Abstract
Heparan sulfate (HS) moieties on cell surfaces are known to provide attachment sites for many viruses including herpes simplex virus type-1 (HSV-1). Here we demonstrate that cells respond to HSV-1 infection by promoting filopodia formation. Filopodia express HS and are subsequently utilized for the transport of HSV-1 virions to cell bodies in a surfing-like phenomenon, which is facilitated by the underlying actin cytoskeleton and is regulated by transient activation of a small Rho GTPase, Cdc42. We also demonstrate that interaction between a highly conserved herpesvirus envelope glycoprotein B (gB) and HS is required for surfing. A HSV-1 mutant that lacks gB fails to surf and quantum-dots conjugated with gB demonstrate surfing-like movements. Our data demonstrates a novel use of a common receptor, HS, which could also be exploited by multiple viruses and quite possibly, many additional ligands for transport along the plasma membrane.
Keywords: Heparan Sulfate, Viral surfing, Herpes Simplex Virus type-1
Introduction
Heparan sulfate (HS) proteoglycans, key components of cell-surfaces and extra-cellular matrix, modulate physiological activities and influence cell growth and differentiation by interacting with a variety of regulatory factors [1]. The versatile ability of HS to interact with a variety of molecules can also be exploited by pathogens including viruses to invade human cells. Herpesviruses, like many other viruses, initiate infection of the host cell by first attaching to HS moieties present on cell surfaces [2,3]. The attachment then initiates the process of viral penetration into host cells. Herpesvirus entry, exemplified by herpes simplex virus type-1 (HSV-1), requires participation from multiple viral glycoproteins (gB, gC, gD, gH, and gL) and cellular receptors [3,4]. Binding of HSV-1 to HS is mediated by gB and gC, followed by interaction of gD with one of its three receptors: HVEM, nectin-1, and 3-O-sulfated heparan sulfate or 3-OS HS [4–7]. Binding of gD to its receptor is essential for viral penetration, which ultimately results in deposition of viral DNA for replication in the nucleus [4].
Although it is well established that HS provides the initial docking sites for the virions, it is not clear where on the host cell surface this interaction occurs first. In this context, projections on the plasma membrane, such as filopodia and retraction fibers, could potentially be important. Both are thin, rod-like, inter-convertible cell surface extensions formed by bundles of parallel actin filaments [8] that grow by the assembly of actin via a process that is signaled by activation of a Rho-family GTPase, Cdc42 [9,10]. Many pathogenic viruses including human herpesviruses such as HSV-1 [11], Epstein-Barr virus [12] and human herpesvirus-8 (HHV-8) [13] also activate Cdc42 during invasion of the host cells.
While filopodia may contribute to viral spread in many instances by facilitating virus particles to bud out [13,14], they could also play a role by facilitating entry of exogenous virions. Recently a phenomenon of viral surfing, whereby virions attach to filopodia/retraction fibers and travel down these extensions to reach the cell body for infection was reported for unrelated retroviruses and papillomavirus [15,16]. Here we provide novel details about HSV-1 surfing and suggest a new role for HS as a mediator of the viral transport phenomenon. The interaction with HS, via HSV-1 gB, results in lateral viral movement along the length of filopodia to bring the virions closer to the cell body. The involvement of HS raises an intriguing possibility that all HS-binding virions may exploit their interactions with HS for targeted transport to cell bodies. It also implicates HS in ligand transport in general.
Material and methods
Plasmids, cell lines & Reagents
The plasmids used in these experiments were pPEP98 (gB), and pPEP99 (gD) [5]. Wild-type CHO-K1, A mutant CHO cell line, pgsA-745 [17] and the gB complementing cell line D6 [18] were used. CHO cells stably expressing nectin-1 were provided by P. Spear (Northwestern University, Chicago IL). Cells were grown in Ham’s F12 medium supplemented with 10% fetal bovine serum (FBS) and containing geneticin (500 μg/ml) for complimenting cell lines. HEK-293 and HeLa cells were passaged with DMEM supplemented with 10% FBS while Vero cells were passaged with DMEM supplemented with 5% FBS. Antibody to Heparan Sulfate (10E4, U.S. biological), gB (Virusys Corporation), and gD (Abcam) were used at predetermined concentrations. Heparinase III was obtained from Sigma. Nectin 1 antibody, PRR1, (Immunotech) was used in a 1:100 dilution. Secondary HRP antibody (Jackson Immunoresearch Laboratories) was used in a 1:5000 dilution. FITC conjugated secondary anti-mouse IgG (SIGMA) was used in 1:200 dilution. Texas Red phalloidan (Invitrogen) was used at a dilution of 1:100.
Virus Stocks and Purification
Wild-type KOS, K26GFP and KO82-K26GFP viruses were used [18–19]. KO82-K26GFP lacks gB, and was grown in a complementing cell line (D6) that stably expresses gB. Virus was propagated on Vero cells and purified using a sucrose gradient as previously described [8,10]. The β-galactosidase-expressing recombinant HSV-1(KOS) gL86 was kindly provided by P. Spear (Northwestern University, Chicago IL). Since the virus lacks gL, it was grown in a complementing Vero cell line that stably expresses gL [6].
Imaging and Analysis
All cell images were taken using a 100x oil objective (plan-apo 1.4) on a Leica SP2 laser scanning confocal microscope (Leica). Cells were grown on a 35-mm glass-bottom dishes (Mattek) coated with collagen (BD Biosciences). Before imaging at 37°C, cells were washed with PBS, and cells were placed in DMEM/10% FBS. In some experiments, cells were serum starved for eight hours. K26GFP virus was imaged at 30 minutes after infection. In some experiments a high multiplicity of infection of 100–500 was used to capture particles in a given plane. For live cell imaging, brightfield and GFP channels were imaged every 30 seconds. Images were acquired using Leica confocal software. All videos, entire images, and analysis were processed using Metamorph (Molecular Devices), Leica confocal software, and Photoshop (Adobe).
Downregulation of Cdc42 and its Effect on Entry
Small interfering (si)-RNA targeting Cdc42, two different RNA duplexes (CDC42 Validated Stealth RNAi) were purchased from Invitrogen. Transfection of siRNA duplexes were performed in HeLa cells with Lipofectamine 2000 (Invitrogen) according to manufacturer’s instruction, and seeded onto glass bottom dishes (Mattek). As a negative control for Cdc42 siRNA low GC RNAi Negative Control Duplexes (Invitrogen) was used. The transfection efficiency of each duplex was confirmed by Block-It Fluorescent Oligo (Invitrogen). Viral Entry assay was described previously [6].
Indirect Immunofluorescence
Indirect immunofluorescence assay was done on Vero and CHO-K1 cells. Cells were adhered to glass bottom dishes, and subsequently blocked in PBS with 3% (w/v) bovine serum albumin, for 1 hour at room temperature. Cells were then incubated with either heparan sulfate antibody (U.S. Biologicals), HVEM antibody, or Nectin 1 antibody (Immunotech) for 1 hour at room temperature. As a negative control, primary antibody was omitted. FITC conjugated secondary anti-mouse IgG (SIGMA) was used in 1:200 dilution.
Cell Lysate and Quantum Dot Preparations
Cells were transfected in 6-well dishes with appropriate plasmid. After 24h post transfection, cells were detached using cell dissociation buffer (Invitrogen) and sonicated on ice for 6 second intervals, and then put on ice for 6 seconds. This was repeated a total of 6 times, and the solution was passed through a 0.8 μm filter. For quantum dot experiments, HeLa cells were blocked in 6% BSA for 1 hour. Anti-gB monoclonal antibody (10B7, Virusys Inc.) was incubated for 1 h with Qdot@605 goat anti-mouse IgG (Invitrogen) and the mixture was then incubated with gB for 1 h before addition to cells with a final end concentration of 0.04 μM, which were pre-blocked with 6% BSA. As a negative control, primary antibody was excluded from quantum dots.
Western Blot
Cdc42 activation and western blot assay was described previously [11]. Time points for Cdc42 activation were set at 0 min, 30 s, 1 and 5 min.
Results and discussion
HSV-1 can induce filopodia
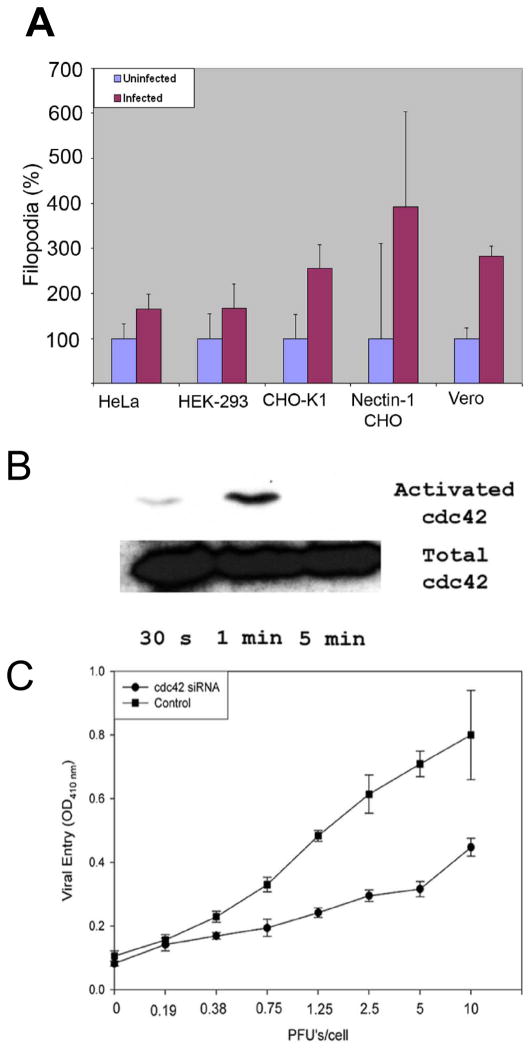
In order to examine the early morphological changes upon HSV-1 infection, a number of cell lines were examined at high magnification on a Leica SP2 laser scanning confocal microscope (Leica). As shown in Fig. 1A, significant (40 to 200%) increase in the number of filopodia per cell was observed at 15 min post virus addition compared to the control cells. The increase in filopodia numbers was observed with all cell lines examined including HeLa, HEK-293, CHO-K1, nectin-1-expressing CHO-K1 and Vero cells. It is noteworthy that CHO-K1 cells allow the virus to attach but not enter due to the lack of any receptors for gD [6]. Thus, it was clear that the increase in the filopodia numbers was not dependent on the presence of a gD receptor. The increase in number was mostly observed with filopodia attached to the substrate. There was only a mild increase in filopodia on the dorsal surface of the cells. Interestingly, the average length of the filopodia was not altered by the infection (data not shown).
Figure 1. Exogenous HSV-1 can induce filopodia formation.
(A) Increase in the number of filopodia upon exposure to HSV-1 were counted for cells indicated. Counting was done 15–30 min before and after the addition of virus. Numbers are represented as percentages to better compare different cell lines. 3 independent experiments were counted for each cell type (n=25). Numbers of filopodia are presented as means with error bars showing standard deviation. (B) Western Blot analysis confirms activation of Cdc42 at the time points indicated. (C) Down-regulation of Cdc42 inhibits HSV-1 entry. HeLa cells were transfected with siRNA against Cdc42 or control siRNA and exposed to a β-galactosidase expressing HSV-1(KOS) virus.
A mechanism for filopodia formation has been attributed to the activation of a RhoA GTPase, Cdc42 [20]. A transient activation of Cdc42 was indeed observed upon addition of HSV-1(KOS) to Vero cells (Fig. 1B). The Cdc42 activation lasted for at least 1 min and became undetectable by 5 min. Of interest is that although detectable Cdc42 activation was transient (within the first 5 min), an increase in the number of filopodia was most noticeable at 15 min after the virus infection. To further understand the role of Cdc42 activation and corresponding increase in filopodia numbers upon HSV-1 infection, Cdc42 expression was down-regulated by transfecting HeLa cells with a commercially validated siRNA against Cdc42 (Invitrogen) or a scrambled siRNA control. At 48 h post transfection, the cells were infected with identical dosages of a reporter HSV-1(gL86) virus and entry was measured at 6 h post infection. A relative decline in viral entry was evident for the cells transfected with the siRNA against Cdc42 and not its scrambled control (Fig. 1C). Taken together, our results suggest that an increase in filopodia numbers regulated by Cdc42 activation may play a supportive role in the entry process.
Virus interacts with filopodia for surfing
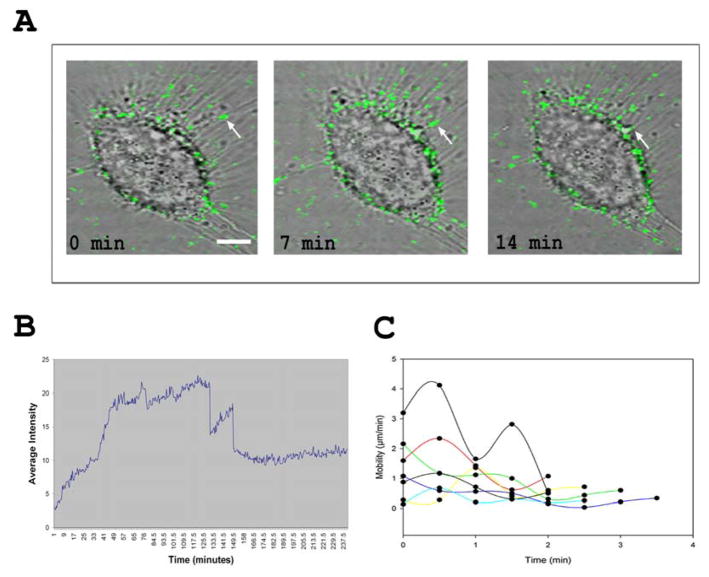
In order to further understand the significance of filopodia induction by HSV-1, cells were infected with GFP-expressing K26GFP virions and the infected cells were examined by laser scanning confocal microscopy. It was observed that the green fluorescent HSV-1 virions showed strong clustering on the filopodia (Fig. 2A). In fact, more virions were observed on the filopodia than on the cell membrane. Time course analysis, using live cell microscopy, indicated an interesting phenomenon. Increase of fluorescence intensity on filopodia, used as a surrogate for HSV-1 binding, was almost instantaneous but then it took about 60 min for the binding to show its peak (Fig. 2B), which continued for another 60 min before showing steady decline. Thus, it appears that the virions kept on finding the filopodia to attach for a relatively long time (about 2.0–2.5h). The occasional sharp declines in the intensity were likely due to large numbers of HSV moving out of the focal plane of the region of interest (ROI). It could happen due to twisting of filopodia and/or rapid movement of particles out of the ROI. Continued live cell imaging of the green HSV-1 virions indicated that bound virions did not remain stationary; instead they demonstrated highly ordered, “surfing-like” [16], lateral movement toward the cell body (Video S1). Virtually all moving particles remained attached to the filopodia and did not show any random motion. To gain additional insight into HSV-1 surfing, the viral movement was quantified by measuring the distance a virion traveled per frame (30 s). It was found that HSV-1 virions on filopodia surfed at variable speeds (Fig. 2C) with a net average speed of 1.5 μm/min where time 0 was when a virion was detected on the filopodia. The speed of travel appears very similar to what has been reported with unrelated retroviruses [16].
Figure 2. HSV-1 binds and surfs along filopodia.
(A) Time course of K26GFP virus (green) surfing (arrow) on a single Vero cell at indicated time points. Cells were infected at an approximate MOI of 200. (B) Representative binding of K26GFP virus to filopodia. To quantify virus binding, the average fluorescent intensity of a selected region of interest on filopodia was plotted over time. (C) Tracking of viral particles demonstrates an average particle speed of 1.5 μm/min. To quantify viral surfing speed, 8 to 10 particles per cell were tracked every 30 s and the distance traveled was plotted over time.
Heparan Sulfate and gB are important for binding and surfing
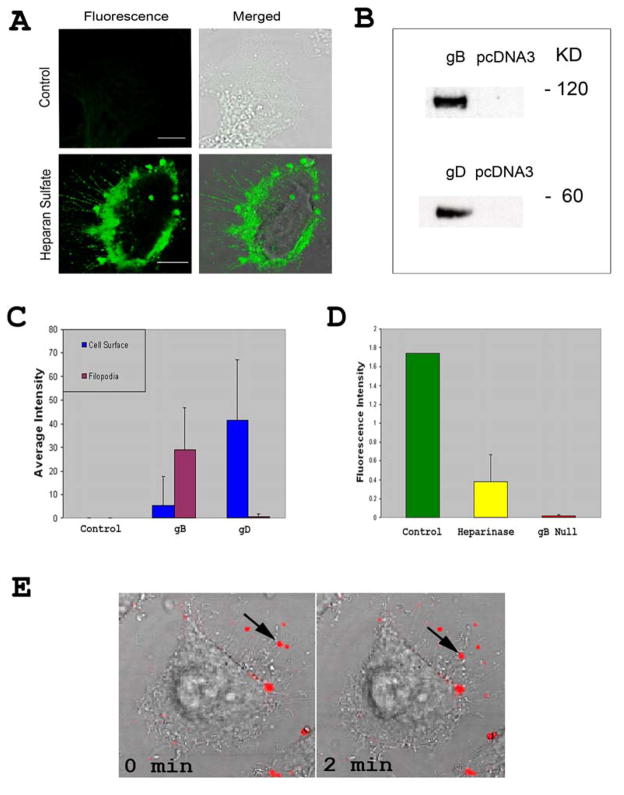
Next, to address the molecular interactions that give rise to surfing we focused on cellular receptors and viral glycoproteins. A previous report had found that trans-interactions of nectin-1 can induce filopodia formation [21]. However, we failed to detect nectin-1 on filopodia of HSV-1 infected cells (data not shown). In contrast, Fig. 3A shows the expression of HS, the attachment receptor for HSV-1, on filopodia, which raised the possibility that HS could be the major filopodial receptor for interaction with HSV-1. To determine a viral ligand for this interaction, we focused on HSV-1 glycoprotein, gB, which is already known to bind HS [3], and found on filopodia of virus infected or gB-expressing cells [13]. Another glycoprotein, gD, was targeted as a control, especially since gD receptors are required for entry and we indirectly wanted to confirm that gD receptors are not present on filopodia. Cell lysates containing gB or gD were prepared and the presence of ~ 60 KD gD and ~120 KD gB proteins was confirmed by Western blot analysis (Fig. 3B). Similar preparations from identically processed mock (pcDNA3.1) transfected cells were used as negative controls. Next, each glycoprotein was separately added to cells to determine the preferential binding locations of gB and gD. As seen in Fig. 3C, gB demonstrates greater binding at the filopodia whereas gD preferentially binds at the cell surface. This observation was determined by calculating green fluorescence intensity measurements at multiple locations on cells. The inability of gD to bind filopodia also indicates that gD receptors are not expressed on filopodia. This may explain why the virions need to travel on filopodia as they need to reach a gD receptor on the cell body for the mediation of entry. A similar finding has been demonstrated with some unrelated retroviruses [16].
Figure 3. Heparan sulfate and gB are important for attachment to filopodia and surfing.
(A) Anti-Heparan sulfate staining of Vero and CHO-K1 cells. As a negative control, primary antibody was omitted. FITC conjugated secondary anti-mouse IgG (SIGMA) was used in 1:200 dilution. (B) Western Blot Analysis confirming gB and gD presence in cell lysates. (C) Quantification of the average green intensity around the cell membrane and filopodia for gB and gD. 20 μg of total protein from gB and gD lysates were taken and incubated with monoclonal antibodies (Virusys Corp and Abcam, respectively) before using as probes on cells. The figure represents 3 independent experiments (n=11). Data are presented as means with error bars showing standard deviations. (D) Quantification of Heparinase treated cells and gB null virus. Average green intensity at filopodia was measured after 1 h of treatment with the enzyme or incubation with the mutant virions. Error bars show standard deviations. (E) Quantum dots (red) conjugated to gB via an antibody were used for demonstration of binding of gB to filopodia and its ability to surf on HeLa cells.
Next, to verify that HSV-1 attachment to filopodia was indeed mediated by interaction of gB with HS, we examined the effects of enzymatic removal of HS from cell surface and the binding and surfing ability of a virus devoid of gB. A gB deletion mutant of HSV-1 was generated in the K26GFP background (KO82-K26GFP) [19]. KO82-K26GFP lacks gB, and was grown in a complementing cell line, D6 [19] that stably expresses gB. Virus was propagated on Vero cells and purified using a sucrose gradient as previously described [18, 22]. Attachment of virions to filopodia was measured by recording fluorescence intensity. As shown (Fig. 3D), about 75% reduction in binding to filopodia was seen with the wild-type K26GFP when HS was removed by heparinase-III treatment (Sigma) (Fig. 3D). Even more, over 95% reduction to filopodial binding was observed when untreated cells were incubated with uncomplemented gB null virus. The fact that the drastically reduced binding was solely due to gB was confirmed by growing KO82-K26GFP virions on HSV-1 gB complementing cells. The complemented virions were able to regain the ability to bind filopodia (data not shown). Next, to demonstrate that gB alone may have the ability to bind and surf along filopodia, we made use of red quantum dots (qdots) (Invitrogen) conjugated to gB. The gB qdots were incubated with cells which were preinfected with HSV-1 to induce filopodia formation. It was evident that gB qdots not only bound filopodia but also a few of them were able to surf along filopodia (Fig. 3E; Video S2). The implication of gB in surfing is very significant given the fact that it is one of the most highly conserved glycoproteins among herpesviruses and virtually all known homologs of it bind HS [3]. It is, therefore, tempting to speculate that other herpesviruses could also surf via a similar gB-mediated mechanism. Likewise, the implication of HS in surfing has even more significance for the viral invasion and ligand transport processes, since many unrelated viruses including retroviruses use HS for attachment to cells [1,2]. Thus, surfing via HS may represent a unique commonality developed and conserved among viruses to help the infection process.
To summarize, the detailed analysis of a surfing-like phenomenon by HSV-1 and the involvement of HS point to the possibility of a common invasion strategy by many unrelated viruses. In an apparent analogy to retroviral surfing, virions in both cases [16; this study] demonstrate a net directional movement on filopodia toward the cell body at an average speed of 1–2 μm/min, require actin cytoskeleton and results in the increase in filopodia numbers. In all cases, surfing lies upstream of cell entry regardless of the mode of entry. Among the major differences lies the fact that the attachment receptor, HS, is an important initiator of surfing for herpesviruses whereas retroviruses have yet to be tested for the use of HS during surfing [16]. In case of papillomavirus the involvement of HS has been speculated but not demonstrated yet [15]. Another possible difference could lie in the motor usage. We have some preliminary evidence to suggest that HS mediated surfing may require myosin X (Oh and Shukla, unpublished observation) whereas myosin II seems to have a role in retrovirus surfing [16]. This difference, however, may not be that critical since the involvement of actin filaments and myosin motors, in general, imply that the cellular mechanism of surfing could be identical, basically involving retrograde F-actin flow [23–25]. Although how HSV-1 engages retrograde F-actin flow remains to be determined, the process itself is a part of the normal cargo delivery to cells [26]. Anchoring of HSV-1 to HS proteoglycans, such as syndecan-2, or EGF receptors, which are linked with filopodial cargo movement, may hold some clues to this process [26–28]. In any case, many identical aspects of retrovirus, papillomavirus and herpesvirus surfing can implicate novel targets for the development of broad-spectrum anti-viral prophylactic agents, a possibility that should also be examined in the near future and so is the role of HS in ligand transport.
Supplementary Material
Video S1 shows HSV-1 particles “surf” down filopodia.
Video S2 shows the movement of gB conjugated quantum dots on filopodia.
Acknowledgments
We thank R. Zelkha for help in image acquisition, editing and analysis, Drs. T. Valyi-Nagy, B. Yue, and members of our laboratory for helpful comments on the manuscript. This work was supported by NIH grants AI057860 (D.S), AI081869 (D.S.), AI33077(PD), and Core Grant EY01792. D.S. is a recipient of Lew Wasserman Merit award from Research to Prevent Blindness, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gorsi B, Stringer SE. Tinkering with heparan sulfate sulfation to steer development. Trends Cell Biol. 2007;17:173–177. doi: 10.1016/j.tcb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Thorp SC. Cell surface heparan sulfate and its roles in assisting viral infections. Med Res Rev. 2002;1:1–25. doi: 10.1002/med.1026. [DOI] [PubMed] [Google Scholar]
- 3.Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spear PG, Longnecker R. Herpesvirus entry: an update. J Virol. 2003;77:10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Donnell CD, Tiwari V, Oh MJ, Shukla D. A role for heparan sulfate 3-O-sulfotranferase isoform 2 in herpes simplex virus type 1 entry and spread. Virology. 2006;346:452–459. doi: 10.1016/j.virol.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 7.Tiwari V, O’Donnell CD, Oh MJ, Valyi-Nagy T, Shukla D. A role for 3-O-sulfotransferase isoform-4 in assisting HSV-1 entry and spread. Biochem Biophys Res Commun. 2005;338:930–937. doi: 10.1016/j.bbrc.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 8.Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caron E, Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282:1717–1721. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- 10.Medalia O, Beck M, Ecke M, Weber I, Neujahr R, Baumeister W, Gerisch G. Organization of actin networks in intact filopodia. Curr Biol. 2007;17:79–84. doi: 10.1016/j.cub.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 11.Clement C, Tiwari V, Scanlan PM, Valyi-Nagy T, Yue BY, Shukla D. A novel role for phagocytosis-like uptake in herpes simplex virus entry. J Cell Biol. 2006;174:1009–1021. doi: 10.1083/jcb.200509155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puls A, Eliopoulos AG, Nobes CD, Bridges T, Young LS, Hall A. Activation of the small GTPase Cdc42 by the inflammatory cytokines TNF(alpha) and IL-1, and by the Epstein-Barr virus transforming protein LMP1. J Cell Sci. 1999;112:2983–2992. doi: 10.1242/jcs.112.17.2983. [DOI] [PubMed] [Google Scholar]
- 13.Sharma-walia N, Naranatt PP, Krishnan HH, Zeng L, Chandran B. Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 envelope glycoprotein gB induces the integrin-dependent focal adhesion kinase-Src-phosphatidylinositol 3-kinase-rho GTPase signal pathways and cytoskeletal rearrangements. J Virol. 2004;78:4207–4223. doi: 10.1128/JVI.78.8.4207-4223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jouvenet N, Windsor M, Rietdorf J, Hawes P, Monaghan P, Way M, Wileman T. African swine fever virus induces filopodia-like projections at the plasma membrane. Cell Microbiol. 2006;8:1803–1811. doi: 10.1111/j.1462-5822.2006.00750.x. [DOI] [PubMed] [Google Scholar]
- 15.Schelhaas M, Ewers H, Rajamäki ML, Day PM, Schiller JT, Helenius A. Human papillomavirus type 16 entry: retrograde cell surface transport along actin-rich protrusions. PLoS Pathog. 2008;4(9):e1000148. doi: 10.1371/journal.ppat.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esko JD, Stewart TE, Taylor WH. Animal cell mutants defective in glycosaminoglycan biosynthesis, Proc. Natl Acad Sci USA. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai P, Person S. Incorporation of the Green Fluorescent Protein into the Herpes Simplex Virus Type 1 Capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai W, Person S, Warner SC, Zhou J, Deluca NA. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1, J. Virol. 1987;61:714–721. doi: 10.1128/jvi.61.3.714-721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 21.Kawaktsu T, Shimizu K, Honda T, Fukuhara T, Hoshino T, Takai Y. Trans-interactions of nectins induce formation of filopodia and Lamellipodia through the respective activation of Cdc42 and Rac small G proteins. J Biol Chem. 2002;277:50749–50755. doi: 10.1074/jbc.M209846200. [DOI] [PubMed] [Google Scholar]
- 22.Pignatti PF, Cassai E. Analysis of herpes simplex virus nucleoprotein complexes extracted from infected cells. J Virol. 1980;36:816–828. doi: 10.1128/jvi.36.3.816-828.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dohner K, Nagel CH, Sodeik B. Viral stop-and-go along microtubles: taking a ride with dynein and kinesins. Trends Microbiol. 2005;13(7):320–327. doi: 10.1016/j.tim.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Fukui Y, Kitanishi-Yumura T, Yumura S. Myosin II independent F-actin flow contributes to cell location in dictyostelium. J Cell Sci. 1999;112:877–886. doi: 10.1242/jcs.112.6.877. [DOI] [PubMed] [Google Scholar]
- 25.Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1:761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- 26.Faix J, Rottner K. The making of filopodia. Curr Opin Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 28.Akhtar J, Tiwari V, Oh MJ, Kovacs M, Jani A, Kovacs SK, Valyi-Nagy T, Shukla D. HVEM and nectin-1 are the major mediators of herpes simplex virus 1 (HSV-1) entry into human conjunctival epithelium. Invest Ophthalmol Vis Sci. 2008;49:4026–35. doi: 10.1167/iovs.08-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1 shows HSV-1 particles “surf” down filopodia.
Video S2 shows the movement of gB conjugated quantum dots on filopodia.