Abstract
The aspirin esterase activity of human plasma is due to butyrylcholinesterase and albumin. Our goal was to identify the amino acid residues involved in the aspirin esterase activity of albumin. Fatty acid free human albumin and human plasma were treated with aspirin for 5 min to 24 h. Acetylated residues were identified by LC/MS/MS and MALDI-TOF/TOF mass spectrometry of tryptic peptides. Treatment with 0.3 mM aspirin resulted in acetylation of Lys-199, Lys-402, Lys-519, and Lys-545. Treatment with 20 mM aspirin resulted in acetylation of 26 lysines. There was no acetylation of Tyr-411, under any conditions. Acetylated lysine was stable for at least 21 days at pH 7.4, 37 °C. Albumin acetylated by aspirin had reduced esterase activity with beta-naphthyl acetate as shown on gels stained for esterase activity. It was concluded that the aspirin esterase activity of albumin is a pseudo-esterase activity in which aspirin stably acetylates lysines and releases salicylate.
Keywords: Albumin, Aspirin, Mass spectrometry, Acetylated lysine, Pseudo-aspirinase activity
1. Introduction
Hawkins et al. reported that aspirin transfers its acetyl to the ε-amino group of lysine residues of albumin both in vitro [1] and in vivo [2]. Later the labeled residue was identified as Lys-199 [3]. The crystal structure of human albumin confirmed that aspirin acetylates Lys-199 [4]. To date no other acetylated residues have been identified, though up to 6 additional radiolabeled peptides were found on the radioautograph of the peptide map of aspirin-treated albumin [2].
Albumin is regarded as an aspirin esterase because salicylic acid is produced by incubation of aspirin with albumin [5]. The esterase activity of albumin with p-nitrophenyl acetate is predominantly a pseudo-esterase activity in which up to 59 lysines are stably acetylated. In addition, Tyr-411 of human albumin is rapidly acetylated by p-nitrophenyl acetate (t1/2 = 0.56 min) and slowly deacetylated (t1/2 = 61 h) [6,7]. The three goals of our study were to determine whether the aspirin esterase activity of albumin is also a pseudo-esterase activity resulting in stable acetylation of many residues; if so, to identify the acetylated residues by mass spectrometry; and to determine whether Tyr-411 is involved in the reaction of albumin with aspirin.
2. Materials and methods
2.1. Materials
A 1 mg/ml solution of fatty acid-free human albumin (Fluka 05418, via Sigma-Aldrich, St. Louis, MO) was prepared in 100 mM potassium phosphate buffer, pH 7.4. The amino acid sequence for the albumin from this source is given in accession number gi:122920512. This albumin has Glu-396 in peptide QNCELFE*QLGEYK where other albumin sequences have Lys-396. A 1 mg/ml solution of porcine pepsin (Sigma-Aldrich P6887) in 10 mM HCl, as well as 20 μg of sequencing grade modified trypsin (Promega V5113, Madison, WI) in 50 μl of 50 mM acetic acid were stored at -80 °C. A saturated solution of α-cyano-4-hydroxycinnamic acid matrix (CHCA) (Applied Biosystems, Foster City, CA) in 50% acetonitrile, 0.1% trifluoroacetic acid was stored at room temperature. Acetylsalicylic acid (aspirin) (Sigma-Aldrich A5376) was dissolved in ethanol to make 0.1 M and 1 M solutions and stored at room temperature. Beta-naphthyl acetate, Fast Blue RR, trifluoroacetic acid and iodoacetamide were from Sigma-Aldrich. Affi-Gel Blue was from Bio-Rad, Hercules, CA. Dithiothreitol, sodium azide, and ammonium bicarbonate were purchased from Fisher Scientific, Fair Lawn, NJ. Acetonitrile was of LC-grade. Water was purified with the Milli-Q system, followed by distillation. Human plasma in heparin anticoagulant was from Innovative Research, Novi, MI.
2.2. Incubation of pure human serum albumin with aspirin
In general, 1 ml of a 1 mg/ml solution of fatty acid-free human albumin (15 μM) in 100 mM phosphate buffer, pH 7.4 was incubated with 0.05 mM to 20 mM aspirin for 24 h at 37 °C. For one experiment, 1 ml of 40 mg/ml solution of human albumin (600 μM) in 100 mM phosphate buffer, pH 7.4 was reacted with 0.3 mM aspirin for 24 h at 37 °C. Control albumin solution was treated with 20 μl ethanol. Treatment with 10-20 mM concentrations of aspirin caused a pH drop, which was corrected by addition of 1 M NaOH.
2.3. Albumin esterase activity staining on nondenaturing PAGE
A 0.75 mm thick, 4-30% polyacrylamide gradient gel was prepared in a Hoefer gel apparatus and run at a constant voltage for 5000 volt-hours (250 V for 20 h) at 4 °C. 5 μl of human plasma, estimated to contain 200 μg albumin, or 20 μl of 10 mg/ml albumin premixed with an equal volume of 50% glycerol, 0.1% bromophenol blue, was loaded per lane. Esterase activity of albumin was visualized by incubating the gel in 100 ml of 50 mM Tris-Cl pH 7.4 solution, containing 50 mg of β-naphthyl acetate dissolved in 1 ml ethanol and 50 mg of Fast Blue RR. Within 10-20 minutes pink bands formed due to the reaction of released β-naphthol with the diazonium salt of Fast Blue RR [8]. The gel was counterstained with Coomassie Blue.
2.4. In vitro modification of human plasma and isolation of albumin
100 μl of human plasma was incubated with 0.3 mM aspirin for 1 h 40 min at 37 °C. Non-treated plasma was used as a control. After the incubation albumin was isolated by affinity chromatography on Blue Affi-Gel. Briefly, 200 μl of affinity media was packed into a microfuge spin column and equilibrated with binding buffer (50 mM potassium phosphate buffer pH 7.0). Plasma samples were diluted 1:20 and 400 μl were applied to the column. Columns were washed with binding buffer until the absorbance of the eluate at 280 nm was zero. Proteins were eluted with 1.5 M KCl in 50 mM potassium phosphate buffer pH 7.0. The absorbance at 280 nm of 1 ml fractions was measured. Albumin concentration was calculated with the formula: A280=ε*l*c where ε is the extinction coefficient of albumin (35700 M-1cm-1) [9], c is the unknown albumin concentration, and l is the pathlength of the cuvette (1 cm). Purified albumin was subjected to tryptic digestion as described in section 2.5.
2.5. Trypsin digestion
Human albumin (1 mg/ml) modified by aspirin was denatured by boiling for 10 min in the presence of 10 mM dithiothreitol, carbamidomethylated with 90 mM iodoacetamide (1 h-incubation in the dark at 37 °C), and dialyzed against 2×4 liters of 10 mM ammonium bicarbonate. Denaturation in urea was avoided because urea adds carbamate (CONH2) to lysine. The added mass of +43 amu from carbamate could be confused with the added mass of +42 amu from acetate (COCH3). A 100-μg aliquot was digested with 2 μg of Promega trypsin overnight at 37 °C. The trypsin digest was subjected to MALDI and Q-TRAP mass spectrometric analyses.
2.6. Pepsin digestion
Pure human serum albumin (15 μM) in 100 mM phosphate pH 7.4 was treated with 0.3, 3, and 20 mM concentrations of aspirin and incubated at 37 °C for 5, 10, 15, 20, 30, 40, 50, 60 min, 3 h, and 24 h. The reaction between aspirin and albumin was stopped by addition of 50 μl of 1% trifluoroacetic acid to 50 μl of the reaction mixture. The samples were digested with 2 μl of 1 mg/ml pepsin at 37 °C for 1.5 h. Samples were diluted 1:10 in water and analyzed in a MALDI-TOF/TOF 4800 mass spectrometer.
2.7. HPLC purification of the acetylated peptide LK*CASLQK and stability assay
1 ml of 1 mg/ml human albumin in 100 mM potassium phosphate pH 7.4 was treated with 0.3 mM aspirin for 24 hours, reduced, carbamidomethylated, dialyzed, and digested with trypsin as described above. The LK*CASLQK peptide was purified on a Waters 625 LC system using a Phenomenex Prodigy 5 micron C18 100×4.60 mm column. Peptides were eluted with a 60-min gradient from 0.1% trifluoroacetic acid in water to 60% acetonitrile, 0.09% trifluoroacetic acid at a flow rate of 1 ml/min. 1 ml fractions were collected. To identify the fraction containing the peptide of interest 1 μl of each fraction was analyzed in the MALDI-TOF/TOF mass spectrometer. The acetylated, carbamidomethylated peptide LK*CASLQK eluted between 16-17 min. This fraction was dried in the SpeedVac, dissolved in 30 μl of 50 mM potassium phosphate buffer, pH 7.4, containing 0.1% (w/v) sodium azide, and incubated at 37 °C. Aliquots were withdrawn over a period of 3 weeks for measurement of acetylated and deacetylated peptide. A 1-μl aliquot was diluted 10-fold in water and analyzed in the MALDI-TOF/TOF 4800 mass spectrometer. The percentage of acetylation was calculated by dividing the cluster area of the acetylated peptide by the sum of the cluster areas for the acetylated and deacetylated peaks [7].
2.8. Analysis in a MALDI-TOF/TOF 4800 (Applied Biosystems, Foster City, CA) mass spectrometer
Essentially salt-free 1-μl samples were spotted on a MALDI target plate, dried in air, and overlaid with 1 μl of saturated CHCA in 50% acetonitrile, 0.1% trifluoroacetic acid. MS spectra were acquired with laser power at 3500 volts in positive reflector mode. For MS/MS spectra laser intensity was 4000 volts. Each spectrum was the average of 500 laser shots. The mass spectrometer was calibrated against bradykinin (904.47 m/z), angiotensin 1 (1296.68 m/z), Glu-fibrinopeptide B (1570.68 m/z), adrenocorticotropic hormone (ACTH) 1-17 clip (2093.09 m/z), ACTH 18-39 clip (2465.20 m/z), and ACTH 7-38 clip (3657.96 m/z) (Cal Mix 5 from Applied Biosystems). Spectra were analyzed with Data Explorer Software.
2.9. LC/MS/MS with the Q-TRAP 2000 mass spectrometer
Tryptic digests were dried in a vacuum centrifuge and dissolved in 5% acetonitrile, 0.1% formic acid to make 6 pmol/μl. A 10-μl aliquot was injected into the HPLC nanocolumn (218MS3.07515 Vydac C18 polymeric rev-phase, 75 μm i.d. × 150 mm long; P.J. Cobert Assoc, St. Louis, MO). Peptides were separated with a 90-min linear gradient from 0 to 60% acetonitrile at a flow rate of 0.3 μl/min and electrosprayed through a fused silica emitter (360 μm o.d., 75 μm i.d., 15 μm taper, New Objective) directly into the Q-TRAP 2000 (Applied Biosystems, Foster City, CA), a hybrid quadrupole linear ion trap mass spectrometer. An ion-spray voltage of 1900 V was maintained between the emitter and the mass spectrometer. Information dependent acquisition was used to collect MS, enhanced MS, and MS/MS spectra for the three most intense peaks in each cycle, having a charge of +1 to +4, a mass between 200 and 1700 m/z, and an intensity >10,000 cps. All spectra were collected in the enhanced mode, using the trap function. Precursor ions were excluded for 30 s after one MS/MS spectrum had been collected. The collision cell was pressurized to 40 μTorr with pure nitrogen and collision energies between 20 and 40 eV were determined automatically by the software based on the mass and charge of the precursor ion. The mass spectrometer was calibrated on selected fragments from the MS/MS spectrum of Glu-fibrinopeptide B. The MS/MS data were processed using Analyst 1.4.1 software and submitted to Mascot for identification of peptide sequences [10].
3. Results
3.1. Residues acetylated by 0.3 mM aspirin
Two different mass spectrometry methods (MALDI-TOF/TOF and Q-TRAP MS/MS) were applied to identify tryptic peptides of albumin acetylated by aspirin. A 24-h reaction of 15 μM human albumin with 0.3 mM aspirin at 37 °C resulted in the acetylation of four albumin residues, namely Lys-199, Lys-402, Lys-519, and Lys-545. No additional acetylation sites were found when the physiological concentration of albumin (600 μM) was used. The masses of labeled tryptic peptides were increased by +42 m/z due to acetylation. MS/MS spectra of candidate peptides were analyzed to confirm the identity of the modified peptide as well as to identify the site of modification.
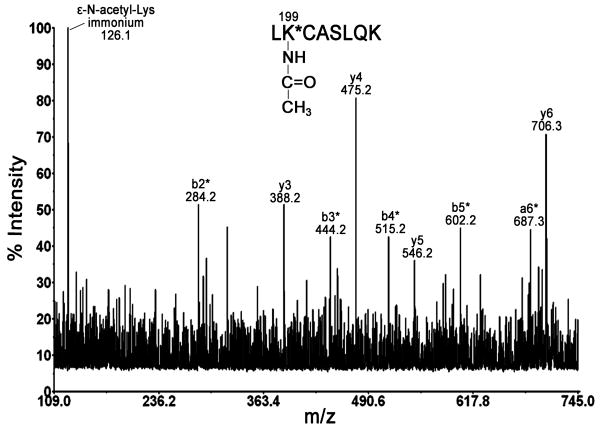
Fig. 1 shows the MALDI MS/MS spectrum of parent ion 989.5 m/z, which corresponds to acetylated peptide LK*CASLQK. Lys-199 was identified as the adduction site by the presence of unacetylated y3, y4, y5, and y6 ions at m/z 388.2, 475.2, 546.2, and 706.3, respectively and by the presence of the acetylated b-ion series b2*, b3*, b4*, and b5* at m/z 284.2, 444.2, 515.2, and 602.2, respectively. The presence of ε-N-acetyllysine immonium ion at m/z 126.1 provides additional evidence for the assignment of acetyllysine-containing peptide [11].
Fig. 1. Identification of acetylated residue Lys-199.
MALDI MS/MS spectrum of parent ion 989.5 m/z shows major y- and b-ions of peptide LK*C(carbamidomethylated)ASLQK. The asterisk * denotes +42 m/z shifted ions as compared to native peptide. Carbamidomethylation of cysteine adds +57 m/z.
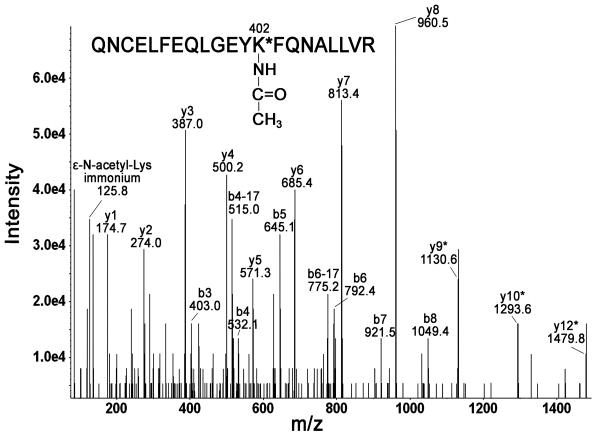
Lys-402 was confirmed to be acetylated by Q-TRAP MS/MS analysis of parent ion [M+3H]3+ at m/z 881.54 (Fig. 2). The acetylated peptide was unambiguously identified as QNCELFEQLGEYK*FQNALLVR, characterized by the partial b-series (b3-b8) and the partial y-series (y1-y8). Lys-402 was identified as the position of acetylation by the presence of the adducted ions y9*, y10*, and y12*. The presence of ε-N-acetyllysine immonium ion at m/z 125.8 served as additional confirmation of acetylation on lysine [11].
Fig. 2. Identification of Lys-402 as an acetylation site.
Q-TRAP MS/MS fragmentation of parent ion [M+3H]3+ at m/z 881.54 yielded y- and b-ion series consistent with acetylated peptide QNC(carbamidomethylated)ELFEQLGEYK*FQNALLVR. The asterisk * indicates +42 m/z shift in ion mass as compared to native peptide ions.
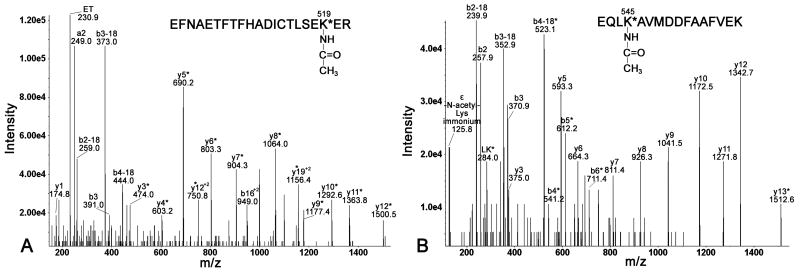
Q-TRAP MS/MS fragmentation of parent ion [M+3H]3+ at m/z 863.46 is shown in Fig. 3A. The spectrum confirms the identity of this acetylated peptide as EFNAETFTFHADICTLSEK*ER and confirms that the adducted residue is Lys-519. Supporting ions are y1, y3*, y4*, y5*, y6*, y7*, y8*, y9*, y10*, y11*, y12*, y12*+2, y19*+2, b3, b16*+2, where the asterisk indicates acetylated ions, as well as b ions that have lost water (b-18). The internal fragment at m/z 230.9 has the sequence ET.
Fig. 3. Identification of Lys-519 and Lys-545 as adduction sites.
Q-TRAP MS/MS spectra of parent ion [M+3H]3+ at m/z 863.46 (panel A) and parent ion [M+2H]2+ at m/z 942.16 (panel B) show major y- and b-ions, as well as internal fragments. Acetylated fragment ions are labeled with an asterisk.
The Q-TRAP MS/MS spectrum of parent ion [M+2H]2+ at m/z 942.16, in Fig. 3B, corresponds to the acetylated peptide EQLK*AVMDDFAAFVEK. The presence of ε-N-acetyllysine immonium ion at m/z 125.8 confirms the presence of acetylated lysine [11]. The MS/MS fragmentation, yielding a partial b-series (b2, b3, b4*-b6*), a partial y-series (y3, y5-y12, y13*) and internal fragment LK* at 284.0 m/z, confirms that Lys-545, but not Lys-557 is acetylated. The ions marked with an asterisk are acetylated.
3.2. Residues acetylated by 20 mM aspirin
Incubation of 15 μM human albumin with 20 mM aspirin at pH 7.4, 37 °C for 24 h resulted in the acetylation of 26 lysines, listed in Table 1. To identify labeled peptides, in silico trypsin digestion of albumin was performed (ProteinProspector v 5.3.0 http://prospector.ucsf.edu), taking into account carbamidomethylation of cysteine and possible oxidation of methionine, as well as possible missed cleavages. Theoretical masses of acetylated tryptic peptides were calculated and used to select candidates for acetylated ions from the MALDI MS spectrum. To confirm peptide sequences and identify adducted residues, peptides were analyzed by MALDI MS/MS.
Table 1.
Tryptic peptides found to be acetylated after 24-h reaction of 15 μM human albumin with 20 mM aspirin at 37 °C. The asterisk * indicates the adducted lysine.
| Sequence position | Peptide sequence | Acetylated lysine | Mass of unlabeled peptide, m/z | Mass of labeled peptide (mass shift 42), m/z | Method of identification |
|---|---|---|---|---|---|
| 11-20 | FK*DLGEENFK | 12 | 1226.6 | 1268.6 | MALDI MSMS, Q-TRAP |
| 65-81 | SLHTLFGDK*LCTVATLR | 73 | 1932.0 | 1974.0 | MALDI MSMS, Q-TRAP |
| 82-98 | ETYGEMADCCAK*QEPER | 93 | 2073.8 | 2115.8 | MALDI MSMS |
| 99-114 | NECFLQHK*DDNPNLPR | 106 | 1996.9 | 2038.9 | Q-TRAP |
| 115-137 | LVRPEVDVMCTAFHDNEETFLK*K | 136 | 2778.3 | 2820.3 | MALDI MSMS |
| 137-144 | K*YLYEIAR | 137 | 1055.6 | 1097.6 | MALDI MSMS, Q-TRAP |
| 146-160 | HPYFYAPELLFFAK*R | 159 | 1899.0 | 1941.0 | MALDI MSMS |
| 161-174 | YK*AAFTECCQAADK | 162 | 1662.7 | 1704.7 | MALDI MSMS, Q-TRAP |
| 163-181 | AAFTECCQAADK*AACLLPK | 174 | 2125.0 | 2167.0 | Q-TRAP |
| 198-205 | LK*CASLQK | 199 | 947.5 | 989.5 | MALDI MSMS |
| 200-209 | CASLQK*FGER | 205 | 1195.6 | 1237.6 | MALDI MSMS, Q-TRAP |
| 210-218 | AFK*AWAVAR | 212 | 1019.6 | 1061.6 | MALDI MSMS, Q-TRAP |
| 223-233 | FPK*AEFAEVSK | 225 | 1252.6 | 1294.6 | Q-TRAP |
| 258-274 | ADLAK*YICENQDSISSK | 262 | 1941.9 | 1983.9 | MALDI MSMS, Q-TRAP |
| 277-286 | ECCEK*PLLEK | 281 | 1305.6 | 1347.6 | Q-TRAP |
| 318-336 | NYAEAK*DVFLGMFLYEYAR | 323 | 2300.1 | 2342.1 | MALDI MSMS |
| 349-359 | LAK*TYETTLEK | 351 | 1296.7 | 1338.7 | Q-TRAP |
| 373-389 | VFDEFK*PLVEEPQNLIK | 378 | 2045.1 | 2087.1 | Q-TRAP |
| 390-410 | QNCELFEQLGEYK*FQNALLVR | 402 | 2599.3 | 2641.3 | MALDI MSMS, Q-TRAP |
| 414-428 | K*VPQVSTPTLVEVSR | 414 | 1639.9 | 1681.9 | Q-TRAP |
| 473-484 | VTK*CCTESLVNR | 475 | 1466.7 | 1508.7 | MALDI MSMS, Q-TRAP |
| 501-521 | EFNAETFTFHADICTLSEK*ER | 519 | 2545.2 | 2587.2 | MALDI MSMS, Q-TRAP |
| 525-534 | K*QTALVELVK | 525 | 1128.7 | 1170.7 | Q-TRAP |
| 539-545 | ATK*EQLK | 541 | 817.5 | 859.5 | MALDI MSMS |
| 542-557 | EQLK*AVMDDFAAFVEK | 545 | 1840.9 | 1882.9 | MALDI MSMS, Q-TRAP |
| 546-560 | AVMDDFAAFVEK*CCK | 557 | 1790.8 | 1832.8 | MALDI MSMS, Q-TRAP |
The albumin sequence is in accession # gi:122920512 in the NCBI protein database. Monoisotopic masses of singly charged ions are given. Cysteines are carbamidomethylated, adding a mass of +57 amu. Acetylated lysines have an added mass of +42 amu.
Q-TRAP MS/MS analysis served as an additional tool for the identification of acetylated peptides. All MS/MS spectra were submitted to Mascot for comparison with the NCBInr human protein database. Peptides corresponding to 60% coverage of the albumin sequence, in accession # gi:122920512, were obtained. MS/MS spectra of candidates for acetylated peptides were manually evaluated and only strong, well-assigned spectra, comparable to those shown in Figs. 1-3, were considered as evidence for peptide acetylation.
All peptides listed in Table 1 have a missed cleavage at the acetylated lysine. This is because trypsin is unable to recognize adducted lysine as a cleavage site, in agreement with previous literature reports [12].
3.3. Mass spectrometric analysis of commercial albumin
Interestingly, acetylation of non-treated pure human albumin was observed. The acetylated residues in commercial albumin were Lys-199, and Lys-525. Less than 0.1 % of commercial albumin was acetylated. In contrast, significant amounts of albumin were acetylated after treatment with aspirin.
3.4. Reaction of aspirin with human plasma
To study the reaction between aspirin and albumin under physiological conditions, 100 μl of human plasma was incubated directly with 0.3 mM aspirin for 1 h 40 min at 37 °C. Albumin was isolated and digested with trypsin as described in materials and methods section. Analysis of the tryptic digest on the MALDI-TOF/TOF mass spectrometer revealed acetylation of Lys-199 and Lys-519.
To model repeated doses of aspirin, 100 μl of human plasma was treated with 0.3 mM aspirin three times. The time between treatments was 1 h 40 min. Two treatments with 0.3 mM aspirin resulted in acetylation of four residues, Lys-199, Lys-402, Lys-519, and Lys-545. Three serial treatments led to the acetylation of 5 residues, the fifth being Lys-136.
Thus, under physiological conditions the most reactive albumin residues were Lys-199 and Lys-519, appearing after the first treatment with 0.3 mM aspirin. Additional “doses” of aspirin led to accumulation of acetylation.
3.5. The role of Tyr-411 in the reaction between aspirin and albumin
To study the involvement of Tyr-411 in the reaction, 15 μM fatty acid-free human albumin was treated with different concentrations of aspirin, digested with pepsin and analyzed on the MALDI mass spectrometer. Pepsin was chosen because experience has shown that peptic peptides containing Tyr-411 are readily observed in both the MALDI and Q-TRAP mass spectrometers. Acetylation of Tyr-411 increases the mass of peptic peptides VRYTKKVPQVSTPTL and LVRYTKKVPQVSTPTL (missed cleavage) at 1717 m/z and 1830 m/z by +42 to 1759 and 1872 m/z [7]. Samples that were incubated with aspirin for 5 min to 3 h gave strong signals for the unlabeled peptides but did not result in the appearance of peptides at m/z 1759 and 1872 (data not shown). However, after 24 h-reaction with 20 mM aspirin, low intensity masses at m/z 1759 and 1872 were observed. Q-TRAP MS/MS analysis showed that the +42 m/z mass shift was due to acetylation of Lys-414, but not Tyr-411 (data not shown).
3.6. The stability of acetylated Lys-199
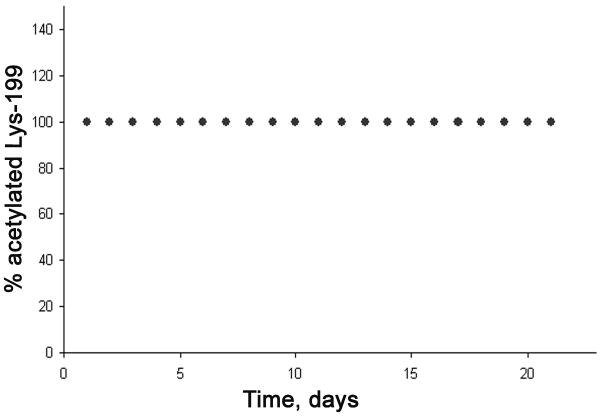
To estimate the stability of acetylation, peptide LK*CASLQK with acetyl on Lys-199 was purified by HPLC and incubated at 37°C in pH 7.4 buffer. At various time intervals a 1 μl aliquot was removed for MALDI-TOF MS analysis to measure the percentage of acetylation. The acetylated peptide at m/z 989.5 would lose 42 amu and shift to a mass of 947.5 m/z if it were deacetylated. Fig. 4 shows that the acetylated peptide was stable for at least 21 days. The absence of non-acetylated peptide after 21 days of incubation leads to the conclusion that acetylation on Lys-199 is stable at pH 7.4, 37 °C.
Fig. 4. The stability of acetylated Lys-199.
Deacetylation was monitored at pH 7.4, 37 °C for 21 days. % acetylation was calculated from cluster area in the MALDI MS spectrum for peptide LK*CASLQK.
3.7. Acetylation sites: order of reactivity
To measure the reactivity of acetylation sites, 15 μM albumin was treated with different concentrations of aspirin for 24 h at 37 °C, digested with trypsin and analyzed in the MALDI mass spectrometer.
Two sites, namely Lys-402 and Lys-519, were acetylated by the lowest aspirin concentration (0.05 mM), when no other sites were labeled. Moreover, peptides containing these acetylated residues rose in abundance with increasing aspirin concentration. Lys-199 was found to be acetylated with 0.1 mM aspirin; however the peak intensity of the peptide containing this lysine was low and did not change with the increase in aspirin concentration.
When albumin was incubated with 0.5 mM aspirin, evidence of acetylation of Lys-73, Lys-136, Lys-137, Lys-159, Lys-212, and Lys-545 appeared in the MS spectrum. Incubation with 20 mM aspirin resulted in acetylation of 26 lysine residues (Table 1).
3.8. Surface location of acetylated residues in human serum albumin
The crystal structure of human albumin is shown in Fig. 5. The 26 acetylated lysines are shown as sticks. Lys-199, Lys-402, Lys-519, and Lys-545 are acetylated by 0.3 mM aspirin. To establish the availability of acetylation sites the solvent accessible surface was analyzed. The solvent accessible surface area is defined as the surface traced out by the center of a water sphere, having a radius of 1.4 angstroms, rolled over the protein atoms [13]. By visual inspection of the molecule, it was concluded that sites Lys-199, Lys-402, Lys-519, as well as Lys-545 are accessible to the solvent.
Fig. 5. Molecular modeling to locate lysines acetylated by aspirin.
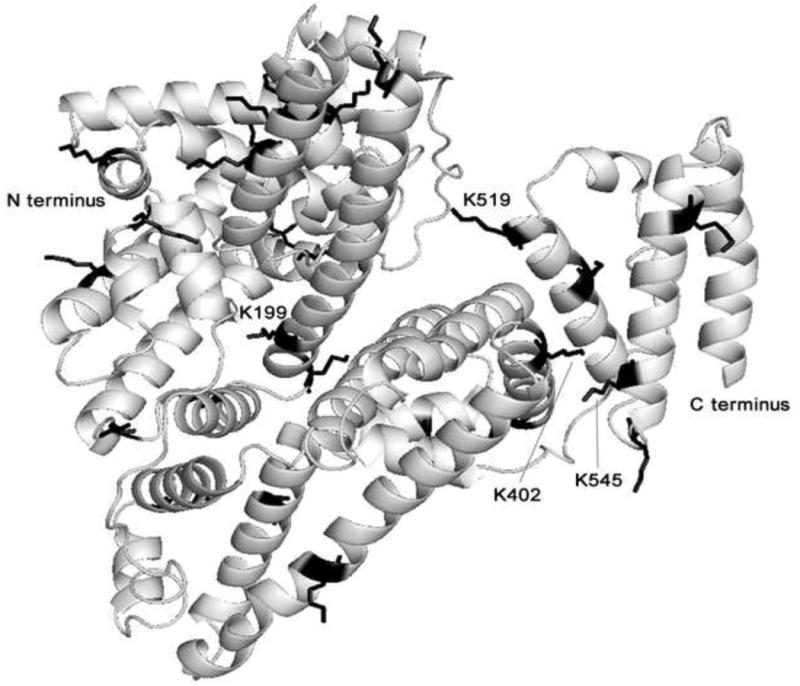
Ribbon model shows the crystal structure of human albumin (Protein Data Bank code 1bmo). The 26 lysines acetylated by 20 mM aspirin are shown in sticks. Lys-199, Lys-402, Lys-519 and Lys-545 are acetylated by 0.3 mM aspirin. The structure was drawn with PyMOL software (DeLano, W.L. The PyMOL Molecular Graphics System (2002) http://www.pymol.org).
3.9. Albumin esterase activity
Staining of a nondenaturing gradient PAGE gel with beta-naphthyl acetate and Fast Blue RR revealed esterase activity of human albumin. The gel in Fig. 6A shows esterase bands of albumin. Fatty acid-free human albumin preincubated for 24 h with different concentrations of aspirin (from 0 to 50 mM) and stained for esterase activity shows decreasing esterase staining with increasing aspirin concentration (lanes 2-8). This result is interpreted to mean that lysines acetylated by aspirin are unavailable for reaction with β-naphthyl acetate. It is concluded that the esterase activity of albumin is due to acetylation of lysines and not to an enzymatic-like hydrolysis. As such albumin exhibits a pseudo-esterase activity. Aspirin by transferring its acetyl group to albumin decreases the number of available esteratic sites.
Fig. 6. Nondenaturing gradient gel stained for esterase activity (A) and counterstained with Coomassie Blue (B).
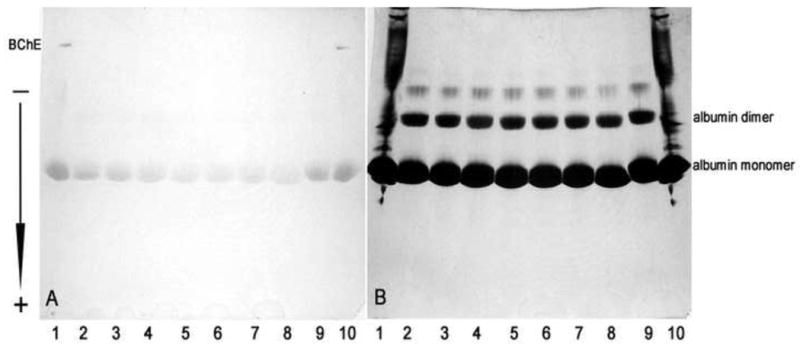
Lanes 1 and 10, control human plasma (5 μl), 0 mM aspirin. Lanes 2-9 contain 200 μg of human albumin treated for 24 h with aspirin. Lane 2, 0 mM aspirin; Lane 3, 1 mM aspirin; Lane 4, 10 mM aspirin; Lane 5, 20 mM aspirin; Lane 6, 30 mM aspirin; Lane 7, 40 mM aspirin; Lane 8, 50 mM aspirin; Lane 9, 0 mM aspirin. Albumin esterase activity in panel A diminishes with increasing aspirin concentrations. Acetylated albumin in lane 8 migrates more rapidly than untreated albumin in lane 9. The arrow shows the direction of migration of proteins on the gel.
To show that equivalent protein concentrations were loaded per lane, the gel was counterstained with Coomassie Blue (Fig. 6B). Pure albumin reveals monomeric forms, as well as albumin dimers and multimers. Albumin treated with 50 mM aspirin (lane 8) migrates slightly further compared to native albumin (lane 9). This fact can be explained by partial elimination of positive charge from the ε-N-amino groups of lysines due to acetylation. Therefore, acetylated protein has a greater net negative charge and migrates more readily to the anode compared to non-treated albumin.
4. Discussion
4.1. Covalent modification of lysines by aspirin
Aspirin transfers its acetyl group to human albumin in vitro. This interaction involves multiple acceptor sites, identified as lysine residues by two different mass spectrometry methods (MALDI TOF/TOF and Q-TRAP). We conclude that the observed adducts are due to covalent binding of the acetyl group to ε-N-amino groups of lysines. Non-covalent adducts do not survive the MALDI process due to the low pH of the matrix as well as the heat generated by high laser energy [14]. Furthermore, non-covalent adducts would not be expected to survive the low pH and high organic solvent conditions that exist in the HPLC step that precedes electrospraying into the Q-TRAP mass spectrometer. Thus, only covalent modifications of albumin are consistent with our experimental conditions.
4.2. Four lysines acetylated by low concentration of aspirin
Aspirin is administered to people in a wide range of doses. Low doses (75-325 mg once a day) are taken to prevent recurrent heart attack and stroke, while high doses (up to 130 mg/kg/day in divided doses) are used to reduce swelling in rheumatoid arthritis patients [15]. The high dose regime amounts to 9100 mg of aspirin per day for a 70 kg (155 lb) individual. Plasma aspirin concentration was found to reach approximately 0.12 mM 20 min after ingestion of 650 mg aspirin [16]. The half-life of aspirin in vivo is approximately 20 min with plasma levels essentially undetectable 1-2 h after ingestion [16,17]. In our study, the 0.3 mM aspirin concentration was comparable to therapeutic blood levels.
The reaction of pure albumin with 0.3 mM aspirin, for 24 h, yielded four modified lysines: Lys-199, Lys-402, Lys-519, and Lys-545. Acetylation of Lys-199 by aspirin has been reported previously [3,4], but the identity of the other acetylated lysines is new in this report. The finding that additional lysines are acetylated is consistent with studies that show the involvement of several sites in the reaction of albumin with low (0.1-0.5 mM) concentrations of aspirin [2,18].
Treatment of human plasma with 0.3 mM aspirin, for 1 h 40 min, resulted in the acetylation of two lysines: Lys-199 and Lys-519. The fact that only two of these lysines were found to be acetylated in plasma may be explained by the shorter incubation period as well as faster hydrolysis of aspirin in plasma compared to phosphate buffer. Aspirin hydrolysis in plasma is catalyzed by butyrylcholinesterase. The half-life of aspirin in 0.1 M phosphate buffer, pH 7.4 at 37 °C is 15.4 h [19] vs. 1.6 h in plasma at 37 °C [20] and vs. 0.8 h in whole blood [20].
To mimic repeated doses of aspirin, plasma was treated with 0.3 mM aspirin 3 times with 1 hour and 40 minutes between treatments. Two treatments led to the acetylation of Lys-199, Lys-402, Lys-519, and Lys-545. The third treatment added Lys-136 to residues found with first two “doses”. The time difference between treatments corresponds to 5 half-lives of aspirin in vivo [17]; consequently about 96% of the aspirin should have been hydrolyzed in 1 h 40 min. However in isolated plasma aspirin was found to survive longer [20,21]. Thus, 1 hour and 40 minutes is not enough for aspirin to break down completely, meaning that the observed increase in the number of acetylation sites may be due to build up of aspirin concentration. The possibility that repeated doses of aspirin would lead to enhancement of acetylation in vivo cannot be ruled out.
4.3. 26 Lysines acetylated by high concentration of aspirin
The reaction of pure albumin with 20 mM aspirin resulted in the acetylation of 26 lysine residues. Six peptides were identified only with MALDI MS/MS, eight only with Q-TRAP MS/MS, and twelve peptides were identified with both methods. This difference between mass spectrometers is not surprising. It is generally accepted that different ionization techniques utilized by MALDI and Q-TRAP mass spectrometers allow identification of more peptides than either method used alone [22,23].
However, this high acetylation burden is not likely to occur in vivo, since a 20 mM aspirin concentration is well beyond the therapeutic blood level. In addition, concentrations of salicylic acid, the metabolite of aspirin, greater than 5.4 mM cause severe toxicity [24].
Lys-199 was proposed to be the preferential site of acetylation in early studies [2]. Our data support this finding: Lys-199 was among the first few sites to be labeled by 0.1 mM aspirin. Moreover, this site together with Lys-519 were the first to react under physiological conditions. Lys-199 reactivity can be explained by the unusually low pKa of its ε-N-amino group (pKa≈8) [9]. This, in turn, may be due to the close proximity of positively charged residues, Lys-195, Arg-281, Arg-222, which would disfavor protonation of Lys-199 [25].
Additional sites with high reactivity towards aspirin were Lys-402 and Lys-545. All of these residues are located on the surface of albumin where they are available to solvent.
4.4. Acetylated lysine is stable
To assess the persistence of acetylation, the stability of acetylated LK*CASLQK peptide was measured. It was found that Lys-199 remains 100% acetylated at pH 7.4, 37 °C for up to 21 days. Assuming an albumin half-life of 20 days in the blood [9], it can be anticipated that once it becomes acetylated, albumin remains acetylated for the remainder of its life-time in the circulation. This idea is supported by the striking finding of a small amount of acetylated albumin in commercial albumin, which indicates that albumin does not lose the acetyl group even under conditions of commercial preparation.
4.5. Tyrosine 411 is not acetylated by aspirin
One of the aims of this study was to establish the role of Tyr-411 in the reaction between aspirin and albumin, since this site was shown to be important in the hydrolysis of other acetyl-containing agents [7,26]. It seemed reasonable to propose that aspirin reacts with albumin by the same mechanism as other phenyl acetates [27]. In which case, labeling of Tyr-411 was expected. In the present study, the reaction with aspirin was stopped after different time points; albumin was digested with pepsin and analyzed by MALDI mass spectrometry to find shifted peptides due to acetylation on Tyr-411. Though the expected mass shift was found, after extended reaction, the labeled residue was Lys-414 not Tyr-411.
Why then does aspirin behave differently from other phenyl esters? Sakurai and coauthors [28] proposed a mechanism by which p-nitrophenyl esters react with Tyr-411. In their model, the side chain of Arg-410 forms a hydrogen bond with the carbonyl oxygen of the ester, thus facilitating nucleophilic attack by the phenolic oxygen of Tyr-411 on the carbonyl carbon of the ester [28]. However, in the case of aspirin the reaction may be complicated by the presence of carboxylic group. Therefore, it is possible that the carboxylic oxygens rather than the carbonyl oxygen of the acetyl group interact with Arg-410, making the acetylation of Tyr-411 impossible due to a wrong orientation of the aspirin molecule.
In conclusion, the aspirin esterase activity of albumin is a half-reaction in which only lysines are stably acetylated. Aspirin molecules are hydrolyzed in a reaction that consumes albumin binding sites, making them unavailable for reaction with other esters. Tyr-411 does not contribute to these reactions.
4.6. Acetylated albumin has altered binding affinity
Albumin function in vivo might be altered by acetylation from aspirin. Interestingly, interaction between aspirin and albumin was first noted because of the increased capacity of aspirin-treated albumin to bind acetrizoate [1]. It was then proposed, that this was a result of structural changes in human albumin induced by acetylation [2]. These early studies generated the idea that albumin affinity for anions that were structurally related to acetrizoate may also be altered by acetylation [1]. Later, acetylation of albumin by aspirin was found to increase the affinity of albumin for phenylbutazone, but decrease its affinity for flufenamic acid [29].
In addition, acetylation of albumin by aspirin was demonstrated to inhibit bilirubin binding [30]. Bilirubin binds to site I located in the IIA subdomain of the albumin molecule [9]. This site also binds aspirin and salicylic acid and includes the reactive residue Lys-199 [4]. Thus, acetylation occurring at binding site I changes the albumin affinity for bilirubin [30].
Finally, binding of aspirin to human albumin was shown to reduce prostaglandins binding to albumin [31]. This was proposed to accelerate clearance of prostaglandins, thus serving as an additional mechanism of the aspirin anti-inflammatory effect [31].
4.7. Summary
Reaction between human albumin and aspirin leads to stable acetylation of multiple lysine residues (see Table 1). Thus the aspirin esterase activity of albumin is more properly classified as a pseudo-esterase activity. Tyr-411 was found not to play a role in this reaction. Considering the widespread use of aspirin by children and adults, investigation into the consequences of the acetylation of albumin on the various functions of albumin in vivo is of interest.
Acknowledgments
Mass spectra were obtained with the support of the Mass Spectrometry and Proteomics core facility at the University of Nebraska Medical Center. This work was supported by a grant from the US Army Medical Research and Materiel Command W81XWH-07-2-0034 and an NCI Cancer Center Support Grant CA36727. M.L. was awarded a Fulbright Russia student grant.
Abbreviations
- PAGE
polyacrylamide gel electrophoresis
- MALDI-TOF/TOF
matrix assisted, laser desorption/ionization tandem mass spectrometer with dual time-of-flight analyzers
- Q-TRAP
hybrid triple quadrupole linear ion trap mass spectrometer
- MS
mass spectrum
- MS/MS
mass spectrum of collision induced dissociation fragments
- LC
liquid chromatography
- CHCA
alpha-cyano-4-hydroxycinnamic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hawkins D, Pinckard RN, Farr RS. Acetylation of human serum albumin by acetylsalicylic acid. Science. 1968;160:780–1. doi: 10.1126/science.160.3829.780. [DOI] [PubMed] [Google Scholar]
- 2.Hawkins D, Pinckard RN, Crawford IP, Farr RS. Structural changes in human serum albumin induced by ingestion of acetylsalicylic acid. J Clin Invest. 1969;48:536–42. doi: 10.1172/JCI106011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker JE. Lysine residue 199 of human serum albumin is modified by acetylsalicyclic acid. FEBS Lett. 1976;66:173–5. doi: 10.1016/0014-5793(76)80496-6. [DOI] [PubMed] [Google Scholar]
- 4.Yang F, Bian C, Zhu L, Zhao G, Huang Z, Huang M. Effect of human serum albumin on drug metabolism: structural evidence of esterase activity of human serum albumin. J Struct Biol. 2007;157:348–55. doi: 10.1016/j.jsb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Morikawa M, Inoue M, Tsuboi M, Sugiura M. Studies on aspirin esterase of human serum. Jpn J Pharmacol. 1979;29:581–6. doi: 10.1254/jjp.29.581. [DOI] [PubMed] [Google Scholar]
- 6.Means GE, Bender ML. Acetylation of human serum albumin by p-nitrophenyl acetate. Biochemistry. 1975;14:4989–94. doi: 10.1021/bi00693a031. [DOI] [PubMed] [Google Scholar]
- 7.Lockridge O, Xue W, Gaydess A, Grigoryan H, Ding SJ, Schopfer LM, et al. Pseudo-esterase activity of human albumin: slow turnover on tyrosine 411 and stable acetylation of 82 residues including 59 lysines. J Biol Chem. 2008;283:22582–90. doi: 10.1074/jbc.M802555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B, Sedlacek M, Manoharan I, Boopathy R, Duysen EG, Masson P, et al. Butyrylcholinesterase, paraoxonase, and albumin esterase, but not carboxylesterase, are present in human plasma. Biochem Pharmacol. 2005;70:1673–84. doi: 10.1016/j.bcp.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Peters T., Jr . All about albumin Biochemistry, genetics, and medical applications. London: Academic Press Ltd.; 1996. [Google Scholar]
- 10.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Trelle MB, Jensen ON. Utility of immonium ions for assignment of epsilon-N-acetyllysine-containing peptides by tandem mass spectrometry. Anal Chem. 2008;80:3422–30. doi: 10.1021/ac800005n. [DOI] [PubMed] [Google Scholar]
- 12.Violand BN, Schlittler MR, Lawson CQ, Kane JF, Siegel NR, Smith CE, et al. Isolation of Escherichia coli synthesized recombinant eukaryotic proteins that contain epsilon-N-acetyllysine. Protein Sci. 1994;3:1089–97. doi: 10.1002/pro.5560030712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.http://www.pymolwiki.org/index.php/Displaying_Biochemical_Properties#Display_solvent_accessible_surface
- 14.Bolbach G. Matrix-assisted laser desorption/ionization analysis of non-covalent complexes: fundamentals and applications. Curr Pharm Des. 2005;11:2535–57. doi: 10.2174/1381612054546923. [DOI] [PubMed] [Google Scholar]
- 15.Bayer Aspirin. http://www.wonderdrug.com/index.html.
- 16.Rowland M, Riegelman S, Harris PA, Sholkoff SD. Absorption kinetics of aspirin in man following oral administration of an aqueous solution. J Pharm Sci. 1972;61:379–85. doi: 10.1002/jps.2600610312. [DOI] [PubMed] [Google Scholar]
- 17.Levy G. Clinical pharmacokinetics of aspirin. Pediatrics. 1978;62:867–72. [PubMed] [Google Scholar]
- 18.Burch JW, Blazer-Yost B. Acetylation of albumin by low doses of aspirin. Thromb Res. 1981;23:447–52. doi: 10.1016/0049-3848(81)90205-x. [DOI] [PubMed] [Google Scholar]
- 19.Bakar SK, Niazi S. Stability of aspirin in different media. J Pharm Sci. 1983;72:1024–6. doi: 10.1002/jps.2600720914. [DOI] [PubMed] [Google Scholar]
- 20.Harthon L, Hedstrom M. Hydrolysis of salicylsalicylic acid in human blood and plasma: a comparison with acetylsalicylic acid. Acta Pharmacol Toxicol(Copenh) 1971;29:155–63. doi: 10.1111/j.1600-0773.1971.tb00602.x. [DOI] [PubMed] [Google Scholar]
- 21.Rylance HJ, Wallace RC. Erythrocyte and plasma aspirin esterase. Br J Clin Pharmacol. 1981;12:436–8. doi: 10.1111/j.1365-2125.1981.tb01244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Zhang S, Howe K, Wilson DB, Moser F, Irwin D, et al. A comparison of nLC-ESI-MS/MS and nLC-MALDI-MS/MS for GeLC-based protein identification and iTRAQ-based shotgun quantitative proteomics. J Biomol Tech. 2007;18:226–37. [PMC free article] [PubMed] [Google Scholar]
- 23.Bodnar WM, Blackburn RK, Krise JM, Moseley MA. Exploiting the complementary nature of LC/MALDI/MS/MS and LC/ESI/MS/MS for increased proteome coverage. J Am Soc Mass Spectrom. 2003;14:971–9. doi: 10.1016/S1044-0305(03)00209-5. [DOI] [PubMed] [Google Scholar]
- 24.Dargan PI, Wallace CI, Jones AL. An evidence based flowchart to guide the management of acute salicylate (aspirin) overdose. Emerg Med J. 2002;19:206–9. doi: 10.1136/emj.19.3.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz N, Suarez D, Sordo TL, Merz KM., Jr Molecular dynamics study of the IIA binding site in human serum albumin: influence of the protonation state of Lys195 and Lys199. J Med Chem. 2001;44:250–60. doi: 10.1021/jm000340v. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe H, Tanase S, Nakajou K, Maruyama T, Kragh-Hansen U, Otagiri M. Role of arg-410 and tyr-411 in human serum albumin for ligand binding and esterase-like activity. Biochem J. 2000;349(Pt 3):813–9. doi: 10.1042/bj3490813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurono Y, Maki T, Yotsuyanagi T, Ikeda K. Esterase-like activity of human serum albumin: structure-activity relationships for the reactions with phenyl acetates and p-nitrophenyl esters. Chem Pharm Bull (Tokyo) 1979;27:2781–6. doi: 10.1248/cpb.27.2781. [DOI] [PubMed] [Google Scholar]
- 28.Sakurai Y, Ma SF, Watanabe H, Yamaotsu N, Hirono S, Kurono Y, et al. Esterase-like activity of serum albumin: characterization of its structural chemistry using p-nitrophenyl esters as substrates. Pharm Res. 2004;21:285–92. doi: 10.1023/b:pham.0000016241.84630.06. [DOI] [PubMed] [Google Scholar]
- 29.Chignell CF, Starkweather DK. Optical studies of drug-protein complexes. V. The interaction of phenylbutazone, flufenamic acid, and dicoumarol with acetylsalicylic acid-treated human serum albumin. Mol Pharmacol. 1971;7:229–37. [PubMed] [Google Scholar]
- 30.Trynda L, Przywarska-Boniecka H, Kosciukiewicz T. Influence of aspirin and iron(III) tetrasulfonated phthalocyanine on bilirubin binding by human serum albumin. J Inorg Biochem. 1990;38:153–67. doi: 10.1016/0162-0134(90)84023-i. [DOI] [PubMed] [Google Scholar]
- 31.Attallah AA, Lee JB. Indomethacin, salicylates and prostaglandin binding. Prostaglandins. 1980;19:311. doi: 10.1016/0090-6980(80)90029-5. [DOI] [PubMed] [Google Scholar]