Abstract
The NF-κB signaling pathways have a critical role in the development and progression of various cancers. In this study, we demonstrated that the small cell lung cancer cell line (SCLC) H69 expressed a unique NF-κB profile as compared to other cancer cell lines. The p105/p50, p100/p52, c-Rel, and RelB protein and mRNA transcripts were absent in H69 cells but these cells expressed RelA/p65. The activation of H69 cells by lipopolysaccharide (LPS) resulted in the induction of RelB and p100 expression. The treatment also induced the nuclear translocation of RelB without the processing of p100 to p52. Furthermore, LPS induced β1 integrin expression and cellular attachment through an NF-κB-dependent mechanism. Blocking RelB expression prevented the increase in the expression of β1 integrin and the attachment of H69. Taken together, the results suggest that RelB was responsible for the LPS-mediated attachment and may play an important role in the progression of some cancers.
Keywords: LPS, SCLC, NF-κB, p65, p52, RelB, Cellular Attachment, ß1 integrin
Introduction
In mammalian cells, there are five nuclear factor kappaB (NF-κB) family members, RelA/p65, RelB, c-Rel, p105/p50, and p100/p52, and different NF-κB complexes are formed as either homo- or heterodimers [1; 2]. In resting cells, inactive NF-κB dimers are sequestered in the cytosol by IκB family members, which include IκBα, IκBβ, and IκBε [3]. Upon stimulation, NF-κB is generally activated by either the canonical or non-canonical pathway. In the canonical pathway, the IκB kinase (IKK) complex phosphorylates the IκB proteins, which leads to IκB degradation and dissociation from the NF-κB complex. The released NF-κB complex translocates into the nucleus and initiates gene transcription. For the non-canonical pathway, RelB is sequestered in the cytosol by p100. After stimulation, the IKK complex phosphorylates and induces the proteolytic processing of p100 to generate p52. The removal of the ankyrin repeats of p100 allows the RelB/p52 complex to translocate into the nucleus. Both the canonical and non-canonical pathways are central coordinators of the innate and adaptive immune response. In addition, it has become clear that the NF-κB pathway contributes to cancer development, progression, chemoresistance, invasiveness, and metastasis [4; 5; 6; 7; 8].
Lipopolysaccharide (LPS) is a complex glycolipid that is a constituent of the outer membrane of Gram-negative bacteria. LPS provides a highly potent stimulus to cells of the immune system through the toll-like receptor-4 (TLR-4)/NF-κB pathway, resulting in an increased production of proinflammatory cytokines and the expression of cell adhesion molecules [9; 10]. TLRs are centrally involved in the initiation of the innate and adaptive immune response. Recent evidence showed that functional TLRs are also expressed on a wide variety of tumors suggesting that TLRs may play an important role in tumor biology [11; 12]. For example, the treatment of tumor cells with LPS induced the production of the proinflammatory factors, nitric oxide, IL-6 and IL-12, which protected these cells against cytotoxic T-lymphocytes and natural killer cells [13]. Moreover, increased resistance to chemotherapy- or tumor necrosis factor-α (TNF-α)-mediated apoptosis by TLR-signaling was observed in myeloma cells and lung cancer cells [14; 15]. Furthermore the activation of the TLR-4 pathway in epithelial ovarian cancer cells resulted in an increase in chemoresistance [16]. In addition, two earlier studies demonstrated that LPS increased the expression of iNOS, MMP2, and β1 integrin through the NF-κB pathway, which contributed to tumor cell invasion and attachment [17; 18]. However, it is still unknown which NF-κB family members play a central role in the LPS-mediated invasiveness and attachment of cancer cells.
Here, we demonstrate that the SCLC cell line H69 expresses a unique NF-κB profile as compared to other cancer cell lines. Protein and mRNA transcript levels of p105/p50, p100/p52, c-Rel and RelB were absent in the SCLC cell line H69. Only p65 was constitutively expressed. Treatment with LPS activated the NF-κB signaling pathway, as seen by the induction of RelB and p100 expression. Furthermore, LPS induced the expression of β1 integrin and cellular attachment through an NF-κB-dependent mechanism. In addition, β1 integrin expression and attachment in H69 were dependent on RelB expression.
Materials and Methods
Cell culture
Human lung cancer cell lines (H69 and A549), human T-cell leukemia cell lines (Jurkat and HUT78), human B-cell lymphoma cell lines (RL and BJAB), and HEK293A cell line were obtained from ATCC. Cells were cultured in RPMI 1640 with 10% fetal bovine serum, 100 U/ml penicillin, 100μg/ml streptomycin, and 2mM glutamine and incubated at 37 °C in a humidified atmosphere of 5% CO2.
Immunoblotting and Immunoprecipitiation
Immunoblot and immunoprecipitation was performed as previously described [19]. Unless indicated otherwise in the figure legend, whole cell lysate (50 μg) was re solved with NuPAGE Novex Bis-Tris Gels (Invitrogen), transferred to PVDF, and probed with the antibodies indicated in the figure legends (see Supplemental Table 1 for antibody information). The antibodies were detected by chemifluorescence and scanned with the Typhoon scanner (Amersham Biosciences). The membranes were stripped and reblotted with antibodies against β-tubulin to monitor loading consistency. The quality of the subcellular fractionation procedure was assessed by stripping and reblotting the membrane with GAPDH (cytoplasmic) and NPM (nuclear) antibodies. For the immunoprecipitation experiments, cytoplasmic and nuclear extract from LPS treated samples were precleared with Protein A/G beads (Santa Cruz Biotechnology) for 1 hour with rotation. The precleared samples were incubated with three microgram of the indicated antibodies. After overnight rotation, 40 μl of Protein A/G beads were added for an additional 4 hours. The beads were washed three times with RIPA buffer and then eluted with sample buffer.
Plasmids and Transfections
Transient transfections were performed with the Lipofectamine-2000 reagent according to the protocol provided by the manufacturer (Invitrogen). Cells were transiently transfected with the dominant negative mutant IκBα-M (Clontech) or RelB (Open Biosystems) expression plasmids.
Quantitative Real-time PCR
Total RNA was isolated with the RNeasy Mini Kit (Qiagen). Complementary DNA (cDNA) was synthesized from 1μg of total RNA using SuperScript III reverse transcriptase kit and random hexamers (Invitrogen) in a total volume of 20μl. One-hundredth of the sample was used in a real-time PCR reaction containing a 0.2μM concentration of both forward and reverse primers and iTaq SYBR Green Supermix with ROX (Bio Rad). Quantitation of fold induction was analyzed by the 2−ΔΔCT method[20]. The quantity of each cDNA was normalized to actin. The primer sequences are listed in the Supplemental Table 2[21].
Attachment Assay
The attachment assay was performed as described with the following modifications [18; 22]. H69 cells (0.5 × 106 cells) were transfected with the pCMV β-galactosidase plasmid with or without IκBα-M, RelB and pEF6 (empty vector) plasmids. After 4 hours incubation, H69 cells were stimulated with LPS (1 μg/ml) for an additional 68 hours. LPS from Escherichia coli serotype 0111:B4 was purchased from DIFCO. Adherent cells were then washed gently with PBS, and analyzed by measuring β-galactosidase activity (Promega). Fibronectin (Sigma) coated-culture flasks were used for this experiment.
NF-κB p65 transcription factor assay
The DNA binding activity of p65 was measured by the TransAM NF-κB p65 transcription factor assay (Active Motif, Carlsbad, CA). The specificity of the DNA binding assay was determined using NF-κB consensus and mutant oligonucleotides (supplemental figure 4).
Statistical analyses
Experiments were carried out a minimum of three times with consistent results. Final results were expressed as ± standard error of mean (SEM). Experimental results were compared by two-tailed, unpaired Student t tests with significance at p<0.05.
Results
H69 has a unique NF-κB profile
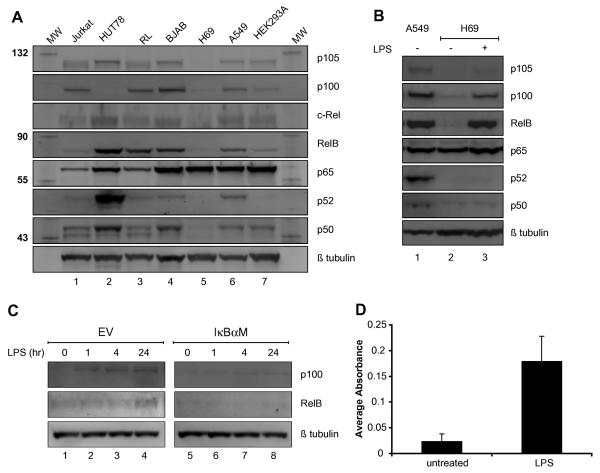
Several studies have shown that the NF-κB proteins may play a role in the progression of lung cancer [23; 24]. Total cell lysates from various cancer cell lines were analyzed by immunoblot for their NF-κB protein profile. As seen in Figure 1a, all of the cancer cell lines analyzed expressed two or more of the various NF-κB proteins except for the human SCLC cell line H69 (see lane 5). Although there were quantitative and qualitative differences among the various cell lines, the amounts of p105, p100, c-Rel, RelB, p52, and p50 were significantly lower or not detectable in H69 (compare lane 5 to other lanes of Fig. 1A). The only NF-κB protein expressed in H69 was p65. The relative amount of p65 mRNA transcripts was comparable in all cell lines (Supplemental Fig. 1). In contrast, the transcripts of the other NF-κB genes were either absent or barely detectable in H69. Furthermore, a similar expression profile of NF-κB proteins, with the exception of c-Rel, was observed with another human SCLC cell line H345 (Supplement Fig. 2).
FIGURE 1. LPS induced RelB and p100 expression in H69.
(A) NF-κB expression profile of H69. Western blot of various NF-κB proteins in seven cell lines. Lane 1, Jurkat (T cell leukemia); Lane 2, HUT78 (cutaneous T cell); Lane 3, RL (diffuse B cell lymphoma); Lane 4, BJAB (Burkitt's lymphoma); Lane 5, H69 (small cell lung cancer); Lane 6, A549 (lung adenocarcinoma); Lane 7, HEK293A (kidney). The molecular weights (KD) of the marker proteins are indicated at left. (B) Induction of RelB and p100 following LPS stimulation. Western blot of whole cell lysates from A549 and H69 cells with or without LPS (1μg/ml) treatment for 24 hours. Lane 1, A549; Lane 2, H69; Lane 3, H69 with LPS. (C) IκBα-M transfection inhibited RelB and p100 induction following LPS treatment. Western blot analysis of whole cell lysates of H69 transfected with either the EV (empty control vector) or IκBaM with LPS (1μg/ml) for indicated time periods (0, 1, 4 and 24 hours). The membranes were probed with the indicated antiserum at right. (D) An increase in p65 DNA binding activity after LPS treatment. The DNA binding activity of p65 in nuclear extracts of H69 that were either untreated or treated with LPS (1 μg/ml) for 2 hours was assessed.
LPS stimulation induced RelB and p100
Based on the NF-κB expression profile, we investigated if H69 could respond to stimuli that activate the NF-κB signaling pathway. To study this, H69 cells were treated with LPS for 24 hours and the induction of NF-κB family proteins was analyzed by immunoblot. In particular, it has been shown that transcription of the RelB gene is regulated by p65 [25]. The treatment of H69 with LPS resulted in the expression of p100 and RelB proteins (Fig. 1B). In accordance, an increase in p100 and RelB transcripts was observed following LPS treatment (Supplemental Fig. 3A). Interestingly, an increase in c-Rel mRNA transcript was also observed; however, the amount of c-Rel protein did not increase after LPS treatment. To determine if the induction of RelB and/or p100 by LPS was dependent on the canonical NF-κB pathway, the non-degradable, dominant negative IκBα mutant (IκBα-M) was expressed in H69. As seen in Figure 1C, the amount of RelB and p100 protein following LPS treatment was reduced in cells transfected with IκBα-M. Likewise, LPS-mediated induction of RelB and p100 transcripts was reduced with the expression of IκBα-M (Supplemental Fig. 3B). In accordance with the activation of the NF-κB signaling pathway, an increase in p65 DNA binding activity was observed in H69 cells following LPS treatment for 2 hours (Fig. 1D and Supplemental Fig. 4). These findings showed that the induction of RelB and p100 by LPS was dependent on the canonical NF-κB signaling pathway, and p65 was sufficient for the NF-κB-mediated induction of RelB and p100 expression by LPS.
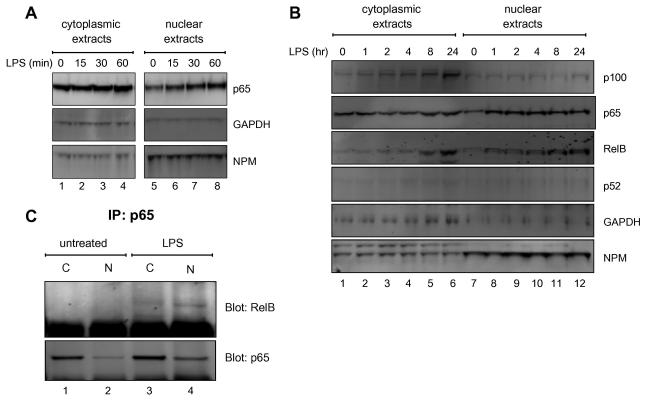
The proteolytic cleavage of p100 to p52 is a pivotal step in the activation of the non-canonical NF-κB pathway, which leads to the nuclear translocation of the RelB/p52 complex. Since RelB and p100 were induced following LPS treatment, it was conceivable that the LPS could activate the non-canonical NF-κB pathway. To further investigate this possibility, the nuclear translocation of RelB and p52 were analyzed in cytoplasmic and nuclear extracts from H69 cells treated with LPS. A rapid translocation of p65 into the nucleus was detected as early as 15 minutes after LPS treatment (Fig. 2A, compare the lanes 5 through 8 of the nuclear p65 panel and supplement Fig. 5). In contrast, LPS gradually induced an increase in the amount of RelB protein in both the cytoplasmic (see lanes 1 to 6 in Fig. 2B) and nuclear extracts (see lanes 7 to 12 in Fig. 2B and supplement Fig. 5). Although LPS induced an increase of cytoplasmic p100, p52 was not detected in either cytoplasmic or nuclear extracts, which indicated that the p100 was not processed to p52. These findings suggested that LPS induced the nuclear translocation of the de novo synthesized RelB in a p52-independent manner.
FIGURE 2. Nuclear translocation of p65 and RelB following LPS stimulation.
Short (A) and long (B) kinetics of p65 and RelB nuclear translocation following LPS stimulation. Western blot analysis of cytoplasmic and nuclear extracts of H69 cells stimulated with LPS (1μg/ml) for the indicated time periods. Twenty five microgram of total protein was loaded into each well. The membrane was probed with the indicated antiserum (right). The levels of GAPDH and NPM (nucleophosmin) are shown as a fractionation control of cytoplasmic and nuclear extracts, respectively. (C) Formation of nuclear RelB/p65 heterodimers following LPS treatment. H69 cells were stimulated with or without LPS (1μg/ml) for 24 hours. Nuclear extracts from cells treated with LPS (1 μg/ml) for 24 hours were immunoprecipitated with anti-p65 immunoprecipitates (IP) and then analyzed by Western blot for the presence of RelB and p65.
Several studies have shown that RelB is unstable and does not form functional homodimers in vivo [13; 26]. Without the processing of p100 to p52, our findings suggested that the nuclear translocation of RelB could be facilitated by p65 following LPS treatment. To investigate this, co-immunoprecipitation experiments of RelB /p65 heterodimers formation were performed after LPS treatment. As seen in Figure 2C, RelB was associated with p65 after LPS treatment. Moreover, the RelB/p65 heterodimer was detected in both cytoplasmic and nuclear extracts only in LPS treated cells. Taken together, these findings suggested that RelB associated with p65 and translocated into the nucleus following LPS treatment in the absence of p100 processing.
LPS-induced β1 integrin and cell attachment
Previous studies have shown that LPS induced cellular attachment in several cancer cell lines [17; 18]. H69 is normally a non-adherent cell line grown in suspension (Supplement Fig. 6). An attachment assay was performed on H69 cells treated with LPS to investigate if LPS could increase cell adhesion in a p65-mediated manner. As seen in Figure 3A and Supplemental Figure 6, treatment with LPS resulted in an increase in cell attachment of H69 cells in wells that were coated with fibronectin. Furthermore, several studies have shown that integrins promote cell attachment to the extracellular matrix (ECM). Accordingly, an increase in β1 integrin transcript and protein were observed following LPS treatment (Fig. 3B & C). To investigate whether the LPS-induced cell attachment and β1 integrin expression were dependent on the NF-κB pathway, H69 cells were transfected with IκBα-M and treated with LPS. A reduction in LPS-mediated cell attachment was observed following the transfection of IκBα-M (Fig. 3A). As expected, the expression of IκBα-M reduced the amount of LPS-induced β1 integrin transcripts (Fig. 3C). Together, these findings indicated that the treatment of H69 with LPS resulted in an increase of β1 integrin expression and cell attachment that was dependent on the NF-κB pathway.
FIGURE 3. LPS induced attachment and β1 integrin up-regulation.
(A) Attachment assay of H69 treated with LPS (1 μg/ml). Cells were transfected with either EV or IκBα-M expression plasmid and ß-galactosidase reporter plasmid, and then plated in fibronectin-coated wells. The β-galactosidase activity reflects the amount of attachment of transfected cells as described in the Material and Methods section. (B) Western blot analysis of H69 with LPS (1μg/ml). The membrane was probed with the indicated antiserum at right. (C) RTq-PCR analysis of H69 cells cultured in uncoated wells. Cells were treated with LPS (1μg/ml) for 24 hours in uncoated wells. The fold induction represents the amount of b1 integrin induction over the H69 without LPS.
RelB expression induced attachment and β1 integrin
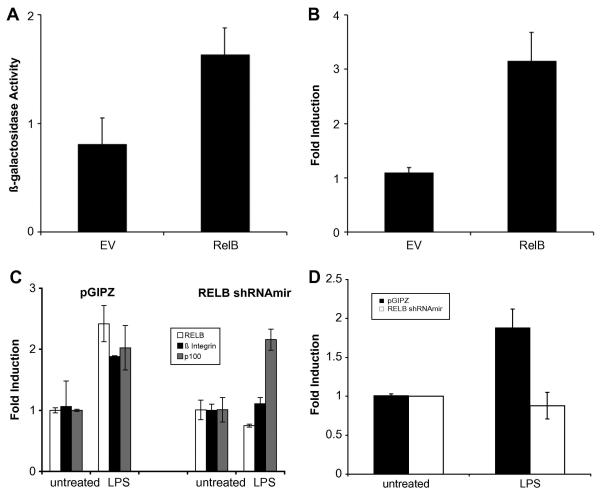
The treatment of H69 with LPS resulted in the nuclear translocation of p65, which induced the expression of RelB. It was conceivable that RelB was responsible for the induction of β1 integrin expression and cellular attachment by LPS. To investigate this, RelB was expressed in H69 without LPS treatment, and cellular attachment and β1 integrin expression were assessed. The expression of RelB resulted in an increase in cell attachment in wells coated with fibronectin (Fig. 4A). Moreover, an increase in β1 integrin transcripts was observed following the expression of RelB (Fig. 4B). These findings demonstrated that the expression of RelB was sufficient to induce an increase in cell attachment and β1 integrin transcription. Furthermore, these results suggested that the action of LPS could be mediated by the expression of RelB. In agreement, LPS did not induce ß1 integrin expression (Fig. 4C) and cell attachment (Suppl. Fig. 7) in H69 cells expressing the inhibitory RelB shRNAmir construct. In contrast, the induction of p100 by LPS, which is p65- but not RelB-dependent, was not affected in these cells. Taken together, the expression of ß1 integrin and cell attachment following LPS treatment were dependent on RelB.
FIGURE 4. RelB expression induced attachment and ß1 integrin expression.
(A) Attachment assay of H69 transfected with either the control vector (EV) or RelB and plated in fibronectin-coated plates. (B) RTq-PCR analysis of EV and RelB transfected H69 cells. Twenty-four hours after transfection (in uncoated plates), cDNA was analyzed by RTq-PCR. The fold induction represents the amount of indicated β1 integrin induction over the H69 without transfection. (C) RTq-PCR analysis of control (pGIPZ) and RELB shRNAmir tranfected H69 cells for RELB, ß1 integrin, and p100 transcripts. Total RNA from transiently transfected cells treated with LPS (1 μg/ml) overnight was used to generate cDNA for RTq-PCR analysis. (D) The relative amount of HIF1-a induction was assessed by RT-qPCR as described in the Materials and Methods section.
It is not clear if RelB directly regulates ß1 integrin expression, although the evidence in this report demonstrated the requirement of RelB for the induction of ß1 integrin by LPS. A recent report has mapped the promoter region of the ß1 integrin (ITGB1) gene and demonstrated the recruitment and binding of Hypoxia-induced factor 1 alpha (HIF-1α) to this region[30]. In addition, several studies have shown that the activation of the NF-κB signaling pathway by LPS resulted in an increase in HIF-1α expression in normoxic conditions [31; 32]. Furthermore, the regulation of HIF-1α by NF-κB transcription factors and the recruitment of RelA and RelB to the HIF-1α promoter were reported [33]. Since the ITGβ1 promoter does not contain NF-κB consensus sequences, it is conceivable that the LPS-mediated induction of ITGβ1 expression could be through the expression of HIF-1a. In support of this hypothesis, an increase in HIF-1α mRNA transcripts was observed in H69 cells following LPS treatment (Fig. 4D). Moreover, the LPS-mediated induction of HIF-1α was abrogated when RelB expression was inhibited by the RelB shRNAmir (Fig. 4D). These findings suggested that RelB indirectly regulated ß1 integrin expression through the induction of HIF-1α.
Discussion
This study has shown that the SCLC line H69 possessed a unique NF-κB protein profile where p65 is the only NF-κB member expressed. Further, the expression of p65 was sufficient for the activation of the NF-κB pathway and the induction of RelB and p100 following LPS treatment. In addition, an increase in β1 integrin expression and cellular attachment was induced by either LPS treatment or RelB expression.
The NF-κB expression profile was not unique to H69 and was observed in another SCLC line H345. In contrast, all of the NF-κB member proteins were observed in the lung adenocarcinoma cell line A549. Lung cancers, in general, can be divided into several histologic subtypes, and it is unclear if these findings are common or are restricted to a subset of small cell lung cancers. The re-examination of the publicly accessible gene expression profile of tissue samples from normal and lung cancer patients diagnosed with the various subtypes [27] has shown that the transcript expression profiles from SCLC samples expressed lower levels of NF-κB family members when compared to other subtypes of lung cancer samples. Although the protein levels were not analyzed, this finding is consistent with the NF-κB profile observed in H69 and H345. It is not clear if other SCLC cell lines or patient samples express a similar NF-κB profile and respond to LPS by expressing RelB and/or p100.
The treatment of H69 with LPS resulted in the translocation of p65 and RelB into the nucleus without the proteolytic processing of p100 to p52. The nuclear translocation of RelB was thought to require the processing of p100 to p52, and this is a hallmark for the activation of the non-canonical NF-κB pathway. Our findings suggested that the nuclear translocation of RelB was independent of p52 and the non-canonical NF-κB pathway. Immunoprecipitation analysis following LPS treatment of H69 revealed that RelB was associated with p65 (Fig 2). It is conceivable that the p65-induced RelB associates with p65 following LPS treatment and this complex translocates into the nucleus and induces transcription. Our findings suggested that LPS induced two waves of NF-κB dimers based on the kinetics of p65 and RelB (Fig. 4). In general, during the initial phase, p65 exists as a homodimer in resting cells (see supplemental Fig. 8), which translocates into the nucleus after LPS treatment. Subsequently, the de novo synthesized RelB associates with cytosolic p65, which results in the nuclear translocation of the RelB/p65 heterodimer. It is noteworthy that the induction of ß1 integrin occurred much later than the expression of RelB and the association between RelB/p65 following LPS treatment. The interconversion of p65 between the p65 homodimer and p65/RelB heterodimer may play a pivotal role in regulating the expression of different subsets of NF-κB responsive genes. It is unclear if the RelB/p65 heterodimer is transcriptionally active. Marienfeld et al.[28] have shown that RelB suppressed p65 by forming transcriptionally inactive RelB/p65 heterodimers in several mouse cell lines. Alternatively, Jacque et al.[29] have shown the RelA (p65) inhibited RelB activity following TNF treatment in mouse fibroblasts. In both reports, the formation of RelB/p65 heterodimers resulted in the inhibition of the NF-κB activity. In contrast, our data showed that the formation of the RelB/p65 heterodimer was associated with an increase in cell attachment and ß1 integrin expression in an NF-κB-dependent manner. The differences between our results and the previous reports could be explained based on the species studied (mouse versus humans). It is more likely that the differences could be explained by the absence of p105/50 in H69 cells. In the prior studies[28; 29], the mode of inhibition by either RelB or RelA was due to the displacement of p50 from the existing p65/p50 or RelB/p50 heterodimers. This was not observed in our study due to the lack of p105/50. It is of interest to determine if the LPS-induced p65/RelB is functional and promotes the transcription of the HIF-1α gene.
Several studies have shown that the NF-κB dimer composition affects the progression of cancer [5; 34]. Here we described the expression of a unique NF-κB profile in the SCLC cell line H69, where p65 was the only member expressed. Furthermore LPS stimulation resulted in expression of RelB and p100 and the subsequent formation of RelB/p65 heterodimers. In addition, LPS treatment or RelB expression resulted in an increase in ß1 integrin expression and cell attachment. These findings suggested that the induction of RelB and ß1 integrin expression were associated with the presence of the p65 homodimer and RelB/p65 heterodimer, respectively, following LPS treatment. Furthermore, it is possilble that the different subclasses of lung cancer can be distinguished from each other based on their NF-κB expression profile.
Supplementary Material
Acknowledgements
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 3.Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH. Negative regulation of toll-like receptor signaling by NF-kappaB p50 ubiquitination blockade. Science. 2007;317:675–8. doi: 10.1126/science.1142953. [DOI] [PubMed] [Google Scholar]
- 4.Stathopoulos GT, Sherrill TP, Han W, Sadikot RT, Yull FE, Blackwell TS, Fingleton B. Host nuclear factor-kappaB activation potentiates lung cancer metastasis. Mol Cancer Res. 2008;6:364–71. doi: 10.1158/1541-7786.MCR-07-0309. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Belguise K, Kersual N, Kirsch KH, Mineva ND, Galtier F, Chalbos D, Sonenshein GE. Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2. Nat Cell Biol. 2007;9:470–8. doi: 10.1038/ncb1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn KS, Sethi G, Aggarwal BB. Reversal of chemoresistance and enhancement of apoptosis by statins through down-regulation of the NF-kappaB pathway. Biochem Pharmacol. 2008;75:907–13. doi: 10.1016/j.bcp.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–47. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 8.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 9.Perera PY, Mayadas TN, Takeuchi O, Akira S, Zaks-Zilberman M, Goyert SM, Vogel SN. CD11b/CD18 acts in concert with CD14 and Toll-like receptor (TLR) 4 to elicit full lipopolysaccharide and taxol-inducible gene expression. J Immunol. 2001;166:574–81. doi: 10.4049/jimmunol.166.1.574. [DOI] [PubMed] [Google Scholar]
- 10.Cross JL, Kott K, Miletic T, Johnson P. CD45 regulates TLR-induced proinflammatory cytokine and IFN-beta secretion in dendritic cells. J Immunol. 2008;180:8020–9. [Google Scholar]
- 11.Huang B, Zhao J, Unkeless JC, Feng ZH, Xiong H. TLR signaling by tumor and immune cells: a double-edged sword. Oncogene. 2008;27:218–24. doi: 10.1038/sj.onc.1210904. [DOI] [PubMed] [Google Scholar]
- 12.Chen R, Alvero AB, Silasi DA, Steffensen KD, Mor G. Cancers take their Toll--the function and regulation of Toll-like receptors in cancer cells. Oncogene. 2008;27:225–33. doi: 10.1038/sj.onc.1210907. [DOI] [PubMed] [Google Scholar]
- 13.Huang B, Zhao J, Li H, He KL, Chen Y, Chen SH, Mayer L, Unkeless JC, Xiong H. Toll-like receptors on tumor cells facilitate evasion of immune surveillance. Cancer Res. 2005;65:5009–14. doi: 10.1158/0008-5472.CAN-05-0784. [DOI] [PubMed] [Google Scholar]
- 14.Jego G, Bataille R, Geffroy-Luseau A, Descamps G, Pellat-Deceunynck C. Pathogen-associated molecular patterns are growth and survival factors for human myeloma cells through Toll-like receptors. Leukemia. 2006;20:1130–7. doi: 10.1038/sj.leu.2404226. [DOI] [PubMed] [Google Scholar]
- 15.He W, Liu Q, Wang L, Chen W, Li N, Cao X. TLR4 signaling promotes immune escape of human lung cancer cells by inducing immunosuppressive cytokines and apoptosis resistance. Mol Immunol. 2007;44:2850–9. doi: 10.1016/j.molimm.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Kelly MG, Alvero AB, Chen R, Silasi DA, Abrahams VM, Chan S, Visintin I, Rutherford T, Mor G. TLR-4 signaling promotes tumor growth and paclitaxel chemoresistance in ovarian cancer. Cancer Res. 2006;66:3859–68. doi: 10.1158/0008-5472.CAN-05-3948. [DOI] [PubMed] [Google Scholar]
- 17.Harmey JH, Bucana CD, Lu W, Byrne AM, McDonnell S, Lynch C, Bouchier-Hayes D, Dong Z. Lipopolysaccharide-induced metastatic growth is associated with increased angiogenesis, vascular permeability and tumor cell invasion. Int J Cancer. 2002;101:415–22. doi: 10.1002/ijc.10632. [DOI] [PubMed] [Google Scholar]
- 18.Wang JH, Manning BJ, Wu QD, Blankson S, Bouchier-Hayes D, Redmond HP. Endotoxin/lipopolysaccharide activates NF-kappa B and enhances tumor cell adhesion and invasion through a beta 1 integrin-dependent mechanism. J Immunol. 2003;170:795–804. doi: 10.4049/jimmunol.170.2.795. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki CY, Barberi TJ, Ghosh P, Longo DL. Phosphorylation of RelA/p65 on serine 536 defines an I{kappa}B{alpha}-independent NF-{kappa}B pathway. J Biol Chem. 2005;280:34538–47. doi: 10.1074/jbc.M504943200. [DOI] [PubMed] [Google Scholar]
- 20.Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 21.Cui W, Taub DD, Gardner K. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007;35:D805–9. doi: 10.1093/nar/gkl767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buller CJ, Zang XP, Howard EW, Pento JT. Measurement of beta-galactosidase tissue levels in a tumor cell xenograft model. Methods Find Exp Clin Pharmacol. 2003;25:713–6. doi: 10.1358/mf.2003.25.9.793338. [DOI] [PubMed] [Google Scholar]
- 23.King KE, Ponnamperuma RM, Allen C, Lu H, Duggal P, Chen Z, Van Waes C, Weinberg WC. The p53 homologue DeltaNp63alpha interacts with the nuclear factor-kappaB pathway to modulate epithelial cell growth. Cancer Res. 2008;68:5122–31. doi: 10.1158/0008-5472.CAN-07-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tew GW, Lorimer EL, Berg TJ, Zhi H, Li R, Williams CL. SmgGDS regulates cell proliferation, migration, and NF-kappaB transcriptional activity in non-small cell lung carcinoma. J Biol Chem. 2008;283:963–76. doi: 10.1074/jbc.M707526200. [DOI] [PubMed] [Google Scholar]
- 25.Bren GD, Solan NJ, Miyoshi H, Pennington KN, Pobst LJ, Paya CV. Transcription of the RelB gene is regulated by NF-kappaB. Oncogene. 2001;20:7722–33. doi: 10.1038/sj.onc.1204868. [DOI] [PubMed] [Google Scholar]
- 26.Ruben SM, Klement JF, Coleman TA, Maher M, Chen CH, Rosen CA. I-Rel: a novel rel-related protein that inhibits NF-kappa B transcriptional activity. Genes Dev. 1992;6:745–60. doi: 10.1101/gad.6.5.745. [DOI] [PubMed] [Google Scholar]
- 27.Virtanen C, Ishikawa Y, Honjoh D, Kimura M, Shimane M, Miyoshi T, Nomura H, Jones MH. Integrated classification of lung tumors and cell lines by expression profiling. Proc Natl Acad Sci U S A. 2002;99:12357–62. doi: 10.1073/pnas.192240599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marienfeld R, May MJ, Berberich I, Serfling E, Ghosh S, Neumann M. RelB forms transcriptionally inactive complexes with RelA/p65. J Biol Chem. 2003;278:19852–60. doi: 10.1074/jbc.M301945200. [DOI] [PubMed] [Google Scholar]
- 29.Jacque E, Tchenio T, Piton G, Romeo PH, Baud V. RelA repression of RelB activity induces selective gene activation downstream of TNF receptors. Proc Natl Acad Sci U S A. 2005;102:14635–40. doi: 10.1073/pnas.0507342102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keely S, Glover LE, MacManus CF, Campbell EL, Scully MM, Furuta GT, Colgan SP. Selective induction of integrin beta1 by hypoxia-inducible factor: implications for wound healing. FASEB J. 2009;23:1338–46. doi: 10.1096/fj.08-125344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–11. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peyssonnaux C, Cejudo-Martin P, Doedens A, Zinkernagel AS, Johnson RS, Nizet V. Cutting edge: Essential role of hypoxia inducible factor-1alpha in development of lipopolysaccharide-induced sepsis. J Immunol. 2007;178:7516–9. doi: 10.4049/jimmunol.178.12.7516. [DOI] [PubMed] [Google Scholar]
- 33.van Uden P, Kenneth NS, Rocha S. Regulation of hypoxia-inducible factor-1alpha by NF-kappaB. Biochem J. 2008;412:477–84. doi: 10.1042/BJ20080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landowski TH, Olashaw NE, Agrawal D, Dalton WS. Cell adhesion-mediated drug resistance (CAM-DR) is associated with activation of NF-kappa B (RelB/p50) in myeloma cells. Oncogene. 2003;22:2417–21. doi: 10.1038/sj.onc.1206315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.