Abstract
Coronins, WD-repeat actin-binding proteins, are known to regulate cell motility by coordinating actin filament turnover in lamellipodia of migrating cell. Here we report a novel mechanism of Coronin 1C-mediated cell motility that involves regulation of cell-matrix adhesion. RNAi silencing of Coronin 1C in intestinal epithelial cells enhanced cell migration and modulated lamellipodia dynamics by increasing the persistence of lamellipodial protrusion. Coronin 1C-depleted cells showed increased cell-matrix adhesions and enhanced cell spreading compared to control cells, while overexpression of Coronin 1C antagonized cell adhesion and spreading. Enhanced cell-matrix adhesion of coronin-deficient cells correlated with hyperphosphorylation of Focal Adhesion Kinase (FAK) and paxillin, and an increase in number of focal adhesions and their redistribution at the cell periphery. siRNA depletion of FAK in coronin-deficient cells rescued the effects of Coronin 1C depletion on motility, cell-matrix adhesion, and spreading. Thus, our findings provide the first evidence that Coronin 1C negatively regulates epithelial cell migration via FAK-mediated inhibition of cell-matrix adhesion.
Keywords: Coronin, FAK, motility, adhesion
Introduction
Migration of epithelial cells plays a vital role in a number of physiological and pathological processes, such as embryogenesis, epithelial renewal, wound healing, and tumor metastasis. Mechanistically, cell migration represents a cyclic process involving extension of lamellipodia at the leading edge, adhesion of protruded lamellipodia to the extracellular matrix, and finally retraction of the trailing edge [1; 2]. The protrusion of lamellipodia is induced by controlled turnover of actin filaments, where the Arp2/3 complex nucleates new branched actin filaments, and existing filaments are disassembled by ADF/cofilin [3]. Cell-matrix adhesion is mediated by focal adhesions (FAs) whose main constituents are integrins and adaptor proteins. The former interact with the extracellular matrix whereas the latter link integrins to the actin cytoskeleton and participate in intracellular signaling [4].
Coronins are evolutionary conserved WD-repeat actin-binding proteins known to regulate various cellular processes involving actin dynamics [5]. Coronin protein family encompasses 7 proteins in mammals [6], divided into three subclasses based on sequence similarity: Type I, II and III [5]. The Type I subclass (Coronins 1A, 1B, and 1C) is the most studied coronin subfamily. Coronins 1A and 1B localize at the leading edge of lamellipodia [7; 8; 9], physically interact with Arp2/3 complex, and regulate the protrusion of lamellipodia and cell migration [7; 8; 10]. Recently, Coronin 1B has been shown to act as a coordinator of filament nucleation and disassembly by bridging together Arp2/3 complex and slingshot 1L, an activator of cofilin, thus controlling actin filament dynamics and architecture at the leading edge of the migrating cell [11].
Coronin 1C is ubiquitously expressed in most tissues [7; 12; 13; 14], localizes at the sites of active actin dynamics, such as lamellipodia and membrane ruffles [15] and co-immunoprecipitates with Arp2/3 complex and cofilin [16]. However, unlike Coronin 1A and 1B, Coronin 1C has not been extensively characterized, and its role in regulating motility of epithelial sheets is not understood. Here we report a novel mechanism for regulation of motility of intestinal epithelial cells (IECs) by Coronin 1C, which involves negative regulation of cell-matrix adhesion through FAK-mediated signaling.
Materials and Methods
Antibodies
Anti-Coronin 1C mouse polyclonal and monoclonal, and anti-Coronin 1B mouse monoclonal antibody were purchased from Abnova (Taipei, Taiwan). Anti-paxillin mouse monoclonal antibodies were obtained from Zymed (Zymed Labs, San Francisco, CA). Anti-FAK, anti-phospho(Y118)paxillin, anti-phospho(T18/S19)RMLC rabbit polyclonal antibodies and anti-phospho(S19)RMLC mouse monoclonal antibodies were from Cell Signaling (Cell Signaling Technology, Beverly, MA). Anti-phospho(Y397)FAK mouse monoclonal antibody were from BD Biosciences (San Jose, CA). Rabbit polyclonal anti-actin antibodies were from Sigma (Sigma Chemical Co., St. Louis, MO). Anti-RMLC rabbit polyclonal antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa 488/546-conjugated goat anti-mouse, goat anti-rabbit and donkey anti-rabbit antibody were purchased from Molecular Probes (Eugene, OR). Horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse secondary antibodies were from Jackson Immunoresearch Labs (West Grove, PA).
Cells, DNA transfection and RNA interference
SK-CO15 (gift of Dr. E. Rodriguez-Boulan, Weill Medical College of Cornell University, NY) and Caco-2 (ATCC, USA) human colonic epithelial cells were grown as described previously [17]. For DNA and RNA transfection cells were plated at ∼75% confluency, transfected the next day and were used in experiments 24 and 72 hours after transfection respectively. Full length Coronin 1C construct in pEGFP-C1 vector were generated as described previously [15] and transfected into cells using Lipofectamine 2000 (Invitrogen) according to manufacturers protocol. Empty pEGFP-C1 vector (Clontech) was used as a control. Human Coronin 1C siGENOME duplex 4 and PTK2 siGENOME SmartPool (Dharmacon, Lafayette, CO) were used to downregulate Coronin 1C and FAK respectively. Scramble duplex 2 SmartPool siRNA (Dharmacon) has been used as a control. RNA transfection was carried out in OPTI-MEM I media (Invitrogen) with 50 nM siRNA using Dharmafect 1 siRNA transfection reagent (Dharmacon) according to standard protocol.
Cell migration assay and live cell microscopy
For microscopy, confluent monolayers growing on collagen-I received a single linear wound using a sterile 20 μL pipette tip attached to low suction. 10×10 wounds per monolayer were created for Western Blot analysis. Live cell microscopy was performed in CO2-independent medium (Invitrogen) using heated cabinet and thermally-controlled microscopy stage (Brook Industries, USA) mounted on Carl Zeiss Axiovert microscope equipped with Zeiss AxioCam MRc5 camera.
Cell adhesion/spreading assay
Confluent cells were trypsinized, washed with Hanks' balanced salt solution (Sigma) containing 0.1% BSA and re-plated on coverslips coated with ECM gel (Sigma) or collagen-I (BD Biosciences, USA) at a density of 105cells/1.9cm2. For cell adhesion assay cells were allowed to adhere for 3 hours, and non-adherent cells were removed by repeated washing with complete media. For cell spreading assay, cells were allowed to spread for additional 8 hours. Cells overexpressing EGFP and EGFP-Coronin 1C were detached from the substrate using Accutase (Innovative Cell Technologies, USA) followed by resuspension in cell sorting buffer (PBS containing 25mM HEPES pH 7.0, 1mM EDTA and 0.1% BSA). EGFP-positive, propidium iodide-negative cells were sorted on a FACSVantage SE (BD Biosciences). The purity of sorting was greater than 95%. Cells were counted manually, and cell surface was measured using MetaMorph software. A total of at least 10 microscopic fields were analyzed per group in each experiment.
Immunofluorescence labeling and confocal microscopy
Cells were fixed in 3.7% paraformaldehyde (PFA) for 15 min and subsequently permeabilized with 0.5% Triton X-100 for 10 minutes at RT. Cells were blocked in Hanks' balanced salt solution (Sigma) containing 1.5 % BSA, and sequentially incubated with primary and Alexa-conjugated secondary antibodies (Molecular Probes) for 1 hour at RT. Nuclei were stained with ToPro-3 iodide (Molecular Probes), followed by mounting on slides using Pro-Long Gold Antifade medium (Molecular Probes). Stained monolayers were analyzed using Zeiss LSM510 laser scanning confocal microscope (Zeiss Microimaging Inc., USA) equipped with Zeiss Plan-apochromat 63×/1.4 Oil lenses. Images shown are representative of at least three independent experiments with multiple images taken per slide.
Image analysis
Wound widths were measured using Scion Image software (Scion Image Corp., USA). Five measurements along the wound length were averaged to determine the wound width. Kymographs were produced using MetaMorph software (Molecular Devices, USA) by taking 1-pixel-wide rectangular regions in the direction of edge movement as shown. For cell adhesion/spreading assay, cells were counted manually, and cell surface was measured using MetaMorph software. A total of at least 10 microscopic fields were analyzed per group in each experiment. For quantification of FAs phospho(Y118)paxillin-positive pattern was thresholded with the same level in all specimens and the integrated surface area of FAs was measured using NIS Elements software (Nikon Inc., USA). To determine the density of FAs along the cell periphery, the total surface of FAs was devided by the cells perimeter length.
Immunoblotting
Cells were scraped in RIPA buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% Na deoxycholate, 2 mM EGTA, 2 mM EDTA, 0.1% SDS) containing protease inhibitor cocktail (1:100, Sigma) and phosphatase inhibitor cocktails 1 and 2 (both at 1:200, Sigma) and homogenized using Wheaton glass homogenizers (Wheaton Industries Inc., USA). Lysates were cleared by centrifugation at 14,000g for 10 minutes at 4°C and equalized for total protein concentration using BCA protein quantification assay (Pierce Biotechnology, USA). Samples were boiled in SDS sample buffer and subjected to SDS-PAGE and Western blotting with 30 μg of total protein per lane. Membranes were blocked with either bovine serum albumin (BSA) (for detection of phosphorylated proteins) or dry milk for 1 hour at room temperature (RT) and incubated with primary antibodies in blocking buffer overnight at 4°C. The results shown are representative immunoblots of three independent experiments.
Statistics
All data shown are representative of at least three independent experiments and expressed as the mean ± standard error of the mean (S.E.). The results were compared by either two-tailed Student's t-test, or a post-hoc Bonferroni test following repeated measures two-way ANOVA with statistical significance assumed at p<0.05.
Results and Discussion
To address the role of Coronin 1C in the epithelial cell motility, we down-regulated its expression in a SK-CO15 intestinal epithelial cells (IEC) using RNA interference. siRNA treatment dramatically (∼85%) and specifically reduced the expression level of Coronin1C, but not the closely related Coronin 1B (Fig. 1A). Coronin 1C-depleted cells demonstrated increased migration rate resulting in more efficient wound closure in scratch-wound assays (1.75±0.15 fold of scramble control) (Fig. 1B). This correlated with dramatic changes in lamellipodia dynamics at the leading edge of migrating IECs. While control cells rapidly extended multiple spiked lamellipodia (Fig. 1C, upper panel) followed by their rapid retraction (Fig. 1C, upper kymograph), Coronin 1C-deficient cells formed flat and rounded lamellipodia (Fig. 1C, bottom panel), that rarely retracted (Fig. 1C, bottom kymograph), thus providing more persistent forward motility of the leading edge and significantly enhancing the overall distance protruded over time (Fig. 1C).
Figure 1. Depletion of Coronin 1C increases motility and modulates lamellipodia dynamics in IECs.
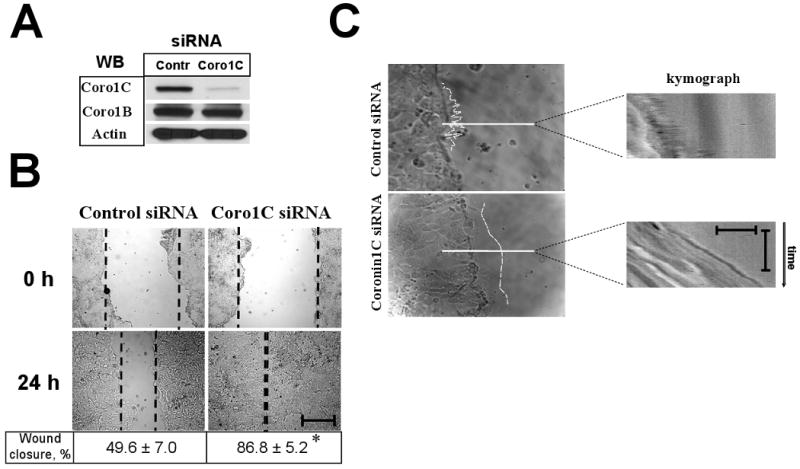
SK-CO15 cells treated with control- or Coronin 1C-specific siRNAs. (A) Cells were lysed and immunobloated for coronins 1C and 1B to ensure efficient and selective Coronin 1C knock-down and for actin to ensure equal protein loading. (B) Representative phase contrast images showing wound closure in control and Coronin 1C-depleted cell monolayers. Bar, 400μm. Wound widths measured 24 hours after wounding and expressed as a % of closure of the initial wound widths at time 0 are indicated under the figure. *, p<0.01 compared to control. (D) Phase contrast images showing the shape of lamellipodia (outlined in white) at the leading edge of the wound 120 minutes after wounding. Kymographs were generated from 180-frame 1 minute interval image sequences using pixel intensities along 1-pixel wide line shown. Vertical bar, 60 seconds; horizontal bar, 20 μm. Note the fuzzy pattern on kymograph reflecting the fast withdrawal rate of the formed protrusions in control cells and smooth pattern representing the persistence of lamellipodial protrusion in Coronin 1C-depleted cells.
The protrusion of lamellipodia is a dynamic process involving both actin-mediated extensions and retractions, and anchoring lamellipodia to the extracellular matrix (ECM) [18]. We therefore reasoned that the increased persistence of lamellipodial protrusion in Coronin 1C-deficient cells might be mediated either through decreased retraction, or enhanced cell-matrix adhesion. Retraction of lamellipodia depends on the activity of the major F-actin motor, non-muscle myosin II [19], which is regulated by phosphorylation of its regulatory light chain (RMLC) on either one (Ser19), or two (Ser19/Thr18) residues [20]. However, down-regulation of Coronin 1C did not significantly alter the levels of mono- and diphosphorylated RMLC in migrating IECs, which suggests unaltered myosin II activity (Fig. 2A) and argues against the role of diminished retraction in the increased lammelipodial protrusion observed in Coronin 1C-deficient IECs.
Figure 2. Coronin 1C negatively regulates cell-matrix adhesion and spreading of IECs.
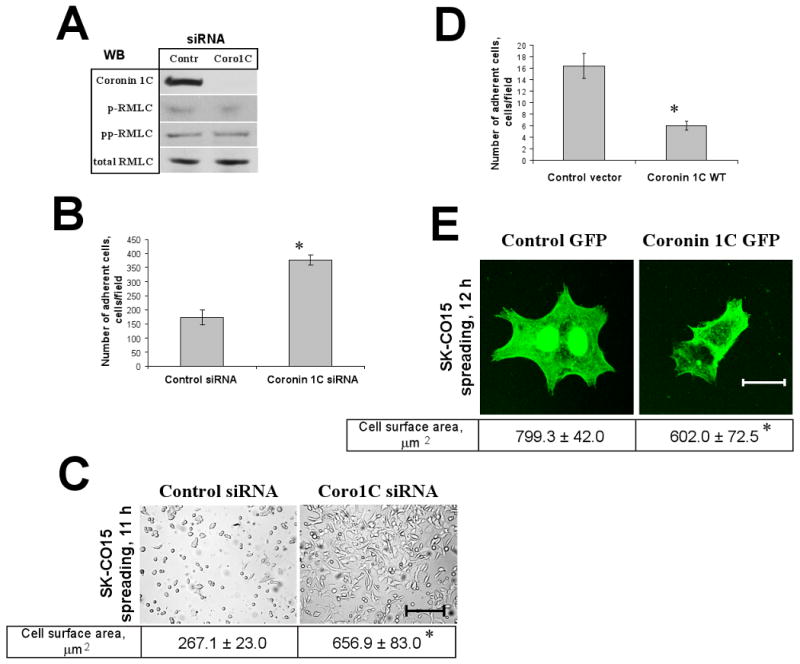
SK-CO15 cells were transfected with the indicated siRNAs. (A) Activation status of myosin II in control and Coronin1C-depleted SK-CO15 cells was compared by immunoblotting analysis of mono- (p) and di-phosphorylated (pp) RMLC expression in cell lysates collected 120 minutes after cell wounding. (B) Cells were plated on ECM gel and non-adherent cells were removed by washing 3 hours after plating. Average count of adhered cells per low power (10×) microscopic field. (C) Adherent cells were allowed to spread for additional 8 hours. Representative phase-contrast images of spreading cells. Bar, 200 μm. The average cell surface measured in spreading cells is shown. *, p<0.01 compared to control. (D,E) SK-CO15 cells were transfected with either GFP-Coronin 1C construct or GFP alone, sorted and plated on ECM as described in Methods. (D) Average number of cells per low power (20×) microscopic field adhered to ECM 12 hours after plating. *, p<0.01 compared to control. (E) Representative fluorescent micrographs of adherent cells. Bar, 20 μm. Average cell surface area of spreading cells is shown under the figure. *, p<0.05 compared to control.
To test whether depletion of Coronin 1C affects cell-matrix adhesion we used two types of ECM: collagen-I and ECM gel (Sigma) composed of laminin, collagen-IV, and entactin. Coronin 1C-depleted cells attached more readily to the ECM gel (2.18±0.16 fold increase) and showed enhanced spreading (2.46±0.15 fold cell surface area increase) following adhesion to ECM compared to control siRNA-transfected cells (Fig. 2B,C). Similar results were obtained on collagen-I and in another intestinal epithelial cell line, Caco-2 (data not shown). Conversely, over-expression of WT Coronin 1C dramatically (2.72±0.18 fold) decreased the number of adherent cells and attenuated their spreading (1.33±0.13 fold decrease in cell surface area) compared to cells transfected with control vector only (Fig. 2D,E). Together, these data suggest that Coronin 1C negatively regulates adhesion and spreading of IECs.
Cells adhere to the ECM by forming specialized multiprotein complexes known as focal adhesions (FAs) that consist of transmembrane integrins, and cytosolic scaffolding and regulatory proteins [4]. Therefore we next examined whether Coronin 1C depletion effects formation of FAs by visualizing the FA marker, Tyr118-phosphorylated (activated) paxillin in spreading IECs. Depletion of Coronin 1C dramatically increased the surface area of FAs and altered their topography towards more dense distribution at the cells periphery (Fig. 3A, Suppl. Fig. 1). These results were consistent with increased levels of total and Tyr118-phosphorylated paxillin in Coronin 1C-deficient IECs (Fig. 3B).
Figure 3. Coronin 1C regulates cell-matrix adhesion through FAK-mediated signaling.
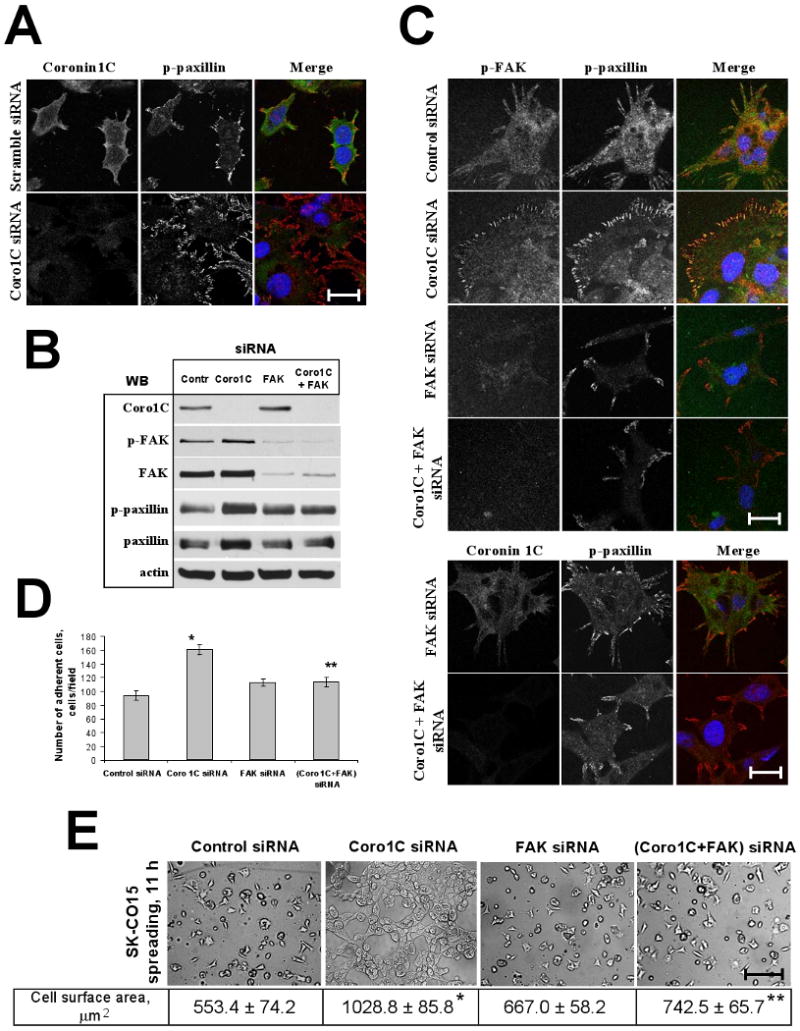
SK-CO15 cells transfected with indicated siRNAs were replated on ECM gel and allowed to spread for 11h. (A) En-face confocal images showing morphology of FAs visualized by immunolabeling for p-paxillin. Note increase in number and density of peripherally located FAs in Coronin 1C-depleted cells. Bar, 25 μm. (B) Cells were lysed and immunoblotted for Coronin 1C, p(397)FAK, FAK, p(118)paxillin, paxillin, and actin (loading control). Note increase in the amount of phosphorylated FAK and paxillin in Coronin 1C-depleted cells (C) En-face confocal images showing localization of phospho-paxillin and phospho-FAK in spreading cells. Note that knock-down of FAK reverses accumulation of peripheral FAs in Coronin 1C-depleted cells. Bar, 25 μm. (D, E) Cell-matrix adhesion and spreading of single and dual Coronin 1C- and FAK knock-downs was analyzed as described in Fig. 2. (D) Average number of adherent cells per microscopic field 3 hours after plaiting. (E) Representative phase contrast image of spreading cells. Bar, 200 μm. Average cell surface area of spreading cells is shown. *, p<0.01 compared to control; ** p<0.05 compared to Coronin 1C siRNA-transfected cells.
Since phosphorylation of paxillin is mediated by focal adhesion kinase (FAK), and FAK signaling plays a central role in regulating both cell-matrix adhesion and cell motility [21], we investigated the role of FAK in adhesiveness and migration of Coronin 1C-depleted IECs. Down-regulation of Coronin 1C resulted in increased activatory phosphorylation of FAK at Tyr397 following attachment to and subsequent spreading on ECM (Fig. 3B).
To determine whether the effects of Coronin 1C depletion on cell-matrix adhesion and migration were FAK-dependent, we performed a single FAK-, and dual FAK/Coronin 1C siRNA-mediated knock-down in SK-CO15 cells. Importantly, down-regulation of FAK protein expression (∼90%) alone did not significantly influence the level of phospho-paxillin (Fig. 3B) and the number or distribution of FAs in adhering/spreading IECs (Fig. 3C, Suppl. Fig. 1). Additionally, depletion of FAK did not significantly affect cell-matrix adhesion (Fig. 3D), cell spreading (Fig. 3E), and IEC migration (Fig 4A). These data suggest that FAK is not required for basal adhesion and motility of IECs and reflect functional redundancy in multiple FAK-mediated signaling pathways [21]. Indeed, the complexity of FAK-mediated signaling is highlighted by a variety of reports showing either negative, or neutral, or positive role of FAK activity in adhesion and migration in different cell types [22; 23; 24].
Figure 4. Coronin 1C regulates motility of IECs via FAK-mediated signaling.
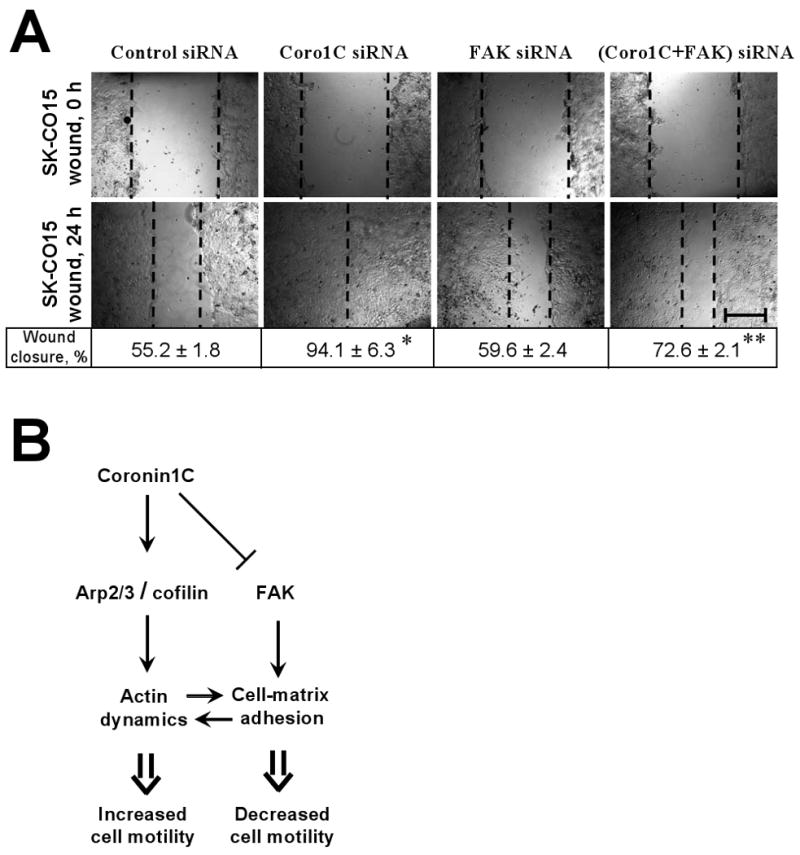
(A) SK-CO15 cells were transfected with the indicated siRNAs. Representative phase contrast images and quantification of wound closure 24 hours after wounding. Bar, 400 μm. *, p<0.01 compared to control; ** p<0.05 compared to Coronin 1C siRNA-transfected cells. (B) A model for the regulation of cell motility by Coronin 1C. Coronin 1C increases the rate of actin filament turnover by regulating Arp2/3/cofilin-dependent actin polymerization/disassembly. Alternatively, Coronin 1C suppresses FAK signaling leading to decreased cell-matrix adhesion. The resulting effect of these two signaling pathways on cell motility would be dependent on the dynamic equilibrium between lamellipodia protrusion and adhesion of protruded lamellipodia to ECM.
However, loss of FAK protein expression effectively reversed major effects of Coronin 1C down-regulation on cell adhesion and migration. Thus, double knock-down of Coronin 1C and FAK efficiently prevented paxillin hyperphosphorylation (Fig. 3B), as well as an increase in number and changes in distribution of focal adhesions (Fig. 3C, Suppl. Fig. 1) caused by Coronin 1C depletion alone. Consistent with these results, depletion of FAK reversed increased cell-matrix adhesion (Fig. 3D), spreading (Fig. 3E) and motility (Fig. 4A) of Coronin 1C-deficient cells. All together, these data suggest that FAK-mediated signaling is a critical downstream component, which mediates negative effects of Coronin 1C on both cell-matrix adhesion and motility of IECs.
The negative regulation of cell migration and cell-matrix adhesion of IECs by Coronin 1C is somewhat unexpected, since Coronin 1B is required for persistent lamellipodia protrusion and efficient motility of fibroblasts and HEK 293 cells [11], and depletion of Coronin 1C negatively regulates wound healing in fibroblasts [16]. Our results can be reconciled with previously published data by proposing a dual mechanism of Coronin 1C-dependent regulation of cell migration. Indeed, a delicate balance between actin-mediated protrusion of lamellipodia and cell-matrix adhesion is required for the efficient cell migration, and both insufficient and excessively strong matrix adhesion can impede cell motility [1; 4; 25; 26], Thus, Coronin 1C can either promote cell migration by increasing the rate of F-actin turnover at the leading edge [11; 27; 28], or attenuate it by decreasing the avidity of cell-matrix adhesions (Fig. 4B). We hypothesize that the prevalence of each regulatory mechanism is likely to be dependent on the cell type and migratory mode, being more pronounced in cells that are characterized by strong adhesion to each other and ECM, such as IECs, and less pronounced in poorly-adherent cells with a fast actin turnover.
To confirm this hypothesis, we studied the effects of Coronin 1C depletion on cell motility, cell-matrix adhesion and spreading in HeLa cells, which have an epithelial origin, but display fibroblast-like behavior. In contrast to IECs, knock down of Coronin 1C (Suppl. Fig. 2A) resulted in significantly (1.4±0.1 fold) attenuated migration of HeLa cells (Suppl. Fig. 2B). This result is in good agreement with previous reports showing a positive role of coronin family members in regulating cell motility of fibroblasts and HEK 293 cells [7; 16]. Importantly, depletion of Coronin 1C had no apparent effect on either cell-matrix adhesion, or spreading of HeLa cells (Suppl. Fig. 2 C,D), which further supports our hypothesis on distinct mechanisms underlying anti-migratory and pro-migratory roles of coronins in different cell types.
Further studies are required to dissect the molecular mechanism(s) responsible for Coronin 1C-dependent inhibition of FAK signaling. One possibility is that Coronin 1C forms a ternary complex with actin, Arp2/3 and FAK. Indeed, a direct interaction between Arp2/3 and FAK has been reported to coordinate integrin signaling and actin dynamics [29], and coronins are binding partners for Arp2/3 complex [7; 10; 16]. However, we failed to co-immunoprecipitate Coronin 1C with FAK, and our immunolocalization studies revealed no co-localization of Coronin 1C with P(397)FAK and P(118)paxillin at focal contacts in both wounded and spreading IECs (our unpublished data). Another possibility is indirect regulation of FAK-mediated signaling via adaptor proteins involved in linking integrin-mediated focal contacts with the cytoskeleton, such as vinculin, which has been shown to co-localize and co-immunoprecipitate with neuronal tissue-restricted Coronin 2B (Clipin C) [30]. Understanding the regulatory mechanisms of Coronin 1C action will provide a deeper insight into the basic mechanisms underlying both cell motility and cell-matrix adhesion.
Supplementary Material
Acknowledgments
We thank Dr. Dr. E. Rodriguez-Boulan for SK-CO15 cells, Drs. A. Noegel and C. Clemen for WT Coronin 1C constructs, Dr. V. Glonty for help with image analysis, and Mr. R.E. Karaffa for the cell sorting. This work was supported by Crohn's and Colitis Foundation of America (Research Fellowship Award to S.S. and S.K., and Carrier Development Award to A.I.I.), National Institute of Health grants DK 61379 and DK 72564 (to C.A.P.), DK 55679, and DK 59888 (to A.N.) and a Digestive Diseases Minicenter grant DK 064399.
Abbreviations
- IEC
intestinal epithelial cell
- Arp
actin-related protein
- ADF
actin-depolymerizing factor
- RMLC
regulatory myosin light chain
- FA
focal adhesion
- FAK
focal adhesion kinase
- ECM
extracellular matrix
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–69. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 2.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 3.Carlier MF, Pantaloni D. Control of actin assembly dynamics in cell motility. J Biol Chem. 2007;282:23005–9. doi: 10.1074/jbc.R700020200. [DOI] [PubMed] [Google Scholar]
- 4.Lock JG, Wehrle-Haller B, Stromblad S. Cell-matrix adhesion complexes: master control machinery of cell migration. Semin Cancer Biol. 2008;18:65–76. doi: 10.1016/j.semcancer.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Uetrecht AC, Bear JE. Coronins: the return of the crown. Trends Cell Biol. 2006;16:421–6. doi: 10.1016/j.tcb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 6.de Hostos EL. The coronin family of actin-associated proteins. Trends Cell Biol. 1999;9:345–50. doi: 10.1016/s0962-8924(99)01620-7. [DOI] [PubMed] [Google Scholar]
- 7.Cai L, Holoweckyj N, Schaller MD, Bear JE. Phosphorylation of coronin 1B by protein kinase C regulates interaction with Arp2/3 and cell motility. J Biol Chem. 2005;280:31913–23. doi: 10.1074/jbc.M504146200. [DOI] [PubMed] [Google Scholar]
- 8.Cai L, Makhov AM, Bear JE. F-actin binding is essential for coronin 1B function in vivo. J Cell Sci. 2007;120:1779–90. doi: 10.1242/jcs.007641. [DOI] [PubMed] [Google Scholar]
- 9.Cai L, Makhov AM, Schafer DA, Bear JE. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 2008;134:828–42. doi: 10.1016/j.cell.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foger N, Rangell L, Danilenko DM, Chan AC. Requirement for coronin 1 in T lymphocyte trafficking and cellular homeostasis. Science. 2006;313:839–42. doi: 10.1126/science.1130563. [DOI] [PubMed] [Google Scholar]
- 11.Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–29. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rybakin V, Clemen CS. Coronin proteins as multifunctional regulators of the cytoskeleton and membrane trafficking. Bioessays. 2005;27:625–32. doi: 10.1002/bies.20235. [DOI] [PubMed] [Google Scholar]
- 13.Iizaka M, Han HJ, Akashi H, Furukawa Y, Nakajima Y, Sugano S, Ogawa M, Nakamura Y. Isolation and chromosomal assignment of a novel human gene, CORO1C, homologous to coronin-like actin-binding proteins. Cytogenet Cell Genet. 2000;88:221–4. doi: 10.1159/000015555. [DOI] [PubMed] [Google Scholar]
- 14.Okumura M, Kung C, Wong S, Rodgers M, Thomas ML. Definition of family of coronin-related proteins conserved between humans and mice: close genetic linkage between coronin-2 and CD45-associated protein. DNA Cell Biol. 1998;17:779–87. doi: 10.1089/dna.1998.17.779. [DOI] [PubMed] [Google Scholar]
- 15.Spoerl Z, Stumpf M, Noegel AA, Hasse A. Oligomerization, F-actin interaction, and membrane association of the ubiquitous mammalian coronin 3 are mediated by its carboxyl terminus. J Biol Chem. 2002;277:48858–67. doi: 10.1074/jbc.M205136200. [DOI] [PubMed] [Google Scholar]
- 16.Rosentreter A, Hofmann A, Xavier CP, Stumpf M, Noegel AA, Clemen CS. Coronin 3 involvement in F-actin-dependent processes at the cell cortex. Exp Cell Res. 2007;313:878–95. doi: 10.1016/j.yexcr.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 17.Ivanov AI, McCall IC, Babbin B, Samarin SN, Nusrat A, Parkos CA. Microtubules regulate disassembly of epithelial apical junctions. BMC Cell Biol. 2006;7:12. doi: 10.1186/1471-2121-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogel V, Sheetz MP. Cell fate regulation by coupling mechanical cycles to biochemical signaling pathways. Curr Opin Cell Biol. 2009;21:38–46. doi: 10.1016/j.ceb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannone G, Dubin-Thaler BJ, Rossier O, Cai Y, Chaga O, Jiang G, Beaver W, Dobereiner HG, Freund Y, Borisy G, Sheetz MP. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128:561–75. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–7. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 22.Lu Z, Jiang G, Blume-Jensen P, Hunter T. Epidermal growth factor-induced tumor cell invasion and metastasis initiated by dephosphorylation and downregulation of focal adhesion kinase. Mol Cell Biol. 2001;21:4016–31. doi: 10.1128/MCB.21.12.4016-4031.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sieg DJ, Hauck CR, Schlaepfer DD. Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J Cell Sci. 1999;112(Pt 16):2677–91. doi: 10.1242/jcs.112.16.2677. [DOI] [PubMed] [Google Scholar]
- 24.Tilghman RW, Slack-Davis JK, Sergina N, Martin KH, Iwanicki M, Hershey ED, Beggs HE, Reichardt LF, Parsons JT. Focal adhesion kinase is required for the spatial organization of the leading edge in migrating cells. J Cell Sci. 2005;118:2613–23. doi: 10.1242/jcs.02380. [DOI] [PubMed] [Google Scholar]
- 25.Csucs G, Quirin K, Danuser G. Locomotion of fish epidermal keratocytes on spatially selective adhesion patterns. Cell Motil Cytoskeleton. 2007;64:856–67. doi: 10.1002/cm.20230. [DOI] [PubMed] [Google Scholar]
- 26.DiMilla PA, Stone JA, Quinn JA, Albelda SM, Lauffenburger DA. Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength. J Cell Biol. 1993;122:729–37. doi: 10.1083/jcb.122.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kueh HY, Charras GT, Mitchison TJ, Brieher WM. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J Cell Biol. 2008;182:341–53. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J Cell Biol. 2006;175:315–24. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serrels B, Serrels A, Brunton VG, Holt M, McLean GW, Gray CH, Jones GE, Frame MC. Focal adhesion kinase controls actin assembly via a FERM-mediated interaction with the Arp2/3 complex. Nat Cell Biol. 2007;9:1046–56. doi: 10.1038/ncb1626. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura T, Takeuchi K, Muraoka S, Takezoe H, Takahashi N, Mori N. A neurally enriched coronin-like protein, ClipinC, is a novel candidate for an actin cytoskeleton-cortical membrane-linking protein. J Biol Chem. 1999;274:13322–7. doi: 10.1074/jbc.274.19.13322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


