Abstract
Protein thiolation by glutathione is a reversible and regulated post-translational modification that is increased in response to oxidants and nitric oxide. Because many mitochondrial enzymes contain critical thiol residues, it has been hypothesized that thiolation reactions regulate cell metabolism and survival. However, it has been difficult to differentiate the biological effects due to protein thiolation from other oxidative protein modifications. In this study, we used diamide to titrate protein glutathiolation and examined its impact on glycolysis, mitochondrial function, and cell death in rat aortic smooth muscle cells. Treatment of cells with diamide increased protein glutathiolation in a concentration-dependent manner and had comparably little effect on protein-protein disulfide formation. Diamide increased mitochondrial proton leak and decreased ATP-linked mitochondrial oxygen consumption and cellular bioenergetic reserve capacity. Concentrations of diamide above 200 µM promoted acute bioenergetic failure and caused cell death, whereas lower concentrations of diamide led to a prolonged increase in glycolytic flux and were not associated with loss of cell viability. Depletion of glutathione using buthionine sulfoximine had no effect on basal protein thiolation or cellular bioenergetics, but decreased diamide-induced protein glutathiolation and sensitized the cells to bioenergetic dysfunction and death. The effects of diamide on cell metabolism and viability were fully reversible upon addition of dithiothreitol. These data suggest that protein thiolation modulates key metabolic processes in both the mitochondria and cytosol.
Keywords: mitochondria, oxidative stress, glutathionylation, glycolysis, extracellular flux, reserve capacity
INTRODUCTION
The formation of mixed disulfides with protein thiols (protein thiolation) is increased in cells and tissues under conditions of oxidative stress. In particular, the most abundant low molecular weight thiol in the cell, glutathione (GSH), readily forms adducts with cysteinyl protein thiols under many pathological conditions [1–4]. Protein glutathiolation (or glutathionylation) is readily reversible and has been shown to be regulated by a number of enzymes including glutaredoxin [5–8], sulfiredoxin [9,10], protein disulfide isomerase (PDI) [11,12], and glutathione-S-transferase pi (GSTP) [13,14]. The abundance of glutathiolated proteins also varies depending on the cell compartment. It has been estimated that up to 50% of the glutathione in the endoplasmic reticulum is protein-bound [15], and that, overall, nearly 3% of all proteins remain bound to glutathione under basal conditions [16–18]. The functionalsignificance of protein thiolation and glutathiolation, however, is not clear. At low levels and under the control of redox regulatory enzymes, it is thought to play a role in cell signaling but effects on cellular metabolism have not been examined. It has also been suggested to be a protective mechanism that prevents deleterious or irreversible protein modifications under conditions of oxidative stress [19].
We hypothesized that bioenergetic systems in the cell are particularly susceptible to protein thiolation for a number of reasons. For example, mitochondria have a high concentration of both glutathione and protein thiols, which act in part as protection against oxidant stress [20]. In addition, solvent-exposed cysteine residues are present in the active sites of many mitochondrial dehydrogenase enzymes and are involved in co-ordinating the structures of enzymes involved in electron transfer for oxidative phosphorylation. For example, alpha-ketoglutarate dehydrogenase [21], isocitrate dehydrogenase [22], and mitochondrial aldehyde dehydrogenase [23] are inhibited by glutathiolation, and electron transport chain complexes I [24,25], II [26], and V [27] can also be modified by glutathione. Key regulatory proteins in the glycolytic pathway have also been shown to be glutathiolated under a number of experimental conditions. In particular, glyceraldehyde-3-phosphate dehydrogenase [28,29] and aldose reductase [30,31] have been shown to be inhibited by glutathiolation, which may redirect glucose flux to other pathways such as the pentose shunt pathway. Therefore, there is a strong rationale for studying the effect of protein thiolation on whole-cell bioenergetic function.
Oxidants such as hydrogen peroxide [21,32,33], nitric oxide (NO) [27,34], and peroxynitrite [2,35] catalyze the formation of protein-glutathione (PSSG) adducts. However, some of these reactive species also promote other oxidative modifications (e.g., protein sulfenic, sulfinic, and sulfonic acids [36,37]) and bind to divalent metals such as the heme centers in guanylate cyclase and cytochrome c oxidase [38]. It has been difficult, therefore, to delineate the bioenergetic effects of protein glutathiolation specifically over other thiol modifications. In this study, we used diamide to titrate protein glutathiolation. The utility of diamide, as opposed to other biochemical reagents or reactive species, is that it reacts preferentially with small acidic thiols (i.e., glutathione) and promotes predominantly the formation of protein-glutathione mixed disulfides (PSSG) [39]. To test these concepts, we examined the effects of diamide on whole-cell bioenergetic function using extracellular flux analysis. This technology allows for the concomitant measurement of glycolytic flux and mitochondrial oxygen consumption in real-time following a treatment such as diamide.
We found that diamide dynamically modulates mitochondrial function and glycolysis. At low concentrations, diamide modestly increased glutathiolation and led to a robust augmentation of mitochondrial proton leak and glycolytic flux. At higher concentrations, it promoted bioenergetic collapse associated with cell death, both of which were fully reversible upon addition of a reducing agent. Depletion of glutathione had no effect on basal protein thiolation or cellular bioenergetic function. However, cells depleted of glutathione that were treated with diamide formed less PSSG adducts at the expense of increased protein-protein dithiol formation; these cells were sensitized to diamide-induced bioenergetic dysfunction and death. Glutathiolation may be a particularly important modification that regulates bioenergetic dysfunction in conditions associated with oxidative stress.
MATERIALS AND METHODS
Materials
Diamide, antimycin A, FCCP (carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone), oligomycin, sodium pyruvate, thiazoyl blue tetrazolium, and buthionine sulfoximine (BSO) were purchased from Sigma (St. Louis, MO). N-ethylmaleimide (NEM) and dithiothreitol (DTT) were from Fisher Scientific (Pittsburgh, PA). Non-fat dry blotting milk was from Biorad (Hercules, CA). Anti-protein-glutathione antibody was from Virogen (Watertown, MA), and horseradish peroxidase-linked anti-mouse secondary antibodies were from Cell Signaling (Danvers, MA).
Cell culture and treatments
Rat aortic smooth muscle cells (RASMC) were harvested from descending thoracic aortas and maintained at 37°C in 5% CO2 in Dulbecco’s Eagle Modified Medium (DMEM) growth medium (Invitrogen; Carlsbad, CA) containing 4 mM GLUTAMAX, 5.5 mM D-glucose, 1 mM pyruvate, 3.7 g/L sodium bicarbonate, 10% fetal bovine serum (FBS, Atlanta Biologicals), 100 U/ml penicillin, and 100 ng/ml streptomycin. Cells of passage 5–12 were used in this study. Extracellular flux, cell viability, and protein thiol modification assays were performed in unbuffered DMEM (Mediatech, Manassas, VA) supplemented with 5.5 mM D-glucose, 4 mM glutamine, and 1 mM pyruvate. The pH of the medium was adjusted to 7.4 with NaOH immediately prior to treatment. For cell treatments, diamide was prepared as a 1M stock solution in DMSO and diluted into assay medium at 0–1 mM experimental concentrations. DMSO served as a vehicle control in all experiments. For in situ reversibility experiments, dithiothreitol (DTT; 1 mM) was added directly to cells treated with vehicle or diamide for the indicated time. In some treatments, buthionine sulfoximine (BSO) was used to deplete glutathione levels. For these experiments, cells were seeded in 24-well plates at 30,000 cells per well and allowed to adhere overnight. The cells were then treated with 0–1 mM BSO for 24 h prior to further treatments with diamide and/or bioenergetic measurements.
Detection of protein-glutathione (PSSG) adducts
Protein glutathiolation was detected by Western blotting using monoclonal anti-protein-glutathione (anti-PSSG) antibodies, essentially as described [27], with minor modifications. Cells treated with vehicle or diamide were lysed in buffer (pH 7.0) containing 20 mM HEPES, 1 mM DTPA, 1% NP-40, 0.1% SDS, 25 mM N-ethylmaleimide (NEM), and protease inhibitor cocktail. Proteins in the cell lysates (10–15 µg) were added to Laemmli sample buffer containing 25 mM NEM, separated by 10% SDS-PAGE, and transferred to PVDF membranes at 40 V overnight or at 100 V for 2 h at 4°C. The membranes were washed briefly in Tris-buffered saline containing 0.1% Tween-20 (TBS-tween) and then blocked in TBS-tween containing 5% milk and 2.5 mM NEM for 2 h at room temperature or overnight at 4°C. In our experience, inclusion of the NEM was critical for preserving PSSG adducts and maximizing the signal on Western blots. After blocking, membranes were washed in TBS-tween, and the membranes were incubated for 2 h at room temperature or overnight at 4°C with the anti-PSSG antibody. Horseradish peroxidase-linked anti-mouse antibodies were used as the secondary antibody. Immunoblots were developed with either ECL Plus chemifluorescent reagent (Invitrogen) or chemiluminescent Super Signal reagent (Pierce). The blots were then imaged on either a Typhoon Variable Mode imager or on a Fluorchem imager, respectively. Adduct signal was quantified by densitometry.
Diagonal electrophoresis
Cells treated with DMSO or diamide (0.25 mM for 40 min) were lysed in NEM-containing lysis buffer, and the proteins were separated by SDS-PAGE as described above. After electrophoresis, the lanes were excised and incubated in warm SDS running buffer (pH 8.3) containing 250 mM DTT for 30 min with constant agitation. The lanes were then laid atop 12% SDS polyacrylamide resolving gels, overlaid with agarose solution containing 100 mM DTT, and separated in the 2nd dimension by electrophoresis. Proteins in the gels were stained using Sypro Ruby and imaged on a Typhoon Variable Mode imager. Proteins below the diagonal were used to assess the relative degree of protein-protein disulfide formation.
Bioenergetic measurements
A Seahorse Bioscience XF24 Extracellular Flux Analyzer (North Billerica, MA) was used to measure mitochondrial function in intact RASMC. The XF24 creates a transient, 7 µl chamber in specialized microplates that allows for the measurement of oxygen and proton concentrations over time [40–42]. These measurements are then used to determine mitochondrial oxygen consumption and glycolytic flux in several samples at once. To allow comparison between different experiments, the data were expressed as the rate of oxygen consumption (OCR) in pmol/min or the rate of extracellular acidification (ECAR) in mpH/min. In some experiments the data were expressed as a percentage of the basal rate of OCR or ECAR. The inclusion of four injection ports per well allow addition of inhibitors or compounds of interest at any time during the assay measurements.
Glutathione measurements
RASMC were seeded in Seahorse® 24-well plates at 40,000 cells per well and allowed to adhere overnight. The cells were then treated with 0–1 mM BSO for 24 h. Total free glutathione was measured by the Tietze recycling assay, essentially as described [43], after lysing the cells in 20 µl of lysis buffer containing phosphate-buffered saline, pH 7.4, 10 µM DTPA, and 0.1% Triton X-100. Total protein was measured by the Bradford assay, and the glutathione content was normalized to total protein.
Cell Viability
Cell viability was measured by MTT assay as described previously [44], with the following modifications. RASMC were seeded as described above into 24-well culture plates at 40,000 cells per well. The cells were allowed to adhere overnight in complete medium. In experiments employing the use of BSO, cells were initially seeded at 30,000 cells per well followed by treatment the following day with BSO in complete medium. The cells were then treated in serum-free, unbuffered DMEM with diamide or DTT, where indicated for 16 h in the continued absence or presence of BSO. The medium was then replaced with media containing 0.4 mg/ml thiazoyl blue tetrazolium. The cells were then incubated at 37°C for an additional 2 h. The media was removed, and the resulting formazan crystals were solubilized in 250 µl DMSO. The absorbance was read at 550 nm and the data are expressed as a percentage normalized to the average absorbance in untreated cells.
Statistical analysis
Data are mean ± standard error of measure (SEM). Unpaired Student's t-tests were used to compare groups. Statistical significance was accepted at p < 0.05.
RESULTS
Induction of protein glutathiolation by diamide
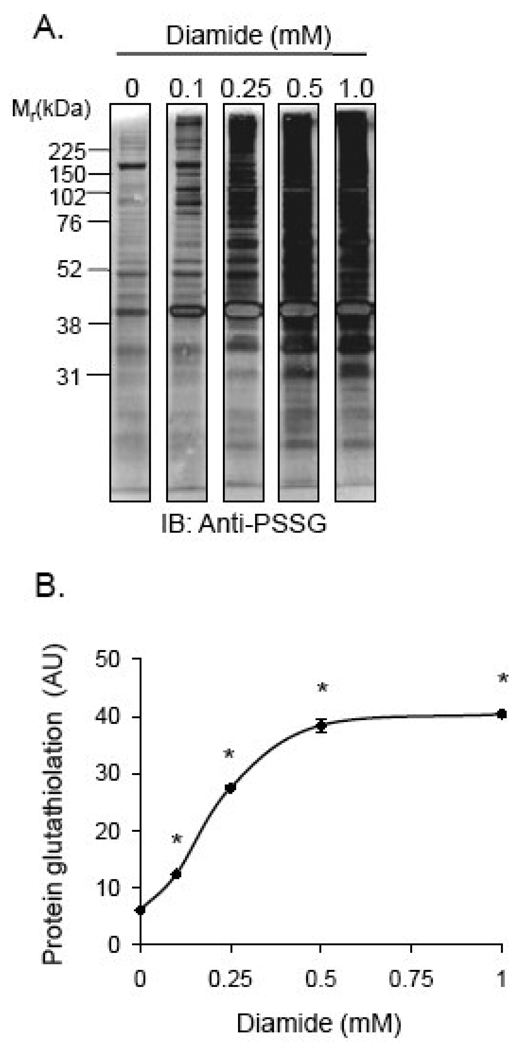
RASMC were treated with 0–1 mM diamide for 60 min, and proteins in the cell lysates were examined for PSSG adducts after separation by SDS-PAGE. As shown in Fig. 1A and B, Western blot analysis with anti-protein-glutathione (anti-PSSG) antibodies showed a diamide-concentration-dependent increase in PSSG adducts that reached a maximal level with 0.5 mM diamide treatment.
Fig. 1. Protein glutathiolation by diamide.
Analysis of protein-glutathione (PSSG) adduct formation by Western blotting: (A) Representative Western blot of PSSG adducts: Cells were treated with diamide (0–1 mM) for 1 h. Cell lysate proteins were then separated by SDS-PAGE followed by chemifluorescent Western blotting using anti-protein-glutathione (anti-PSSG) antibodies. (B) Group data from panel A. n = 3 per group; *p<0.05 vs. cells not treated with diamide.
Effect of protein thiolation on cellular bioenergetics
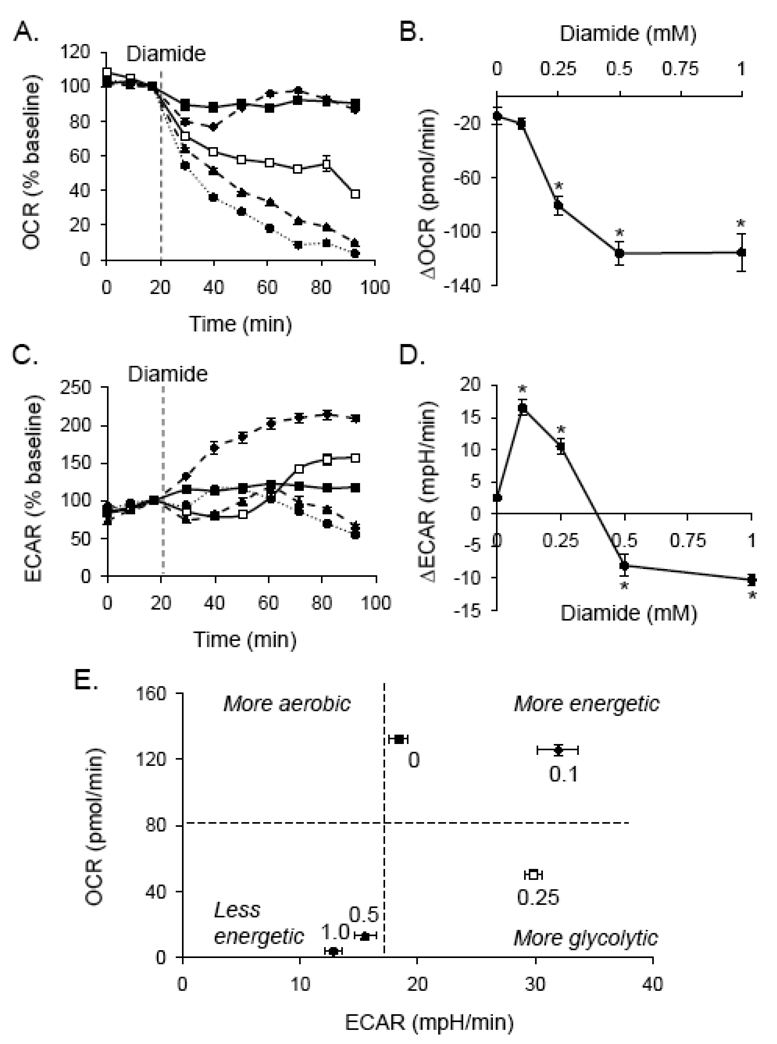
To determine whether the protein thiolation described in Fig. 1 affects cellular bioenergetics, cells were treated with diamide, and the effects on oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were examined by extracellular flux analysis. After measurement of basal OCR and ECAR, diamide (0–1 mM) was injected. As shown in Fig. 2A and B, diamide decreased OCR over time in a concentration-dependent manner. The effects of diamide treatment on glycolysis were also examined. Low concentrations of diamide (100 µM) consistently increased ECAR to ≥ 200% of the control level (Fig. 2C). Interestingly, higher concentrations of diamide (e.g., 0.25 mM) caused an initial depression of ECAR followed by recovery. As shown in Fig. 2D, higher concentrations of diamide decreased ECAR below basal levels.
Fig. 2. Bioenergetic changes induced by protein thiolation.
Extracellular flux analyses of diamide treated cells: Baseline measurements of oxygen consumption rate (OCR; panel A) and extracellular acidification rate (ECAR; panel C) were measured. Diamide was then injected where indicated to 0 (closed squares), 0.1 (diamonds; dashed line), 0.25 (open squares), 0.5 (triangles; dashed line), or 1.0 (circles; dotted line) mM final concentrations. Shown in panels A and C are the OCR and ECAR expressed as a percentage of baseline. (B and D) The changes in OCR (panel B) and ECAR (panel D) at the last time point measured are shown as a function of diamide concentration. N = 4 per group, *p < 0.05 vs. cells not treated with diamide. (E) Metabolic profile of diamide-treated cells: The last time point of OCR and ECAR measurements in panels A and C, respectively, were used to construct a 2D plot of bioenergetic function. The numbers above each point indicate the concentration of diamide (in mM).
The bioenergetic changes in ECAR and OCR were then plotted against one another at the final timepoint indicated in Fig. 2A and C. This analysis yields a metabolic profile of the cells after diamide treatment at a defined time of exposure. As shown in Fig. 2E, the lowest concentration of diamide (0.1 mM) appeared to stimulate glycolysis in the cell; i.e., it increased the glycolytic rate but did not significantly affect the consumption of oxygen at this timepoint. However, higher concentrations of diamide (0.5–1.0 mM) resulted in a less energetic cellular phenotype due to a decrease in both the aerobic and glycolytic flux in the cell.
Protein thiolation and mitochondrial function
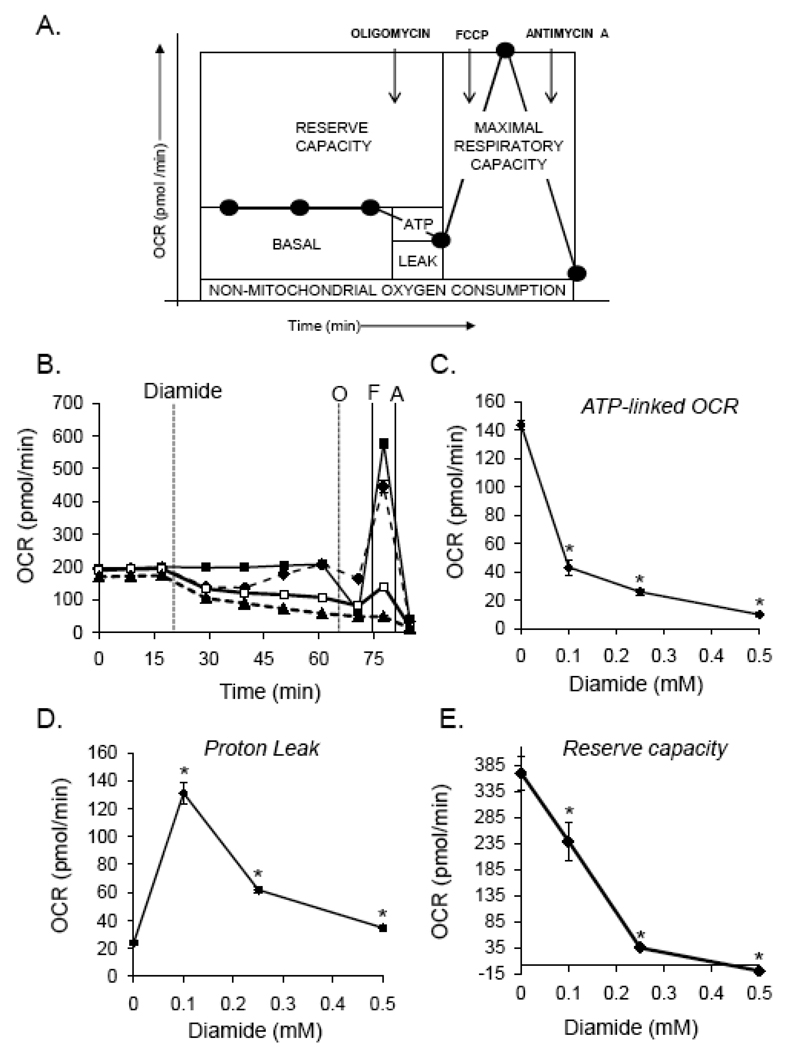
To determine the effect of diamide on specific indices of mitochondrial function, three basal oxygen consumption rates (OCR) were recorded, followed by the sequential injection of oligomycin (1 µg/ml), FCCP (1 µM), and antimycin A (10 µM). OCR measurements were recorded after each injection and used to determine how intracellular mitochondria responded to treatment. As shown in Fig. 3A, this assay was used to determine the level of oxygen consumption related to ATP production and proton leak; i.e., the oligomycin-sensitive OCR was ascribed to oxygen consumption linked to ATP production, and the oligomycin-insensitive OCR was attributed to non-ATP-linked oxygen consumption which may be due to proton leak across the mitochondrial inner membrane. As shown in Fig. 3A, the antimycin A-insensitive OCR was subtracted from the basal rate and the oligomycin-insensitive rate to estimate the mitochondrial contribution to these rates of oxygen consumption. The FCCP-stimulated OCR was similarly used to calculate the maximal respiratory capacity, and the bioenergetic reserve capacity was calculated by subtracting the pre-oligomycin OCR from the FCCP-stimulated OCR[45].
Fig. 3. Characterization of mitochondrial dysfunction induced by protein thiolation.
Delineation of changes in ATP-linked oxygen consumption, proton leak, and respiratory capacity in cells treated with diamide. (A) Extracellular flux analysis was used to measure mitochondrial function in intact, adherent smooth muscle cells. To probe individual components of respiration that contributed to the consumption of oxygen, oligomycin (1 µg/ml), FCCP (1 µM), and antimycin A (10 µM) were injected sequentially. This allowed for estimation of the contribution of proton leak (Leak) and ATP demand (ATP) to mitochondrial oxygen consumption. The maximal respiratory capacity was determined using the FCCP-stimulated rate. The residual oxygen consumption that occurred after addition of antimycin A was ascribed to non-mitochondrial sources. (B) After baseline measurements of oxygen consumption rate (OCR), diamide was injected to 0 (closed squares), 0.1 (diamonds; dotted line), 0.25 (open squares), or 0.5 (triangles; dotted line) mM final concentrations. At the indicated time, oligomycin (O; 1 µg/ml) was injected and OCR was measured. FCCP (F; 1 µM) was then injected followed by another OCR measurement. Last, antimycin A (A; 10 µM) was injected and OCR was again measured. (C and D) The oligomycin-sensitive and –insensitive rates of oxygen consumption were used to calculate the amount of oxygen consumption that was linked to ATP production (panel C) and proton leak (panel D). (E) The reserve or spare respiratory capacity was calculated in control and diamide-treated cells by subtracting the maximal (FCCP-stimulated) rate of oxygen consumption from the basal OCR. N = 5 per group, *p < 0.05 vs. cells not treated with diamide.
To examine the effect of protein thiolation on each of these indices of mitochondrial function, diamide was injected after a basal OCR was established. As shown in Fig. 3B, 0–0.5 mM diamide decreased the OCR; however, as also shown in Fig. 2A, cells treated with 0.1 mM diamide quickly recovered to their pre-diamide rate of oxygen consumption. Sequential injection of oligomycin, FCCP, and antimycin A then allowed for the determination of the effects of diamide on ATP-linked OCR, proton leak, maximal respiratory capacity, and the mitochondrial reserve capacity. As shown in Fig. 3C and D, oxygen consumption in cells treated with vehicle was largely linked to ATP production and proton leak was minimal. However, treatment of cells with 0.1 mM diamide led to an approximately 6-fold increase in proton leak and decreased ATP-linked oxygen consumption by > 3-fold. Nevertheless, at this concentration of diamide the response to FCCP was largely conserved and the cells retained a substantial reserve capacity (Fig. 3B and E). Treatment of the cells with 0.25 or 0.5 mM diamide further increased the loss of ATP-linked OCR, led to less of an increase in proton leak, and abrogated the maximal respiratory capacity and the mitochondrial reserve capacity.
Effect of glutathione on mitochondrial function and cell viability
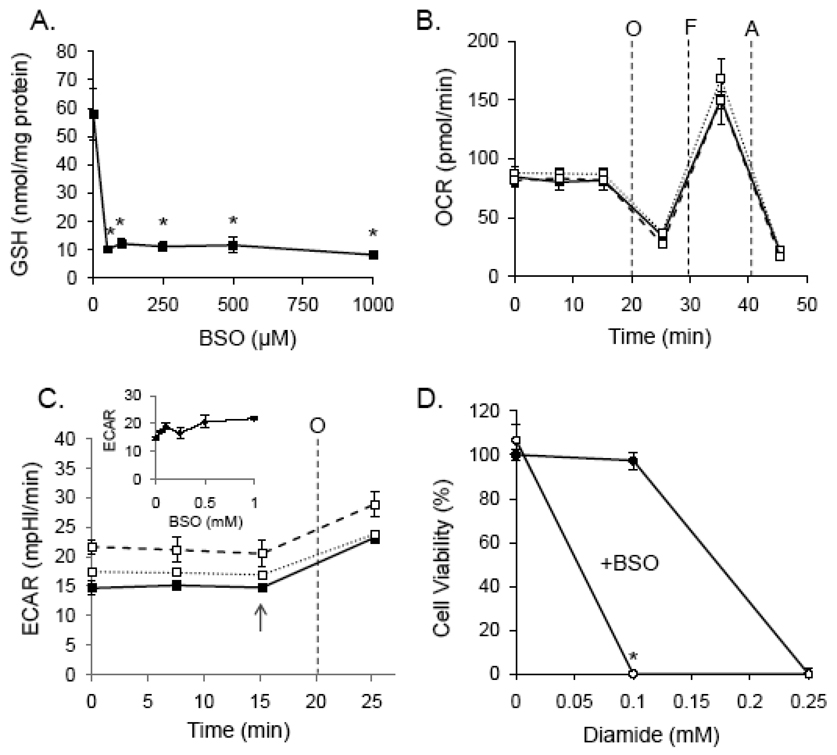
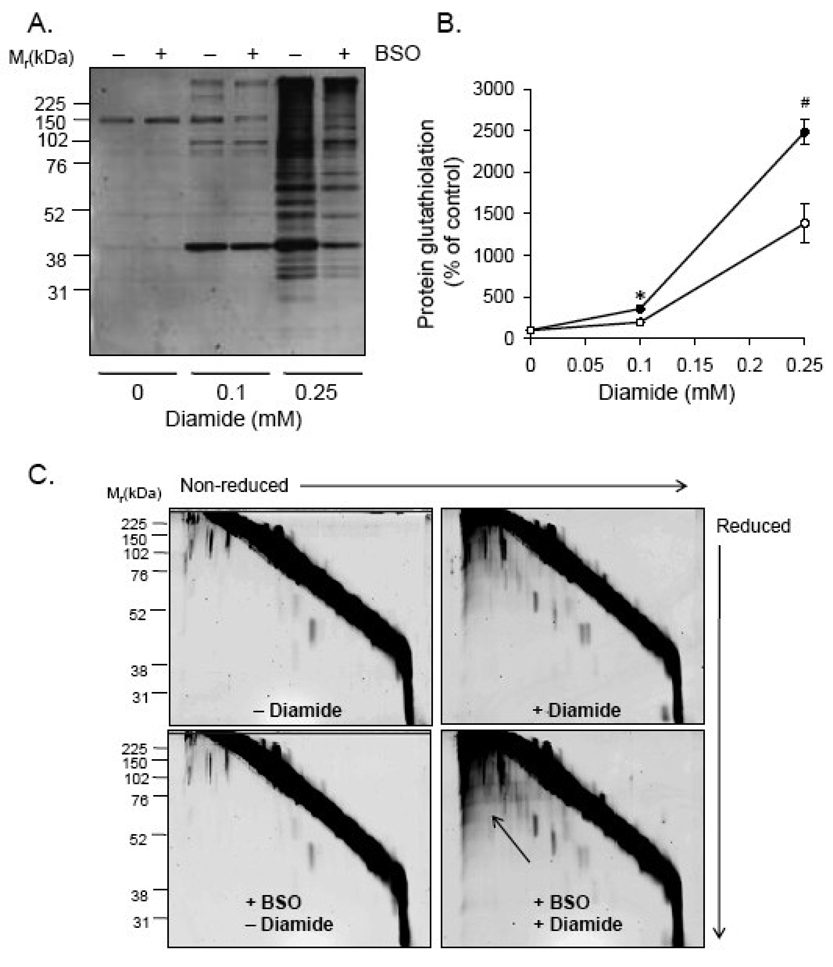
Because diamide increases protein-bound glutathione and decreases free GSH levels [39,46], we tested whether the effects of diamide on bioenergetics could be simply be the indirect effects of GSH depletion. Buthionine sulfoximine (BSO) was used to inhibit GSH synthesis in RASMC. As shown in Fig. 4A, 50 µM BSO depleted GSH maximally within a 24 h period and as shown in Fig. 4B, 0–0.5 mM BSO treatment had no effect on ATP-linked oxygen consumption, proton leak, maximal respiratory capacity, mitochondrial reserve capacity, or the non-mitochondrial rates of oxygen consumption. Similarly, ECAR was not significantly affected by GSH depletion. However, GSH depletion sensitized cells to diamide-induced cell death (Fig. 4D). These data suggest that protein thiolation reactions, and not GSH depletion per se, are responsible for diamide-induced changes in bioenergetics and cell viability.
Fig. 4. Glutathione depletion does not affect mitochondrial function but potentiates diamide-induced cell death.
(A) Cells were treated with buthionine sulfoximine (BSO; 0 – 1 mM) for 24 h and free glutathioine was measured by glutathione recycling assay. N ≥ 3 per group, *p < 0.05 vs. cells not treated with BSO. (B) Mitochondrial function in glutathione-depleted cells: Cells were treated with 0 (closed squares), 0.05 (open squares; dashed line), or 0.5 (open squares; dotted line) mM BSO for 24 h followed by mitochondrial function assay. After three baseline measurements of oxygen consumption rate (OCR), oligomyin (O; 1 µg/ml), FCCP (F; 1 µM), and antimycin A (A; 10 µM) were injected sequentially, and rates of oxygen consumption were recorded after each injection. (C) Glycolytic flux in glutathione-depleted cells: After 24 h of 0 (closed squares), 0.05 (open squares; dashed line), or 0.5 (open squares; dotted line) mM BSO treatment, baseline and oligomycin-stimulated extracellular acidification rates (ECAR) were measured. Inset: At the point shown by the arrow, the rate of extracellular was plotted for cells treated with 0–1 mM BSO. (D) Viability of normal (closed circles) and glutathione-depleted (open circles) cells treated with 0–0.25 mM diamide. N ≥ 3, and *p < 0.05 vs. diamide-treated cells not treated with BSO.
Protein thiolation reactions in glutathione-depleted cells
To determine whether glutathione depletion affected PSSG adduct formation, cells treated without and with BSO were exposed to diamide for 40 min, and the glutathione adducts were examined by Western blotting with anti-PSSG antibodies. As expected, GSH depletion significantly decreased protein glutathiolation by diamide, but had no effect on basal levels of GSH adducts (Fig. 5A and B).
Fig. 5. Glutathione depletion decreases diamide-induced protein-glutathione adduct formation.
Protein-glutathione adducts and protein-protein dithiol formation in glutathione-depleted cells treated with diamide. (A) Protein-glutathione adducts in cells depleted of glutathione: Cells were treated with 100 µM BSO for 24 h followed by diamide (0–0.25 mM) for 40 min. Equal amounts of protein (10 µg) from cell lysates were separated by SDS-PAGE, and glutathiolated proteins were detected by chemifluorescent Western blotting with anti-PSSG antibodies. (B) Group data from panel A. n = 3 per group; *p < 0.05 vs. glutathione-depleted cells treated with 0.1 mM diamide; #p<0.05 vs. glutathione-depleted cells treated with 0.25 mM diamide. (C) Diagonal electrophoresis gels in control cells (upper left panel), cells treated with 0.25 mM diamide (upper right panel), glutathione-depleted cells (lower left panel), and glutathione-depleted cells treated with 0.25 mM diamide. The arrow points to the area of the gel showing increased protein-protein dithiol formation.
Diagonal electrophoresis was then used to determine whether treatment with diamide also promoted protein-protein disulfide formation. After treatment with 0.25 mM diamide, the cells were collected in NEM-containing buffer and separated by SDS-PAGE. In the 2nd dimension, the proteins were reduced with DTT and again separated by size. Protein signal below the diagonal therefore represents proteins that were covalently disulfide-linked to another protein. As shown in Fig. 5C, treatment of cells with 0.25 mM diamide promoted little protein-protein thiolation compared to the large PSSG adduct signal shown in Fig. 1A and Fig. 5A. These results are consistent with the preference of diamide for oxidizing low molecular weight thiols in preference to protein thiols and suggest that glutathiolation was the predominant modification occurring during diamide treatment.
Treatment of GSH-depleted cells with diamide, however, resulted in a smaller increase in glutathiolated proteins (Fig. 5A and B) and a relatively larger increase in protein-protein linked proteins (Fig. 5C, lower right panel). BSO treatment alone had no effect on protein-protein disulfide formation (Fig. 5C, lower left panel). These results show that GSH levels regulate the degree to which proteins can be glutathiolated in the cell under conditions oxidant stress. Last, the relative increase in diamide-induced, protein-protein disulfides in glutathione-depleted cells was associated with a loss of viability (Fig. 4D).
Mitochondrial function in glutathione-depleted cells treated with diamide
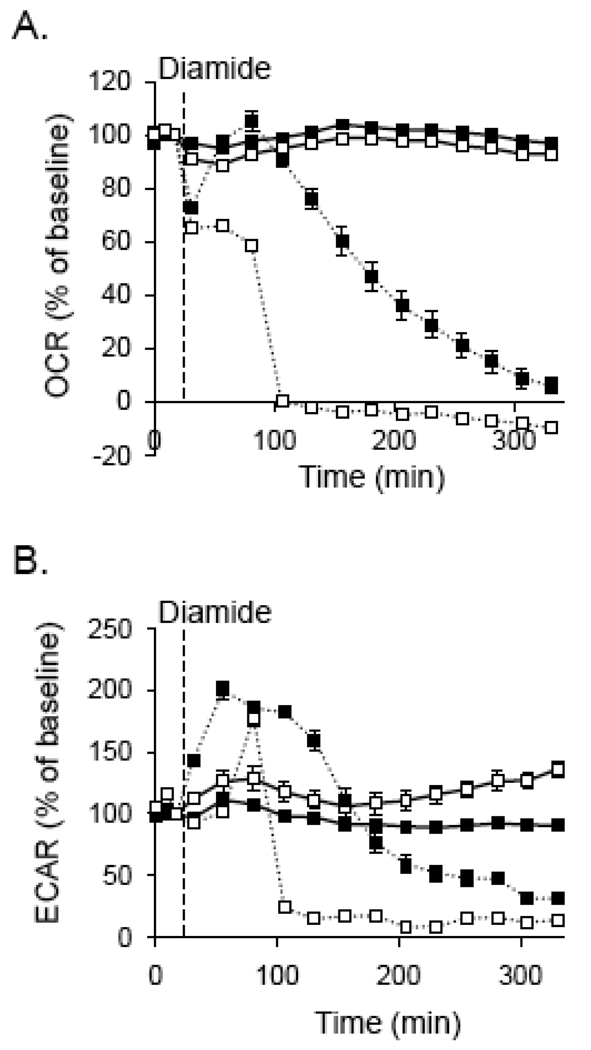
Mitochondrial function was also significantly affected in glutathione-depleted cells that were treated with diamide. As shown in Fig. 6A, BSO treatment by itself had no effect on oxygen consumption; however, diamide treatment of glutathione-depleted cells resulted in a rapid decline in oxygen consumption. Although there was a significant decrease in oxygen consumption over a longer period of time in cells treated with diamide only, these cells were attached and still viable after 16 h as assessed by the MTT assay (Fig. 4D and data not shown).
Fig. 6. Bioenergetic function in glutathione-depleted cells treated with diamide.
Extracellular flux analysis plots of oxygen consumption and extracellular acidification: (A) Cells were treated without or with 100 µM BSO for 24 h (open squares), and baseline oxygen consumption rates (OCR) were recorded. Diamide (dotted lines) was then injected to 0.1 mM final concentrations where indicated, and the OCR were followed for approximately 300 min. (B) Glycolytic flux in glutathione-depleted cells treated with diamide: Cells were treated as described in panel A and rates of extracellular acidification were measured. N = 5 per group.
The glycolytic response of the cells to diamide was also significantly affected by glutathione depletion. As shown in Fig. 6B, diamide by itself increased glycolytic flux for the first 150 min after treatment and then decreased the ECAR to below control levels. In BSO-treated cells, however, the cells demonstrated a much abbreviated enhancement of ECAR followed by an abrupt decrease in glycolytic flux.
Reversibility of diamide-induced protein thiolation and bioenergetic function
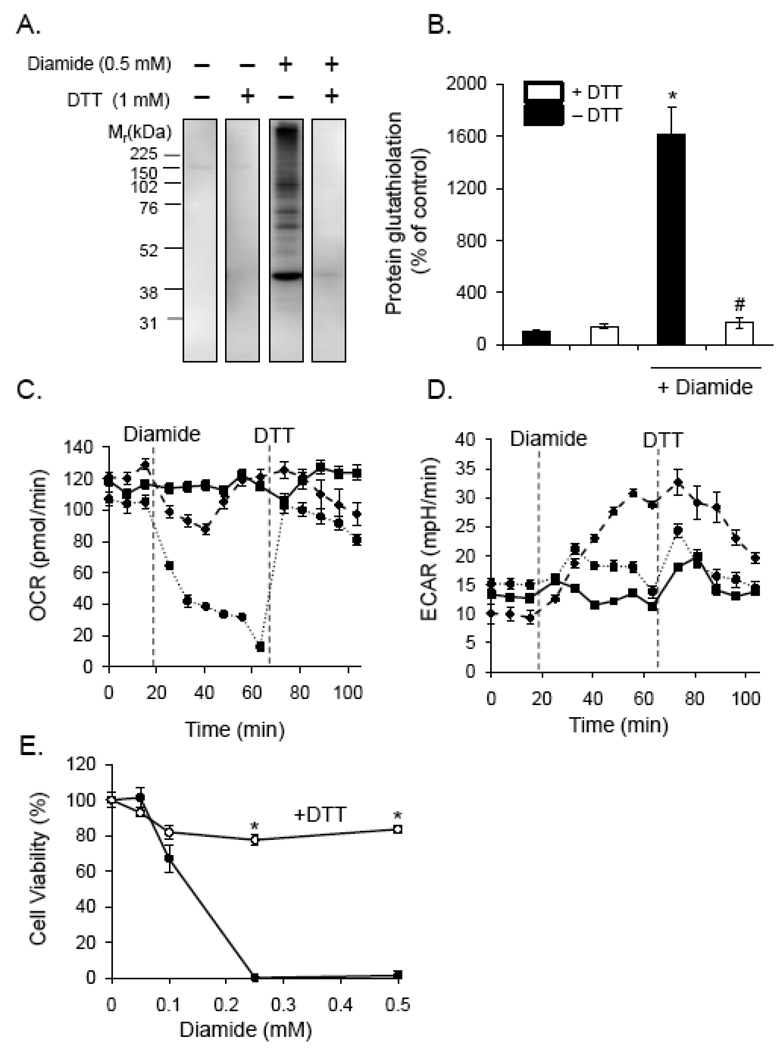
We next tested whether reduction of the protein thiol adducts would result in recovery of bioenergetic function and cell viability. To determine the degree of reversibility of the adducts, cells were treated with 0.5 mM diamide for 40 min followed by addition of 1 mM DTT for 10 min. The cells were then lysed in NEM-containing buffer, and PSSG adduct formation was examined by Western blotting. As shown in Fig. 7A and B, PSSG adducts formed upon diamide addition were completely reduced after incubation of the cells with 1 mM DTT.
Fig. 7. De-thiolation promotes bioenergetic recovery and prevents cell death.
Protein-glutathione adducts, bioenergetics, and cell viability in diamide-treated cells exposed to the reducing agent, dithiothreitol. (A) Dithiothreitol (DTT) reverses protein glutathiolation induced by diamide: Cells were treated without or with diamide for 40 min. DTT was then added to the cells to a final concentration of 1 mM for 10 min. The medium was then removed, and the cells were lysed in buffer containing N-ethylmaleimide. Protein-glutathione (PSSG) adducts were assessed by chemiluminescent Western blotting with anti-PSSG antibodies. (B) Group data from panel A. n = 3 per group; *p<0.05 vs. cells not treated with diamide; #p<0.05 vs. diamide-treated cells not exposed to DTT. (C and D) Bioenergetic measurements after diamide and DTT addition to cells: After measurement of the baseline oxygen consumption (panel C) and extracellular acidification (panel D) rates (OCR and ECAR, respectively), diamide was injected to 0 (closed squares), 0.1 (triangles; dashed line), or 0.5 (circles; dotted line) mM concentrations. At the indicated time, DTT was injected to a final concentration of 1 mM, and OCR and ECAR measurements were recorded. N = 5 per group. (E) Cell viability in diamide and DTT-treated cells. Cells were treated with diamide in the absence or presence of 1 mM DTT for 16 h. Cell viability was then measured by MTT assay. N = 4 per group; *p < 0.05 vs. cells treated with diamide alone.
To determine if removal of adduct formation was sufficient to reverse the effect of diamide on bioenergetic function, cells were treated with 0, 0.1, or 0.5 mM diamide for 40 min, and OCR was monitored over time. DTT was then injected to a final concentration of 1 mM, and the OCR was examined. As shown in Fig. 7C, 0.1 mM diamide had little effect on OCR in this time period, but 0.5 mM diamide led to a collapse of mitochondrial function. DTT addition led to a nearly instantaneous and complete recovery of the OCR. The effect of diamide on glycolytic flux was also reversed by DTT treatment. Interestingly, the increase in ECAR by 0.1 mM diamide was more slowly reversed by DTT treatment, and DTT treatment by itself had a small but abbreviated effect on glycolytic flux in the 0 and 0.5 mM diamide treatment groups.
The effects of diamide on cell viability were also reversed by DTT treatment. Cells were treated with 0–0.5 mM diamide in the absence or presence of DTT. As shown in Fig. 7E, cells treated with 0.25 or 0.5 mM diamide were not viable after 16 h of treatment, and the presence of DTT almost completely prevented death due to diamide as assessed by MTT assay.
DISCUSSION
Protein glutathiolation is an important post-translational protein modification that is increased under conditions of oxidative and nitrosative stress. It regulates the function of a variety of enzymes and is implicated in redox signaling, metabolic function, and apoptosis. Nevertheless, an understanding of how glutathiolation itself modulates cell function has been difficult to achieve. This is largely due to the many types of oxidative modifications induced by reactive species, which confound our ability to discern the contribution of protein-glutathione adducts to functional readouts. For instance, NO has been shown to increase protein glutathiolation in a number of in vitro and in vivo settings [27]; however, NO also interacts with metal centers in proteins and can react with oxidants and amino acids [37,38]. In this study, we examined how diamide modulates protein thiolation and cellular bioenergetics. There are several advantages to using diamide for examining the overall impact of glutathiolation-induced changes on bioenergetics. Reduced glutathione reacts rapidly with diamide, with a rate constant of 300 M−1 sec−1 at pH 7.3 [39]. Furthermore, glutathione is 3–10 times more reactive with diamide compared with other small thiols such as cysteine, and lipoic acid [39]. Protein glutathiolation by diamide proceeds via a sulfenylhydrazine intermediate during its reaction with GS– or protein thiolate anion (PS–) and therefore yields the glutathiolating agent GSSG or PSSG directly [39,47]. Oxygen is not required for diamide reactivity, and no radical intermediates are involved in diamide-induced thiolation reactions [39]. Therefore, the perturbation of thiol status in the cell by diamide primarily involves the GSH/GSSG couple and the protein thiol redox state, thereby minimizing subsidiary protein modifications.
We found that diamide promoted primarily protein-glutathione (PSSG) adducts that were associated with robust effects on glycolysis, mitochondrial function, and cell viability. Low levels of diamide did not induce significant toxicity, yet changed both glycolysis and mitochondrial function; whereas higher levels of diamide caused bioenergetic failure and cell death. In support of the hypothesis that these effects were indeed due to protein glutathiolation/thiolation, we found that: a) depletion of intracellular GSH with BSO prevented protein glutathiolation caused by diamide (Fig. 5); b) GSH depletion sensitized cells to diamide-induced bioenergetic collapse (Fig. 6) and death (Fig. 4D); and c) the thiol reducing agent DTT reversed the metabolic effects and cell death caused by diamide while having no significant effects on control cells (Fig. 7). These data therefore appear to rule out any non-specific effects of diamide (e.g., with reduced pyridine nucleotides, etc. [48]). Collectively, these results point to a role for thiolation reactions in the modulation of bioenergetic function in the cell as well as suggest that protein glutathiolation at low levels could be protective under conditions where thiol oxidation is increased.
Mitochondria figure predominantly in bioenergetic function and redox signaling. They are sites of reactive oxygen species generation [49], and mitochondrial metabolic flux can be controlled by NO through its binding to cytochrome c oxidase [38]. Mitochondria are also potential hotspots for oxidative post-translational protein modifications. Several types of modifications have been shown to occur on mitochondrial proteins, including, but not limited to, protein sulfenic, sulfinic, and sulfonic acids [50,51], S-nitrosation [52,53], and protein thiolation [20,26,27]. Of these, protein thiolation-type modifications appear to be abundant, increased under conditions of oxidative/nitrosative stress, and reversible, suggesting that they could potentially regulate mitochondrial function. Nevertheless, it has been difficult to discern the whole-cell functional effects caused by glutathiolation from those of other oxidative modifications.
In our treatments, protein-protein disulfide formation was ancillary to protein glutathiolation. From diagonal electrophoresis gels (Fig. 5), it was apparent that diamide induced comparatively little protein-protein thiol modifications compared to PSSG adducts. The inhibitory effects of diamide on oxygen consumption and glycolysis were maximal at 0.5 mM diamide. These changes were fully reversible with DTT treatment; however, longer durations of treatment with diamide (i.e., > 60 min) resulted in less DTT reversibility of the diamide-induced bioenergetic changes (data not shown). Interestingly, lower concentrations of diamide (100 µM) had biphasic effects on glycolysis and oxygen consumption. Diamide at this concentration consistently increased ECAR, suggesting that it regulated glycolytic flux. This may be due to either modification of enzymes that regulate glycolysis or a response to loss of mitochondrial ATP production. Evidence for the former is shown by several studies, where enzymes regulating glucose utilization branch points are regulated by thiolation. For instance, aldose reductase, which metabolizes glucose to sorbitol, has been shown to be inactivated by glutathiolation [30,31]. Therefore, glutathiolation may reroute carbohydrate flux by regulating enzymes controlling glucose metabolism pathways. Glyceraldehyde-3-phosphate dehydrogenase has also been shown to be modified and inactivated by glutathiolation [5,28], and this may underlie the loss of ECAR at higher concentrations or longer exposures to diamide. The latter possibility, i.e., an increase in glycolysis due to loss of respiratory activity, is supported by the fact that diamide, even at low concentrations, decreased the apparent oxygen consumption linked to ATP production and made oxygen utilization less efficient (Fig. 3). However, the changes in glycolysis due to diamide exceeded the increase in ECAR due to oligomycin treatment alone (see Fig. 4C), suggesting that loss of respiration cannot be wholly responsible for increased glycolytic flux during diamide treatment.
Our studies support the notion that protein glutathiolation causes mitochondrial uncoupling. It should be noted that the measurements of proton leak described here are reflective of what occurs in state 4 respiration, i.e., the measurements were performed in the presence of oligomycin; this is known to increase the mitochondrial membrane potential which maximizes changes in proton leak [54]. Nevertheless, these measurements show that glutathiolation uncouples mitochondria and decreases the capacity of the cell to utilize oxygen efficiently for ATP production. This is consistent with a previous study showing that diamide enhances proton conductivity through the Fo portion of the mitochondrial ATPase [55]; however, it has also been shown in submitochondrial particles that oligomycin inhibits this increase in conductivity [56], suggesting that the diamide-induced proton leak occurring in mitochondria in intact cells occurs via a different mechanism. At higher concentrations, diamide decreased the FCCP-stimulated rate of oxygen consumption and diminished the reserve capacity (Fig. 3), consistent with inhibition of mitochondrial electron transfer. Collectively, these results suggest that diamide, and therefore protein thiolation, decreases mitochondrial efficiency and the bioenergetic reserve capacity.
Interestingly, low concentrations of diamide had little effect on cell viability. Diamide at the 0.1 mM concentration caused at most a 20% loss in cell viability (Fig. 7E). In most experiments, however, no increase in cell death was observed after 16 h of treatment with this concentration of diamide (Fig. 4D). However, 0.1 mM diamide caused complete loss of viability in glutathione-depleted cells. This is consistent with the hypothesis that glutathiolation, at low levels such as that induced by 0.1 mM diamide (see Fig. 1 and Fig. 5), could be protective under thiol-oxidizing conditions. For example, protein glutathiolation, at low concentrations or when specifically regulated by enzyme systems, could form a molecular cap that prevents other deleterious modifications, e.g., protein-protein dithiol formation [19]. Glutathione depletion, by itself, had no effect on cell viability or basal mitochondrial function and little effect on glycolysis. However, loss of glutathione due to BSO treatment decreased protein glutathiolation due to diamide, and, under the same conditions, increased protein-protein dithiol formation, as assessed qualitatively by diagonal electrophoresis. These results support the widely held view that glutathione levels regulate the degree of protein glutathiolation that can be achieved in the cell under conditions of oxidant stress.
The reversibility of the PSSG adducts with DTT was associated with a rapid restoration of oxygen consumption, normalization of glycolytic flux, and protection from cell death due to diamide treatment. These findings are consistent with glutathiolation being a reversible and regulated thiol modification that controls cell function. Candidate proteins that can enzymatically modulate PSSG adducts include the thiolation/de-thiolation enzymes—glutaredoxin, sulfiredoxin, PDI, and GSTP [14,47]. An important limitation of these studies is that they do not take into account such enzymatic control of glutathiolation, especially within defined domains in the cell such as the mitochondrion. Furthermore, it can be difficult to relate the thiolation reactions caused by diamide to those that could be caused by endogenous or exogenous NO or ROS. However, many of the same mitochondrial proteins or proteins involved in glucose metabolism have been shown to be S-nitrosated [30,57], glutathiolated [4,27], and S-oxidized [36,58], suggesting that S-nitrosated and sulfenic acid-modified proteins could precede their glutathiolated forms. Moreover, receptor agonists such as angiotensin II have been shown to promote protein glutathiolation in smooth muscle cells [59], and TNF-α promotes glutathiolation of GAPDH [60]. Our results indicate that thiolation of these subproteomes could impact cellular bioenergetics and that even small increases in protein thiolation can affect glycolysis and aerobic respiration. Taken as a whole, glutathiolation appears to be an important modification regulating bioenergetic function and cell death. Interventions that modulate protein thiolation levels in specific sub-domains could therefore have therapeutic potential in pathological conditions characterized by increased oxidative or nitrosative stress.
ACKNOWLEDGEMENTS
This work was supported in part by Seahorse Biosciences (V.D.U.) and NIH grants ES10167 (V.D.U.), T32 HL07457 (B.G.H.), T32 HL007918 (B.P.D.), and an American Heart Association predoctoral fellowship 0815177E (A.N.H).
ABBREVIATIONS
- RASMC
rat aortic smooth muscle cells
- PSSG
protein-glutathione
- XF
extracellular flux
- OCR
oxygen consumption rate
- ECAR
extracellular acidification rate
- ECL
enhanced chemiluminescence
- ROS
reactive oxygen species
- FCCP
carbonyl cyanide p-[trifluoromethoxy]-phenyl-hydrazone
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- NO
nitric oxide
- NP-40
noniodet P40
- SDS
sodium dodecyl sulfate
- PVDF
polyvinylidene fluoride
- DMEM
Dulbecco’s Eagle Modified Medium
- HRP
horseradish peroxidase
- DMSO
dimethylsulfoxide
- FBS
fetal bovine serum
- PDI
protein disulfide isomerase
- DTT
dithiothreitol
- BSO
buthionine sulfoximine
- NEM
N-ethylmaleimide
- GSTP
glutathione-S-transferase pi
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Diotte NM, Xiong Y, Gao J, Chua BH, Ho YS. Attenuation of doxorubicin-induced cardiac injury by mitochondrial glutaredoxin 2. Biochim. Biophys. Acta. 2009;1793:427–438. doi: 10.1016/j.bbamcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, Cohen RA. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat. Med. 2004;10:1200–1207. doi: 10.1038/nm1119. [DOI] [PubMed] [Google Scholar]
- 3.Ghezzi P, Romines B, Fratelli M, Eberini I, Gianazza E, Casagrande S, Laragione T, Mengozzi M, Herzenberg LA, Herzenberg LA. Protein glutathionylation: coupling and uncoupling of glutathione to protein thiol groups in lymphocytes under oxidative stress and HIV infection. Mol. Immunol. 2002;38:773–780. doi: 10.1016/s0161-5890(01)00114-6. [DOI] [PubMed] [Google Scholar]
- 4.Eaton P, Byers HL, Leeds N, Ward MA, Shattock MJ. Detection, quantitation, purification, and identification of cardiac proteins S-thiolated during ischemia and reperfusion. J. Biol. Chem. 2002;277:9806–9811. doi: 10.1074/jbc.M111454200. [DOI] [PubMed] [Google Scholar]
- 5.Lind C, Gerdes R, Schuppe-Koistinen I, Cotgreave IA. Studies on the mechanism of oxidative modification of human glyceraldehyde-3-phosphate dehydrogenase by glutathione: catalysis by glutaredoxin. Biochem. Biophys. Res. Commun. 1998;247:481–486. doi: 10.1006/bbrc.1998.8695. [DOI] [PubMed] [Google Scholar]
- 6.Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, Murphy MP. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant DEFENSE. J. Biol. Chem. 2004;279:47939–47951. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 7.Gallogly MM, Starke DW, Mieyal JJ. Mechanistic and Kinetic Details of Thiol-Disulfide Exchange by Glutaredoxins and Potential Mechanisms of Regulation. Antioxid. Redox. Signal. 2009 doi: 10.1089/ars.2008.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan S, Berk BC. Glutathiolation regulates tumor necrosis factor-alpha-induced caspase-3 cleavage and apoptosis: key role for glutaredoxin in the death pathway. Circ. Res. 2007;100:213–219. doi: 10.1161/01.RES.0000256089.30318.20. [DOI] [PubMed] [Google Scholar]
- 9.Findlay VJ, Townsend DM, Morris TE, Fraser JP, He L, Tew KD. A novel role for human sulfiredoxin in the reversal of glutathionylation. Cancer Res. 2006;66:6800–6806. doi: 10.1158/0008-5472.CAN-06-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JW, Mieyal JJ, Rhee SG, Chock PB. Deglutathionylation of 2-Cys peroxiredoxin is specifically catalyzed by sulfiredoxin. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peltoniemi MJ, Karala AR, Jurvansuu JK, Kinnula VL, Ruddock LW. Insights into deglutathionylation reactions: Different intermediates in the glutaredoxin and protein disulphide isomerase catalysed reactions are defined by the gamma-linkage present in glutathione. J. Biol. Chem. 2006 doi: 10.1074/jbc.M605602200. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura S, Matsushima M, Song H, Kikuchi M. A role of PDI in the reductive cleavage of mixed disulfides. J. Biochem. 1996;120:525–530. doi: 10.1093/oxfordjournals.jbchem.a021445. [DOI] [PubMed] [Google Scholar]
- 13.Manevich Y, Feinstein SI, Fisher AB. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3780–3785. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Townsend DM, Manevich Y, He L, Hutchens S, Pazoles CJ, Tew KD. Novel role for glutathione S-transferase pi. Regulator of protein S-Glutathionylation following oxidative and nitrosative stress. J. Biol. Chem. 2009;284:436–445. doi: 10.1074/jbc.M805586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bass R, Ruddock LW, Klappa P, Freedman RB. A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J. Biol. Chem. 2004;279:5257–5262. doi: 10.1074/jbc.M304951200. [DOI] [PubMed] [Google Scholar]
- 16.Klatt P, Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur. J. Biochem. 2000;267:4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- 17.Thomas JA, Poland B, Honzatko R. Protein sulfhydryls and their role in the antioxidant function of protein S-thiolation. Arch. Biochem. Biophys. 1995;319:1–9. doi: 10.1006/abbi.1995.1261. [DOI] [PubMed] [Google Scholar]
- 18.Ziegler DM. Role of reversible oxidation-reduction of enzyme thiols-disulfides in metabolic regulation. Annu. Rev. Biochem. 1985;54:305–329. doi: 10.1146/annurev.bi.54.070185.001513. [DOI] [PubMed] [Google Scholar]
- 19.Hill BG, Darley-Usmar VM. S-nitrosation and thiol switching in the mitochondrion: a new paradigm for cardioprotection in ischaemic preconditioning. Biochem. J. 2008;412:e11–e13. doi: 10.1042/BJ20080716. [DOI] [PubMed] [Google Scholar]
- 20.Hurd TR, Costa NJ, Dahm CC, Beer SM, Brown SE, Filipovska A, Murphy MP. Glutathionylation of mitochondrial proteins. Antioxid. Redox. Signal. 2005;7:999–1010. doi: 10.1089/ars.2005.7.999. [DOI] [PubMed] [Google Scholar]
- 21.Nulton-Persson AC, Starke DW, Mieyal JJ, Szweda LI. Reversible inactivation of alpha-ketoglutarate dehydrogenase in response to alterations in the mitochondrial glutathione status. Biochemistry. 2003;42:4235–4242. doi: 10.1021/bi027370f. [DOI] [PubMed] [Google Scholar]
- 22.Kil IS, Park JW. Regulation of mitochondrial NADP+dependent isocitrate dehydrogenase activity by glutathionylation. J. Biol. Chem. 2005;280:10846–10854. doi: 10.1074/jbc.M411306200. [DOI] [PubMed] [Google Scholar]
- 23.Wenzel P, Hink U, Oelze M, Schuppan S, Schaeuble K, Schildknecht S, Ho KK, Weiner H, Bachschmid M, Munzel T, Daiber A. Role of reduced lipoic acid in the redox regulation of mitochondrial aldehyde dehydrogenase (ALDH-2) activity. Implications for mitochondrial oxidative stress and nitrate tolerance. J. Biol. Chem. 2007;282:792–799. doi: 10.1074/jbc.M606477200. [DOI] [PubMed] [Google Scholar]
- 24.Taylor ER, Hurrell F, Shannon RJ, Lin TK, Hirst J, Murphy MP. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J. Biol. Chem. 2003;278:19603–19610. doi: 10.1074/jbc.M209359200. [DOI] [PubMed] [Google Scholar]
- 25.Chen CL, Zhang L, Yeh A, Chen CA, Green-Church KB, Zweier JL, Chen YR. Site-specific S-glutathiolation of mitochondrial NADH ubiquinone reductase. Biochemistry. 2007;46:5754–5765. doi: 10.1021/bi602580c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YR, Chen CL, Pfeiffer DR, Zweier JL. Mitochondrial complex II in the post-ischemic heart: oxidative injury and the role of protein S-glutathionylation. J. Biol. Chem. 2007;282:32640–32654. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 27.West MB, Hill BG, Xuan YT, Bhatnagar A. Protein glutathiolation by nitric oxide: an intracellular mechanism regulating redox protein modification. FASEB J. 2006;20:1715–1717. doi: 10.1096/fj.06-5843fje. [DOI] [PubMed] [Google Scholar]
- 28.Ravichandran V, Seres T, Moriguchi T, Thomas JA, Johnston RB., Jr S-thiolation of glyceraldehyde-3-phosphate dehydrogenase induced by the phagocytosis-associated respiratory burst in blood monocytes. J. Biol. Chem. 1994;269:25010–25015. [PubMed] [Google Scholar]
- 29.Schuppe-Koistinen I, Moldeus P, Bergman T, Cotgreave IA. S-thiolation of human endothelial cell glyceraldehyde-3-phosphate dehydrogenase after hydrogen peroxide treatment. Eur. J. Biochem. 1994;221:1033–1037. doi: 10.1111/j.1432-1033.1994.tb18821.x. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava S, Dixit BL, Ramana KV, Chandra A, Chandra D, Zacarias A, Petrash JM, Bhatnagar A, Srivastava SK. Structural and kinetic modifications of aldose reductase by S-nitrosothiols. Biochem. J. 2001;358:111–118. doi: 10.1042/0264-6021:3580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandra A, Srivastava S, Petrash JM, Bhatnagar A, Srivastava SK. Modification of aldose reductase by S-nitrosoglutathione. Biochemistry. 1997;36:15801–15809. doi: 10.1021/bi9714722. [DOI] [PubMed] [Google Scholar]
- 32.Fratelli M, Demol H, Puype M, Casagrande S, Eberini I, Salmona M, Bonetto V, Mengozzi M, Duffieux F, Miclet E, Bachi A, Vandekerckhove J, Gianazza E, Ghezzi P. Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3505–3510. doi: 10.1073/pnas.052592699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brennan JP, Wait R, Begum S, Bell JR, Dunn MJ, Eaton P. Detection and mapping of widespread intermolecular protein disulfide formation during cardiac oxidative stress using proteomics with diagonal electrophoresis. J. Biol. Chem. 2004;279:41352–41360. doi: 10.1074/jbc.M403827200. [DOI] [PubMed] [Google Scholar]
- 34.Reinartz M, Ding Z, Flogel U, Godecke A, Schrader J. Nitrosative stress leads to protein glutathiolation, increased s-nitrosation, and up-regulation of peroxiredoxins in the heart. J. Biol. Chem. 2008;283:17440–17449. doi: 10.1074/jbc.M800126200. [DOI] [PubMed] [Google Scholar]
- 35.Clavreul N, Adachi T, Pimental DR, Ido Y, Schoneich C, Cohen RA. S-glutathiolation by peroxynitrite of p21ras at cysteine-118 mediates its direct activation and downstream signaling in endothelial cells. FASEB J. 2006;20:518–520. doi: 10.1096/fj.05-4875fje. [DOI] [PubMed] [Google Scholar]
- 36.Saurin AT, Neubert H, Brennan JP, Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17982–17987. doi: 10.1073/pnas.0404762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J. Biol. Chem. 1991;266:4244–4250. [PubMed] [Google Scholar]
- 38.Landar A, Darley-Usmar VM. Evidence for oxygen as the master regulator of the responsiveness of soluble guanylate cyclase and cytochrome c oxidase to nitric oxide. Biochem. J. 2007;405:e3–e4. doi: 10.1042/BJ20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosower NS, Kosower EM. Diamide: an oxidant probe for thiols. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- 40.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov. Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Choi SW, Gerencser AA, Nicholls DG. Bioenergetic analysis of isolated cerebrocortical nerve terminals on a microgram scale: spare respiratory capacity and stochastic mitochondrial failure. J. Neurochem. 2009;109:1179–1191. doi: 10.1111/j.1471-4159.2009.06055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, Chomicz S, Ferrick DA. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am. J. Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 43.Tietze F. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal. Biochem. 1969;27:502–522. doi: 10.1016/0003-2697(69)90064-5. [DOI] [PubMed] [Google Scholar]
- 44.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 45.Hill BG, Dranka BP, Zou L, Chatham JC, Darley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem. J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilge JL, Fisher M, Chai YC. The effect of oxidant and the non-oxidant alteration of cellular thiol concentration on the formation of protein mixed-disulfides in HEK 293 cells. PLoS. One. 2008;3:e4015. doi: 10.1371/journal.pone.0004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill BG, Bhatnagar A. Role of glutathiolation in preservation, restoration and regulation of protein function. IUBMB. Life. 2007;59:21–26. doi: 10.1080/15216540701196944. [DOI] [PubMed] [Google Scholar]
- 48.Harris JW, Biaglow JE. Non-specific reactions of the glutathione oxidant "diamide" with mammalian cells. Biochem. Biophys. Res. Commun. 1972;46:1743–1749. doi: 10.1016/0006-291x(72)90045-9. [DOI] [PubMed] [Google Scholar]
- 49.Murphy MP. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo MS, Kang SW, Kim K, Baines IC, Lee TH, Rhee SG. Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. J. Biol. Chem. 2000;275:20346–20354. doi: 10.1074/jbc.M001943200. [DOI] [PubMed] [Google Scholar]
- 51.Woo HA, Kang SW, Kim HK, Yang KS, Chae HZ, Rhee SG. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J. Biol. Chem. 2003;278:47361–47364. doi: 10.1074/jbc.C300428200. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y, Loscalzo J. S-nitrosoprotein formation and localization in endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 2005;102:117–122. doi: 10.1073/pnas.0405989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burwell LS, Nadtochiy SM, Tompkins AJ, Young S, Brookes PS. Direct evidence for S-nitrosation of mitochondrial complex I. Biochem. J. 2006;394:627–634. doi: 10.1042/BJ20051435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brand MD, Chien LF, Ainscow EK, Rolfe DF, Porter RK. The causes and functions of mitochondrial proton leak. Biochim. Biophys. Acta. 1994;1187:132–139. doi: 10.1016/0005-2728(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 55.Zanotti F, Guerrieri F, Scarfo R, Berden J, Papa S. Effect of diamide on proton translocation by the mitochondrial H+ATPase. Biochem. Biophys. Res. Commun. 1985;132:985–990. doi: 10.1016/0006-291x(85)91904-7. [DOI] [PubMed] [Google Scholar]
- 56.Yagi T, Hatefi Y. Thiols in oxidative phosphorylation: inhibition and energy-potentiated uncoupling by monothiol and dithiol modifiers. Biochemistry. 1984;23:2449–2455. doi: 10.1021/bi00306a020. [DOI] [PubMed] [Google Scholar]
- 57.Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ. Res. 2007;101:1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 58.Kaiserova K, Srivastava S, Hoetker JD, Awe SO, Tang XL, Cai J, Bhatnagar A. Redox activation of aldose reductase in the ischemic heart. J. Biol. Chem. 2006;281:15110–15120. doi: 10.1074/jbc.M600837200. [DOI] [PubMed] [Google Scholar]
- 59.Adachi T, Pimentel DR, Heibeck T, Hou X, Lee YJ, Jiang B, Ido Y, Cohen RA. S-glutathiolation of Ras mediates redox-sensitive signaling by angiotensin II in vascular smooth muscle cells. J. Biol. Chem. 2004;279:29857–29862. doi: 10.1074/jbc.M313320200. [DOI] [PubMed] [Google Scholar]
- 60.Sullivan DM, Wehr NB, Fergusson MM, Levine RL, Finkel T. Identification of oxidant-sensitive proteins: TNF-alpha induces protein glutathiolation. Biochemistry. 2000;39:11121–11128. doi: 10.1021/bi0007674. [DOI] [PubMed] [Google Scholar]