Abstract
Proteasome inhibition has become synonymous with inhibition of NF-κB activity. However, hyperactive NF-κB responses often accompany physiological conditions marked by proteasomal defects, i.e. advancing age, geriatric diseases, and bortezomib resistance. These paradoxical NF-κB responses are likely to be impervious to proteasomal defects because they stem from atypical NF-κB signaling induced by upstream mechanisms which are proteasome-independent. While this atypical pathway does not require proteasome for NF-κB nuclear translocation, a role for proteasome in regulating nuclear NF-κB remains unexplored. We now demonstrate that proteasome stringently controls transcription of inflammatory mediators regulated by this atypical NF-κB pathway. Proteolytic activity of the proteasome mediates the removal of the NF-κB subunit, p65/RelA, from inflammatory genes, thereby terminating atypical NF-κB-dependent transcriptional responses. For the first time, we demonstrate that both 19S and 20S components of the 26S proteasome complex are recruited to an inflammatory gene promoter; additionally, the 19S and 20S complexes appear to play distinct roles in the negative regulation of NF-κB-dependent transcription. By demonstrating that proteasome regulates the termination of atypical NF-κB-dependent transcriptional responses, these studies clearly indicate a novel, regulatory role for proteasome in atypical NF-κB signaling. Moreover, these results signal a potential interplay between lowered proteasomal function and increased inflammation and may explain why inflammation accompanies physiological conditions under which proteasomal function is compromised, such as during advancing age or following bortezomib treatment. Given this role for proteasome in inflammation resolution, restoration of proteasome function may constitute a novel mechanism for intervening in chronic inflammatory diseases.
Keywords: Proteasome, NF-κB, pervanadate, inflammation, IL-6
1. Introduction
As a principal regulator of inflammatory and immune response genes, the NF-κB/Rel family encompasses five transcription factors, p65/RelA, cRel, RelB, p50, and p52 [1]. NF-κB/Rel proteins are sequestered in the cytoplasm of unstimulated cells in an inactive form through its association with a member of the inhibitory protein (IκB) family. In the canonical pathway of NF-κB induction, IκBs are phosphorylated at two amino-terminal serines, thus targeting them for polyubiquitination and subsequent proteasomal degradation. IκB degradation enables NF-κB to translocate to the nucleus and bind target genes, including IκBα. NF-κB-dependent resynthesis of IκBα promotes a negative feedback loop which is designed to terminate NF-κB signaling. Additionally, proteasomal degradation of transcriptionally-active p65/RelA promotes the prompt termination of NF-κB responses [2]. Collectively, these mechanisms comprise the dominant mechanisms for shutting off canonical NF-κB-dependent transcriptional responses. Presently, it remains unclear if these negative regulatory mechanisms contribute to signal inactivation in additional NF-κB induction pathways.
Although it is lesser-known, the atypical NF-κB pathway is a phospho-tyrosine dependent pathway that is generally associated with redox stimuli such as H2O2, pervanadate and hypoxia/reoxygenation, as well as various growth factors, including ciliary neurotrophic factor (CNTF), epidermal growth factor (EGF), and nerve growth factor (NGF) [3–11]. Upstream signaling within this pathway culminates in tyrosine phosphorylation of IκBα followed by its dissociation from NF-κB without the involvement of the proteasome [3,4]. While mechanisms associated with tyrosine phosphorylation of IκBα have been studied extensively, few studies have focused on the mechanisms which terminate atypical NF-κB-dependent transcription. Recent studies have highlighted a role for IκBα resynthesis but further clarification is required to determine if it has the same significance in the atypical pathway as it has in the canonical pathway [12,13]. Additionally, it is unclear if the atypical pathway employs proteasomal degradation of nuclear p65/RelA as a mechanism for down-regulating NF-κB activity.
Based primarily on in vitro studies, NF-κB induction by tyrosine phosphorylation of IκBα appears to be a vital cellular response to acute events, such as ischemia/reperfusion and hypoxia/reoxygenation [3,5,14,15]. As for its role in chronic, pathological conditions, studies have linked tyrosine phosphorylation of IκBα to the development of inflammatory-osteolytic arthritis and HCV-related hepatocellular carcinoma [16,17]. Furthermore, atypical pathways of NF-κB induction have recently been implicated in the chronic activation of NF-κB which accompanies aging [18]. However, given the paucity of information on this pathway’s negative regulatory mechanisms, it is unclear if these chronic conditions are the result of excessive induction of NF-κB and/or insufficient termination of NF-κB activity.
Distinct from its tyrosine-phosphorylation dependent mechanisms, the atypical pathway has the hallmark of being a proteasome-independent mechanism of NF-κB induction. This is quite significant because proteasome inhibition has become synonymous with inhibition of NF-κB activity. For example, the proteasome inhibitor PS-341 (Velcade/bortezomib) is currently being used in the treatment of multiple myeloma, with the therapeutic benefit of bortezomib partly attributed to its ability to block NF-κB induction [19,20]. Paradoxically, many reports have described the development of inflammatory symptoms and increases in serum levels of pro-inflammatory cytokines following administration of bortezomib [21–23]. As suggested recently, complications and resistance to bortezomib treatment may stem from NF-κB signaling induced by proteasome-independent mechanisms [24]. As yet, there have been few studies, if any, which have proposed that proteasome inhibition might further impact the termination of these atypical NF-κB responses. Hence, we explored the effects of proteasome inhibition on NF-κB-mediated, inflammatory gene transcription induced by upstream mechanisms which are both proteasome-independent and phospho-tyrosine dependent.
We now demonstrate that proteasome stringently controls transcription of inflammatory mediators regulated by the atypical NF-κB pathway. Proteolytic activity of the proteasome mediates the removal of the NF-κB subunit, p65/RelA, from inflammatory genes, thereby terminating atypical NF-κB-dependent transcriptional responses. Both the 19S and 20S components of the 26S proteasome complex are recruited to an inflammatory gene promoter, but appear to play distinct roles in the negative regulation of NF-κB-dependent transcription. Our results uncover a novel role for the proteasome in NF-κB signaling induced by upstream mechanisms which are both proteasome-independent and phospho-tyrosine dependent. Finally, our study provides a molecular basis for the hyperactive NF-κB responses accompanying physiological conditions marked by proteasomal dysfunction, i.e. advancing age, geriatric diseases, and bortezomib resistance.
2. Materials and methods
2.1. Reagents
Phospho-Tyrosine antibody was obtained from Cell Signaling Technology (Danvers, MA). FK2 (Anti-Ubiquitin) and Sug1 and 20S proteasome antibodies were purchased from Biomol (Plymouth Meeting, PA). Phospho-specific IκBα (Tyr-42) was purchased from ECM Biosciences (Versailles, KY). All other antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Enhanced Chemiluminescence reagents were from Amersham (Arlington Heights, IL). All fine chemicals, unless otherwise mentioned, were obtained from Sigma Chemical Company (St. Louis, MO). Electrophoresis supplies and Molecular weight standards were from BioRad (Richmond, CA). Aclacinomycin (Acla), Lactacystin (Lacta), and PSI-1 were from Calbiochem (La Jolla, CA). MG132 was obtained from Sigma Chemical Company (St. Louis, MO). Murine TNFα was purchased from R&D Systems (Minneapolis, MN). SN50 peptide was obtained from Biomol (Plymouth Meeting, PA).
2.2. Cell Culture
Bone marrow derived stromal cell line +/+LDA.11 were maintained in DMEM medium supplemented with 2mM glutamine, 100units/mL penicillin, 100μg/mL streptomycin, and 10% fetal bovine serum. ILU-18 clone was derived from +/+LDA.11 parental line which was stably transfected with a murine IL-6 minigene designated ILU, containing the entire IL-6 promoter and first intron, including the 3′UTR [25]. Both +/+LDA.11 and ILU-18 cells were kindly provided by Dr. Charles O’Brien (UAMS, Little Rock, AR). Murine embryonic fibroblasts (MEF) were obtained from p65−/− and p50−/− mice and were maintained in DMEM supplemented with 10% FBS.
2.3. RNA interference
siRNA duplexes targeting murine Sug1 or β5 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). ILU-18 cells were seeded one day before transfection at a density of 75,000 cells per well. Cells were transfected with either control siRNA or Sug1/β5 siRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Cells underwent transfection twice in 72 hours; thereafter cells were either harvested for protein extraction or appropriately stimulated for analysis by luciferase reporter assay.
2.4. Hypoxia/Reoxygenation
ILU-18 cells were seeded at a density of 1×106 cells/well and plated in medium supplemented with 10mM Hepes. Cells were subjected to hypoxia (H) by incubating at 37°C for 6h in an airtight chamber equilibrated with 5% CO2 and 95% N2. Cells then underwent reoxygenation (R) by exposure to atmospheric oxygen (21% O2) at 37°C for 18 hours. Control cells were maintained at 5% CO2 in atmospheric oxygen.
2.5. Western Blotting
Cytosolic lysates were prepared as described previously [26]. Protein content of cytosolic extracts was determined using BioRad protein assay. Cell lysates equalized for protein (30μg) were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, immuno-blotted with specific antibody/s, and detected using anti-IgG coupled to horseradish peroxidase followed by Enhanced Chemi-luminescence, per manufacturer’s instructions (ECL, Amersham).
2.6. RT-PCR
Total RNA was isolated using TRIzol Reagent (Invitrogen Corporation, Carlsbad, CA). Four microgram of RNA per sample was used in the reverse transcription reaction. The resultant cDNA was amplified using PCR. The resulting PCR products were resolved by 2% agarose gel electrophoresis and visualized by ethidium bromide staining. Gene-specific primers for murine IL-6 and β-ACTIN were designed by Primer3 employing sequences obtained from GenBank [27]. β-actin was used as a loading control.
2.7. Quantitative PCR
Four micrograms of RNA per sample was used in reverse transcription reaction; resulting cDNA was diluted to 100μL and used in subsequent real-time PCR reactions. Real-time fluorescence detection was carried out using BioRad iCycler PCR system. Reactions were performed in micro 96-well reaction plates using 12.5μL of SYBR Green Master Mix (SuperArray Biosciences Corporation), forward and reverse primers (0.3nmol each), and cDNA (3μL) in a final volume of 25μL. Amplification parameters were denaturation at 95°C for 10 min followed by 40 cycles at 95°C for 30s and 60°C for 70s. No template containing wells served as negative control. Samples were analyzed in duplicate, and murine β-actin was used as a housekeeping gene control. Fold induction was calculated after normalization to murine β-actin using the ΔΔCt method. Dissociation curves indicated that each reaction consisted of a single reaction product. Primer sequences are available upon request.
2.8. Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation (ChIP) assay was performed with the ChIP assay kit (Upstate Biotechnology, Lake Placid, NY) according to the manufacturer’s protocol. Briefly, untreated or treated ILU-18 cells were subjected to cross-linking with 1% formaldehyde at 37°C for 10 minutes, followed by cell lysis using SDS Lysis buffer (1% SDS, 10mM EDTA, 50mM Tris-HCl - pH 8.1, and protease inhibitors). Sonication was used to shear the genomic DNA into fragments with an average size of ~500bp which were then diluted 10-fold in ChIP dilution buffer. After removing an aliquot for evaluation as input DNA, the lysates were pre-cleared with Salmon Sperm DNA/Protein G Agarose. The pre-cleared lysates were immunoprecipitated with the appropriate antibody by overnight incubation and immune complexes were precipitated with Salmon Sperm DNA/Protein G Agarose. The precipitates were washed sequentially with buffers, including low salt wash buffer, high salt wash buffer, LiCl wash buffer, and TE buffer. Antibody-DNA complexes were eluted twice with extraction buffer (100mM NaHCO3, 1% SDS, 20μg yeast tRNA). The eluates were heated overnight at 65°C to dissociate the cross-linking. Genomic DNA fragments were extracted and purified by phenol:chloroform extraction and Ethanol precipitation before proceeding to detection of specific chromatin fragments by PCR. Using primer sequences previously published, the amplification conditions were 94°C for 45 seconds, 60°C for 1 minute, and 72°C for 1 minute for 30–36 Cycles. PCR products were analyzed by 3% agarose gel electrophoresis and stained with ethidium bromide. ChIP assay employing quantitative real-time PCR (using the SYBR green Master mix in the Bio-Rad iCycler PCR system) was performed on the immunoprecipitated DNA which was obtained as described above. Fold differences represent Anti-ubiquitin (FK2) ChIP relative to control ChIP, where control ChIP was performed on untreated cells using Anti-Ubiquitin antibody for the immunoprecipitation. Fold differences were calculated as follows: 2[ΔCT(CONTROL) − ΔCT(FK2)], where ΔCT = CT(immunoprecipitated sample) − CT(input). Each treatment condition was quantitated in duplicate on at least two separate occasions, and from at least two independent immunoprecipitations, to give a minimum of four independent quantitations.
2.9. Luciferase Reporter Assay
ILU-18 cells, with or without pretreatment, were activated with pervanadate (100μM) for 18 hours at 37°C. Utilizing a Luciferase Reporter assay kit (Promega, Madison, WI), lysates were prepared with Reporter lysis buffer followed by repeated freezing and thawing. Protein content was determined by Bio-Rad assay. Per the supplier’s protocol, 50μg cell extract was combined with 100μL of the luciferase assay reagent at room temperature and analyzed by a luminometer.
2.10. IL-6 ELISA
After addition of the stimuli, cells were incubated for 4, 8, or 24 hours and then the culture supernatants were collected and frozen. Assays for murine IL-6 were performed with an enzyme-linked immunosorbent assay kit (eBioscience, San Diego, CA) according to the manufacturer’s instructions.
2.11. Statistical Analyses
Data were analyzed using student’s t-test. Differences were considered significant, if p < 0.05.
3. Results
3.1. Proteasome function is required for down-regulating pro-inflammatory IL-6 secretion induced by phospho-tyrosine dependent mechanisms
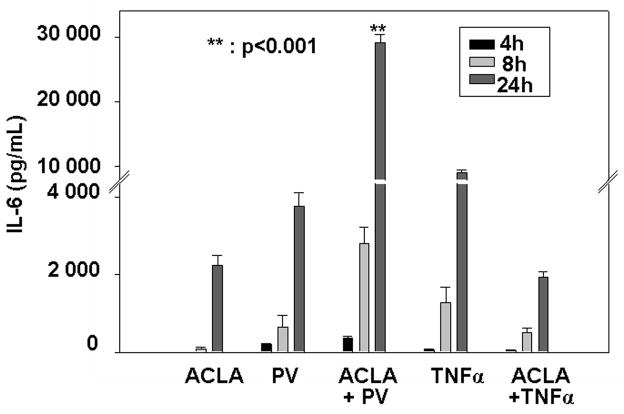
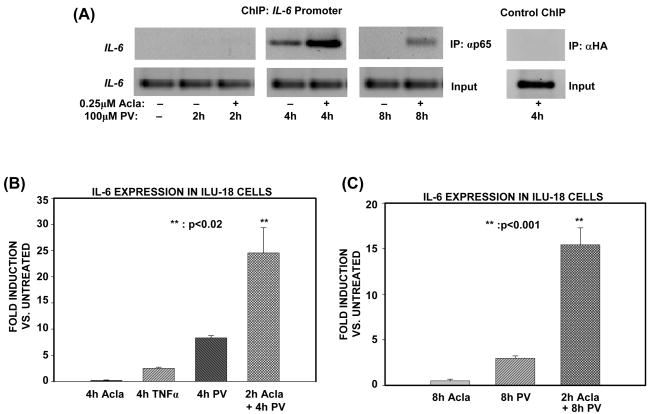
Our fundamental understanding of the interplay between tyrosine phosphorylation and NF-κB activation is principally derived from studies employing pervanadate, a tyrosine phosphatase inhibitor [4,5]. Therefore, we utilized pervanadate (PV) and the proteasome inhibitor, Aclacinomycin, to analyze the effects of proteasome inhibition on IL-6 secretion induced by the activation of cellular tyrosine kinases. We focused on IL-6 secretion by murine stromal cells because flow cytometric bead array analysis highlighted IL-6 as a pro-inflammatory cytokine that is highly sensitive to the combined treatment of proteasome inhibitor pretreatment and PV stimulation. As depicted in Figure 1, pretreatment with proteasome inhibitor significantly increased IL-6 production induced by PV, regardless of treatment duration. As a control, we measured IL-6 in the supernatant of cells undergoing pretreatment with proteasome inhibitor followed by TNF-α treatment. As expected, pretreatment with proteasome inhibitor significantly blocked TNF-α-mediated IL-6 production. Based on these results, proteasome appears to function as a negative regulator of pro-inflammatory IL-6 secretion induced by the activation of cellular tyrosine kinases.
Figure 1. Proteasome function negatively regulates pro-inflammatory IL-6 secretion induced by PV.
ILU-18 cells were either left untreated or pretreated with 0.25μM Aclacinomycin for 2h. At the end of 2h, cells were activated with TNFα (20ng/mL) or Pervanadate (100μM) for 4, 8, or 24 hours and supernatants were collected. Supernatants were analyzed by murine IL-6 ELISA kit per manufacturer’s protocol. Values presented are mean ± standard error from a minimum of three independent experiments and were adjusted for basal (control unstimulated cells) IL-6 secretion (338 pg/mL). Statistically significant differences are denoted by **.
3.2. NF-κB subunit, p65, mediates IL-6 transcription induced by phospho-tyrosine dependent mechanisms and may be a target of the 26S proteasome
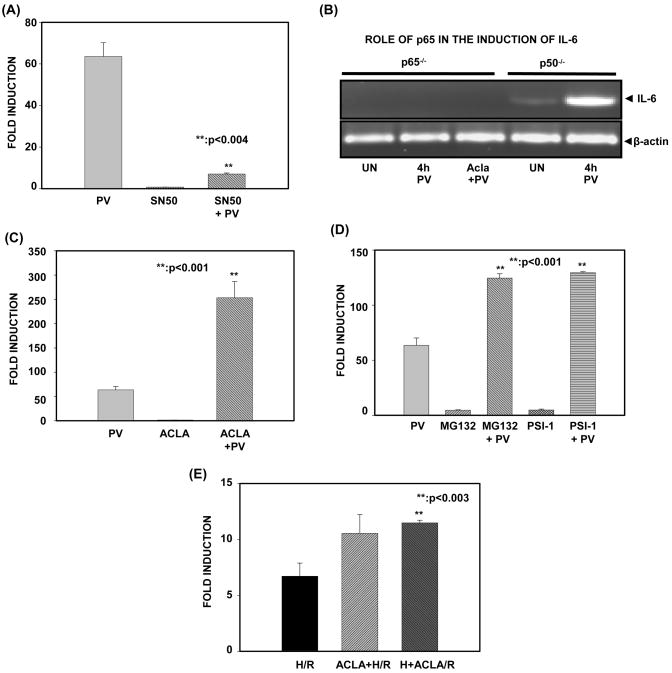
To establish a role for NF-κB in PV-mediated IL-6 expression, we again utilized the ILU-18 cell line because it is stably transfected with a murine IL-6 luciferase minigene. To evaluate this, cells were pretreated with the NF-κB-specific inhibitor, SN50 (inhibits nuclear translocation of NF-κB), before activation with PV. As depicted in Fig. 2A, pretreatment with SN50 inhibited PV-induced IL-6 reporter activity, indicating that PV-induced IL-6 expression is dependent upon nuclear translocation of NF-κB. To show conclusively that NF-κB is instrumental in PV-mediated transcription of IL-6, we employed murine embryonic fibroblasts lacking either the NF-κB subunit, p65, or the NF-κB subunit, p50 (Fig. 2B). In p65−/− MEFs, IL-6 expression was not induced by PV, regardless of proteasome inhibition. In contrast, p50−/− MEFs responded to PV by inducing transcription of IL-6. Thus, nuclear translocation and activity of the NF-κB subunit, p65, is necessary for PV-mediated expression of IL-6.
Figure 2. Proteasome limits IL-6 gene transcription mediated by NF-κB subunit, p65.
(A). ILU-18 cells were either pretreated with SN50 (18μM) for 1 hour or left untreated. At the end of 1 hour, these cells were either exposed to PV (100μM) or left untreated for 18 hours. At the end of 18 hours, cell lysates were obtained and luciferase activity was determined employing the Promega luciferase assay kit. For panels (A, C, D and E), fold induction represents a ratio of luciferase activity obtained in treatment-induced to that of untreated cells. Data represents mean ± standard deviation from four independent experiments, following normalization. Statistically significant differences are denoted by **.
(B). Murine Embryonic Fibroblasts (p50−/− and p65−/−), either untreated (UN) or treated with PV (100μM) for 4 hours with or without pretreatment with Acla (0.25μM) for 2 hours were used to isolate total RNA. Equal amounts of RNA were analyzed by RT-PCR. β-actin was used as a loading control.
(C). ILU-18 cells were either pretreated with Aclacinomycin (0.25μM) or left untreated. At the end of 2 hours, cells were stimulated with PV (100μM) for 18 hours or left untreated. Lysates obtained were evaluated for luciferase activity.
(D). ILU-18 cells were pretreated with either MG132 (1μM) or PSI-1 (20μM) for 2 hours or left untreated. At the end of 2h, cells were activated with PV (100μM) for 18 hours. Cells treated with MG132 or PSI-1 for 20 hours served as controls. Lysates obtained were evaluated for luciferase activity.
(E). ILU-18 cells were either pretreated with 0.25μM Acla or left untreated. At the end of 2 hours, cells were subjected to hypoxia (H). Cells underwent reoxygenation (R) for 18 hours, either in the presence or absence of 0.25μM Acla. This resulted in the following samples: H/R, Acla+H/R, and H+Acla/R. Lysates obtained were evaluated for luciferase activity.
To further address the possibility that proteasome activity negatively regulates the role of nuclear NF-κB, we then determined the effect of proteasome inhibition on IL-6 reporter activity induced by atypical, NF-κB stimuli. Regardless of the proteasome inhibitor employed in the pretreatment, a significant increase in PV-induced IL-6 reporter activity was observed (Fig. 2C & 2D). Similar results were obtained when siRNA to proteasome catalytic subunit, β5, was employed (Fig. 6C); this further confirms that catalytic activity of the proteasome plays a vital role in down-regulating PV-induced IL-6 reporter activity. To determine whether proteasome inhibition accentuated IL-6 reporter activity induced by yet another atypical stimulus, we carried out hypoxia/reoxygenation (H/R) in ILU-18 cells (Fig. 2E). Data presented in Fig. 2E demonstrates that similar to PV, H/R-induced IL-6 is up-regulated by proteasome inhibition, irrespective of the timing of inhibition (i.e. either before or after hypoxia). A priori, these results suggest that the proteasome limits NF-κB-mediated, inflammatory gene transcription induced by phospho-tyrosine dependent mechanisms.
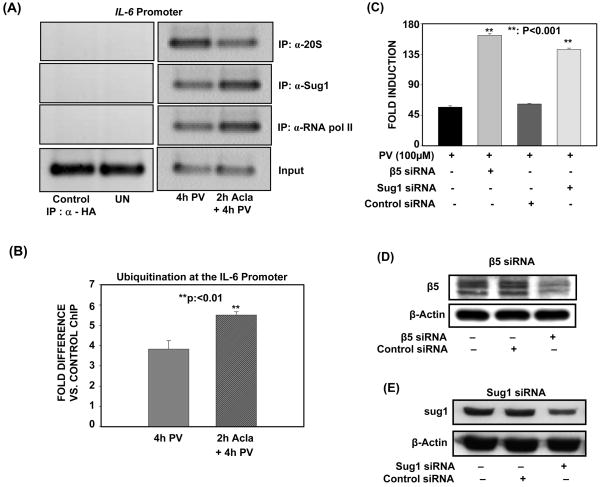
Figure 6. Catalytic core and regulatory subunits of the proteasome are recruited to the IL-6 promoter to negatively regulate PV-induced IL-6 transcription.
(A). ILU-18 cells were either untreated or treated with 100μM PV for 4h, with or without pretreatment with 0.25μM Acla for 2 hours. ChIP assays employing anti-20S, anti-Sug1, and anti-RNA pol II were then performed using immunoprecipitated DNA amplified with primers specific for the IL-6 promoter. ChIP assay employing α-HA (irrelevant antibody) in cells treated with Acla+PV served as a specificity control.
(B). ILU-18 cells were either untreated or treated with 100μM PV for 4h, with or without pretreatment with 0.25μM Acla for 2 hours. ChIP assay employing Anti-Ubiquitin (FK2) was then performed. Immunoprecipitated DNA was analyzed by quantitative real-time PCR analysis using primers amplifying the promoter region of IL-6. Fold difference is Anti-Ubiquitin (FK2) ChIP relative to control ChIP (immunoprecipitated DNA from untreated cells). Statistically significant differences are denoted by **.
(C). ILU-18 cells were either left untreated or transfected twice in 72 hours with either scrambled, β5-specific, or Sug1-specific siRNA. Cells were treated with 100μM PV for 18 hours, following transfection. Fold induction represents normalization of treatment-induced luciferase activity to basal luciferase activity obtained in untreated cells. Data obtained from at least four replicates are presented as mean ± standard error. Statistically significant differences are denoted by **.
(D), (E). ILU-18 cells were either left untreated or transfected twice in 72 hours with either scrambled, β5-specific, or Sug1-specific siRNA. Cells were lysed following transfection and 30μg of each sample was analyzed by immunoblotting using an antibody specific to β5 (D) or Sug1 (E). After stripping, the blot was reprobed with anti-β-actin to demonstrate equal loading.
3.3. Aclacinomycin specifically targets the catalytic activity of the proteasome without disrupting PV-induced tyrosine phosphorylation of cellular proteins, including IκBα
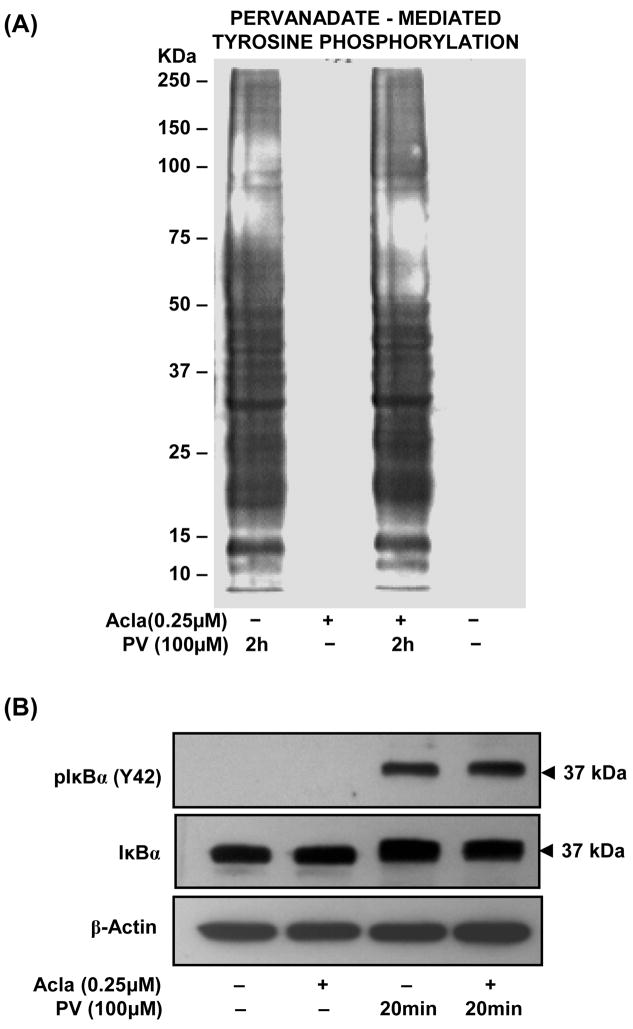
Before examining the precise mechanisms by which proteasome down-regulates atypical NF-κB transcriptional responses, we verified the cellular effects of the proteasome inhibitor, Aclacinomycin. By employing an active-site directed chemical probe (Biomol), we confirmed that Acla specifically inhibits chymotryptic activity by targeting β5 subunits of the 26S proteasome (Supplementary Fig. S1). Additionally, PV treatment, even in the presence of Acla, resulted in robust tyrosine phosphorylation of cytosolic proteins (Fig. 3A). Correspondingly, tyrosine phosphorylation of IκBα (Tyr-42) induced by PV was unaffected by pretreatment with proteasome inhibitor (Fig. 3B). Thus, in the process of targeting the chymotryptic activity of the proteasome, Acla does not interfere with the PV-mediated inhibition of tyrosine phosphatases and/or activation of tyrosine kinases which regulates tyrosine phosphorylation of IκBα.
Figure 3. Pretreatment with proteasome inhibitor, Aclacinomycin, does not modulate PV-induced tyrosine phosphorylation.
(A). ILU-18 cells were treated with PV (100μM) for 2 hours, with or without prior treatment with Acla (0.25μM) for 2 hours. At the end of incubation, cells were washed and cell lysates prepared. As controls, cell lysates were made from ILU-18 cells left untreated or treated for 4 hours with 0.25μM Aclacinomycin alone. Lysates, equalized for protein, were analyzed by immunoblotting using an antibody specific to Phospho-Tyrosine residues. Molecular weights derived from standards are indicated in kDa.
(B). ILU-18 cells were treated with PV (100μM) for 20 min, with or without prior treatment with Aclacinomycin (0.25μM) for 2 hours. At the end of incubation, cells were washed and cytosolic lysates prepared. As controls, cell lysates were made from ILU-18 cells either left untreated or treated for 140 minutes with 0.25μM Aclacinomycin alone. Lysates, equalized for 30μg protein, were resolved using SDS-PAGE, followed by western blotting using both antibodies to IκBα and phospho-IκBα (Tyr-42) and ECL. After stripping, the blot was reprobed with anti-β-actin to demonstrate equal loading.
3.4. Proteasome limits association of p65/RelA with the IL-6 promoter to terminate atypical, NF-κB-dependent transcriptional responses
In the canonical pathway, proteasome terminates NF-κB transcription by mediating the timely removal of p65/RelA from the promoters of target genes [2]. To delineate if proteasome exploits a similar mechanism to terminate atypical, NF-κB transcriptional responses, NF-κB was induced with PV in cells deficient or sufficient in proteasome activity, and then ChIP assay was performed with an antibody to p65. At the IL-6 promoter, inhibition of the proteasome results in the enhanced retention of p65 at 4h post-activation (Fig. 4A). Pretreatment with proteasome inhibitor does not appear to alter the kinetics with which NF-κB is recruited to the IL-6 promoter because, both in the absence and presence of proteasome inhibitor, p65 was not detected at the promoter upon 2h PV activation (Fig. 4A). Furthermore, p65 is found at the IL-6 promoter as late as 8h post-activation when proteasome is inhibited (Fig. 4A). Since p65 is persistently bound only when proteasome is inhibited, these results indicate that proteasome regulates the timely removal of p65 from the promoter.
Figure 4. Proteasome limits association of p65/RelA with the IL-6 promoter to terminate atypical, NF-κB-dependent transcriptional responses.
(A). ILU-18 cells were either untreated or treated with PV (100μM) for 2, 4, and 8 hours, with (+) or without (−) pretreatment with Acla (0.25μM) for 2 hours. ChIP assay employing anti-p65 was performed using immunoprecipitated DNA amplified with primers specific for the IL-6 promoter region. ChIP assay employing α-HA (irrelevant antibody) in cells treated with Acla+PV served as a specificity control. Input samples are representative data generated from sonicated DNA fragments not subjected to immunoprecipitation and analyzed by PCR amplification with IL-6 promoter primers.
(B), (C). Equal amounts of RNA were analyzed by real-time PCR detection, after isolation of total RNA from ILU-18 cells treated with TNFα (20ng/mL) or Acla (0.25μM) for 4 hours, or PV (100μM) for 4 hours with or without pretreatment with Acla for 2 hours (B). Data obtained following 8h treatment regimen with PV (100μM), with or without pretreatment with Acla (0.25μM) for 2 hours is presented as fold induction in (C). Fold induction represents treatment-induced IL-6 expression relative to basal IL-6 expression by untreated cells; data represents results from two independent experiments. Statistically significant differences are denoted by **.
An inability to remove promoter-associated p65 is likely to impact IL-6 expression because, as determined by mRNA analysis of p65−/− and p50−/− MEFs, the NF-κB subunit, p65, is absolutely required for PV-induced IL-6 expression (Fig. 2B). Consequently, we deduced that IL-6 expression would be significantly affected by the dysregulation in p65 transactivation accompanying proteasome inhibition. As predicted, IL-6 transcript levels analyzed by qPCR were enhanced and more persistently up-regulated by PV in ILU-18 cells deficient in proteasome activity (Fig. 4B & 4C). Moreover, the enhanced expression of IL-6 detected in the presence of proteasome inhibitor was not attributable to post-transcriptional mechanisms (Supplementary Fig. S2). Collectively, these data demonstrate that removal of p65 from target promoters by the proteasome results in the termination of atypical NF-κB transcriptional responses.
3.5. Proteasomal regulation of promoter-associated p65/RelA limits inflammatory gene transcription induced by PV
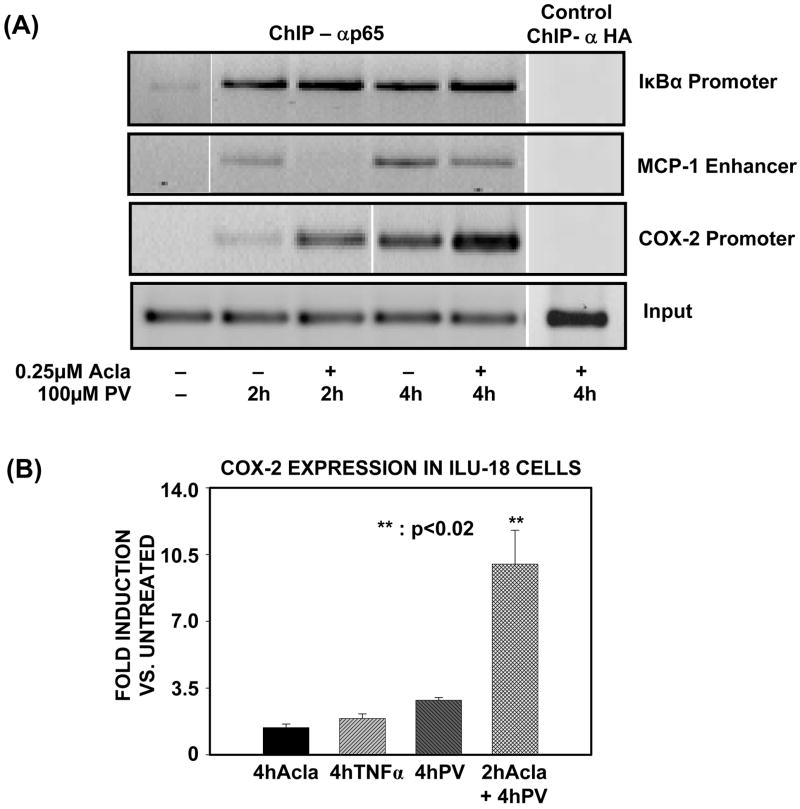
Having established a distinct role for the proteasome in enhanced and persistent retention of p65 at the IL-6 promoter, we next tested the role for proteasome at additional NF-κB target genes, i.e. MCP-1, IκBα, and COX-2. At the κB site in the MCP-1 enhancer, proteasome inhibition prior to PV-treatment negatively impacted p65 binding by decreasing p65 association with the MCP-1 enhancer (Fig. 5A). In contrast, proteasome inhibition prior to PV-treatment modestly enhanced p65 associated with the IκBα promoter (Fig. 5A). Most important, pretreatment with proteasome inhibitor elicited results at the COX-2 promoter which are similar to those observed at the IL-6 promoter, as inhibition of the proteasome significantly enhanced p65 association with the COX-2 promoter (Fig. 5A). Accordingly, this increase in retention of p65 at the COX-2 promoter led to an up-regulation in COX-2 transcript levels, as confirmed by qPCR (Fig. 5B). Thus, despite the selectivity with which proteasome targets p65/RelA at NF-κB-regulated promoters, proteasomal regulation of promoter-associated p65 clearly limits inflammatory transcription induced by PV.
Figure 5. Proteasomal regulation of promoter-associated p65/RelA limits inflammatory gene transcription induced by PV.
(A). ILU-18 cells were either untreated or treated with PV (100μM) for 2 and 4 hours, with or without pretreatment with Acla (0.25μM) for 2 hours. ChIP assay employing anti-p65 was performed using immunoprecipitated DNA amplified with primers specific for the promoter region of the genes, MCP-1, IκBα, and COX-2. ChIP assay employing α-HA (irrelevant antibody) in cells treated with Acla+PV served as a specificity control.
(B). Total RNA was extracted from ILU-18 cells treated with TNFα (20ng/mL) or Acla (0.25μM) for 4 hours or PV (100μM) for 4 hours with or without pretreatment with Acla for 2 hours. Equal amounts of RNA were analyzed by real-time PCR. Fold induction represents treatment-induced COX-2 expression relative to basal COX-2 expression in untreated cells; representative data from two independent experiments are presented. Statistically significant differences are denoted by **.
3.6. To negatively regulate PV-induced, IL-6 transcription, both 19S and 20S components of the 26S proteasome are recruited to the IL-6 promoter
To provide additional evidence for proteasomal involvement in the transcriptional regulation of NF-κB, we next determined if 19S and 20S components of the 26S proteasome directly associate with the IL-6 promoter, and compared their recruitment with that of p65, by employing ChIP assays on the IL-6 promoter in vivo. As depicted in Figure 6A, enhanced recruitment of 20S occurred upon 4h PV activation, and coincided with the observation of decreased p65 binding at the IL-6 promoter (Fig. 4A). Unexpectedly, inhibition of proteasome catalytic activity interfered with the recruitment of the 20S catalytic core to the IL-6 promoter (Fig. 6A). Given this defective recruitment of the 20S catalytic core, we determined if an accumulation of ubiquitinated proteins are associated with the promoter when proteasome is inhibited. For these analyses, we performed qChIP assay using an immunoprecipitating antibody specifically recognizing mono- and poly-ubiquitinated proteins, but not free ubiquitin. As depicted in Fig. 6B, proteasome inhibition is accompanied by a significant increase in mono- and poly-ubiquitinated proteins at the IL-6 promoter upon 4h PV-activation (Fig. 6B). Re-ChIP analysis, employing first an antibody directed at p65 followed by an antibody which recognizes mono- and poly-ubiquitinated proteins, revealed that p65 is among the promoter-associated factors which are retained when proteasome is inhibited (Supplementary Fig. S3). As added proof that p65 undergoes signal-induced ubiquitination, immunoprecipitation of p65 confirmed that ubiquitinated p65 accumulates following combined exposure to PV and Aclacinomycin (proteasome inhibition) (Supplementary Fig. S4). Altogether, these results suggest that the activity of the 20S catalytic core must be intact if it is to be properly recruited to degrade ubiquitinated, promoter-associated proteins, such as RelA/p65.
In contrast to the 20S catalytic core, enhanced association of the 19S proteasome subunit, Sug1, occurred when proteasome was inhibited and this recruitment corresponds with the increased p65 binding observed upon proteasomal inhibition (Fig. 6A). Since the 19S and 20S components appeared to fulfill distinct roles at the IL-6 promoter, we determined if both components negatively regulate IL-6 transcription induced by PV, by employing siRNA. Transient transfection of either β5 or Sug1 siRNA led to up-regulation of PV-induced IL-6 reporter activity (Fig. 6B & 6C). Thus, both 20S and 19S components of the 26S proteasome function as negative regulators of PV-induced IL-6 transcription.
3.7. To effectively terminate NF-κB activity, proteasome activity must be intact upon PV-stimulation
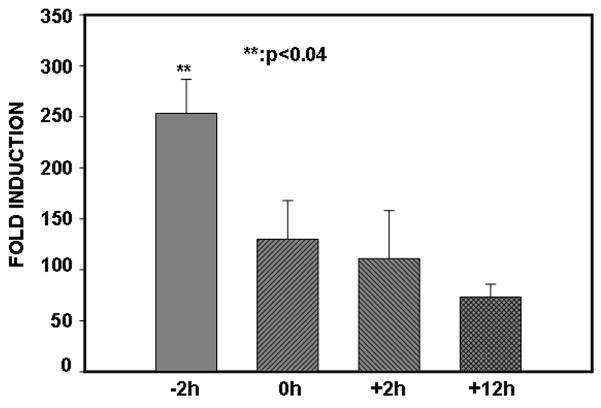
To gain further insight into proteasomal inhibition of NF-κB activity, we performed IL-6 reporter activity assay in which the addition of Acla was staggered relative to the addition of PV. Results depicted in Fig. 7 demonstrate that the maximum up-regulation of NF-κB responses is observed only if inhibition of proteasome activity precedes exposure to PV.
Figure 7. To effectively terminate NF-κB activity, proteasome activity must be intact upon PV-stimulation.
ILU-18 cells were treated with Aclacinomycin and PV in the following order: simultaneous addition of Acla & PV (0h); pretreated with Acla followed by PV (−2h); pretreated with PV followed by Acla (+2h); pretreated with PV followed by Acla (+12h); (− and + denote time in hours when Acla addition occurred, before (−) or after (+) addition of PV.) Eighteen hours after the addition of PV (100μM), cells were lysed and analyzed for luciferase activity, as previously described. Data obtained from at least four independent experiments are presented; values are data means ± standard deviation from the mean. Statistically significant differences are denoted by **.
4. Discussion
Proteasomal regulation of NF-κB induction has been a subject of intense investigation, however, the molecular link between proteasome and NF-κB down-regulation remains to be resolved, particularly as it relates to the dynamics and prevalence of proteasomal regulation. Unlike IκBα resynthesis, proteasome has not been recognized as a global mechanism for the termination of NF-κB-dependent transcription because its role has only been examined in studies employing activators of the NF-κB canonical pathway, i.e. TNFα and LPS [2,28]. Consequently, this study sought to advance our knowledge of proteasome and NF-κB down-regulation by exploring the role of proteasome in the termination of NF-κB-mediated transcription induced by atypical stimuli.
The results of our study reveal that pervanadate, a protein tyrosine phosphatase inhibitor, potently induces NF-κB-dependent genes, particularly IL-6. Additionally, we now demonstrate that the inhibition of proteasome catalytic function leads to a significant enhancement in PV-induced IL-6 transcription and secretion. Moreover, these sustained IL-6 responses obtained with PV following proteasome inhibition occur as a result of transcriptional up-regulation mediated by the NF-κB subunit, p65. Finally, we show for the first time that the catalytic activity of the 20S proteasome complex directly terminates NF-κB transcription induced by tyrosine phosphorylation-dependent mechanisms.
Our identification of a molecular link between lowered proteasomal function and increased inflammatory IL-6 induction has particular significance from an aging perspective because it suggests a possible connection between two well-established aging phenomena: proteasome inhibition and up-regulation of pro-inflammatory cytokine induction. During advancing age, low-grade inflammation is implicated in a spectrum of age-associated diseases whose onset and course are influenced by pro-inflammatory cytokines, i.e. cardiovascular disease, osteoporosis, arthritis, Alzheimer’s disease, and frailty [29]. Amongst the pro-inflammatory cytokines elevated with age, IL-6 has gained notoriety as the “cytokine for gerontologists” because studies consistently describe an age-related increase in IL-6 serum concentration [30]. Concomitant to up-regulation in levels of IL-6, an age-related decline in proteasomal function has been reported in myriad mammalian tissues, including liver, heart, lymphocytes, muscle, and neuronal and epidermal cells [31,32].
While there is no evidence of a direct molecular link between proteasomal defects and increased IL-6 expression during aging, proteasome inhibition has been reported to induce a pro-inflammatory response by activating transcription of pro-inflammatory mediators [33,34]. By employing antioxidants, these studies concluded that proteasome inhibition elevated levels of reactive oxygen species that in turn functioned as the regulatory mechanism underlying inflammation. Contrary to these reports, in our system, proteasome inhibition alone did not induce a pro-inflammatory response, i.e. marked up-regulation of IL-6. Correspondingly, we did not observe an increase in reactive oxygen species following treatment with Aclacinomycin (data not shown). Therefore, we propose that dysregulation in proteasome function does not represent the driving force in age-associated inflammation but is instead a contributing factor.
The predominant mechanism underlying induction of inflammatory genes during advancing age remains unclear, however possible mechanisms can be deduced by virtue of NF-κB-dependent regulation of these inflammatory genes. A role for NF-κB in these processes is supported by evidence of age-associated increases in constitutive nuclear NF-κB activity in rat liver, cardiac muscle, and hippocampus, as well as epidermis and major lymphoid organs, such as spleen and bone marrow, of aged mice [35–38]. As for the mechanism by which NF-κB becomes activated, redox regulation is considered as a major determinant of constitutive NF-κB activity in aged rodents. For example, a murine model of aging established that endogenous IL-6 promoter activity increases with age and that altered redox regulation profoundly influenced the endogenous IL-6 gene transcription [39]. Additionally, studies have attempted to determine if the constitutive activation of NF-κB accompanying aging is also derived from alterations in NF-κB response termination. Thus far, IκB synthesis and translation appear unaffected by age [40], indicating that dysregulation in IκB-mediated down-regulation of NF-κB activity is probably not significantly contributing to age-related alterations in NF-κB activity.
To our knowledge, there have been few studies, if any, which have reasoned that age-associated defects in proteasomal proteolysis might impact down-regulation of NF-κB signaling. However, given the significant up-regulation in IL-6 induction which we obtained with proteasome inhibition, we propose that alterations in proteasome-mediated termination of NF-κB responses are linked to age-related inflammation. Furthermore, consistent with the role for redox imbalance in the activation of NF-κB, we hypothesize that age-associated increases in IL-6 are due to defects in the proteasome’s ability to terminate oxidant-induced NF-κB transactivation. Substantiating this hypothesis with in vivo data presents a considerable challenge. However, as we gain further insight into genetic and chemical means for restoration of proteasome function [41], future studies have the potential to provide direct in vivo evidence that proteasome dysfunction indeed plays a role in age-associated chronic inflammation.
Similar to aging, multiple myeloma represents a pathological condition in which hyperactive NF-κB responses accompany proteasomal inhibition. Based on its resistance to bortezomib, constitutive NF-κB activity in primary multiple myeloma cells has been attributed to proteasome-independent mechanisms of NF-κB activation [24]. As yet, there is no evidence that atypical NF-κB responses are involved in additional, bortezomib-related complications. However, despite its purported anti-inflammatory effects, bortezomib treatment has been reported to induce inflammatory symptoms, such as neuropathy and increases in serum levels of pro-inflammatory cytokines [21–23]. In accordance with our findings, these symptoms suggest that proteasome inhibitors may serve to amplify rather than prevent inflammation. Therefore, in the long term, increased inflammation may be an unintended result of proteasome inhibition.
Our study highlights several areas for future studies. Firstly, the precise function and mode of interaction between the proteolytic and regulatory components of the 26S proteasome at inflammatory promoters will require further clarification. Secondly, the signal(s) mediating recruitment of proteasome to the promoter remain unknown. Clearly, promoter specificities underlie proteasomal regulation of NFκB-mediated transcriptional responses induced by both atypical and canonical pathways [2]. Thus, as the effects of proteasome inhibition appear to be gene specific, understanding promoter-specific differences in proteasomal recruitment may further elucidate the underlying mechanisms which govern its recruitment to NF-κB target genes.
In summary, these studies reinforce the importance of proteasome in the negative regulation of nuclear NF-κB because we demonstrate that proteasome regulates the termination of NF-κB transcription induced by upstream mechanisms which are both phospho-tyrosine dependent and proteasome-independent. Correspondingly, our study provides a molecular basis for the hyperactive NF-κB responses accompanying physiological conditions marked by proteasomal defects, i.e. advancing age, geriatric diseases, and bortezomib resistance.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Charles O’Brien for providing us with ILU-18 and its parental cell line. This work was supported by NIH grants AG13081, AG030599 and AG025220 to UP and in part by the UAMS Graduate Student Research Fund to SJC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 2.Saccani S, Marazzi I, Beg AA, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J Exp Med. 2004;200:107–13. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canty TG, Jr, Boyle EM, Jr, Farr A, Morgan EN, Verrier ED, Pohlman TH. Oxidative stress induces NF-kappaB nuclear translocation without degradation of IkappaBalpha. Circulation. 1999;100:II361–4. doi: 10.1161/01.cir.100.suppl_2.ii-361. [DOI] [PubMed] [Google Scholar]
- 4.Imbert V, Rupec RA, Livolsi A, Pahl HL, Traenckner EB, Mueller-Dieckmann C, et al. Tyrosine phosphorylation of I kappa B-alpha activates NF-kappa B without proteolytic degradation of I kappa B-alpha. Cell. 1996;86:787–98. doi: 10.1016/s0092-8674(00)80153-1. [DOI] [PubMed] [Google Scholar]
- 5.Fan C, Li Q, Ross D, Engelhardt JF. Tyrosine phosphorylation of I kappa B alpha activates NF kappa B through a redox-regulated and c-Src-dependent mechanism following hypoxia/reoxygenation. J Biol Chem. 2003;278:2072–80. doi: 10.1074/jbc.M206718200. [DOI] [PubMed] [Google Scholar]
- 6.Natarajan R, Fisher BJ, Jones DG, Fowler AA., III Atypical mechanism of NF-kappaB activation during reoxygenation stress in microvascular endothelium: a role for tyrosine kinases. Free Radic Biol Med. 2002;33:962–73. doi: 10.1016/s0891-5849(02)00990-5. [DOI] [PubMed] [Google Scholar]
- 7.Gallagher D, Gutierrez H, Gavalda N, O’Keeffe G, Hay R, Davies AM. Nuclear factor-kappaB activation via tyrosine phosphorylation of inhibitor kappaB-alpha is crucial for ciliary neurotrophic factor-promoted neurite growth from developing neurons. J Neurosci. 2007;27:9664–9. doi: 10.1523/JNEUROSCI.0608-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sethi G, Ahn KS, Chaturvedi MM, Aggarwal BB. Epidermal growth factor (EGF) activates nuclear factor-kappaB through IkappaBalpha kinase-independent but EGF receptor-kinase dependent tyrosine 42 phosphorylation of IkappaBalpha. Oncogene. 2007;26:7324–32. doi: 10.1038/sj.onc.1210544. [DOI] [PubMed] [Google Scholar]
- 9.Bui NT, Livolsi A, Peyron JF, Prehn JH. Activation of nuclear factor kappaB and Bcl-x survival gene expression by nerve growth factor requires tyrosine phosphorylation of IkappaBalpha. J Cell Biol. 2001;152:753–64. doi: 10.1083/jcb.152.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gloire G, Legrand-Poels S, Piette J. NF-kappaB activation by reactive oxygen species: fifteen years later. Biochem Pharmacol. 2006;72:1493–505. doi: 10.1016/j.bcp.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Schoonbroodt S, Ferreira V, Best-Belpomme M, Boelaert JR, Legrand-Poels S, Korner M, et al. Crucial role of the amino-terminal tyrosine residue 42 and the carboxyl-terminal PEST domain of I kappa B alpha in NF-kappa B activation by an oxidative stress. J Immunol. 2000;164:4292–300. doi: 10.4049/jimmunol.164.8.4292. [DOI] [PubMed] [Google Scholar]
- 12.Crevecoeur J, Merville MP, Piette J, Gloire G. Geldanamycin inhibits tyrosine phosphorylation-dependent NF-kappaB activation. Biochem Pharmacol. 2008;75:2183–91. doi: 10.1016/j.bcp.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Horion J, Gloire G, El MN, Quivy V, Vermeulen L, Vanden BW, et al. Histone deacetylase inhibitor trichostatin A sustains sodium pervanadate-induced NF-kappaB activation by delaying IkappaBalpha mRNA resynthesis: comparison with tumor necrosis factor alpha. J Biol Chem. 2007;282:15383–93. doi: 10.1074/jbc.M609166200. [DOI] [PubMed] [Google Scholar]
- 14.Okaya T, Lentsch AB. Hepatic expression of S32A/S36A IkappaBalpha does not reduce postischemic liver injury. J Surg Res. 2005;124:244–9. doi: 10.1016/j.jss.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 15.Fan C, Li Q, Zhang Y, Liu X, Luo M, Abbott D, et al. IkappaBalpha and IkappaBbeta possess injury context-specific functions that uniquely influence hepatic NF-kappaB induction and inflammation. J Clin Invest. 2004;113:746–55. doi: 10.1172/JCI17337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clohisy JC, Roy BC, Biondo C, Frazier E, Willis D, Teitelbaum SL, et al. Direct inhibition of NF-kappa B blocks bone erosion associated with inflammatory arthritis. J Immunol. 2003;171:5547–53. doi: 10.4049/jimmunol.171.10.5547. [DOI] [PubMed] [Google Scholar]
- 17.Waris G, Livolsi A, Imbert V, Peyron JF, Siddiqui A. Hepatitis C virus NS5A and subgenomic replicon activate NF-kappaB via tyrosine phosphorylation of IkappaBalpha and its degradation by calpain protease. J Biol Chem. 2003;278:40778–87. doi: 10.1074/jbc.M303248200. [DOI] [PubMed] [Google Scholar]
- 18.Kriete A, Mayo KL. Atypical pathways of NF-kappaB activation and aging. Exp Gerontol. 2009;44:250–5. doi: 10.1016/j.exger.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14:1649–57. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- 20.Hideshima T, Chauhan D, Richardson P, Mitsiades C, Mitsiades N, Hayashi T, et al. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem. 2002;277:16639–47. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- 21.Ravaglia S, Corso A, Piccolo G, Lozza A, Alfonsi E, Mangiacavalli S, et al. Immune-mediated neuropathies in myeloma patients treated with bortezomib. Clin Neurophysiol. 2008;119:2507–12. doi: 10.1016/j.clinph.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Min CK, Lee S, Kim YJ, Eom KS, Lee JW, Min WS, et al. Cutaneous leucoclastic vasculitis (LV) following bortezomib therapy in a myeloma patient; association with pro-inflammatory cytokines. Eur J Haematol. 2006;76:265–8. doi: 10.1111/j.0902-4441.2005.t01-1-EJH2437.x. [DOI] [PubMed] [Google Scholar]
- 23.Maruyama D, Watanabe T, Heike Y, Nagase K, Takahashi N, Yamasaki S, et al. Stromal cells in bone marrow play important roles in pro-inflammatory cytokine secretion causing fever following bortezomib administration in patients with multiple myeloma. Int J Hematol. 2008;88:396–402. doi: 10.1007/s12185-008-0194-0. [DOI] [PubMed] [Google Scholar]
- 24.Markovina S, Callander NS, O’Connor SL, Kim J, Werndli JE, Raschko M, et al. Bortezomib-resistant nuclear factor-kappaB activity in multiple myeloma cells. Mol Cancer Res. 2008;6:1356–64. doi: 10.1158/1541-7786.MCR-08-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin SC, Yamate T, Taguchi Y, Borba VZ, Girasole G, O’Brien CA, et al. Regulation of the gp80 and gp130 subunits of the IL-6 receptor by sex steroids in the murine bone marrow. J Clin Invest. 1997;100:1980–90. doi: 10.1172/JCI119729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiernan R, Bres V, Ng RW, Coudart MP, El MS, Sardet C, et al. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem. 2003;278:2758–66. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 27.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–43. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 29.Ershler WB, Keller ET. Age-associated increased interleukin-6 gene expression, late-life diseases, and frailty. Annu Rev Med. 2000;51:245–70. doi: 10.1146/annurev.med.51.1.245. [DOI] [PubMed] [Google Scholar]
- 30.Maggio M, Guralnik JM, Longo DL, Ferrucci L. Interleukin-6 in aging and chronic disease: a magnificent pathway. J Gerontol A Biol Sci Med Sci. 2006;61:575–84. doi: 10.1093/gerona/61.6.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chondrogianni N, Gonos ES. Proteasome dysfunction in mammalian aging: steps and factors involved. Exp Gerontol. 2005;40:931–8. doi: 10.1016/j.exger.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Ponnappan U, Zhong M, Trebilcock GU. Decreased proteasome-mediated degradation in T cells from the elderly: A role in immune senescence. Cell Immunol. 1999;192:167–74. doi: 10.1006/cimm.1998.1418. [DOI] [PubMed] [Google Scholar]
- 33.Rockwell P, Yuan H, Magnusson R, Figueiredo-Pereira ME. Proteasome Inhibition in Neuronal Cells Induces a Proinflammatory Response Manifested by Upregulation of Cyclooxygenase-2, Its Accumulation as Ubiquitin Conjugates, and Production of the Prostaglandin PGE2. Arch Biochem Biophys. 2000;374:325–33. doi: 10.1006/abbi.1999.1646. [DOI] [PubMed] [Google Scholar]
- 34.Chen JJ, Huang WC, Chen CC. Transcriptional regulation of cyclooxygenase-2 in response to proteasome inhibitors involves reactive oxygen species-mediated signaling pathway and recruitment of CCAAT/enhancer-binding protein delta and CREB-binding protein. Mol Biol Cell. 2005;16:5579–91. doi: 10.1091/mbc.E05-08-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helenius M, Hanninen M, Lehtinen SK, Salminen A. Changes associated with aging and replicative senescence in the regulation of transcription factor nuclear factor-kappa B. Biochem J. 1996;318:603–8. doi: 10.1042/bj3180603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufmann JA, Bickford PC, Taglialatela G. Free radical-dependent changes in constitutive Nuclear factor kappa B in the aged hippocampus. Neuroreport. 2002;13:1917–20. doi: 10.1097/00001756-200210280-00017. [DOI] [PubMed] [Google Scholar]
- 37.Spencer NF, Poynter ME, Im SY, Daynes RA. Constitutive activation of NF-kappa B in an animal model of aging. Int Immunol. 1997;9:1581–8. doi: 10.1093/intimm/9.10.1581. [DOI] [PubMed] [Google Scholar]
- 38.Adler AS, Sinha S, Kawahara TLA, Zhang JY, Segal E, Chang HY. Motif module map reveals enforcement of aging by continual NF-{kappa}B activity. Genes Dev. 2007;21:3244–57. doi: 10.1101/gad.1588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Dai J, Lu Y, Yao Z, O’Brien CA, Murtha JM, et al. In vivo visualization of aging-associated gene transcription: evidence for free radical theory of aging. Exp Gerontol. 2004;39:239–47. doi: 10.1016/j.exger.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 40.Helenius M, Kyrylenko S, Vehvilainen P, Salminen A. Characterization of Aging-Associated Up-Regulation of Constitutive Nuclear Factor-kappaB Binding Activity. Antioxid Redox Sign. 2001;3:147–56. doi: 10.1089/152308601750100669. [DOI] [PubMed] [Google Scholar]
- 41.Chondrogianni N, Gonos ES. Proteasome activation as a novel antiaging strategy. IUBMB Life. 2008;60:651–5. doi: 10.1002/iub.99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.