) and (D) (PBP/MED1 immunohistochemical staining) of third generation PBP/MED1fl/fl HCC transplants following intratumoral injection of adeno-Cre-GFP. Areas of necrosis (N) represent Cre-mediated deletion of the PBP/MED1fl/fl gene. (E and F) Represent soft agar colony formation assay. Treatment of PBP/MED1fl/flHCC cells with adeno-Cre-GFP caused degeneration and reduction of anchorage-independent colony in soft agar (F) compared with control adeno-EGFP-treated controls (E). (G) Genotyping of HCC lines (HCC-1 and HCC-2) were done with the primers flanking exon 8–10 of PBP/MED1 allele to confirm the deletion of floxed DNA. Infection with Ad/Cre-GFP excised floxed region which resulted into the amplification of mutated band of ∼1.5 kb whereas infection with Ad/EGFP has an intact PBP/MED1 gene which could not be amplified due to large size of amplicon (∼5.2 kb). Liver genomic DNA from wild-type (C57BL6/J), PBP/MED1fl/fl and PBP/MED1ΔLiv mice were used for control. The bar graph panels on the right show the qPCR expression levels of PBP/MED1, insulin-like growth factor binding protein 1, cyclin D and cyclin B1 in PBP/MED1fl/fl (1), PBP/MED1ΔLiv (2) mouse livers and liver tumors transplanted in nude mice (3). Standard error bar represents three independent experiments.
Deletion of PBP/MED1gene in transplanted HCC causes tumor necrosis in vivo and suppresses colony formation in vitro
The failure of PBP/MED1-deleted hepatocytes to give rise to liver tumors by PPARα ligand Wy-14,643 (32), or following initiation by genotoxic carcinogen DEN and promotion by PB as described here, prompted us to explore the possible biological significance of this coactivator in tumor survival. Since HCC transplanted in nude mice express PBP/MED1 and the PBP/MED1fl/fl gene in these cells can be disrupted by adeno-Cre, we injected adeno-Cre-GFP (2 × 1011 virus particles) intratumorally in a volume of 100 μl and the tumor harvested on the third day for histological analysis (Figure 7C). Extensive areas of tumor necrosis were observed (Figure 7C and D) and the viable tumor tissue was immunohistochemically positive for PBP/MED1 (Figure 7D). Deletion of PBP/MED1fl/fl alleles in PBP/MED1fl/flHCC cell lines by adeno-Cre-GFP resulted in marked suppression of colony formation in soft agar (Figure 7F) as compared with cells with intact PBP/MED1fl/fl alleles (Figure 7E), further implying the importance of this coactivator in the maintenance of neoplastic change. PCR-based genotyping confirmed the deletion of floxed PBP/MED1 allele in HCC-1 and HCC-2 cell lines after infection with Ad/Cre-GFP virus resulting into a short band of ∼1.5 kb after the deletion of floxed exons 8–10. However, HCC cell lines infected with Ad/EGFP has intact PBP/MED1 that could not be amplified due to large size of the amplicon (∼5.2 kb) causing PCR constraint (Figure 7G).
Discussion
In the present study, we show that hepatocyte-specific deletion of PBP/MED1fl/fl targeted allele with the Alb-Cre recombinase transgene decreases the susceptibility of PBP/MED1-deficient hepatocytes to genotoxic carcinogen DEN-induced and PB promoted hepatocarcinogenesis. In these PBP/MED1ΔLiv mice, hepatocytes that escaped Cre-mediated deletion of PBP/MED1fl/fl-targeted allele exhibited profound proliferative potential and these cells are responsible for liver tumor development in this DEN-initiation and PB promotion liver tumor model. Accordingly, all liver tumors expressed PBP/MED1 and none were PBP/MED1 negative. Previously, we reported that PBP/MED1-deficient hepatocytes failed to develop liver tumors when chronically exposed to a non-genotoxic hepatocarcinogen that acts by enhancing PPARα signaling (27,32). Absence of PBP/MED1 abrogates PPARα signaling in hepatocytes and accordingly abolishes the PPARα-ligand-induced pleiotropic responses including peroxisome proliferation, induction of fatty acid oxidation enzymes and liver tumor development (27,32). The data presented here extend these observations to demonstrate that PBP/MED1-deficient hepatocytes are also resistant to tumorigenesis initiated by a potent genotoxic hepatocarcinogen DEN and promoted by PB.
The mechanisms underlying the inability of PBP/MED1-deficient hepatocytes to give rise to HCC in response to DEN-initiation and PB promotion can reside at two or more levels. First, the metabolism of the genotoxic carcinogen DEN, which was used for initiation, could be defective in hepatocytes with PBP/MED1 deficiency as compared with PBP/MED1fl/fl hepatocytes in the same PBP/MED1ΔLiv mouse liver. Metabolic activation of DEN occurs in liver within hours after administration via cytochrome P450 (CYP)-dependent α-hydroxylation and oxidation to form reactive products capable of generating mutagenic DNA adducts (40–42). CYPs involved in DEN metabolism include CYP1A1, CYP1A2, CYP2B1, CYP2B2 and CYP2E1 and several of these are regulated by the nuclear receptor CAR (42,43). In PBP/MED1ΔLiv mouse liver, the basal mRNA levels of these CYPs were lower than in PBP/MED1fl/fl livers (30). Parenthetically, it should be noted that these genes are also not inducible in PBP/MED1ΔLiv mouse liver by CAR activators such as PB and the pesticide contaminant 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (30). The diminished basal levels of CYPs may account for a reduction in DEN metabolism and DNA alkylation contributing to lower number of ‘initiated’ cells among PBP/MED1-deficient cells in comparison with PBP/MED1fl/fl hepatocytes in the same liver. This can explain the absence of liver tumors with PBP/MED1-deficient genotype in PBP/MED1ΔLiv mouse liver. However, given that the mice received only a single injection of DEN while they were 14 day old, enzyme induction may not play a major role. Future studies on CYP levels and DEN-related DNA adducts may provide insights into the role of DEN metabolism in HCC development in PBP/MED1ΔLiv mouse liver. Second, it has been suggested that the process of initiation, which follows the primary DNA alkylation reaction, may take several weeks because two or more rounds of cell division are needed to fix the ‘initiation’ (44). Fixation of critical mutation may not occur in PBP/MED1-deficient hepatocytes because of their diminished ability to undergo cell proliferation and this could account for a possible reduction in the initiated cell population (27,30,32). Third, initiated PBP/MED1-deficient cells may be highly susceptible to apoptotic cell death and extinction. We noted a higher incidence of apoptosis in PBP/MED1-deficient cells and attributed this to compressive pressure on hepatocytes with inherent defect to divide slowly. Fourth, in this DEN-initiation–PB promotion liver carcinogenesis model, promotion of initiated cells is an integral part toward liver tumor development and evidence suggests that this may be defective in PBP/MED1-deficient hepatocytes. Promotion of mouse hepatocarcinogenesis by PB involves the selection of hepatocytes carrying activating mutations and such a population of initiated cells serves as a sensitive target for PB tumor promotion. Available evidence indicates that CAR is the molecular target of promotion by PB and that activation of this nuclear receptor is an essential requirement for liver tumor development (38).
The DEN-initiation and PB promotion studies in CAR knockout mice have established that CAR is necessary for liver tumor promotion by PB (39). CAR activation by PB and 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene increases hepatocellular proliferation and upregulates drug-metabolizing enzymes (39,43). For CAR to exert its effects on gene expression in liver, PB and other activators of this receptor have to induce its translocation into hepatocyte nucleus (45). Of interest is studies using PBP/MED1ΔLiv mouse livers have clearly established that PBP/MED1 is essential for the nuclear translocation of CAR into hepatocytes and for the proper functioning of this xenobiotic receptor (30,46,47). In the absence of this coactivator, CAR remains in the cytoplasm and fails to induce CYP1A2, CYP2B10 and other CAR-regulated genes in liver (30,46). Absence of PBP/MED1 in liver cells results in the abrogation of hypertrophic and hyperplastic influences mediated by CAR ligands (30). DEN is a genotoxic chemical carcinogen capable of generating DNA adducts that contribute to mutational events necessary for initiation. In contrast, carcinogenesis by non-genotoxic mechanisms, for example PPARα-and CAR-mediated carcinogenesis, generally involves receptor-mediated cell proliferation and other metabolic events. The DEN-initiated cells can be influenced by a variety of mechanisms that enhance the proliferative capacity of initiated cells. The studies with PBP/MED1 strongly suggest that this coactivator plays a major role in liver tumorigenesis, whether it is genotoxic and non-genotoxic carcinogenesis.
Finally, we have established a transplantable HCC in athymic nude mice that expresses high levels of insulin-like growth factor binding protein 1, cyclin D and cyclin B1 and show that deletion of PBP/MED1fl/fl allele using adeno-Cre results in tumor cell necrosis. Two HCC cell lines with PBP/MED1fl/fl allele have also been generated and preliminary studies suggest that adeno-Cre-mediated disruption of PBP/MED1 gene in these cells inhibits colony formation in soft agar and tumor formation in nude mice. We have previously reported that PBP/MED1 (PPARBP) gene, which is localized to chromosome 17q12, a region known to contain the oncogene erbB-2/HER-2, is amplified and overexpressed in breast cancer (48). Recently, it has been shown that the loss of this coactivator inhibits prostate cancer cell proliferation and survival (49). The available data indicate that PBP/MED1 is important for cell proliferation and neoplastic growth and the absence of this molecule adversely affects cell growth. In contrast, a recent study shows that downregulation of the expression of this coactivator in fact enhances the tumorigenic phenotype of human melanoma cells (50). It is possible that PBP/MED1 may exert different effects in different tumor cell types. Additional studies are needed to understand the function of PBP/MED1 and other essential coactivators such as PRIP (51) in tumor formation, growth and survival in liver and other types of cancers.
Funding
National Institutes of Health (GM23750, DK083163 to J.K.R.).
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BrdUrd
bromodexoyuridine
- CAR
constitutive androstane receptor
- CYP
cytochrome p450
- DEN
diethylnitrosamine
- DMEM
Dulbecco's modified Eagles medium
- EGFP
enhanced green fluorescent protein
- HCC
hepatocellular carcinoma
- MED1
mediator subunit 1
- mRNA
messenger RNA
- PB
phenobarbital
- PBP
peroxisome proliferator-activated receptor-binding protein
- PBP/MED1ΔLiv
PBP/MED1 liver conditional null
- PCR
polymerase chain reaction
- PPAR
peroxisome proliferator-activated receptor
- PRIP
peroxisome proliferator-activated receptor interacting protein
References
- 1.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonoda J, et al. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582:2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKenna NJ, et al. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 4.Lonard DM, et al. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 5.Hermanson O, et al. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- 6.Kornberg RD. Mediator and the mechanisms of transcriptional activation. Trends Biochem. Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Onate SA, et al. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 8.Hörlein AJ, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:387–388. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 9.Chen JD, et al. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 10.Reddy JK, et al. Nuclear receptor transcriptional coactivators in development and metabolism. Adv. Develop. Biol. 2006;16:389–420. [Google Scholar]
- 11.Zhu Y, et al. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 12.Yuan CX, et al. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl Acad. Sci. USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rachez C, et al. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y, et al. Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. J. Biol. Chem. 2000;275:14779–14782. doi: 10.1074/jbc.C000121200. [DOI] [PubMed] [Google Scholar]
- 15.Ito M, et al. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 16.Kuang SQ, et al. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. J. Biol. Chem. 2002;277:45356–45360. doi: 10.1074/jbc.C200509200. [DOI] [PubMed] [Google Scholar]
- 17.Zhu YJ, et al. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J. Biol. Chem. 2003;278:1986–1990. doi: 10.1074/jbc.C200634200. [DOI] [PubMed] [Google Scholar]
- 18.Mahajan MA, et al. Nuclear receptor coactivator/coregulator NCoA6 (NRC) is a pleiotropic coregulator involved in transcription, cell survival, growth and development. Nucl. Recept. Signal. 2008;6:1–19. doi: 10.1621/nrs.06002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonson P, et al. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol. Cell. Biol. 2003;23:1260–1268. doi: 10.1128/MCB.23.4.1260-1268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, et al. Partial hormone resistance in mice with disruption of the steroid receptor coactivator -1 (SRC-1) gene. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 21.Qi C, et al. Mouse steroid receptor coactivator-1 is not essential for peroxisome proliferator-activated receptor α-regulated gene expression. Proc. Natl Acad. Sci. USA. 1999;96:1585–1590. doi: 10.1073/pnas.96.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gehin M, et al. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol. Cell. Biol. 2002;22:5923–5937. doi: 10.1128/MCB.22.16.5923-5937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, et al. Regulation of somatic growth by the p160 coactivator p/CIP. Proc. Natl Acad. Sci. USA. 2000;97:13549–13554. doi: 10.1073/pnas.260463097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, et al. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc. Natl Acad. Sci. USA. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Yadav N, et al. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine mehyltransferase 1-deficient mice. Proc. Natl Acad. Sci. USA. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia Y, et al. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARα-regulated gene expression in liver. J. Biol. Chem. 2004;279:24427–24434. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- 28.Sarkar J, et al. Transcription coactivator PRIP, the peroxisome proliferator-activating receptor (PPAR)-interacting protein, is redundant for the function of nuclear receptors PPARα and CAR, the constitutive androstane receptor, in mouse liver. Gene Expr. 2007;13:255–269. doi: 10.3727/000000006780666948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yakar S, et al. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc. Natl Acad. Sci. USA. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia Y, et al. Transcription coactivator peroxisome proliferator-activated receptor-binding protein/mediator 1 deficiency abrogates acetaminophen hepatotoxicity. Proc. Natl Acad. Sci. USA. 2005;102:12531–12536. doi: 10.1073/pnas.0506000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu S, et al. Transcription coactivators for peroxisome proliferator-activated receptors. Biochim. Biophys. Acta. 2007;1771:936–951. doi: 10.1016/j.bbalip.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto K, et al. Critical role for transcription coactivator peroxisome proliferator-activated receptor PPAR)-binding protein TRAP/220 in liver regeneration and PPARα ligand-induced liver tumor development. J. Biol. Chem. 2007;282:17053–17060. doi: 10.1074/jbc.M701956200. [DOI] [PubMed] [Google Scholar]
- 33.Reddy JK, et al. Hypolipidaemic hepatic peroxisome proliferators form a novel class of chemical carcinogens. Nature. 1980;283:397–398. doi: 10.1038/283397a0. [DOI] [PubMed] [Google Scholar]
- 34.Neumann H-G. Risk assessment of chemical carcinogens and thresholds. Crit. Rev. Toxicol. 2009;39:449–461. doi: 10.1080/10408440902810329. [DOI] [PubMed] [Google Scholar]
- 35.Diwan BA, et al. Interstrain differences in susceptibility to liver carcinogenesis initiated by N-nitrosodiethylamine and its promotions by Phenobarbital in C57BL/6NCr, C3H/HeNCrMTV- and DBA/2NCr mice. Carcinogenesis. 1986;7:215–220. doi: 10.1093/carcin/7.2.215. [DOI] [PubMed] [Google Scholar]
- 36.Tamano S, et al. Rapid development of hepatic tumors in transforming growth factor alpha transgenic mice associated with increased cell proliferation in precancerous hepatocellular lesions initiated by N-nitrosodiethylamine and promoted by phenobarbital. Carcinogenesis. 1994;15:1791–1798. doi: 10.1093/carcin/15.9.1791. [DOI] [PubMed] [Google Scholar]
- 37.Kalinichenko VV, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takami T, et al. Loss of hepatocyte growth factor/c-Met signaling pathway accelerates early stages of N-nitrosodiethylamine-induced hepatocarcinogenesis. Cancer Res. 2007;67:9844–9851. doi: 10.1158/0008-5472.CAN-07-1905. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto Y, et al. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004;64:7197–7200. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
- 40.Verna L, et al. N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol. Ther. 1996;71:57–81. doi: 10.1016/0163-7258(96)00062-9. [DOI] [PubMed] [Google Scholar]
- 41.Kang JS, et al. Role of CYP2E1 in diethylnitrosamine-induced hepatocarcinogenesis in vivo. Cancer Res. 2007;67:11141–11146. doi: 10.1158/0008-5472.CAN-07-1369. [DOI] [PubMed] [Google Scholar]
- 42.Wei P, et al. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- 43.Swales K, et al. CAR, driving into the future. Mol. Endocrinol. 2004;18:1589–1598. doi: 10.1210/me.2003-0397. [DOI] [PubMed] [Google Scholar]
- 44.Marx-Stoelting P, et al. Hepatocarcinogenesis in mice with a conditional knockout of the hepatocyte growth factor receptor c-Met. Int. J. Cancer. 2009;124:1767–1772. doi: 10.1002/ijc.24167. [DOI] [PubMed] [Google Scholar]
- 45.Kawamoto T, et al. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol. Cell. Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo D, et al. Peroxisome proliferator-activated receptor (PPAR)-binding protein (PBP) but not PPAR-interacting protein (PRIP) is required for nuclear translocation of constitutive androstane receptor in mouse liver. Biochem. Biophys. Res. Commun. 2006;347:485–495. doi: 10.1016/j.bbrc.2006.06.129. [DOI] [PubMed] [Google Scholar]
- 47.Jia Y, et al. Conditional ablation of mediator subunit MED1 (MED1/PPARBP) gene in mouse liver attenuates glucocorticoid receptor agonist dexamethasone-induced hepatic steatosis. Gene Expr. 2009;14:291–306. doi: 10.3727/105221609788681213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Y, et al. Amplification and overexpression of peroxisome proliferator-activated receptor binding protein (PBP/PPARBP) gene in breast cancer. Proc. Natl Acad. Sci. USA. 1999;96:10848–10853. doi: 10.1073/pnas.96.19.10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vijayvargia R, et al. A coregulatory role for the mediator complex in prostate cancer cell proliferation and gene expression. Cancer Res. 2007;67:4034–4041. doi: 10.1158/0008-5472.CAN-06-3039. [DOI] [PubMed] [Google Scholar]
- 50.Ndong Jde L, et al. Down-regulation of RB18A/MED1, a cofactor of transcription, triggers strong tumorigenic phenotype of human melanoma cells. Int. J. Cancer. 2009;124:2597–2606. doi: 10.1002/ijc.24253. [DOI] [PubMed] [Google Scholar]
- 51.Zhu YT, et al. PRIP promotes tumor formation through enhancing serum-responsive factor-mediated FOS expression. J. Biol. Chem. 2009;284:14485–14492. doi: 10.1074/jbc.M900935200. [DOI] [PMC free article] [PubMed] [Google Scholar]