Abstract
The Hippo (Hpo) signaling pathway controls cell growth, proliferation and apoptosis in both Drosophila and vertebrates. In Drosophila, Hpo signaling regulates gene expression by inhibiting a transcription complex consisting of the transcriptional coactivator Yorkie (Yki) and the TEAD/TEF family of transcription factor Scalloped (Sd). Here we provide genetic evidence that both isoforms of 14-3-3, 14-3-3ε and 14-3-3ζ, regulate Yki activity through modulating its subcellular localization. Inactivation of 14-3-3 by RNAi or genetic mutations enhanced whereas overexpression of 14-3-3 suppressed tissue overgrowth induced by Yki overexpression. Loss of 14-3-3 resulted in the accumulation of Yki in the nucleus. We found that regulation of Yki by 14-3-3 was mediated by phosphorylation of Yki at S168. In addition, we found that Hpo signaling also inhibited Yki nuclear localization and activity by phosphorylating Yki at S111 and S250, and this inhibition appears to be independent of 14-3-3. Finally, we provided evidence that Hpo signaling restricted Yki nuclear localization depending on CRM1-mediated nuclear export.
Introduction
How the size of an organ is controlled has remained an unsolved mystery in development biology. The organ size is mainly determined by the number and size of its constituent cells, and both extrinsic environmental factors such as hormones and nutrients and intrinsic growth-control mechanisms contribute to the control of tissue growth by coordinately regulating cell growth, cell proliferation and cell apoptosis (Bryant and Simpson, 1984; Conlon and Raff, 1999). Genetic screens in Drosophila have identified a large number of tumor suppressor genes, mutations of which result in overgrowth of imaginal disc derivatives (Hariharan and Bilder, 2006). Several tumor suppressor genes, including warts(wts)(Justice et al., 1995; Xu et al., 1995), salvador(sav)(Tapon et al., 2002) and hpo(also called dMST)(Harvey et al., 2003; Jia et al., 2003; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003), fat (Bennett and Harvey, 2006; Cho et al., 2006; Mahoney et al., 1991; Silva et al., 2006; Willecke et al., 2006), and expanded (ex) (Cho et al., 2006; Hamaratoglu et al., 2006; Maitra et al., 2006; Pellock et al., 2007), define a common tumor suppressor pathway, the so called Hpo pathway (Pan, 2007; Zhang et al., 2009).
The Hpo pathway regulates tissue growth and organ size by inhibiting cell growth and proliferation as well as by promoting apoptosis through modulating the expression of genes including cyclin E, diap1 and bantam that are involved in these cellular processes (Harvey et al., 2003; Jia et al., 2003; Pantalacci et al., 2003; Tapon et al., 2002; Thompson and Cohen, 2006; Udan et al., 2003; Wu et al., 2003). The core components of the Hpo pathway, Hpo, Sav, Wts and Mats, constitute a kinase cascade. The upstream kinase Hpo, the Drosophila homolog of mammalian Ste20 family kinases MST1 and MST2, associates with the WW-domain containing scaffolding protein Sav to phosphorylate and activate the downstream kinase Wts, which belongs to the Nuclear Dbf-2-related (NDR) kinase family (Harvey et al., 2003; Jia et al., 2003; Justice et al., 1995; Pantalacci et al., 2003; Tapon et al., 2002; Udan et al., 2003; Wu et al., 2003; Xu et al., 1995). The phosphorylated Wts interacts with a cofactor Mats, which is also phosphorylated by Hpo (Lai et al., 2005; Wei et al., 2007), to regulate the expression of Hpo pathway target genes by phosphorylating a transcriptional coactivator Yki, the Drosophila homolog of Yap (Huang et al., 2005).
Recently, the TEAD/TEF family transcription factor Scalloped (Sd) has been implicated as a transcription factor for the Hpo pathway. Sd both genetically and physically interacts with Yki and is essential for tissue overgrowth induced by Yki overexpression or by loss of Hpo or Wts (Goulev et al., 2008; Wu et al., 2008; Zhang et al., 2008). Sd binds to the enhancer elements of diap1 (Wu et al., 2008; Zhang et al., 2008). In addition, Sd promotes nuclear localization of Yki and recruits Yki to the diap1 promoter (Zhang et al., 2008). On the other hand, phosphorylation of Yki at Ser168 by Hpo signaling restricts Yki nuclear localization (Dong et al., 2007; Oh and Irvine, 2008; Zhang et al., 2008). Mutating Ser168 to Ala promotes Yki nuclear localization and its ability to induce tissue overgrowth when overexpressed in vivo and rendered Yki more resistant to Hpo-mediated repression (Dong et al., 2007; Oh and Irvine, 2008; Zhang et al., 2008). Similarly, mutating the corresponding Ser residue in Yap (Ser127) promoted Yap nuclear localization and activity and rendered Yap more resistant to Lats-mediated inhibition (Hao et al., 2008; Zhao et al., 2007). In response to Hpo signaling, Yki/Yap interacted with 14-3-3 whereas YkiS168A/YapS127A failed to bind 14-3-3 (Dong et al., 2007; Oh and Irvine, 2008; Zhao et al., 2007). As 14-3-3 binding often regulate protein subcellular localization including nuclear/cytoplasmic shuttling (Fu et al., 2000), these observations imply that phosphorylation of Yki/Yap at S168/S127 restrict their nuclear localization by promoting 14-3-3 binding. However, genetic evidence for the involvement of 14-3-3 in the Hpo pathway and in regulating Yki/Yap nuclear localization is still lacking.
14-3-3 proteins are highly conserved and ubiquitously expressed phosphor-S/T binding proteins. 14-3-3 proteins form homo- and hetero-dimers and participate in many cellular processes including signal transduction through binding to specific phosphorylated sites on their target partners (Mackintosh, 2004; Morrison, 2009). There are two isoforms of 14-3-3 in Drosophila, 14-3-3ε and 14-3-3ζ/Leonardo that often act redundantly in many cellular processes (Acevedo et al., 2007; Benton and St Johnston, 2003). Here we provide evidence that both 14-3-3 isoforms are involved in modulating Yki activity through regulating its subcellular localization. We showed that inactivating 14-3-3 either by RNAi knockdown or genetic mutations enhanced tissue overgrowth induced by Yki overexpression whereas excess amounts of 14-3-3 suppressed Yki-induced overgrowth phenotype. In addition, 14-3-3 RNAi resulted in the accumulation of Yki in the nucleus. We provided both genetic and biochemical evidence that regulation of Yki by 14-3-3 is through phosphorylation of Yki at Ser168. Furthermore, we provided evidence that Hpo signaling inhibited Yki nuclear localization and activity through two additional phosphorylation sites, Ser111 and Ser250, and this regulation appeared to be 14-3-3 independent. Finally, we provided evidence that Yki subcellular localization is regulated by CRM1-mediated nuclear export.
Materials and Methods
Mutants and Transgenes
The 14-3-3- ζ -RNAi transgene was obtained from VDRC (http://www.vdrc.at/). 14-3-3εj2B10 and 14-3-3ζ12BL are strong loss-of-function alleles (http://flybase.org/). To construct UAS-14-3-3ε-RNAi transgenes, genomic DNA fragment corresponding to 14-3-3ε aa45-108 was amplified by PCR and subcloned between the BglII and KpnI sites of the pUAST vector. The corresponding cDNA fragment was inserted in a reverse orientation between KpnI and XbaI sites. Yki point mutations were generated by PCR-based site directed mutagenesis and verified by DNA sequence. The Yki coding sequence was amplified by PCR and subcloned in frame into the pUAST-6Myc vector (Zhang et al., 2008). To construct wild type and mutant forms of attB-UAS-Myc-Yki transgenes, a pUAST vector with attB sequence inserted upstream of the UAS-binding sites was used (Liu et al., 2007). The vas-phi-zh2A-VK5 flies were used to generate Yki transformants inserted at the 75B1 attP locus. To construct HA-14-3-3, 14-3-3 coding sequences were amplified by PCR and subcloned in frame into the pUAST-3HA vector (Zhang et al., 2008).
Cell Culture, Transfection, Immunoprecipitation, Western Blot Analysis and Luciferase Reporter Assay
S2 cells were cultured in Drosophila Schneider’s Medium (Invitrogen) with 10% fetal bovine serum, 100U/ml of penicillin, and 100ug/ml of Streptomycin. Transfection was carried out using Calcium Phosphate Transfection Kit (Specialty Media) according to manufacturer’s instructions. A ubiquitin-Gal4 construct was cotransfected with pUAST expression vectors for all the transfection experiments. Immunoprecipitation and western blot analysis were performed using standard protocols as previously described (Zhang et al., 2008). Antibodies used were mouse anti-HA (Santa Cruz), mouse anti-Myc (Santa Cruz). For Luciferase reporter assays, S2 cells were transfected with 3×Sd2-Luc and copia-renilla luciferase reporter constructs (Zhang et al., 2008) in 12 well plates together with constructs expressing Sd and different mutant forms of Yki. Cells were incubated for 48 hr after transfection and the luciferase reporter assay was performed using the Dual-Luciferase reporter assay system (Promega). Dual-Luciferase measurements were performed in triplicate using FLUOstar OPTIMA (BMG LABETCH).
Immunostaining
Immunostaining of imaginal discs and cultured cells was carried out as described (Jia et al., 2004; Jiang and Struhl, 1995). Antibodies or dyes used in this study were as follows: mouse anti-Myc (Santa Cruz), rabbit anti-HA (Santa Cruz), rabbit anti-Yki (Oh and Irvine, 2008) and 7-AAD (Molecular Probes). For LMB treatment of S2 cells, LMB was added at a concentration of 20 nM for 4 hrs before cells were collected.
Results
Knockdown of 14-3-3 enhances Yki-induced overgrowth
To investigate whether 14-3-3 proteins regulate Yki activity, we used transgenic RNAi technique (Kalidas and Smith, 2002; Kennerdell and Carthew, 2000) to knockdown either 14-3-3ε or 14-3-3ζ and examined whether reduction in 14-3-3 activity would modify Yki-induced overgrowth phenotype. Overexpression of UAS-Yki posterior to the morphogenetic furrow (MF) using the GMR-Gal4 driver (referred to as GMR-Yki) resulted in enlarged eyes (Fig. 1B, B′)(Wu et al., 2008; Zhang et al., 2008). Coexpression of either UAS-14-3-3ε-RNAi or UAS-14-3-3ζ-RNAi transgene enhanced the eye overgrowth phenotype (Fig. 1C–D′), whereas expression of neither RNAi transgene with GMR-Gal4 in otherwise wild type eyes caused any visible phenotypes (data not shown). The effect of 14-3-3 RNAi on GMR-Yki is specific as 14-3-3 RNAi did not modify the overgrowth phenotype caused by expressing an active form of insulin receptor (data not shown).
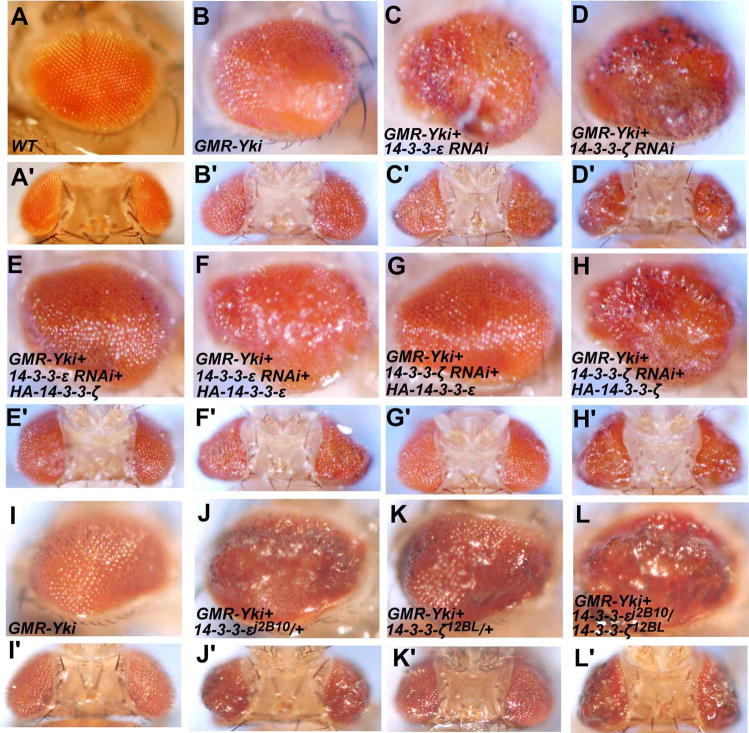
Figure 1. Loss of function of either 14-3-3 isoform can enhance Yki-induced eye overgrowth.
(A-L′) Side views (A-L) or dorsal views (A′-L′) of adult eyes of GMR-Gal4 (A and A′), GMR-Gal4 UAS-Yki (B and B′), GMR-Gal4 UAS-Yki; UAS-14-3-3εRNAi (C and C′), GMR-Gal4 UAS-Yki; UAS-14-3-3ζRNAi (D and D′), GMR-Gal4 UAS-Yki; UAS-14-3-3
εRNAi/UAS-HA-14-3-3ζ(E and E′), GMR-Gal4 UAS-Yki; UAS-14-3-3ε
RNAi/UAS-HA-14-3-3ε(F and F′), GMR-Gal4 UAS-Yki; UAS-14-3-3ζ
RNAi/UAS-HA-14-3-3ε(G and G′), GMR-Gal4 UAS-Yki; UAS-14-3-3ζ
RNAi/UAS-HA-14-3-3ζ(H and H′), GMR-Gal4 UAS-Yki (I and I′), GMR-Gal4 UAS-Yki; 14-3-3εj2B10/+ (J and J′), GMR-Gal4 UAS-Yki; 14-3-3ζ12BL/+ (K and K′) and GMR-Gal4 UAS-Yki; 14-3-3εj2B10/+ 14-3-3ζ12BL/+ (L and L′). Reduction of 14-3-3 either by RNAi knockdown or loss of function mutations enhanced the overgrown phenotype caused by Yki overexpression.
To determine whether modification of the GMR-Yki phenotype was due to inactivation of 14-3-3 rather than off-target effects, we generated transgenes that express either 14-3-3ε or 14-3-3ζ under the control of the UAS promoter. As shown in Fig. 1, enhancement of the GMR-Yki phenotype by 14-3-3ε RNAi was significantly suppressed by coexpression of UAS-14-3-3ζ (Fig. 1E and E′) but not by coexpression of UAS-14-3-3ε (Fig. 1F and 1F′). Similarly, coexpression of UAS-14-3-3ε but not UAS-14-3-3ζ suppressed the enhancement of GMR-Yki phenotype caused by 14-3-3ζ RNAi (Fig. 1G–H′). The isoform-specific rescue suggests that the transgenic RNAi for each isoform did not significantly interfere with the other isoform and that increasing the level of one 14-3-3 isoform could compensate for the loss of the other.
As an independent approach to determine whether loss of 14-3-3 would modulate Yki activity, we turned to loss-of-function mutations of 14-3-3. 14-3-3ε j2B10 and 14-3-3ζ12BL are strong loss-of-function alleles of 14-3-3ε and 14-3-3ζ, respectively (flybase). We found that either 14-3-3ε j2B10 or 14-3-3ζ12BL heterozygosity could enhance the GMR-Yki phenotype (Fig. 1J–K′). 14-3-3ε j2B10 and 14-3-3ζ12BL double heterozygosity enhanced the GMR-Yki phenotype more profoundly (Fig. 1L and L′). Taken together with the results from the RNAi experiments, these observations suggest that both 14-3-3 isoforms are involved in restricting Yki activity.
Overexpression of 14-3-3 suppresses Yki-induced overgrowth
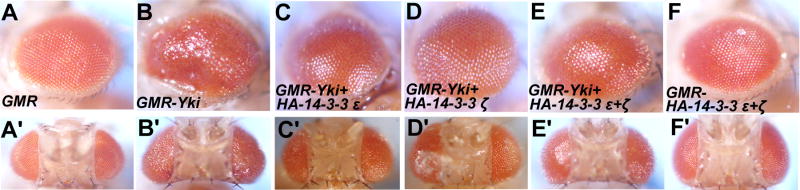
We next investigated whether increasing the level of 14-3-3 could suppress the phenotype induced by GMR-Yki. UAS-Yki was coexpressed with two copies of either UAS-14-3-3ε, UAS-14-3-3ζ or both. We observed dose dependent suppression of the GMR-Yki phenotype by 14-3-3 overexpression (Fig. 2B–E′). Overexpression of UAS-14-3-3ε and UAS-14-3-3ζ with GMR-Gal4 did not significantly affect eye development (Fig. 2F, F′). Thus, both loss-of-function and gain-of-function studies suggested that both 14-3-3 isoforms participate in restricting Yki activity.
Figure 2. Overexpression of either 14-3-3 isoform can suppress Yki-induced eye overgrowth.
(A-E) Side views (A-E) or dorsal views (A′-E′) of adult eyes of GMR-Gal4 (A and A′), GMR-Gal4 UAS-Yki (B and B′), GMR-Gal4 UAS-Yki; UAS-HA-14-3-3ε (C and C′), GMR-Gal4 UAS-Yki; UAS-HA-14-3-3ζ(D and D′), GMR-Gal4 UAS-Yki; UAS-HA-14-3-3ε+ UAS-HA-14-3-3ζ(E and E′) and GMR-Gal4; UAS-HA-14-3-3ε+ UAS-HA-14-3-3ζ(F and F′).
Loss of 14-3-3 results in nuclear accumulation of Yki
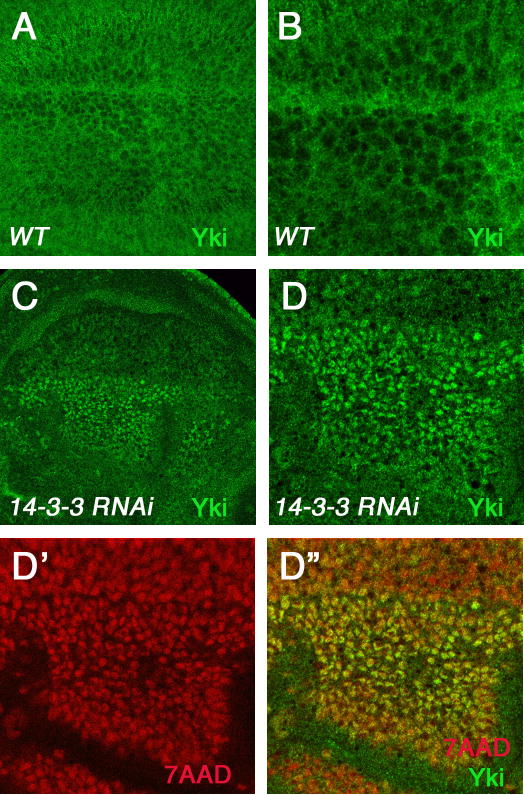
To determine whether 14-3-3 regulates Yki activity by inhibiting its nuclear localization, we examined the subcellular localization of endogenous Yki in either wild type wing discs or wing discs expressing both 14-3-3ε and 14-3-3ζ RNAi transgenes under the control of the wing specific Gal4 driver MS1096. As 14-3-3 function is required for many cellular processes, a more complete loss of 14-3-3 could result in cell lethality. We therefore coexpressed a cell death inhibitor P35 (Hay et al., 1994) together with 14-3-3 RNAi transgenes to increase the survival of cells lacking 14-3-3 activity. Consistent with previous observations (Dong et al., 2007; Oh and Irvine, 2008), the endogenous Yki was predominantly localized in the cytoplasm (Fig. 3A and B). In 14-3-3 knockdown wing discs, Yki was accumulated in the nucleus (Fig. 3C–D″), particularly in the dorsal compartment cells where MS1096-gal4 expression is higher (Wang et al., 1999). These results demonstrated that 14-3-3 is required for the nuclear exclusion of Yki.
Figure 3. Loss of 14-3-3 results in nuclear accumulation of Yki.
(A and B) Low (A) and high (B) magnification view of a wild type wing disc immunostained with an anti-Yki (green) antibody. Yki is primarily localized in the cytoplasm.
(C-D″) Low (C) and high (D-D″) magnification view of a wing disc expressing both UAS-14-3-3ε RNAi and UAS-14-3-3ζ RNAi together with UAS-P35 using MS1096 and immunostained with anti-Yki antibody (green in C, D and D″) and 7-AAD (red in D′ and D″) to label the nuclei. The wing disc is oriented with anterior to the left and ventral up. Severe knockdown of both 14-3-3 isoforms in the dorsal region of the wing pouch resulted in nuclear accumulation of endogenous Yki.
Yki activity is regulated by multiple phosphorylation events
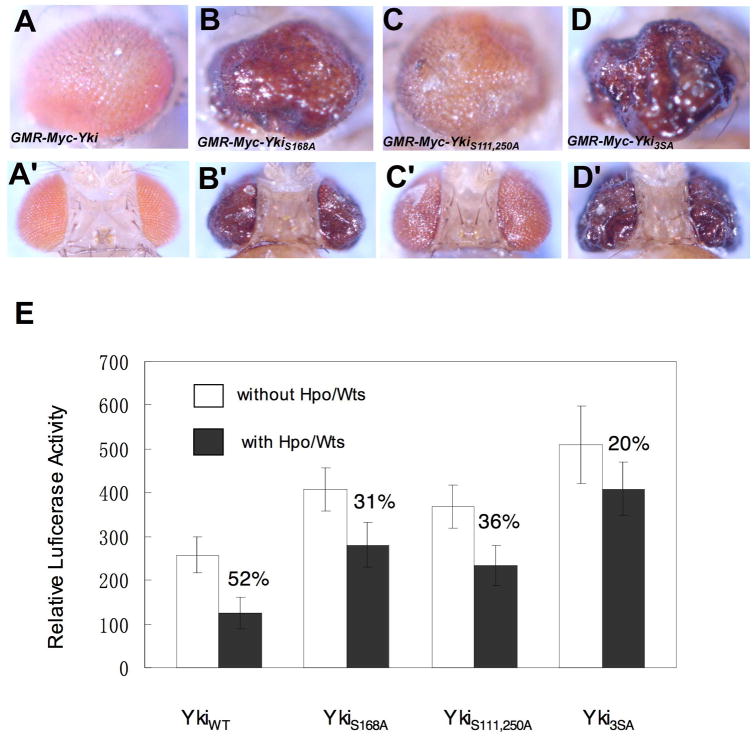
It has been shown that Yap is regulated by phosphorylation at multiple sites that fall into the consensus sequence: HXRXXS (Hao et al., 2008; Zhao et al., 2007). Besides Yki Ser168, which is equivalent to Yap Ser127, two additional sites (Ser111 and Ser250) fall into the same consensus (Fig. 6A). To determine whether phosphorylation at Ser111 and Ser250 modulates Yki activity, we mutated them into Ala in otherwise wild type background (YkiS111,250A) or in the background where Ser168 was also mutated into Ala (Yki3SA). To directly compare their activities with those of wild type Yki and YkiS168A without evoking the difference in the levels of transgene expression, transformants for Myc-tagged wild type Yki (Myc-Yki) or Yki variants (Myc-YkiS168A, Myc-YkiS111, 250A and Myc-Yki3SA) were generated using the phiC31 integration system to ensure that the transgenes were expressed at the same level (Bischof et al., 2007). Consistent with our previous observation (Zhang et al., 2008), Myc-Yki induced only modest overgrowth when expressed posterior to the MF using the GMR-Gal4 driver (Fig. 4A, A′). In contrast, GMR-Myc-YkiS168A induced more dramatic eye overgrowth (Fig. 4B, B′). Interestingly, GMR-Myc-YkiS111,250A also induced enlarged eyes although the phenotype was weaker than that of GMR-Myc-YkiS168A but stronger than GMR-Myc-Yki (Fig. 4C, C′). Finally, Myc-Yki3SA appeared to be more potent than Myc-YkiS168A as it induced more profound overgrowth than Myc-YkiS168A when expressed using GMR-Gal4 (Fig. 4D, D′). As all the transgenes were inserted into the same chromosomal location and expected to express at comparable levels, the difference in the overgrowth phenotypes induced by different forms of Yki reflected the difference in their activities.
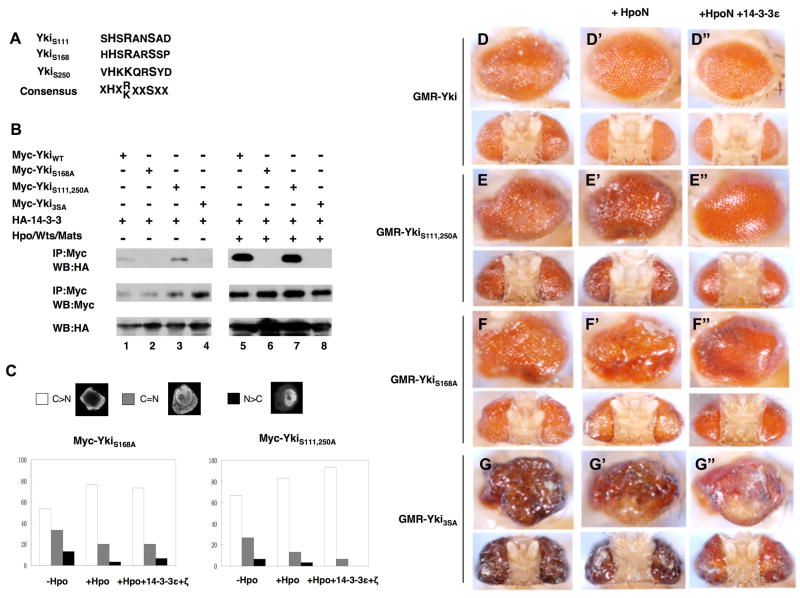
Figure 6. Phosphorylation at S111 and S250 regulates Yki independent of 14-3-3.
(A) The alignment of Yki sequences surrounding S111, S168 or S250 with the consensus sequence for Wts phosphorylation site indicated underneath. X: any amino acid.
(B) Mutating S111 and S250 does not affect 14-3-3 binding but mutating S168 abolishes 14-3-3 binding. S2 cells were transfected with indicated Myc-tagged wild type and mutant Yki constructs and HA-14-3-3 constructs with or without Hpo/Wts/Mats coexpression, followed by immunoprecipitation and western blot analyses with indicated antibodies.
(C) 14-3-3 regulates nuclear localization of YkiS111, 250A but not YkiS168A. S2 cells were transfected with HA-SD, Myc-YkiS111, 250A or Myc-YkiS168A, without or with coexpression of Hpo or Hpo plus 14-3-3. Cells were immunostained with Myc and HA antibodies, and the subcellular localization of Myc-YkiS111, 250A or Myc-YkiS168A was monitored using confocal microscopy. Cells with different nucleocytoplasmic distributions of Myc tagged Yki were counted. A total of 100 cells were counted for each Yki construct. C>N: cells contain higher levels of Yki in the cytoplasm than in the nucleus. C=N: cells contain Yki equally distributed in cytoplasm and nucleus. N>C: cells contain higher levels of Yki in the nucleus than in the cytoplasm. The y axis indicates the percentage of cells in each category.
(D-G″) Side (top) or (bottom) dorsal views of adult eyes expressing GMR-Gal4 UAS-Yki (D), GMR-Gal4 UAS-YkiS111, 250A (E), GMR-Gal4 UAS-YkiS168A (F) or GMR-Gal4 UAS-Yki3SA (G). HpoN (D′-G′) or HpoN and 14-3-3 (D″-G″) were coexpressed with the wild type or mutant Yki as indicated.
Figure 4. Multiple phosphorylation events contribute to Yki regulation.
(A-D′) Side views (A-D) or dorsal views (A′-D′) of adult eyes expressing UAS-Myc-Yki (A and A′), UAS-Myc-YkiS168A (B and B′), UAS-Myc-YkiS111,250A (C and C′) or UAS-Myc-Yki3SA (D and D′) with GMR-Gal4.
(E) S2 cells were transfected with the indicated Yki constructs together with Sd and an Sd-luciferase reporter gene, with or without Hpo and Wts expressing constructs. Cell lysates were subjected to dual luciferase assay. Error bars indicate standard deviation (triplicate wells). Numbers indicate degrees of suppression of Yki/Sd activity by Hpo/Wts.
To further confirm that Yki activity is regulated by multiple phosphorylation events, we directly measured the activity of Yki variants using the sd-luciferase reporter assay (Zhang et al., 2008). As shown in Fig. 4E, Yki variants exhibited higher activity than the wild type Yki with the order of Yki3SA > YkiS168A > YkiS111,250A >YkiWT, which is consistent with the in vivo activity exhibited by these Yki variants. In addition, Yki variants were more resistant to Hpo-mediated inhibition. Taken together, these results demonstrated that, besides phosphorylation at S168, phosphorylation at S111 and S250 also inhibited Yki activity, albeit to a lesser extent than S168 phosphorylation.
Regulation of Yki subcellular localization by phosphorylation at S111 and S250
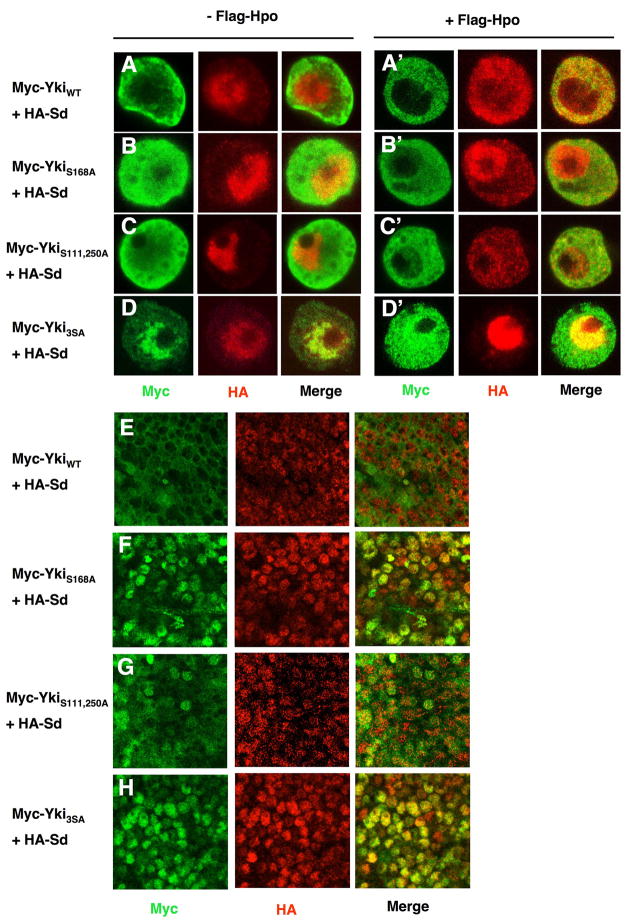
It has been shown that Hpo signaling inhibits Yki nuclear localization by phosphorylating Yki at S168 (Dong et al., 2007; Oh and Irvine, 2008; Zhang et al., 2008). To investigate whether phosphorylation at S111 and S250 also modulated Yki nuclear localization, we examined the subcellular localization of YkiS111,250A in S2 cells and wing disc cells. We have shown previously that Sd facilitates Yki nuclear localization in both S2 cells and wing imaginal discs (Zhang et al., 2008). In agree with these previous findings, Myc-YkiWT exhibited significant nuclear signal when coexpressed with HA-Sd but was excluded from the nucleus in the presence of Flag-Hpo coexpression (Fig. 5A, A′). In contrast, Myc-YkiS168A exhibited more profound nuclear localization than Myc-YkiWT when coexpressed with HA-Sd and a significant portion of Myc-YkiS168A remained in the nucleus when Flag-Hpo was coexpressed (Fig. 5B, B′). Myc-YkiS111,250A also exhibited higher levels of nuclear signal than Myc-YkiWT and nuclear YkiS111,250A signal was readily detectable when Flag-Hpo was coexpressed (Fig. 5C, C′). However, the levels of nuclear Myc-YkiS111,250A appeared to be lower than those of Myc-YkiS168A (cf. Fig. 5C with 5B, also see Fig. 6C and Fig. 7 for quantification). Finally, Myc-Yki3SA was predominantly localized in the nucleus when coexpressed with HA-Sd and the majority of Myc-Yki3SA remained in the nucleus even when Flag-Hpo was coexpressed (Fig. 5D, D′).
Figure 5. Phosphorylation at S111 and S250 restricts Yki nuclear localization.
(A-D′) S2 cells expressing HA-Sd together with Myc-YkiWT (A and A′), Myc-YkiS168A (B and B′), Myc-YkiS111,250A (C and C′) or Myc-Yki3SA (D and D′) with (A′-D′) or without Fg-Hpo (A-D) were immunostained with anti-Myc (green) and anti-HA (red) antibodies.
(E-H) High-magnification views of wing discs expressing HA-Sd and Myc- YkiWT (E), Myc-YkiS168A(F), Myc-YkiS111,250A(G) or Myc-Yki3SA (H) using MS1096 and immunostained with anti-Myc (green) and anti-HA (red) antibodies.
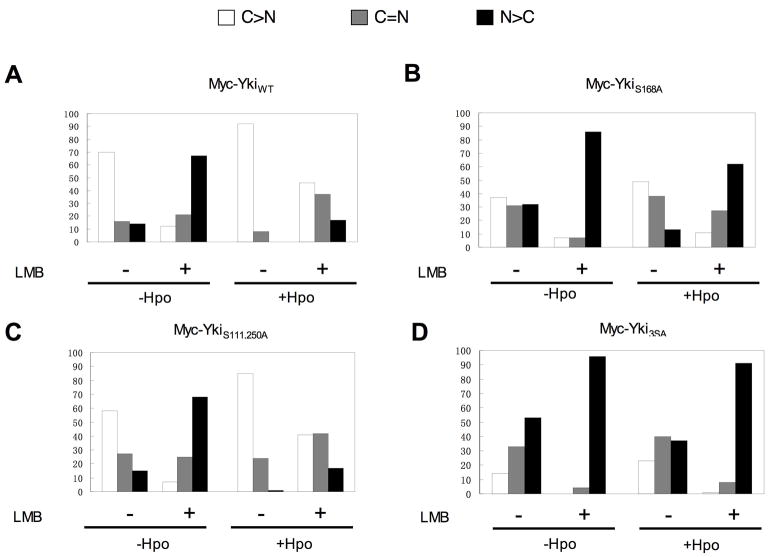
Figure 7. Restriction of Yki nuclear localization by Hpo signaling depends on nuclear export.
S2 cells were transfected with Myc-tagged wild type or mutant forms of Yki together with HA-Sd and with or without a Flag-Hpo expression construct. Cells were treated without or with LMB at a concentration of 20 nM for 4 hours, followed by immunostaining with Myc and HA antibodies. For each Yki construct, 100 cells were randomly selected and Yki subcellular localization was determined using confocal microscopy. C>N: cells contain higher levels of Yki in the cytoplasm than in the nucleus. C=N: cells contain Yki equally distributed in cytoplasm and nucleus. N>C: cells contain higher levels of Yki in the nucleus than in the cytoplasm.
We observed a similar trend of nuclear localization of Yki variants in wing imaginal discs. When coexpressed with HA-Sd using the MS1096 Gal4 driver, Myc-YkiS111,250A exhibited increased nuclear localization as compare with Myc-YkiWT (cf. Fig. 5G with 5E) whereas both Myc-YkiS168A and Myc-Yki3SA were predominantly nuclear (Fig. 5F, H). Thus, mutating S111 and S250 facilitated Yki nuclear localization, albeit less dramatically compared with S168A mutation.
Regulation of Yki subcellular localization by 14-3-3 is mediated by phosphorylation at S168 but is independent of phosphorylation at Ser111 and S250
Phosphorylation of YkiS168/Yap127 generates a 14-3-3 binding site that fall into the consensus RXXSpXP (Dong et al., 2007; Oh and Irvine, 2008; Zhao et al., 2007). In contrast, phosphorylation at S111 or S250 does not generate a 14-3-3 binding consensus site because a critical Pro residue C-terminal to the phospho-Ser residue is missing (Fig. 6A), implying that regulation of Yki by these two phosphorylation sites is 14-3-3 independent. To confirm that phosphorylation at Ser111 and 250 does not render 14-3-3 binding, we carried out immunoprecipitation experiments to determine the interaction between 14-3-3 with various Yki mutants. As shown in Fig. 6, HA-14-3-3 was coimmunoprecipitated with Myc-YkiWT and the interaction between 14-3-3 and Myc-YkiWT increased dramatically when the Hpo/Wts/Mats complex was coexpressed (Fig. 6B, lanes 1 and 5). Mutating S111 and S250 did not affect either the basal or Hpo signaling-induced 14-3-3 binding (Fig. 6B, lanes 3 and 7). In contrast, mutating S168 completely abolished both the basal and Hpo signaling-induced 14-3-3 binding (Fig. 6B, lanes 2 and 6). Myc-Yki3SA and Myc-YkiS168A behaved similarly with respect to 14-3-3 binding (cf. lanes 4 and 8 with lanes 2 and 6). Consistent with the observations that phosphorylation at S168 but not at S111 and S250 promotes 14-3-3 binding, Hpo-mediated inhibition of Myc-YkiS111,250A nuclear localization was enhanced by coexpression of 14-3-3 in S2 cells (Fig. 6C). In contrast, Hpo-mediated inhibition of Myc-YkiS168A nuclear localization was not significantly affected by 14-3-3 coexpression. Thus, phosphorylation at S111 and S250 does not regulate Yki/14-3-3 association and nuclear restriction of Yki by 14-3-3.
In corroboration with the observations obtained in S2 cells, 14-3-3 cooperated with the Hpo kinase domain (HpoN) to inhibit the overgrowth phenotypes caused by overexpression of Yki or YkiS111,250A (Fig. 6D–D″ and 6E–E″). In contrast, 14-3-3 did not collaborate with HpoN to suppress the overgrowth phenotypes caused by overexpression of YkiS168A or Yki3SA (Fig. 6F–F″ and 6G–G″). Furthermore, the overgrowth phenotype induced by YkiS111,250A overexpression was consistently enhanced by 14-3-3 knockdown (Fig. S1C–D′). In contrast, the overgrowth phenotype caused by YkiS168A or Yki3SA was not significantly enhanced by 14-3-3 knockdown (Fig. S1A–B′ and E–F′). These observations strength the view that 14-3-3 regulates Yki activity through phosphorylation at S168 whereas phosphorylation at S111 and S250 regulates Yki activity independent of 14-3-3.
Phosphorylation of Yki does not regulate Ex-mediated inhibition of Yki
It has been shown recently that Ex directly binds Yki and inhibits its nuclear localization and activity (Badouel et al., 2009). We found that Ex inhibited the activity of Myc-YkiWT, Myc-YkiS111,250A, Myc-YkiS168A and Myc-Yki3SA in the sd-luciferase reporter assay (Supplementary Fig. S2A). In addition, mutating S111, S168 and S250 did not affect cytoplasmic sequestration of Yki by Ex (Supplementary Fig. S2B), suggesting that phosphorylation of Yki at S111, S168 and S250 as well as 14-3-3 binding is not required for Ex-mediated inhibition of Yki.
Regulation of Yki subcellular localization by CRM1-mediated nuclear export
Hpo signaling could prevent Yki nuclear localization by impeding its nuclear import or by promoting its nuclear export. To investigated whether Yki is regulated by nuclear export, we treated cells transfected with Yki and Sd expression constructs with Leptomycin B (LMB), a drug that blocks CRM1-dependent nuclear export (Kudo et al., 1998), and determined if blocking nuclear export affect Yki subcellular localization. In the absence of LMB treatment, the majority of cells (~70%, N=100) exhibited higher levels of Myc-YkiWT in the cytoplasm whereas only a small fraction of cells (~30%, N=100) exhibited equal distribution of Myc-YkiWT in the cytoplasm and nucleus or higher levels of Myc-YkiWT in the nucleus. Strikingly, after LMB treatment, the majority of cells (>80%, N=100) exhibited equal distribution of Myc-YkiWT in the cytoplasm and nucleus or higher levels of nuclear Myc-YkiWT (Fig. 7A). In the presence of Flag-Hpo, >90% (N=100) of cells exhibited exclusive cytoplasmic Myc-YkiWT in the absence of LMB, whereas after LMB treatment, about 50% of cells (N=100) exhibited higher nuclear signals or equal distribution of nuclear and cytoplasmic signals of Myc-YkiWT (Fig. 7A). LMB treatment also promoted nuclear localization of Myc-YkiS111,250A, Myc-YkiS168A and Myc-Yki3SA, and interfered with Hpo-mediated cytoplasmic retention of these Yki variants (Fig. 7B–D). In contrast, Ex sequestered both Myc-YkiWT and Myc-Yki3SA in the cytoplasm efficiently in the presence of LMB treatment (Fig. S3). Thus, phosphorylation-mediated inhibition of Yki nuclear localization critically depends on nuclear export whereas Ex/Yki interaction inhibits Yki nuclear localization is independent of Yki nuclear export.
Discussion
The Hpo pathway is an evolutionarily conserved pathway that regulates cell growth, cell proliferation and apoptosis. The transcriptional coactivator Yki/YAP is the critical downstream regulatory target of the Hpo kinase cascade. It has been suggested that regulation of the subcellular localization of Yki/Yap is the primary mechanism by which the Hpo pathway influences gene expression (Pan, 2007; Zhang et al., 2009). The observation that Hpo signaling regulates Yki nuclear localization by phosphorylating S168 and phosphorylation at S168 promotes 14-3-3 binding led to the proposal that Hpo signaling-mediated phosphorylation of Yki at S168 sequesters Yki in the cytoplasm through 14-3-3 binding. Here we provide genetic evidence that 14-3-3 inhibits Yki nuclear localization and restricts Yki activity.
As complete loss of 14-3-3 leads to cell lethality due to the essential roles of 14-3-3 in many cellular processes, we applied RNAi to partially inactivate 14-3-3 and asked whether Yki activity was potentiated. Indeed, we found that knockdown of either 14-3-3ε or 14-3-3ζ enhanced the overgrowth phenotype induced by GMR-Yki (Fig. 1C–D′). In addition, removal of one copy of either 14-3-3ε or 14-3-3ζ slightly enhanced the GMR-Yki phenotype whereas removal of one copy of both 14-3-3ε and 14-3-3ζ resulted in a more dramatic enhancement of the GMR-Yki phenotype (Fig. 1J–L′). On the other hand, overexpressing either 14-3-3ε or 14-3-3ζ can suppress the GMR-Yki phenotype (Fig. 2). Thus, both loss of function and gain of function studies suggest that both 14-3-3 isoforms participate in restricting Yki activity. Furthermore, we found that knockdown of 14-3-3 led to nuclear accumulation of endogenous Yki in wing imaginal disc cells (Fig. 3), suggesting that 14-3-3 inhibits Yki activity by restricting its nuclear localization.
Hpo signaling restricts Yki nuclear localization and activity by phosphorylating Yki at S168, which is equivalent to S127 of Yap. Phosphorylation at YkiS168/Yap S127 generates 14-3-3 binding site. Consistent with phosphorylation at this site inhibits Yki nuclear localization and activity through 14-3-3 binding, we found that the activity of YkiS168A was not sensitive to changes in 14-3-3 activity as RNAi knockdown or overexpression of 14-3-3 did not significantly modulate the overgrowth phenotype caused by GMR-YkiS168A (Fig. 6F–F″; Fig. S1A–B′). Moreover, the subcellular localization of YkiS168A was not influenced by overexpression of 14-3-3 (Fig. 6C). Taken together with the biochemical data, these observations suggest that 14-3-3 regulates Yki nuclear localization and activity through interaction with phosphorylated Yki at S168.
Although phosphorylation at YkiS168 is a major regulatory mechanism of Yki nuclear localization and activity, it is not the only mechanism. Two previous studies revealed that Yap was phosphorylated at 4 additional sites that conform the consensus sequence, HXRXXS, and that phosphorylation at these sites also inhibited Yap activity (Hao et al., 2008; Zhao et al., 2007). We found that Yki has two additional sites that match the same consensus, HSRANS111AD and HKKQRS250YN (Fig. 6A). Interestingly, mutating Ser 111 and S250 to A (YkiS111.250A) increased Yki activity (Fig. 4C cf. 4A) and rendered Yki less sensitive to Hpo/Wts mediated inhibition (Fig. 4E). Compared with wild type Yki, YkiS111.250A exhibited increased levels of nuclear localization in both S2 cells and wing discs (Fig. 5C and G cf. 5A and E). YkiS111.250A appears to be less potent in inducing overgrowth as compared YkiS168A (Fig. 4). Consistently, nuclear localization of Yki S111.250A was less profound than that of YkiS168A (Fig. 5). Finally, Ser 111 and S250 to A mutation had an additive effect with S168 mutation as Yki3SA exhibited the highest levels of activity and more profound nuclear localization than YkiS168A or Yki S111,250A, suggesting that these phosphorylation events act in parallel to inhibit Yki nuclear localization and activity. In agreement with our findings, Oh and Irvine also observed that mutating Yki S111 and S250 enhanced Yki subcellular localization and activity, and rendered Yki less sensitive to Hpo/Wts-mediated inhibition (Oh and Irvine, 2009).
Mutating Yki S111 and S250 (Yki S111,250A) did not affect 14-3-3 binding to Yki and phosphorylation at these two sites in the absence of S168 (YkiS168A) failed to confer any detectable Yki/14-3-3 association (Fig. 6B; Oh and Irvine, 2009). Furthermore, when S127 of Yap was mutated, phosphorylation at other 4 sites failed to recruit 14-3-3 (Hao et al., 2008; Zhao et al., 2007). Consistent with these biochemical data, we found that YkiS168A, which has intact S111 and S250, was insensitive to either gain or loss of 14-3-3 activity. On the other hand, Yki lacking Ser 111 and S250 (Yki S111,250A) was still inhibited by 14-3-3. These observations support the view that phosphorylation at Ser 111 and S250 may regulate Yki nuclear localization and activity independent of 14-3-3 association. A recent study has revealed that Ex can physically associate with Yki and this interaction appears to sequester Yki in the cytoplasm. We found that Ex not only sequestered the wild type Yki but also the phosphorylation deficient forms of Yki (Fig. S2), suggesting that regulation of Yki nuclear localization by phosphorylation at Ser 111 and S250 is unlikely mediated by Ex.
How phosphorylation and 14-3-3 binding regulate Yki nuclear localization is not clear but it appears that nuclear export is required for restricting Yki nuclear localization in response to Hpo signaling (Fig. 7). Bind of 14-3-3 to target proteins has been suggested to regulate nucleocytoplasmic shuttling by impeding nuclear import and/or promoting nuclear export (Brunet et al., 2002; Kumagai and Dunphy, 1999). Yki does not have an intrinsic NLS (Nuclear Localization Signal) and its nuclear import appears to depend on its binding partner Sd (Goulev et al., 2008; Zhang et al., 2008). It is not impossible that binding of 14-3-3 to Yki could mask the NLS of Sd. An alternative but not mutually exclusive model is that phosphorylation and 14-3-3 binding may facilitate CRM1-mediated nuclear export. Consistent with this model, we found that blocking CRM1-mediated nuclear export by LMB treatment effectively prevented cytoplasmic localization in response to Hpo signaling. However, since the nuclear levels of YkiS168A and Yki3SA increased after LMB treatment, Yki can be exported out of the nucleus even in the absence of Yki phosphorylation and 14-3-3 binding. Thus, phosphorylation and 14-3-3 binding is not absolutely required for but may facilitate Yki nuclear export. In contrast, Ex-mediated cytoplasmic localization of Yki was not effectively blocked by LMB treatment (Fig. S3), suggesting that Ex/Yki interaction may sequester Yki in the cytoplasm independent of nuclear export. Since Ex can effectively retain YkiS168A and Yki3SA in the cytoplasm and both YkiS168A and Yki3SA no longer bind 14-3-3, the regulation of Yki by Ex is likely to be 14-3-3 independent.
CRM1 mediates nuclear export of target proteins through NES (Nuclear Export Signal) (Fornerod et al., 1997; Fukuda et al., 1997). Inspection of Yki and Yap sequences identified multiple NES like sequences (our unpublished observation). It will be interesting to determine the roles of these putative NES sequences in regulating Yki/Yap subcellular localization and activity and how 14-3-3 proteins control the nucleocytoplasmic shuttling of Yki/Yap. We also note that the subcellular localization and activity of Yki3SA is still regulated by Hpo signaling, albeit much less effectively as compared with the wild type Yki. Similar observation has been made for the phosphorylation deficient form of Yap that lacks all the Wts phosphorylation sites (Zhao et al., 2007). Thus, Hpo signaling may regulate Yki through additional mechanism(s) independent of its phosphorylation.
Supplementary Material
Acknowledgments
We thank Drs. Ken Irvine, Jianhang Jia, VDRC and Bloomington stock centers for reagents. This work was supported by grants from NIH (GM061269 and GM067045) and Welch Foundation (I-1603) to J. Jiang (J. J). J. J is a Eugene McDermott Endowed Scholar in Biomedical Science at UTSW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acevedo SF, Tsigkari KK, Grammenoudi S, Skoulakis EM. In vivo functional specificity and homeostasis of Drosophila 14-3-3 proteins. Genetics. 2007;177:239–53. doi: 10.1534/genetics.107.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, McNeill H. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–20. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–10. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–7. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Kanai F, Stehn J, Xu J, Sarbassova D, Frangioni JV, Dalal SN, DeCaprio JA, Greenberg ME, Yaffe MB. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol. 2002;156:817–28. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant PJ, Simpson P. Intrinsic and extrinsic control of growth in developing organs. Q Rev Biol. 1984;59:387–415. doi: 10.1086/414040. [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–50. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–44. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–33. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–60. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–47. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–11. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–41. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- Hariharan IK, Bilder D. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annu Rev Genet. 2006;40:335–61. doi: 10.1146/annurev.genet.39.073003.100738. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–67. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–9. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–34. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jia J, Tong C, Wang B, Luo L, Jiang J. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;432:1045–50. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–9. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Protein kinase A and hedgehog signaling in Drosophila limb development. Cell. 1995;80:563–72. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–46. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kalidas S, Smith DP. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron. 2002;33:177–84. doi: 10.1016/s0896-6273(02)00560-3. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–8. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–7. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 1999;13:1067–72. doi: 10.1101/gad.13.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–85. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cao X, Jiang J, Jia J. Fused-Costal2 protein complex regulates Hedgehog-induced Smo phosphorylation and cell-surface accumulation. Genes Dev. 2007;21:1949–63. doi: 10.1101/gad.1557407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J. 2004;381:329–42. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney PA, Weber U, Onofrechuk P, Biessmann H, Bryant PJ, Goodman CS. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell. 1991;67:853–68. doi: 10.1016/0092-8674(91)90359-7. [DOI] [PubMed] [Google Scholar]
- Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16:702–9. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–8. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–27. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–97. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–7. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Pellock BJ, Buff E, White K, Hariharan IK. The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Dev Biol. 2007;304:102–15. doi: 10.1016/j.ydbio.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–9. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–78. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–74. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–20. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes & Dev. 1999;13:2828–37. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. Embo J. 2007;26:1772–81. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–56. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–98. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–63. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–87. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yue T, Jiang J. Hippo signaling pathway and organ size control. Fly. 2009;3:68–73. doi: 10.4161/fly.3.1.7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.