Abstract
As shown by X-ray crystallography, horse liver alcohol dehydrogenase undergoes a global conformational change upon binding of NAD+ or NADH, involving a rotation of the catalytic domain relative to the coenzyme binding domain and the closing up of the active site to produce a catalytically efficient enzyme. The conformational change requires a complete coenzyme and is affected by various chemical or mutational substitutions that can increase the catalytic turnover by altering the kinetics of the isomerization and rate of dissociation of coenzymes. The binding of NAD+ is kinetically limited by a unimolecular isomerization (corresponding to the conformational change) that is controlled by deprotonation of the catalytic zinc-water to produce a negatively-charged zinc-hydroxide, which can attract the positively-charged nicotinamide ring. The deprotonation is facilitated by His-51 acting through a hydrogen-bonded network to relay the proton to solvent. Binding of NADH also involves a conformational change, but the rate is very fast. After the enzyme binds NAD+ and closes up, the substrate displaces the hydroxide bound to the catalytic zinc; this exchange may involve a double displacement reaction where the carboxylate group of a glutamate residue first displaces the hydroxide (inverting the tetrahedral coordination of the zinc), and then the exogenous ligand displaces the glutamate. The resulting enzyme-NAD+-alcoholate complex is poised for hydrogen transfer, and small conformational fluctuations may bring the reactants together so that the hydride ion is transferred by quantum mechanical tunneling. In the process, the nicotinamide ring may become puckered, as seen in structures of complexes of the enzyme with NADH. The conformational changes of alcohol dehydrogenase demonstrate the importance of protein dynamics in catalysis.
Keywords: Enzyme mechanism, Enzyme activation, Protein structure, Crystallography, Kinetic simulation Isomerization, Proton relay, Zinc coordination
Conformational flexibility has long been recognized as an important feature of enzyme catalysis and has been invoked to explain substrate specificity (“induced fit”), catalytic efficiency, and allosteric effects. X-ray crystallography has demonstrated that many enzymes change conformation upon binding substrates, but it is more problematic to explain how and why the changes are important for catalysis. Some binding energy must be used to bring about the conformational changes, and in principle this would reduce the catalytic efficiency, but it could be a small cost for creating the environment required for catalysis. In this review, we will focus on aspects of three conformational changes that occur during catalysis by liver alcohol dehydrogenase. The major emphasis will be on the global domain motion and the associated loop rearrangement that occur when coenzyme binds (producing a more efficient catalyst). We will also discuss a potential local conformational change during exchange of ligands at the catalytic zinc and the subtle puckering of the nicotinamide ring during hydride transfer.
Structural foundations
Liver alcohol dehydrogenase was one of the first enzymes for which different conformational states for apo- and holo-enzymes were determined by X-ray crystallography [1, 2]. A recent review summarizes several different aspects of the structures of the alcohol dehydrogenases [3]. The dimeric horse liver apoenzyme has two identical protein subunits, each of which has two domains. The conformational change can be described as a rigid body rotation of about 10° around an axis between the catalytic and coenzyme binding domains (Fig. 1), which closes up the active site [4, 5]. The apoenzyme typically crystallizes in the orthorhombic C2221 space group with one subunit in the “open” conformation in the asymmetric unit, whereas the holoenzyme complexes crystallize in the triclinic P1 or monoclinic P21 space groups with one or two molecules in the “closed” conformation in the asymmetric unit [6]. The loop containing residues 292-299 of the coenzyme binding domain also rearranges so that the catalytic domain can move, and this results in the isopropyl group of Val-294 flipping from a position buried in the interface between the coenzyme binding domains to a position in the active site under the nicotinamide ribose (Fig. 2). Some amino acid residues from the catalytic domain move about 5 Å closer to the coenzyme binding domain, and with substrate bound in the active site, water is excluded from the site of hydride transfer (Fig. 3). The proton relay system involving Ser-48, the 2′-hydroxyl group of the coenzyme and His-51 is also installed, which can function to relay a proton to solvent from the water or alcohol bound to the catalytic zinc (Fig. 4) [7]. Since the catalytic mechanism is ordered, with coenzyme binding before the substrate, the conformational change could be required to set up the enzyme for catalysis. Nevertheless, it is not clear that the conformational change is absolutely required, as the catalytic residues are in relatively similar positions in both the open and closed conformations. How much activity might the enzyme have in an open conformation?
Fig. 1.
Global conformational change when ADH binds coenzyme. The dimer in red is the apoenzyme conformation (1JU9.pdb [65]), and the dimer in green is the holoenzyme with the zincs and coenzyme shown in space-filling representation (1HLD.pdb [20]). The coenzyme binding domains (Coe) are superimposed. The major change is apparent from the shifts in the catalytic domains (Cat). The loop containing residues 292 to 299 (290's) is indicated. The figures were prepared with Molray [109].
Fig. 2.
Conformational change of the flexible loop. Residues 291 to 300 from the apoenzyme (1YE3.pdb) are shown in red, and those from holoenzyme (1HLD.pdb) are shown in standard colors. Note that the rearrangement flips the position of Val-294, which forms a floor under the nicotinamide ribose and protrudes into the substrate binding site (PFB, 2,3,4,5,6-pentafluorobenzyl alcohol).
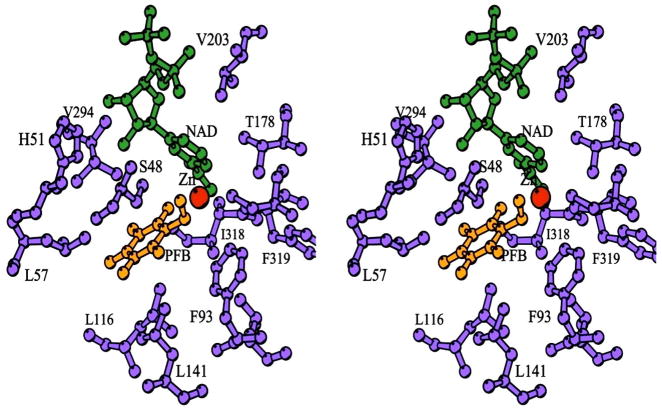
Fig. 3.
Binding of 2,3,4,5,6-pentafluorobenzyl alcohol in the active site of horse liver alcohol dehydrogenase. Water is excluded from the site of hydride transfer. This complex has an orientation that resembles the presumed Michaelis complex. The figure is based on 1HLD.pdb [20]. (Stereo image is for cross-eyed viewing.)
Fig. 4.
Hydrogen bonded system in the active site. The hydroxyl group of the alcohol is connected (red dashed lines) via Ser-48 and the 2′-hydroxyl group of the coenzyme to His-51, which contacts a solvent water on the surface of the protein (based on 1HLD.pdb). This system could function as a relay system to allow the alcohol to deprotonate and transfer the proton to solvent. See Scheme 2. The black dotted line connects C4 of the nicotinamide ring to C7 of the benzyl alcohol to indicate the hydride transfer (3.4 Å).
Binding of the complete coenzyme is critical for producing the conformational change in horse liver alcohol dehydrogenase. The apoenzyme binds adenosine diphosphoribose and remains in the open conformation, but binding of NAD+ or NADH along with a substrate or substrate analog produces enzyme that crystallizes in the closed conformation [6, 8, 9]. Co-crystallization with NAD+ or NADH alone also gives the closed conformation, but it is not known which form of coenzyme is present in such crystals, as NAD+ can be reduced by the precipitant, 2-methyl-2,4-pentanediol or contaminating alcohols, and NADH can be oxidized over the days required for crystallization [7, 10, 11]. Crystallization with NADH in the presence of imidazole, which binds to the catalytic zinc and hinders binding of the nicotinamide ring, yields the apoenzyme conformation [12]. Enzyme crystallized with the tetrahydronicotinamide adenine dinucleotide (H2NADH) was in the open conformation, but enzyme with H2NADH and the substrate N,N-dimethylcinnamaldehyde was closed [13]. NAD analogs with a thiazole-4-carboxamide or its selenium derivative in place of the nicotinamide ring formed crystals with the open conformation [14]. Isosteric C-glycosidic analogs of NAD, where the pyridine nitrogen is moved to the 2 or 5 position of the ring, crystallized with enzyme in the closed form [15]. However, the F93W:V203A doubly mutated enzyme crystallized with the analog with the N in the 2 position in the open form, apparently due to some subtle effect of the mutations [16]. Thus, it appears that the nicotinamide ring must bind properly to allow the conformational change and that the conformational equilibrium is sensitive to small alterations in structures.
A variety of ligands can form ternary complexes that crystallize in the closed form. Complexes with NAD+ and pyrazoles, 2,2,2-trifluoroethanol, 2,3,4,5,6-pentafluorobenzyl alcohol, 4-bromobenzyl alcohol and cholic acid, or with NADH and dimethyl sulfoxide, tetramethylene sulfoxides, or formamides (R2S=O or RNHCHO, analogs of the carbonyl substrates) are in the closed conformation [4, 10, 16-23]). It is worth noting that the nicotinamide rings in complexes with NAD+ and a pyrazole or with NADH and (R)-N-1-methylhexylformamide are not planar, but rather are puckered, boat-like conformations [18, 23]. One N of the pyrazole forms a partial covalent bond (1.7 Å) with C4 of the nicotinamide ring, and the complex absorbs at 292 nm, in between the maximum of 260 nm for the oxidized nicotinamide ring and the maximum of 325 nm for enzyme-bound NADH [18, 24]. This complex may resemble the transition state for the catalytic reaction. The complex with NADH and the formamide was the first example of a puckered, reduced nicotinamide ring in a dehydrogenase where there was no evidence of a chemically modified ring [23]. The ring puckering may be relevant for catalysis as it can decrease the distance between C4 of the nicotinamide ring and the reacting C of the substrate (Fig. 5). In order to properly refine the puckered structures with the CCP4 Refmac5 program, we removed some restraints on the CCP4 Refmac5 dictionary for the NAD. Careful analysis of structures of other enzymes determined by X-ray crystallography shows other examples of puckered nicotinamide rings [25]. These ternary complexes form the basis of our understanding of structures of ternary complexes that should resemble the Michaelis complexes and be relevant for catalysis.
Fig. 5.
Structure of the horse liver alcohol dehydrogenase complexed with NADH and N-isopropylformamide. Note that the nicotinamide ring is puckered. The dotted black line connects C4 of the nicotinamide ring to C1 of the formamide to illustrate hydride transfer (3.6 Å). The figure is based on 1P1R.pdb [23].
Since the catalytic zinc ion is close to C5 of the nicotinamide ring (3.34 Å) and binds the N or O of the ligands in the complexes, it would seem that the zinc would also be critical for the conformational change. The catalytic zinc is tightly bound to the protein by the sulfhydryl groups of Cys-46 and Cys-174 and the imidazole of His-67, in a distorted tetrahedral coordination where the exogenous ligand (water, substrate, inhibitor) occupies the fourth position. Surprisingly, removal of the zinc ion or substitution with Cd(II) or Cu(II) produces enzymes that form the closed conformation with NADH [26-28]. Perhaps the sulfhydryl groups of the cysteines that ligate the catalytic zinc or the substituted metals can replace the functionality of the zinc, at least with respect to the conformational change.
The conformational change is a general property of the dimeric alcohol dehydrogenases. The homologous human and cod class I liver alcohol dehydrogenases also crystallize in the closed conformation when complexed with coenzyme, but the extents of closure can differ somewhat as compared to the horse liver enzyme [29-33].
Kinetic mechanism
Steady-state kinetics
The minimal kinetic mechanism for horse liver alcohol dehydrogenase is an Ordered Bi Bi reaction, with the addition of an isomerization step (step 2) for the enzyme-NAD+ complex, which probably reflects the conformational change shown by X-ray crystallography (Scheme 1). (We use “isomerization” to refer to a kinetic event, whereas “conformation” refers to a structural change.)
Scheme 1.
In this mechanism, the dissociation of the product coenzyme in either direction is a major rate-limiting step for turnover, as shown originally by Theorell and Chance [34]. The Theorell-Chance mechanism, however, assumed that formation of the central, ternary complexes was not kinetically significant. Careful initial velocity studies (varying substrate concentrations) supported the Theorell-Chance mechanism, but raised the possibility that the enzyme-coenzyme complexes isomerized [35]. These studies found that the rate constant for dissociation of NAD+ calculated from steady-state results was somewhat slower than the observed turnover number for the reaction of NADH and acetaldehyde, but it was concluded that it was not possible to distinguish between mechanisms that had an isomerization or an inactive (dead-end) enzyme-coenzyme complex. Product inhibition studies demonstrated the kinetic significance of the ternary (enzyme-coenzyme-substrate) complexes and suggested that the enzyme-NAD+ and maybe the enzyme-NADH complexes isomerized, again on the basis of the inconsistency between the turnover number and calculated rate constants [36]. With the isomerization step as shown in Scheme 1, the rate constant calculated from steady-state data for NAD+ dissociation is koff = V1Kia/EtKa = k−1k2k−2/(k−1 + k2)(k2 + k−2), which is less than k−1, and the rate constant for binding of NAD+ is kon = V1/EtKa = k1k2/(k−1 + k2), where V1/Et is the turnover number for reaction of NAD+ and alcohol, Ka is the Michaelis constant for NAD+, and Kia is the “inhibition” (dissociation) constant for NAD+. If the enzyme-NAD+ complex isomerizes, isomerization of the enzyme-NADH complex would not be detected by the inconsistency in observed and calculated rate constants [36].
More complicated mechanisms are possible, as steady-state kinetic studies also show that abortive enzyme-NAD+-aldehyde and enzyme-NADH-alcohol complexes can form when the concentrations of substrates are very high [36-38]. NADH dissociates more slowly from the abortive enzyme-NADH-alcohol complex when the alcohol is ethanol (producing substrate inhibition), but faster (giving substrate activation) when the alcohol is cyclohexanol [37, 38]. Benzyl alcohol also forms an abortive enzyme-NADH-alcohol complex and inhibits dissociation of NADH [11]. Human ADH1C also forms these abortive complexes, resulting in kinetic cooperativity [39]. In these studies, the possibility was raised that the mechanism might be random, with either alcohol or NAD+ binding to free enzyme, but it appears that the binding of NAD+ to the enzyme-alcohol complex is much slower than to free enzyme [11, 39, 40]. In contrast, binding of an aldehyde does not inhibit binding of NADH, and the mechanism of reaction of NADH and aldehyde may be random, but conforming to an ordered mechanism because dissociation of NADH from the ternary enzyme-NADH-aldehyde complex is slower than the dissociation of the aldehyde [41]. For the purposes of the present discussion, it is sufficient to note that the minimal ordered mechanism given in Scheme 1 explains all of the steady-state kinetic data collected with concentrations of substrates that do not greatly exceed the Km values. Nevertheless, justification for the mechanism, in particular the isomerization of the enzyme-NAD+ complex, requires additional information that transient kinetics provides.
Transient kinetics
The binding of NAD+ was studied with a stopped-flow instrument by following the increase in absorbance at 290 nm due to trapping of the enzyme-NAD+ complex with pyrazole [42]. By varying the concentration of NAD+, a limiting first order reaction with a rate constant of about 100 s−1 was observed, which was assigned at that time to an isomerization of the enzyme-NAD+-pyrazole complex [43]. Subsequent investigation, using a stopped-flow instrument with a shorter dead-time and binding experiments that did not include pyrazole, demonstrated that the isomerization of the enzyme-NAD+ complex has a limiting first order rate constant of about 500 s−1 [44]. Kinetic simulations using KINSIM and FITSIM [45] of a family of progress curves provided estimates of the rate constants for the initial bimolecular binding step and the isomerization, thus defining rate constants for steps 1 and 2 in the mechanism in Scheme 1 (Table 1). A previous study, using pressure relaxation, obtained similar rate constants [46]. In contrast, transient kinetic studies on the binding of NADH (monitoring quenching of protein fluorescence or the shift in spectrum of bound NADH) showed a linear dependence on the concentration of NADH up to an apparent rate constant of 1000 s−1 (i.e., no limiting rate or saturation), and the bimolecular association rate constant is 1.1 × 107 M−1s−1 [47]. Apparently, the binding of NADH is followed by a rapid isomerization step. (However, later work suggested the binding of NADH is limited by an isomerization with a rate constant of about 1300 s−1 [48]). The rate constant for dissociation of the enzyme-NADH complex was also determined by mixing preformed enzyme-NADH complex with varied concentrations of NAD+ and pyrazole, which traps the free enzyme, and following the change in absorption at 293 or 355 nm [49]. These transient studies characterize the binding of coenzyme and the isomerization of the enzyme-NAD+ complex for the mechanism in Scheme 1, with the rate constants given in Table 1. The rate constant for the reverse isomerization, k−2, is about the same magnitude as the turnover number for acetaldehyde reduction (Table 2), suggesting that the isomerization is the major rate limiting step for wild-type enzyme. The ratio of rate constants for step 2 indicates that the equilibrium position favors the isomerized form of the complex by a factor of 10.
Table 1.
Estimated rate constants for ethanol oxidation and acetaldehyde reduction for the mechanism shown in Scheme 1a
| Rate constant | ethanol/acetaldehyde |
|---|---|
| k1 (M−1s−1) | 4.5 × 107 |
| k−1 (s−1) | 2.3 × 104 |
| k2 (s−1) | 620 |
| k−2 (s−1) | 64 |
| k3 (M−1s−1) | 2.4 × 105 |
| k−3 (s−1) | 560 |
| k4 (s−1) | 490 (150) |
| k−4 (s−1) | 610 (360) |
| k5 (s−1) | 64 |
| k−5 (M−1s−1) | 9.2 × 104 |
| k6 (s−1) | 5.5 |
| k−6 (M−1s−1) | 1.1 × 107 |
Data are from ref. [47]. Values for step 4 in parentheses are for deuterio substrates where the rate constants for the other steps were fixed for the simulation. From these values, the overall rate constant for binding of NAD+ is given by kon = k1k2/(k−1 + k2), and for dissociation by koff = k−1k2k−2/(k−1 + k2)(k2 + k−2), and the dissociation constant, Kia = k−1k−2/k1(k2 + k−2). Note that the values for k1 and k−1 are highly correlated, but the ratio is well determined.
Table 2.
Steady state kinetic constants for wild-type and modified liver alcohol dehydrogenases acting on ethanol and acetaldehyde a
| Kinetic constant | WTb | PI-AIc | I269Sd | V292Se | G293A/P295Tf |
|---|---|---|---|---|---|
| Ka (μM) | 3.9 | 420 | 1000 | 130 | 1900 |
| Kb (mM) | 0.35 | 18 | 11 | 26 | 710 |
| Kp (mM) | 0.40 | 5.7 | 11 | 3.3 | 110 |
| Kq (μM) | 5.8 | 125 | 570 | 95 | 510 |
| Kia (μM) | 27 | 640 | 9500 | 1000 | 1100 |
| Kiq (μM) | 0.50 | 32 | 180 | 50 | 320 |
| V1/Et (s−1) | 3.5 | 32 | 90 | 44 | 5.2 |
| V2/Et (s−1) | 47 | 560 | 1500 | 480 | 260 |
| V1/EtKb (mM−1s−1) | 10 | 1.8 | 8.4 | 1.7 | 0.0073 |
| V2/EtKp (mM−1s−1) | 120 | 97 | 140 | 150 | 2.4 |
| k4, apparent (s−1) | 180g | 32 | 90 | 44 | 5.2 |
| H/D isotope effect on k4 | 3.8g | 4.8 | 4.0 | 2.8 | 4.8 (30 °C) |
| Ki AMP (μM) | 41d | 300 | 790 | 86 | |
| Ki CF3CH2OH (μM)h | 8.4 | - | 7.8 | - | 4100 |
| kon, NAD+ (μM−1s−1)i | 1.2 | 0.21 | 0.047 | 0.11 | 0.027 |
Kinetic constants were determined at 25 °C in 33 mM sodium phosphate and 0.25 mM EDTA buffer, pH 8.0. Ka, Kb, Kp, Kq are the Michaelis constants for NAD+, alcohol, aldehyde, and NADH, respectively. Kia and Kiq are the inhibition constants for NAD+ and NADH, respectively. V1/Et is the turnover number for alcohol oxidation, and V2/Et is the turnover number for aldehyde reduction. The standard errors for the fitted values were 10-25% of the values.
Wild-type enzyme [58].
Picolinimidylated, acetimidylated enzyme [43].
I269S enzyme [63].
V292S enzyme, unpublished data, but see [65]
Doubly-mutated enzyme [66]
Apparent rate constant for transient oxidation of alcohol and the observed isotope effect [47]. For the other enzymes, no burst phase was observed and the steady-state turnover was governed by hydride transfer as shown by the isotope effects.
2,2,2-CF3CH2OH was tested as competitive inhibitor against ethanol.
Apparent rate constant for binding of NAD+, determined by transient kinetics by trapping with pyrazole.
The rate constants for binding and reaction of alcohols and aldehydes, including the interconversion of the ternary complexes, were estimated by simulations and global fitting of a series of transient reactions of NAD+ and alcohol or NADH and aldehyde where the substrate concentrations were varied [47]. In such simulations, the rate constants for coenzyme binding steps are fixed. These simulations give small standard errors of the fits, but some rate constants are correlated and may represent lower limits of the constants. Estimates for the rate constants for the hydride transfer steps were validated by using the deuterated substrates and showing that isotope effects were observed on the transients and reflected in the rate constants. The rate constants (Table 1) also were used to calculate the steady-state kinetic constants (Table 2) and the overall equilibrium constant (Haldane) determined independently, with good agreement. Rate constants describing the ethanol/acetaldehyde substrate pair are given in Table 1. Comparable values have been estimated previously for different conditions [50].
Modifications of amino acid residues that affect coenzyme binding and the conformational states
Lys-228 in the adenosine monophosphate binding site
Before the advent of site-directed mutagenesis, chemical modification was often used to identify amino acid residues that participated in enzyme catalysis. Typically, these modifications decreased or eliminated enzyme activity and led to a classification of a residue as “essential” for catalysis [51]. Of course, it was recognized that simply blocking the active site could inactivate an enzyme, and that modification of an amino acid residue adjacent to, or even distant from the active site, could modulate activity. Nevertheless, it was a surprise when amidination of amino groups of horse liver alcohol with methyl picolinimidate increased catalytic activity by 20-fold! Further work showed that the mechanism remained Ordered Bi Bi and that the enzyme was activated because coenzymes were bound less tightly and released much faster than for wild-type enzyme, so that hydride transfer became rate-limiting for alcohol oxidation [43, 52]. Differential labeling (first modify amino groups in the presence of coenzyme, then remove the coenzyme and modify with a different reagent) showed that modification of Lys-228 in the adenosine ribose phosphate binding site (Fig. 6) was responsible for the change in activity [53-55]. Various modifications of Lys-228 could affect the activity to different extents, and some modified enzymes were used for studies on the mechanism and pH dependencies of hydride transfer [56-60]. An X-ray structure determined at 3.2 Å for enzyme in which 23 of the 30 lysine residues per subunit were isonicotinimidylated showed that the apoenzyme was in the open conformation, and NADH could only be diffused into the crystals with low occupancy in the active site without causing the crystals to shatter [61]. Co-crystals with NADH were not obtained. These structural results show that modification of the lysine in the active site would interfere with binding of the coenzyme, as deduced from examining a structure of a ternary complex of ADH at high resolution [4, 20].
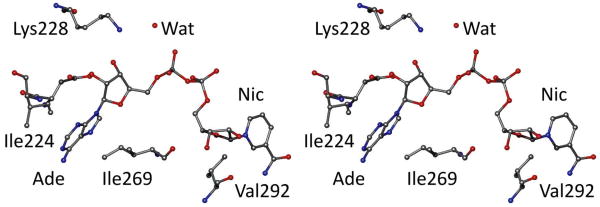
Fig. 6.
Selected amino acid residues in the NAD binding site. Isoleucine residues 224 and 269 make a sandwich of the adenine ring. Lys-228 interacts with the 3′-hydroxyl group of the adenosine ribose and via a water molecule with a phosphate oxygen. Val-292 is near the nicotinamide ring. The figure is based on 1HLD.pdb.
Kinetic constants for a picolinimidylated enzyme are given in Table 2. Affinity for coenzymes decreased 23 to 64-fold, and part of this is accounted for by a 7-fold decrease in affinity for AMP. Because the turnover number for acetaldehyde reduction (V2/Et) increased by 12-fold as compared to wild-type enzyme, it is clear that the reverse isomerization step (k−2) has increased in this enzyme (to at least 560 s−1). Nevertheless, catalytic efficiency for ethanol oxidation only decreased 5-fold, and this indicates that the enzyme can adopt a catalytically active conformation that is about the same as that of wild-type enzyme. The overall rate constant for the binding of NAD+ was determined by stopped-flow experiments to be 2.1 × 105 M−1s−1, with a limiting rate constant (k2) of 155 s−1, which is slower than the constant of 620 s−1 for wild-type enzyme (Table 1). Although simulations of the mechanism with isomerization of the enzyme-NAD+ complex were not done at that time, we can now interpret the data in terms of the mechanism in Scheme 1, by using the catalytic efficiency for ethanol as a measure of the fraction of enzyme in the E*-NAD+ form that can bind and react with ethanol. Since V1/EtKb is decreased by about 5-fold, it appears that the enzyme-NAD+ complex is about 20% E*-NAD+ and 80 % E-NAD+, and from this equilibrium position, we estimate that k−2 is 650 s−1. This is a reasonable value as the turnover number for acetaldehyde reduction is 560 s−1, and thus k−2 may be a major rate-limiting step in turnover. The microscopic rate constants for k1 and k−1 were not calculated, but the ratio can be estimated to be 0.77 mM (see equations for kon and Kia in Table 1). This work shows that modifications of the AMP binding site that weaken binding of NAD+ can affect modestly the conformational change, which nevertheless still occurs during catalysis.
Ile-269 in the adenine binding site
When site-directed mutagenesis became feasible [62], we could design substitutions that specifically probed coenzyme binding and catalysis. Substitutions of either of the two leucine residues that make a sandwich of the adenine ring of NAD in horse ADH (Fig. 6), with the glycine and serine residues found in yeast ADH, decrease affinity for coenzymes and increase catalytic activity, but not to the extent found in the yeast enzyme [63]. The turnover numbers for the I269S enzyme are about 30-fold higher than those for wild-type enzyme, and affinity for coenzymes decreases 400-fold, but the mechanism remains ordered, and an enzyme was produced for which the rate-limiting step for the oxidation of ethanol is hydride transfer (Table 2). Part (about 50-fold) of the decreased affinity for coenzymes is explained by the altered affinity for the adenine nucleotide, but the isomerization step (k−2) substantially increases as the turnover number for acetaldehyde reduction increases 30-fold, to 1500 s−1. The overall rate constant for binding of NAD+, determined by transient kinetics, decreases about 25-fold. Remarkably, catalytic efficiency for ethanol (V1/EtKb) is the about the same as that for wild-type enzyme, and furthermore, the binding constant for 2,2,2-trifluoroethanol is the same. These results show again that binding of NAD+ is coupled to the isomerization, but the binding may not affect the position of the isomerization as long as the complete coenzyme with the nicotinamide ring is binding. Saturation of the enzyme with coenzyme will lead to predominantly E*-NAD+. It is interesting that the I224G enzyme, which has decreased affinity for coenzymes and catalytic efficiency for ethanol that is about the same as that for wild-type enzyme, has isotope effects that are about the same as those for the parent enzyme, showing that distal substitutions may have little effect on the quantum mechanical hydrogen tunneling of liver alcohol dehydrogenase [64].
Val-292 in the nicotinamide binding site
Val-292 is about 4 Å away from the nicotinamide ring in ternary complexes, on the face opposite to the one that reacts with substrates (Fig. 6). It is also in the loop of the coenzyme binding domain that rearranges when the enzyme changes conformation (Fig. 2) [65]. The V292S enzyme is similar in many respects to the picolinimidylated enzyme in that affinity for coenzymes decreases 15 to 110-fold, turnover numbers increase 10 to 12-fold, catalytic efficiency for alcohol oxidation decreases 6-fold, and affinity for pentafluorobenzyl alcohol decreases 4-fold relative to wild-type enzyme (Table 2) [65]. There is no transient burst phase for ethanol oxidation, and turnover of ethanol exhibits an H/D isotope effect of 2.8, indicating that hydride transfer is a major rate-limiting step. The kinetic mechanism fits the ordered reaction. The observed rate constant for binding of NAD+ is 1.1 × 105 M−1s−1, indicating a kinetically significant isomerization step, but no limiting rate at high concentrations of NAD+ was observed. The turnover number with acetaldehyde indicates that k−2 is at least of 480 s−1. By the same reasoning applied for the picolinimidylated enzyme, we can estimate the equilibrium position for the isomerization favors E-NAD+ over E*-NAD+ by about 5:1. In other words, about 20% of the enzyme is in the catalytically active form. Supporting this conclusion, X-ray crystallography showed that enzyme saturated with NAD+ and pentafluorobenzyl alcohol was in the open conformation. However, only the ADP portion of the NAD was observed in the electron density map, and no defined density was observed in the active site for pentafluorobenzyl alcohol. Apparently, the nicotinamide ribose and benzyl alcohol are not bound in one predominant position. The structural basis for the altered conformational equilibrium is not obvious, but the hydrophobic interactions near the nicotinamide ring may be less favorable in the mutated enzyme. In any case, it appears from the kinetic data that some fraction of the enzyme can adopt the active, closed conformation. The question arises, then, if enzyme that is locked in the open conformation can be active.
290s loop in the coenzyme binding domain
This issue was explored by doing partially random mutagenesis of amino acid residues in the loop of the coenzyme binding domain that rearranges during the conformational change (Figs. 1 and 2). Residues 293 and 295 were targeted, and the doubly mutated G293A/P295T enzyme was found to have enhanced activity [66]. As for the other activated enzymes in Table 1, this enzyme has decreased affinity for coenzymes and increased turnover numbers, but catalytic efficiency for ethanol is decreased by 1400-fold and affinity for trifluoroethanol is decreased by 490-fold. The rate constant for binding of NAD+ is 2.7 × 104 M−1s−1. The mechanism remains Ordered Bi Bi, as determined with dead-end inhibition studies, but substantial H/D isotope effects are observed on V1/Et, V1/EtKb, and V1/EtKa. Crystals were prepared with concentration of trifluoroethanol sufficiently high to saturate the enzyme (400 mM), but the structure of the complex was in the open conformation, with only the ADP portion of NAD visible in the electron density maps. Inspection of the structure of the loop region shows that the structures of the open conformations of the wild-type and mutated enzyme are very similar, but the P295T substitution allows a new water molecule to bind. If the mutated enzyme were to change conformation as does wild-type enzyme, steric conflicts would be generated with Ala-293 and Thr-295, and therefore it seems reasonable to conclude that the double mutant is “locked” in an open conformation. Additional evidence in support of this conclusion is that the pH dependence for V1/Et shows maximal activity above a pK value of 8.4, two units higher than that observed for wild-type enzyme. This result appears to indicate that His-51 is not installed in the hydrogen-bonded proton relay system that is observed in the closed conformation (Fig. 4). A catalytic role for His-51 in wild-type enzyme is consistent with the observation that the H51Q enzyme has 35-fold decreased catalytic efficiency (V1/EtKb) at pH 7 and maximal activity above a pK value of 8.4 [67]. In the H51Q and G293A/P295T enzymes, the pK of the zinc-alcohol appears to control the reaction. In the G293A/P295T enzyme, we propose that a failure of the enzyme to change conformation and put His-51 in its proper place produces a handicapped enzyme with a 1400-fold decrease in V1/EtKb. However, the observed rate constant for hydride transfer from the alcohol only decreases from 180 s− 1 to 5.2 s−1 (35-fold). The enzyme may have very low activity in the open state, but direct evidence for the activity of the open conformation is lacking.
pH Dependence of NAD+ Binding and Isomerization
His-51 in the proton relay system
Since the binding of NAD+ results in the conformational change, and His-51 is involved in the process, it was of interest to investigate which groups control the binding of NAD+. The equilibrium dissociation constant (equivalent to Kia) shows highest affinity at high pH and lower affinity at low pH [68]. The rate constants for binding of NAD+ show a bell-shaped pH dependence with pK values of about 7 and 9 [35, 44]. One interpretation of these results is that a group on the free enzyme, perhaps the zinc-water, with a pK of 9 is shifted to a pK of 7 in the enzyme-NAD+ complex [69-71]. However, X-ray crystallography shows that His-51 and Lys-228 participate directly in binding of NAD+, and their contributions to the pH dependence were therefore investigated by using the H51Q and K228R enzymes [72]. The H51Q substitution causes a large effect, decreasing the rate constant for binding of NAD+ at pH 8 from 1.2 μM−1s−1 to 0.09 μM−1s−1 and producing a pH dependence with maximal rates above a pK of 8.0. The doubly mutated enzyme, H51Q/K228R, is similar, but the rate constant decreases to 0.011 μM−1s−1 and the pK value increases to 9.0. In the doubly mutated enzyme, it seems likely that the ionization of the catalytic zinc-water is controlling NAD+ binding. His-51 appears to facilitate the deprotonation as shown in Scheme 2. The ligand to the zinc can be water or alcohol, and in the absence of NAD(H) in the apoenzyme, a water molecule substitutes for the 2′-hydroxyl group (1YE3.pdb). This mechanism is in principle the same as the one proposed by Brändén et al., except for the additional hydroxyl group in the relay [9]. Interestingly, the rate constants and pH dependence for binding of NADH are only modestly affected by these substitutions. The steady-state kinetic constants for the H51Q enzyme are very similar to those of wild-type enzyme [72], and the three dimensional structures of ternary complexes with NAD+ and difluorobenzyl alcohols are essentially superimposable with corresponding structures of wild-type enzyme [67]. These studies show that the proton relay system (Scheme 2) is important for controlling the overall binding of NAD+, but leave open the question of how the conformational change is controlled.
Scheme 2.
Catalytic zinc-water and proton release
Since a macroscopic pK value of about 9.2 in apoenzyme is shifted to a pK value of about 7.6 in the enzyme-NAD+ complex, accompanied by release of about one proton per subunit [73, 74], proton release could control the conformational change or vice versa. The sequence of events was investigated by transient kinetic studies of proton release and protein fluorescence or UV absorption during the binding of NAD+ and formation of ternary complexes [75]. The critical experiments illustrated in Fig. 7 show a biphasic proton release and uptake when caprate and NAD+ bind, with varied concentrations of ligands. The families of transient progress curves were fitted by kinetic simulation to provide estimates of the rate constants for each step in the mechanism. Several different kinetic mechanisms were examined, and the simplest mechanism that fits the data is shown in Scheme 3, where a rate-limiting deprotonation of the HE-NAD+ complex (k2, about 230 s−1) is coupled to the conformational change and occurs before the caprate binds. The initial deprotonation is coincident with changes in protein fluorescence and absorption at 280 nm, which seem to report on the protein conformation [60, 76, 77]. In this case, the kinetic “isomerization” is concluded to be equivalent to the structural “conformational change”. The binding of NAD+ appears to be in rapid equilibrium (K1 = 0.67 mM), and pK2 for the HE-NAD+ complex is 7.3. The initial proton release phase is followed by proton uptake (equivalent to hydroxide dissociation) associated with caprate binding, demonstrating that the conformational change occurs before caprate binds. A similar mechanism was obtained for binding of 2,2,2-trifluorethanol and pyrazole, except that they only bind preferentially (about 10 times faster) to the deprotonated E-NAD+ complex as compared to HE-NAD+, and only a proton release phase is associated with their binding. The simplest interpretation is that the E-NAD+ complex has a zinc-hydroxide, and trifluoroethanol and pyrazole deprotonate upon binding to produce a water and the zinc alkoxide or pyrazolate.
Fig. 7.
Transient and simulated progress curves for reaction of ADH with NAD+ and caprate. Points are experimental and lines are simulated progress curves for absorbance at 559 nm due to proton release and uptake with phenol red as indicator (A, B), protein fluorescence (C, D), and absorbance at 280 nm (E, F). Enzyme was allowed to react with two different concentrations of caprate and three or four different concentrations of NAD+ in the three different exxperiments. The lines are calculated using estimated microscopic rate constants for the mechanism in Scheme 3. The reactions were carried out in 33 mM Na2SO4 and 0.25 mM EDTA, pH 8.0 at 25° C. (Reprinted with permission from Kovaleva, E. G., and Plapp, B. V. (2005) Biochemistry 44, 12797-12808 [75]. Copyright 2005 American Chemical Society.)
Scheme 3.
Although we propose that the proton is released from the zinc-water, His-51 facilitates the process as the H51Q enzyme only binds NAD+ about 1/12 as fast as wild-type enzyme does, and the substitution dramatically shifts the pH dependencies. A simple structural explanation is that His-51 facilitates the relay of the proton from the zinc-water to solvent, and the formation of the zinc-hydroxide attracts the positively-charged nicotinamide ring to a position near the zinc in a reaction coupled to the conformational change (Scheme 4). Whether or not the conformational change or the deprotonation occurs first is an open question, but it does not appear likely that the H51Q substitution would hinder the conformational change, whereas it would affect proton transfer. Thus it appears that deprotonation occurs first and controls the global conformational change. The structural change proposed in Scheme 4 is consistent with the observation that for those complexes of modified enzymes with NAD that crystallize in the open conformation, the position of the nicotinamide ring is not apparent in the electron density maps, but in complexes with the closed conformation the nicotinamide ring is next to the zinc (e.g.,refs. [65, 66].
Scheme 4.
Conformational changes during exchange of zinc ligands
After the deprotonation and the global conformational change, it appears that the ligands, caprate, pyrazole or trifluoroethanol, bind most rapidly to enzyme with the zinc-hydroxide [75]. The mechanism of this exchange is uncertain, however. Perhaps it involves the formation of a transient pentacoordinated zinc, which was proposed before a three-dimensional structure for a ternary complex was determined [59]. Some flexibility in zinc ligation is possible, as the complex of apoenzyme with 1,10-phenanthroline is pentacoordinate [78]. However, in ternary complexes with coenzyme and substrate analogs, there is very little room to move a water past a zinc-bound ligand (e.g., see Fig. 3, ref. [4, 7, 20]. Nevertheless, spectroscopic studies suggest that pentacoordinated species can form during the transient binding of alcohol (in Thermoanaerobacter brockii ADH [79]).
An alternative mechanism involves a double displacement in which a glutamate residue that is located on the opposite side of the zinc from which the substrate approaches, moves into ligation with the zinc and displaces the hydroxide, and then is subsequently displaced by the incoming ligand. Such a mechanism was suggested on the basis of theoretical calculations [80] and is illustrated in Scheme 5. (This scheme is based on the residue numbering for yeast alcohol dehydrogenase.) Note that the water and the alcohol are shown in a protonated state, but the water would be deprotonated through the proton relay system before displacement by the glutamate carboxyl group, and after an alcohol binds it would deprotonate to form the alkoxide before transfer of the hydride ion to NAD+. This mechanism is supported by X-ray crystallography, where for instance, the homologous dimeric human ADH3 (glutathione-dependent formaldehyde dehydrogenase) shows two different ligation states, which depend upon the ligands [81]. In subunits of the enzyme where the exogenous ligand is water or 12-hydroxydodecanoic acid (a substrate), or NADH and hydroxymethylglutathione (a physiological substrate), the ligand is bound to the tetracoordinated zinc and the glutamate is 4.8 Å away from the zinc. In contrast, in subunits with NAD(H) or NAD+ and dodecanoic acid, the glutamate is ligated to the tetracoordinated zinc. It appears that when the ligand is neutral, the glutamate will coordinate to the zinc. In the homologous, but tetrameric, ADH from Escherichia coli the situation is more complicated, as the apoenzyme has three of the four subunits with the alternative coordination with glutamate, whereas the holoenzyme with NAD(H) has 3 of the 4 subunits with the classical coordination with water as a ligand. The fourth subunit has no bound coenzyme and the alternative coordination [3, 82]. In these complexes, the zinc has moved about 2 Å during the conformational change. Tetrameric alcohol dehydrogenases from Clostridium beijerinckii, Sulfolobous solfataricus, and Saccharomyces cerevisiae also have some subunits in which the glutamate is ligated to the zinc [83-85][2hcy.pdb, Plapp, unpublished]. Some experimental support for a role for the glutamate in catalysis comes from a study of yeast alcohol dehydrogenase, in which the substitution of the glutamate with glutamine decreases catalytic efficiency by 100-fold [86]. However, in the Thermoanaerobacter brockii enzyme, substituting the glutamate with alanine or aspartic acid residues only decreases activity by 4 to 6-fold [87]. Although the mechanism of ligand exchange is unknown at this time, it is clear that there is considerable flexibility in the zinc coordination that would make the mechanism shown in Scheme 5 plausible.
Scheme 5.
Further understanding of the role of the conformational changes in ADH catalysis may come from computational studies. An early study suggested that the energetic barrier for the global change is relatively small and would readily allow the large 10° relative rotation [5]. It was also noted that the flexible loop including residues 292-299 would have to rearrange to allow the catalytic and coenzyme binding domains to move together (to form the “closed” conformation). A more recent study using 10 ns free molecular dynamics simulations suggested that when the loop is in the conformation found in the open enzyme it could hinder the global conformational change, but when the conformations of loops in both subunits are as observed in the closed conformation, NAD+ binding in one subunit might cooperatively facilitate the global conformational change or the change in the other subunit even it did not have bound NAD+ [88]. Since the loop is at the interface of the subunits in the dimeric molecule (see also ref. [89]), it is reasonable to ask if coenzyme binding is cooperative. Even though half-of-the-sites reactivity was proposed [90], contrary evidence abounds [91-93]. It may be noted that the complexes of the horse liver enzyme with coenzyme and substrate usually crystallize with both subunits occupied and in a closed conformation, but the dimeric molecule is in the asymmetric unit and the subunits are not identical even if they have very similar subunit structures. Furthermore, the structures of the tetrameric enzymes often have a mixed population of states. More critical tests of the potential cooperativity are warranted.
Molecular dynamics simulations of the holoenzyme complexes have identified some anticorrelated motions that may be important for driving the substrates together during the hydride transfer [94-96]. These motions, which generally reflect the domain closure, could orient and move the reacting carbons of the nicotinamide ring and the alcohol some 0.5 Å closer together. Such motions would be consistent with the evidence for quantum mechanical hydrogen tunneling in alcohol dehydrogenases [65, 97], where the distance between reacting carbons would be about 2.7 Å [98-100]. Normal mode analysis also identifies some motions of the domains that seem to be relevant for catalysis [101]. On the atomic level, these motions could also result in the puckering of the nicotinamide ring, which would bring the reacting carbons together [102]. Quantum mechanical calculations support the proposal that puckering of the nicotinamide ring strongly impacts the reaction [99, 103]. As shown in Fig. 5, the reduced nicotinamide ring is puckered in a structure with a substrate analog [23]. (However, secondary 15N isotope effects are negligible (with the 3-acetylpyridine analog of NAD), suggesting that the nicotinamide ring nitrogen remains planar during the hydrogen transfer step [104]). Additional computations suggest that very fast motions of amino acid residues near the active site may contribute to protein promoting vibrations that facilitate the hydrogen tunneling [105-108]. The temperature independent H/D isotope effects for benzyl alcohol oxidation and benzaldehyde reduction catalyzed by the V292S enzyme may be explained by such motions [65]. Analysis of atomic displacement parameters derived from atomic resolution X-ray crystallography may also provide experimental evidence for the directions and amplitudes of relevant motions in the holoenzyme complexes [29]. The involvement of protein dynamics and hydrogen tunneling in catalysis remains an active area of investigation.
Summary and future directions
In this review, we have shown that the global conformational change is important for most efficient, but not necessarily absolutely required, for catalysis by horse liver alcohol dehydrogenase. The enzyme is able to accommodate various substitutions and retain catalytic activity, indicating that the enzyme is adaptable and resilient. The available data provide a foundation for further analyses of this enzyme and raise some questions to be addressed in other alcohol dehydrogenases. A combination of kinetics, mutagenesis, crystallography, computations and other techniques may provide the relevant information. The kinetics of the conformational change of the apoenzyme needs to be determined in order to complete the thermodynamic cycle during binding of NAD+. The rate constants for the conformational change that occurs when NADH binds need to be determined with appropriate methods and conditions. The structural features of the protein that control the motions around the “hinge” regions need examination. Further study of the exchange of ligands at the catalytic zinc is needed to clarify the sequence of events as the alternative coordination appears in apo- and holo-enzymes. The tetrameric alcohol dehydrogenases may provide interesting subjects for such studies. Cooperativity between subunits during the conformational change is not fully understood. The role of electrostatics in binding of coenzymes needs exploration. We still need to account for the catalytic power of alcohol dehydrogenases in a quantitative manner, which may come from computations and correlations with enzymes with different rates of catalysis. Although horse liver alcohol dehydrogenase has been extensively studied, we do not understand the quantitative correlations between structure and function.
Abbreviation used
- ADH
alcohol dehydrogenase
Footnotes
This work was supported by Grant GM078446 from the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brändén CI. Arch Biochem Biophys. 1965;112:215–217. [Google Scholar]
- 2.Brändén CI, Eklund H. Ciba Found Symp. 1978;60:63–80. doi: 10.1002/9780470720424.ch5. [DOI] [PubMed] [Google Scholar]
- 3.Eklund H, Ramaswamy S. Cell Mol Life Sci. 2008;65:3907–3917. doi: 10.1007/s00018-008-8589-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eklund H, Samama JP, Wallén L, Brändén CI, Åkeson Å, Jones TA. J Mol Biol. 1981;146:561–587. doi: 10.1016/0022-2836(81)90047-4. [DOI] [PubMed] [Google Scholar]
- 5.Colonna-Cesari F, Perahia D, Karplus M, Eklund H, Brändén CI, Tapia O. J Biol Chem. 1986;261:15273–15280. [PubMed] [Google Scholar]
- 6.Eklund H, Brändén CI. In: Biological Macromolecules and Assemblies: Volume 3-Active Sites of Enzymes. Jurnak FA, McPherson A, editors. Wiley; 1987. pp. 73–141. [Google Scholar]
- 7.Eklund H, Plapp BV, Samama JP, Brändén CI. J Biol Chem. 1982;257:14349–14358. [PubMed] [Google Scholar]
- 8.Nordström B, Brändén CI. In: Structure and Conformation of Nucleic Acids and Protein-Nucleic Acid Interactions. Sundaralingam M, Rao ST, editors. University Park Press; Baltimore: 1975. pp. 387–395. [Google Scholar]
- 9.Brändén CI, Jörnvall H, Eklund H, Furugren B. The Enzymes. 3rd. Vol. 11. 1975. pp. 103–190. [Google Scholar]
- 10.Plapp BV, Eklund H, Brändén CI. J Mol Biol. 1978;122:23–32. doi: 10.1016/0022-2836(78)90105-5. [DOI] [PubMed] [Google Scholar]
- 11.Shearer GL, Kim K, Lee KM, Wang CK, Plapp BV. Biochemistry. 1993;32:11186–11194. doi: 10.1021/bi00092a031. [DOI] [PubMed] [Google Scholar]
- 12.Cedergren-Zeppezauer E. Biochemistry. 1983;22:5761–5772. doi: 10.1021/bi00294a013. [DOI] [PubMed] [Google Scholar]
- 13.Cedergren-Zeppezauer E, Samama JP, Eklund H. Biochemistry. 1982;21:4895–4908. doi: 10.1021/bi00263a011. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Hallows WH, Punzi JS, Marquez VE, Carrell HL, Pankiewicz KW, Watanabe KA, Goldstein BM. Biochemistry. 1994;33:23–32. doi: 10.1021/bi00167a004. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Hallows WH, Punzi JS, Pankiewicz KW, Watanabe KA, Goldstein BM. Biochemistry. 1994;33:11734–11744. doi: 10.1021/bi00205a009. [DOI] [PubMed] [Google Scholar]
- 16.Colby TD, Bahnson BJ, Chin JK, Klinman JP, Goldstein BM. Biochemistry. 1998;37:9295–9304. doi: 10.1021/bi973184b. [DOI] [PubMed] [Google Scholar]
- 17.Eklund H, Samama JP, Wallen L. Biochemistry. 1982;21:4858–4866. doi: 10.1021/bi00263a005. [DOI] [PubMed] [Google Scholar]
- 18.Rubach JK, Plapp BV. Biochemistry. 2003;42:2907–2915. doi: 10.1021/bi0272656. [DOI] [PubMed] [Google Scholar]
- 19.Ramaswamy S, Scholze M, Plapp BV. Biochemistry. 1997;36:3522–3527. doi: 10.1021/bi962491z. [DOI] [PubMed] [Google Scholar]
- 20.Ramaswamy S, Eklund H, Plapp BV. Biochemistry. 1994;33:5230–5237. doi: 10.1021/bi00183a028. [DOI] [PubMed] [Google Scholar]
- 21.Adolph HW, Zwart P, Meijers R, Hubatsch I, Kiefer M, Lamzin V, Cedergren-Zeppezauer E. Biochemistry. 2000;39:12885–12897. doi: 10.1021/bi001376s. [DOI] [PubMed] [Google Scholar]
- 22.Cho H, Ramaswamy S, Plapp BV. Biochemistry. 1997;36:382–389. doi: 10.1021/bi9624604. [DOI] [PubMed] [Google Scholar]
- 23.Venkataramaiah TH, Plapp BV. J Biol Chem. 2003;278:36699–36706. doi: 10.1074/jbc.M305419200. [DOI] [PubMed] [Google Scholar]
- 24.Theorell H, Yonetani T. Biochem Z. 1963;338:537–553. [PubMed] [Google Scholar]
- 25.Meijers R, Cedergren-Zeppezauer E. Chem Biol Interact. 2009;178:24–28. doi: 10.1016/j.cbi.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 26.Schneider G, Eklund H, Cedergren-Zeppezauer E, Zeppezauer M. EMBO J. 1983;2:685–689. doi: 10.1002/j.1460-2075.1983.tb01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider G, Cedergren-Zeppezauer E, Knight S, Eklund H, Zeppezauer M. Biochemistry. 1985;24:7503–7510. doi: 10.1021/bi00346a070. [DOI] [PubMed] [Google Scholar]
- 28.Al-Karadaghi S, Cedergren-Zeppezauer ES, Dauter Z, Wilson KS. Acta Crystallogr D Biol Crystallogr. 1995;51:805–813. doi: 10.1107/S090744499500045X. [DOI] [PubMed] [Google Scholar]
- 29.Ramaswamy S. In: Enzymology and Molecular Biology of Carbonyl Metabolism 7. Weiner H, Maser E, Crabb DW, Lindahl R, editors. Kluwer Academic; New York: 1999. pp. 275–284. [Google Scholar]
- 30.Ramaswamy S, el-Ahmad M, Danielsson O, Jörnvall H, Eklund H. Protein Sci. 1996;5:663–671. doi: 10.1002/pro.5560050410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurley TD, Bosron WF, Stone CL, Amzel LM. J Mol Biol. 1994;239:415–429. doi: 10.1006/jmbi.1994.1382. [DOI] [PubMed] [Google Scholar]
- 32.Niederhut MS, Gibbons BJ, Perez-Miller S, Hurley TD. Protein Sci. 2001;10:697–706. doi: 10.1110/ps.45001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibbons BJ, Hurley TD. Biochemistry. 2004;43:12555–12562. doi: 10.1021/bi0489107. [DOI] [PubMed] [Google Scholar]
- 34.Theorell H, Chance B. Acta Chem Scand. 1951;5:1127–1144. [Google Scholar]
- 35.Dalziel K. J Biol Chem. 1963;238:2850–2858. [PubMed] [Google Scholar]
- 36.Wratten CC, Cleland WW. Biochemistry. 1963;2:935–941. doi: 10.1021/bi00905a007. [DOI] [PubMed] [Google Scholar]
- 37.Dalziel K, Dickinson FM. Biochem J. 1966;100:34–46. doi: 10.1042/bj1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalziel K, Dickinson FM. Biochem J. 1966;100:491–500. doi: 10.1042/bj1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charlier HA, Jr, Plapp BV. J Biol Chem. 2000;275:11569–11575. doi: 10.1074/jbc.275.16.11569. [DOI] [PubMed] [Google Scholar]
- 40.Andersson P, Kvassman J, Olden B, Pettersson G. Eur J Biochem. 1984;144:317–324. doi: 10.1111/j.1432-1033.1984.tb08466.x. [DOI] [PubMed] [Google Scholar]
- 41.Andersson P, Kvassman J, Olden B, Pettersson G. Eur J Biochem. 1984;139:519–527. doi: 10.1111/j.1432-1033.1984.tb08036.x. [DOI] [PubMed] [Google Scholar]
- 42.Shore JD, Gilleland MJ. J Biol Chem. 1970;245:3422–3425. [PubMed] [Google Scholar]
- 43.Plapp BV, Brooks RL, Shore JD. J Biol Chem. 1973;248:3470–3475. [PubMed] [Google Scholar]
- 44.Sekhar VC, Plapp BV. Biochemistry. 1988;27:5082–5088. doi: 10.1021/bi00414a020. [DOI] [PubMed] [Google Scholar]
- 45.Frieden C. Methods Enzymol. 1994;240:311–322. doi: 10.1016/s0076-6879(94)40053-9. [DOI] [PubMed] [Google Scholar]
- 46.Coates JH, Hardman MJ, Shore JD, Gutfreund H. FEBS Lett. 1977;84:25–28. doi: 10.1016/0014-5793(77)81049-1. [DOI] [PubMed] [Google Scholar]
- 47.Sekhar VC, Plapp BV. Biochemistry. 1990;29:4289–4295. doi: 10.1021/bi00470a005. [DOI] [PubMed] [Google Scholar]
- 48.Adolph HW, Kiefer M, Cedergren-Zeppezauer E. Biochemistry. 1997;36:8743–8754. doi: 10.1021/bi970398k. [DOI] [PubMed] [Google Scholar]
- 49.DeTraglia MC, Schmidt J, Dunn MF, McFarland JT. J Biol Chem. 1977;252:3493–3500. [PubMed] [Google Scholar]
- 50.Kvassman J, Pettersson G. Eur J Biochem. 1978;87:417–427. doi: 10.1111/j.1432-1033.1978.tb12391.x. [DOI] [PubMed] [Google Scholar]
- 51.Plapp BV. Methods Enzymol. 1982;87:469–499. doi: 10.1016/s0076-6879(82)87027-4. [DOI] [PubMed] [Google Scholar]
- 52.Plapp BV. J Biol Chem. 1970;245:1727–1735. [PubMed] [Google Scholar]
- 53.Sogin DC, Plapp BV. J Biol Chem. 1975;250:205–210. [PubMed] [Google Scholar]
- 54.Dworschack R, Tarr G, Plapp BV. Biochemistry. 1975;14:200–203. doi: 10.1021/bi00673a002. [DOI] [PubMed] [Google Scholar]
- 55.Eklund H, Nordström B, Zeppezauer E, Söderlund G, Ohlsson I, Boiwe T, Söderberg BO, Tapia O, Brändén CI, Åkeson Å. J Mol Biol. 1976;102:27–59. doi: 10.1016/0022-2836(76)90072-3. [DOI] [PubMed] [Google Scholar]
- 56.Zoltobrocki M, Kim JC, Plapp BV. Biochemistry. 1974;13:899–903. doi: 10.1021/bi00702a011. [DOI] [PubMed] [Google Scholar]
- 57.Fries RW, Bohlken DP, Blakley RT, Plapp BV. Biochemistry. 1975;14:5233–5238. doi: 10.1021/bi00694a034. [DOI] [PubMed] [Google Scholar]
- 58.Dworschack RT, Plapp BV. Biochemistry. 1977;16:111–116. doi: 10.1021/bi00620a018. [DOI] [PubMed] [Google Scholar]
- 59.Dworschack RT, Plapp BV. Biochemistry. 1977;16:2716–2725. doi: 10.1021/bi00631a020. [DOI] [PubMed] [Google Scholar]
- 60.Parker DM, Hardman MJ, Plapp BV, Holbrook JJ, Shore JD. Biochem J. 1978;173:269–275. doi: 10.1042/bj1730269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plapp BV, Eklund H, Jones TA, Brändén CI. J Biol Chem. 1983;258:5537–5547. [PubMed] [Google Scholar]
- 62.Plapp BV. Methods Enzymol. 1995;249:91–119. doi: 10.1016/0076-6879(95)49032-9. [DOI] [PubMed] [Google Scholar]
- 63.Fan F, Plapp BV. Biochemistry. 1995;34:4709–4713. doi: 10.1021/bi00014a027. [DOI] [PubMed] [Google Scholar]
- 64.Chin JK, Klinman JP. Biochemistry. 2000;39:1278–1284. doi: 10.1021/bi9920331. [DOI] [PubMed] [Google Scholar]
- 65.Rubach JK, Ramaswamy S, Plapp BV. Biochemistry. 2001;40:12686–12694. doi: 10.1021/bi011540r. [DOI] [PubMed] [Google Scholar]
- 66.Ramaswamy S, Park DH, Plapp BV. Biochemistry. 1999;38:13951–13959. doi: 10.1021/bi991731i. [DOI] [PubMed] [Google Scholar]
- 67.LeBrun LA, Park DH, Ramaswamy S, Plapp BV. Biochemistry. 2004;43:3014–3026. doi: 10.1021/bi036103m. [DOI] [PubMed] [Google Scholar]
- 68.Taniguchi S, Theorell H, Åkeson Å. Acta Chem Scand. 1967;21:1903–1920. doi: 10.3891/acta.chem.scand.21-1903. [DOI] [PubMed] [Google Scholar]
- 69.Kvassman J, Pettersson G. Eur J Biochem. 1979;100:115–123. doi: 10.1111/j.1432-1033.1979.tb02039.x. [DOI] [PubMed] [Google Scholar]
- 70.Evans SA, Shore JD. J Biol Chem. 1980;255:1509–1514. [PubMed] [Google Scholar]
- 71.Pettersson G. CRC Crit Rev Biochem. 1987;21:349–389. [PubMed] [Google Scholar]
- 72.LeBrun LA, Plapp BV. Biochemistry. 1999;38:12387–12393. doi: 10.1021/bi991306p. [DOI] [PubMed] [Google Scholar]
- 73.Shore JD, Gutfreund H, Brooks RL, Santiago D, Santiago P. Biochemistry. 1974;13:4185–4191. doi: 10.1021/bi00717a019. [DOI] [PubMed] [Google Scholar]
- 74.Eftink MR, Bystrom K. Biochemistry. 1986;25:6624–6630. doi: 10.1021/bi00369a044. [DOI] [PubMed] [Google Scholar]
- 75.Kovaleva EG, Plapp BV. Biochemistry. 2005;44:12797–12808. doi: 10.1021/bi050865v. [DOI] [PubMed] [Google Scholar]
- 76.Shore JD, Gutfreund H, Yates D. J Biol Chem. 1975;250:5276–5277. [PubMed] [Google Scholar]
- 77.Laws WR, Shore JD. J Biol Chem. 1979;254:2582–2584. [PubMed] [Google Scholar]
- 78.Boiwe T, Brändén CI. Eur J Biochem. 1977;77:173–179. doi: 10.1111/j.1432-1033.1977.tb11655.x. [DOI] [PubMed] [Google Scholar]
- 79.Kleifeld O, Frenkel A, Martin JM, Sagi I. Nat Struct Biol. 2003;10:98–103. doi: 10.1038/nsb889. [DOI] [PubMed] [Google Scholar]
- 80.Ryde U. Protein Sci. 1995;4:1124–1132. doi: 10.1002/pro.5560040611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanghani PC, Bosron WF, Hurley TD. Biochemistry. 2002;41:15189–15194. doi: 10.1021/bi026705q. [DOI] [PubMed] [Google Scholar]
- 82.Karlsson A, el-Ahmad M, Johansson K, Shafqat J, Jörnvall H, Eklund H, Ramaswamy S. Chem Biol Interact. 2003;143-144:239–245. doi: 10.1016/s0009-2797(02)00222-3. [DOI] [PubMed] [Google Scholar]
- 83.Korkhin Y, Kalb G, Peretz M, Bogin O, Burstein Y, Frolow F. J Mol Biol. 1998;278:967–981. doi: 10.1006/jmbi.1998.1750. [DOI] [PubMed] [Google Scholar]
- 84.Esposito L, Sica F, Raia CA, Giordano A, Rossi M, Mazzarella L, Zagari A. J Mol Biol. 2002;318:463–477. doi: 10.1016/S0022-2836(02)00088-8. [DOI] [PubMed] [Google Scholar]
- 85.Esposito L, Bruno I, Sica F, Raia CA, Giordano A, Rossi M, Mazzarella L, Zagari A. Biochemistry. 2003;42:14397–14407. doi: 10.1021/bi035271b. [DOI] [PubMed] [Google Scholar]
- 86.Ganzhorn AJ, Plapp BV. J Biol Chem. 1988;263:5446–5454. [PubMed] [Google Scholar]
- 87.Kleifeld O, Shi SP, Zarivach R, Eisenstein M, Sagi I. Protein Sci. 2003;12:468–479. doi: 10.1110/ps.0221603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hayward S, Kitao A. Biophys J. 2006;91:1823–1831. doi: 10.1529/biophysj.106.085910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Strasser F, Dey J, Eftink MR, Plapp BV. Arch Biochem Biophys. 1998;358:369–376. doi: 10.1006/abbi.1998.0882. [DOI] [PubMed] [Google Scholar]
- 90.Dunn MF, Bernhard SA, Anderson D, Copeland A, Morris RG, Roque JP. Biochemistry. 1979;18:2346–2354. doi: 10.1021/bi00578a033. [DOI] [PubMed] [Google Scholar]
- 91.Hadorn M, John VA, Meier FK, Dutler H. Eur J Biochem. 1975;54:65–73. doi: 10.1111/j.1432-1033.1975.tb04114.x. [DOI] [PubMed] [Google Scholar]
- 92.Weidig CF, Halvorson HR, Shore JD. Biochemistry. 1977;16:2916–2922. doi: 10.1021/bi00632a018. [DOI] [PubMed] [Google Scholar]
- 93.Andersson P, Pettersson G. Eur J Biochem. 1982;122:559–568. doi: 10.1111/j.1432-1033.1982.tb06475.x. [DOI] [PubMed] [Google Scholar]
- 94.Luo J, Bruice TC. Proc Natl Acad Sci USA. 2002;99:16597–16600. doi: 10.1073/pnas.262667599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luo J, Bruice TC. Proc Natl Acad Sci USA. 2004;101:13152–13156. doi: 10.1073/pnas.0405502101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bruice TC. Chem Rev. 2006;106:3119–3139. doi: 10.1021/cr050283j. [DOI] [PubMed] [Google Scholar]
- 97.Bahnson BJ, Park DH, Kim K, Plapp BV, Klinman JP. Biochemistry. 1993;32:5503–5507. doi: 10.1021/bi00072a003. [DOI] [PubMed] [Google Scholar]
- 98.Klinman JP. Trends Biochem Sci. 1989;14:368–373. doi: 10.1016/0968-0004(89)90010-8. [DOI] [PubMed] [Google Scholar]
- 99.Billeter SR, Webb SP, Agarwal PK, Iordanov T, Hammes-Schiffer S. J Am Chem Soc. 2001;123:11262–11272. doi: 10.1021/ja011384b. [DOI] [PubMed] [Google Scholar]
- 100.Nagel ZD, Klinman JP. Chem Rev. 2006;106:3095–3118. doi: 10.1021/cr050301x. [DOI] [PubMed] [Google Scholar]
- 101.Luo J, Bruice TC. Biophys Chem. 2007;126:80–85. doi: 10.1016/j.bpc.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 102.Luo J, Bruice TC. J Am Chem Soc. 2001;123:11952–11959. doi: 10.1021/ja0109747. [DOI] [PubMed] [Google Scholar]
- 103.Webb SP, Agarwal PK, Hammes-Schiffer S. J Phys Chem B. 2000;104:8884–8894. [Google Scholar]
- 104.Rotberg NS, Cleland WW. Biochemistry. 1991;30:4068–4071. doi: 10.1021/bi00230a035. [DOI] [PubMed] [Google Scholar]
- 105.Caratzoulas S, Mincer JS, Schwartz SD. J Am Chem Soc. 2002;124:3270–3276. doi: 10.1021/ja017146y. [DOI] [PubMed] [Google Scholar]
- 106.Mincer JS, Schwartz SD. J Phys Chem B. 2003;107:366–371. [Google Scholar]
- 107.Mincer JS, Schwartz SD. J Proteome Res. 2003;2:437–439. doi: 10.1021/pr025590+. [DOI] [PubMed] [Google Scholar]
- 108.Antoniou D, Basner J, Nunez S, Schwartz SD. Chem Rev. 2006;106:3170–3187. doi: 10.1021/cr0503052. [DOI] [PubMed] [Google Scholar]
- 109.Harris M, Jones TA. Acta Crystallogr. 2001;D57:1201–1203. doi: 10.1107/s0907444901007697. [DOI] [PubMed] [Google Scholar]