Abstract
While high-resolution 3D structures reveal the locations of all atoms in a molecule, it is the dynamics that correlates the structure with the function of a biological molecule. The complete characterization of dynamics of a membrane protein is in general complex. In this study, we report the influence of dynamics on the channel-forming function of pardaxin using chemical shifts and dipolar couplings measured from 2D broadband-PISEMA experiments on mechanically aligned phospholipids bilayers. Pardaxin is a 33-residue antimicrobial peptide originally isolated from the Red Sea Moses sole, pardachirus marmoratus, which functions via either a carpet-type or barrel-stave mechanism depending on the membrane composition. Our results reveal that the presence of cholesterol significantly reduces the backbone motion and the tilt angle of the C-terminal amphipathic helix of pardaxin. In addition, a correlation between the dynamics-induced heterogeneity in the tilt of the C-terminal helix and the membrane disrupting activity of pardaxin by the barrel-stave mechanism is established. This correlation is in excellent agreement with the absence of hemolytic activity for the derivatives of pardaxin. These results explain the role of cholesterol in the selectivity of the broad-spectrum of antimicrobial activities of pardaxin.
Introduction
Design of novel antibiotic compounds to overcome the increasing bacterial resistance towards conventional antibiotics is of considerable current importance. [1] Numerous studies have reported the isolation, characterization, biological functions, and potential pharmaceutical applications of naturally occurring AMPs. [2, 3] A variety of biochemical and biophysical approaches have reported mechanisms of membrane permeation/disruption, bioavailability, and synergistic activities of AMPs. [4–7] High-resolution cutting-edge techniques have been used to obtain atomic-level insights into the structure, dynamics, folding, oligomerization, and membrane orientation of AMPs and their interactions with lipid membrane. [8] These studies have been utilized in the design of potent peptide antibiotics. [9–11] In spite of these valuable studies, unfortunately, only a very few peptides have been found to be suitable for pharmaceutical applications. [5, 12–15] One of the major difficulties in designing potent AMPs is determining the role of membrane components that play important roles in the selectivity of AMPs. Therefore, there is a significant need for biophysical studies to identify the roles of membrane components on the function of AMPs. In this study, we report atomic-level insights into the role of cholesterol on the function of pardaxin using solid-state NMR experiments on mechanically aligned model phospholipid membranes. Cholesterol is abundantly present in mammalian membranes, but not in bacterial membranes, and has been thought to be crucial in reducing the toxicity of AMPs.
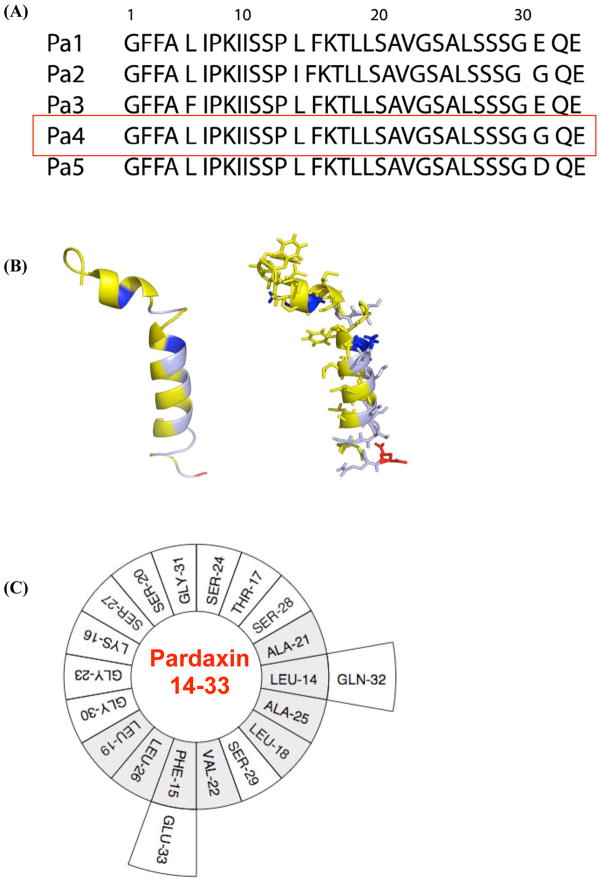
Pardaxin is a 33-residue antimicrobial peptide originally isolated from the secretions from the Red Sea Moses sole, pardachirus marmoratus. Pardaxin has been shown to exhibit a broad-spectrum of antimicrobial activities at very low concentrations, to act as a channel-forming neurotoxin, and also to cause cell death by necrosis at slightly higher concentrations. [16–21] Amino acid sequences of pardaxin peptides are given in Figure 1 along with the 3D NMR structure of the peptide. An early NMR study on pardaxin in a TFE:water solution reported a helix-hinge-helix structure similar to that of melittin. [22] But a recent NMR study reported the more biologically-relevant structure of the peptide in a membrane environment, where the structure of the N-terminal domain significantly differs from that obtained from TFE:water structure (Figure 1). [23] Recent NMR studies have also reported the membrane orientation, role of membrane composition, and potential mechanisms of the membrane permeation of pardaxin. [23, 24] The antimicrobial activities of peptide fragments corresponding to 1–11, 1–18, 1–22, 10–33, and 12–33 residues of pardaxin and several mutants have also been reported. The amidated forms of the 22-residues C-terminal fragment and the 11 amino acid N-terminal domain exhibit antimicrobial activity only against Gram-positive bacteria,[17] whereas the fragment 1–18 has been shown to be active against E.coli but not against Gram-positive bacteria. [25] These differences in the activities of the derivatives of pardaxin are not well understood.
Figure 1. Pardaxin - a cationic antimicrobial peptide originally isolated from the Red Sea Moses sole, pardachirus marmoratus.
(A) Amino acid sequences of pardaxin peptides. Pa4 was used in this study. (B) NMR structure of pardaxin determined from DPC micelles [23]. (C) A helical wheel representation of the C-terminal amphipathic domain of pardaxin.
In this study, we show that the presence of cholesterol reduces the dynamics of pardaxin but does not influence the membrane orientation of pardaxin. Influences of temperature and cholesterol on the backbone dynamics of pardaxin are investigated using a combination of 2D broadband-PISEMA (polarization inversion spin exchange at the magic angle) experiments and mechanically aligned phospholipids bilayers to accurately measure 15N chemical shifts and 1H-15N dipolar couplings. [26–28] Implications of solid-state NMR results on the mechanisms of membrane disruption and selectivity by pardaxin are discussed.
Materials and Methods
Materials
Methanol and chloroform were purchased from Aldrich Chemical Inc. (Milwaukee, WI). Phospholipids, cholesterol, and naphthalene were purchased from Avanti Polar Lipids (Alabaster, AL), Sigma (St. Louis, MO), and Fisher Scientific (Pittsburg, PA) respectively. All these chemicals were used without further purification. All peptides were synthesized and purified as explained in our earlier publication. [24] Mechanically aligned bilayer samples were prepared using the naphthalene procedure as explained in our previous publications. [29]
Solid-state NMR
All of the solid-state NMR experiments were performed on a Chemagnetics/Varian Infinity 400 MHz spectrometer operating at resonance frequencies of 400.138, 161.979, and 40.55 MHz for 1H, 31P, and 15N nuclei, respectively using home-built flat-coil probes. 31P chemical shift spectra obtained using a spin-echo sequence (90°–τ–180°–τ–acquisition; t = 70 μs) with a 90° pulse length of 3.5 μs under 40 kHz proton decoupling were used to confirm the alignment of each mechanically aligned bilayer samples after an equilibration for about 30 minutes at a chosen temperature before recording the spectrum. All reported solid-state NMR experiments were performed with the bilayer normal of the sample oriented parallel to the external magnetic field. After optimization of the experimental conditions for sensitivity, resolution, and sample stability using a series of 1D NMR experiments as explained elsewhere [28], 2D BB-PISEMA experiments were performed as explained in our previous publication. [26] A ramp cross-polarization sequence with a 1H π/2 pulse length of 3 μs, 50 kHz cross-polarization power, a 1H decoupling of 75 kHz during acquisition, and a 2 s recycle delay were used in 1D experiments. An RF field strength of 62.5 kHz was used for the Lee-Goldburg off-resonance pulses during the t1 period of BB-PISEMA. 32 t1 increments each with 3600 scans were recorded for 2D BB-PISEMA spectra. All experimental data were processed using the Spinsight (Chemagnetics/Varian) software on a Sun SPARC workstation.
Results
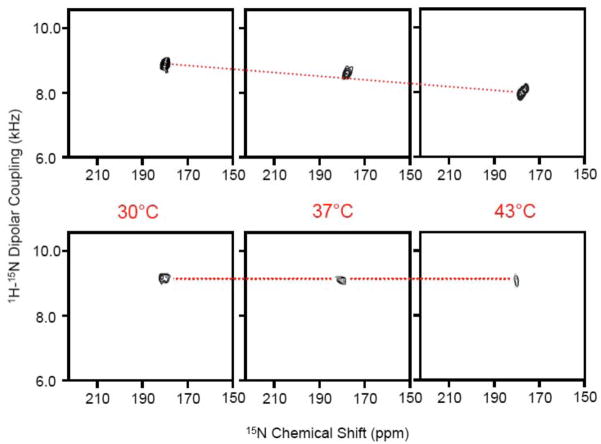
A previous solid-state NMR study reported a transmembrane orientation for the C-terminal helix of pardaxin in DMPC and a membrane surface orientation in POPC bilayers. [24] Since we are interested in probing the role of cholesterol on the dynamics of the peptide and its implications for the barrel-stave mechanism of membrane disruption by pardaxin, we chose to use DMPC and DMPG lipids for this study in which the peptide is in a transmembrane orientation. Two types of model membrane compositions were used in this study: 3:1 DMPC:DMPG bilayers and DMPC bilayers containing 15 mole % cholesterol to understand the role of cholesterol present in mammalian membranes. 2D BB-PISEMA spectra of these samples at various temperatures are given in Figure 2. A single peak observed in the 15N chemical shift region of 175 to 183 ppm and a 15N-1H dipolar coupling frequency of 8–9.3 kHz confirms the transmembrane orientation of the C-terminal helix of pardaxin under this condition. 2D experiments were performed to measure the influence of temperature in the presence and absence of cholesterol. A decrease in both the 15N chemical shift and 15N-1H dipolar coupling frequencies were observed with increasing temperature in the absence of cholesterol (Figure 3A). On the other hand, no significant changes were observed in the presence of cholesterol (Figure 3B). A model explaining the influence of cholesterol on lipid bilayers and on pardaxin’s backbone dynamics is depicted in Figure 4 and discussed below.
Figure 2. The C-terminal helical segment of pardaxin has a transmembrane orientation.
2D BB-PISEMA spectra of mechanically aligned lipid bilayers containing 2 mole % 15N-Ala21-pardaxin: (top) 3:1 DMPC:DMPG bilayers and (bottom) DMPC bilayers with 15 mole % cholesterol at 30, 37 and 43°C. A single peak observed in each spectrum, even in the absence of cholesterol, suggests an average of dynamically disordered tilts of the C-terminal helical domain of pardaxin.
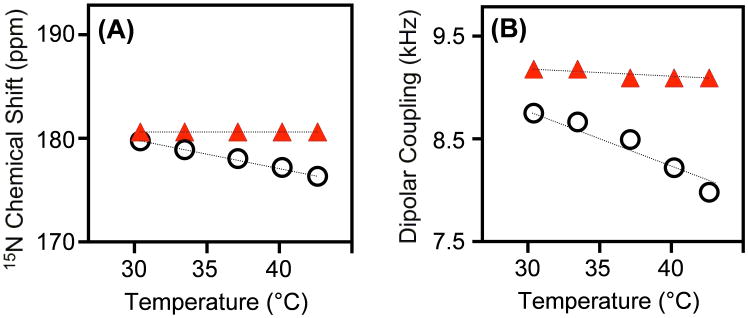
Figure 3. Cholesterol reduces pardaxin’s dynamics in lipid bilayers.
Changes in 15N chemical shift (A) and 1H-15N dipolar coupling (B) frequencies measured from 2D BB-PISEMA spectra of 3:1 DMPC:DMPG bilayers (open circles) and DMPC bilayers with 15 mole % cholesterol (filled triangles).
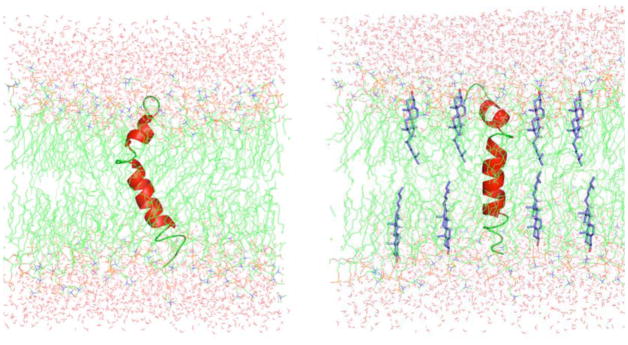
Figure 4. A model depicting the dynamics-driven membrane orientation of pardaxin.
(A) Disordered lipid acyl chains thin the bilayer and therefore disordered tilted orientations are expected for the C-terminal helix of pardaxin. (B) The presence of cholesterol thickens the lipid bilayer, reduces the dynamical disorder in the transmembrane region, and could reduce the channel activity and hemolytic activity of pardaxin.
Discussion
Previous solid-state NMR studies have shown that pardaxin has a transmembrane orientation in DMPC bilayers while it is oriented close to the bilayer surface of POPC bilayers due to the hydrophobic mismatch between its hydrophobic length and the hydrophobic thickness of the lipid bilayer. [23, 24] In this study we measured the backbone dynamics of the C-terminal amphipathic helix of pardaxin using solid-state NMR experiments on mechanically aligned lipid bilayers at various temperatures. Dipolar couplings between 15N and 1H nuclei at the amide site of pardaxin selectively labeled with 15N isotope were accurately measured using 2D broadband-PISEMA experiments. The influence of cholesterol embedded in lipid bilayers on the dynamics of pardaxin was also investigated.
PISEMA experiments on mechanically aligned lipid bilayers provide piercing insights into pardaxin’s function
A number of studies have well-utilized a combination of PISEMA and mechanically aligned lipid bilayers to investigate folding and topology of membrane-associated peptides and proteins. [30–36] This approach has also revealed mechanisms of membrane disruption by antimicrobial peptides [37–40] and amyloid peptides [41]; and in fact the geometry of toroidal pores induced by antimicrobial peptides was also determined at high-resolution. [42, 43] Previous studies have shown that the absence of the overall motion is an advantage in the structural studies of transmembrane proteins. [30] Lack of bulk water in mechanically aligned bilayers enabled the measurement of backbone dynamics only from membrane-bound peptide populations. Therefore, in this study, we used this approach to determine the influence of the backbone dynamics on the function of pardaxin. Pardaxin was labeled with 15N at the amide site of Ala21 residue, which is located in the helical region of the peptide (Figure 1B) [23]. A single peak observed near the parallel edge of 15N chemical shift anisotropy with an N-H dipolar coupling ~8 kHz in 2D PISEMA (Figure 2) spectra suggest that the C-terminal helix has a transmembrane orientation confirming previously reported channel-forming mechanism of Pardaxin. [23, 24] The variation of NMR parameters with temperature is given in Figure 3 and discussed below.
Cholesterol reduces pardaxin’s dynamics and disorder in its transmembrane orientation
Our previous solid-state NMR study could not differentiate the effects of the tilt of the C-terminal amphipathic helix from the bilayer normal and the peptide’s dynamics on the observed 15N chemical shift frequency from mechanically aligned bilayers. [24] In this study, we measured 15N chemical shift and 1H-15N dipolar coupling values by varying the temperature. As seen from 2D PISEMA data in Figure 2, both chemical shift and dipolar coupling values decrease as the temperature of the sample is increased. These results suggest that the transmembrane orientation of the peptide is disordered due to dynamics. Increasing temperature increases the mobility of the transmembrane C-terminal helix rendering the peptide to orient heterogeneously relative to the bilayer normal. On the other hand, a decreasing temperature reduces the mobility of the helix and also the disorder in the tilt of the helix. Our results in Figures 2 and 3 indicate that the presence of cholesterol decreases the disorder in the tilt of the C-terminal helix and also its mobility even at 43°C. This is mainly because cholesterol increases the order of acyl chains of the lipid bilayer, which restricts the mobility of the C-terminal helix of the peptide (Figure 4). These observed changes in the mobility of the channel-forming helix and also in its transmembrane orientation due to the presence of cholesterol or because of a change in the temperature suggest that peptide-peptide interactions are not sufficiently strong to retain the same tilt angle of the helix. This is in excellent agreement with the previously reported difference in the membrane orientation of the C-terminal helix in DMPC vs. POPC bilayers due to hydrophobic mismatch [23, 24]. These results therefore imply that cholesterol significantly influences the barrel-stave mechanism of pardaxin; similar effects have been reported for other channel-forming peptides like melittin. [24, 44–50] Since cholesterol is abundantly present in mammalian cell membranes, it would be interesting to investigate the effect of higher concentration of cholesterol on the barrel-stave activities of pardaxin. A higher concentration of cholesterol could prevent the membrane insertion of pardaxin. [16] It is also be possible that the direct interaction between cholesterol and pardaxin could play some role in the observed dynamical changes or tilt of the helix, particularly at a concentration of cholesterol higher than used in this study. More solid-state NMR experimental measurements are needed to confirm this effect. Such studies will be useful in understanding the role of cholesterol on channel-forming peptides and proteins in general, and possibly on the fusiogenic activities of viral proteins. [51, 52] Cholesterol is also known to play important roles in the membrane toxicity of channel-forming amyloid peptides. [53–57]
Cholesterol effectively suppresses carpet-type and barrel-stave mechanisms of pardaxin
Solid-state NMR experiments on mechanically aligned POPC and POPC:POPG bilayers revealed the membrane surface orientation of pardaxin and peptide-induced disorder near the lipid head group region and in the hydrophobic core of the bilayer. [23, 24] These studies also reported that pardaxin disrupts membrane via a carpet-type mechanism which was found to be inhibited by the presence of cholesterol. Since cholesterol decreases lipid acyl chain disorder in the hydrophobic region of bilayers, it readily suppresses membrane disruption by those AMPs that functioning via a carpet-type mechanism. [37, 58, 59]. As a consequence the concentration of an AMP required to disrupt a mammalian cell membrane is higher than MIC. This also explains the presence of unperturbed cholesterol-rich domains in lipid bilayers even at a concentration as high as 5 mole % pardaxin as reported from our previous solid-state NMR and DSC studies. [24, 60] The decrease in the backbone dynamics is expected to suppress the channel-forming activity of pardaxin and therefore would play a role in the selectivity of this antimicrobial peptide. This prediction from our studies enables us to hypothesize that any mutation of pardaxin that is accompanied by a reduction in the dynamical disorder of the C-terminal helix would reduce the hemolytic activity of the mutant peptide. For example, a substitution of Ala for Pro-13 should extend the helicity towards the N-terminal domain by removing the hinge and therefore should significantly suppress the dynamical disorder in the C-terminal transmembrane helical region of the mutant. Such Pro to Ala mutation has indeed been reported to abolish the toxicity of the peptide. [7] It is also interesting to note that similar behavior has been reported for a derivative of pardaxin that lacks the first 9 N-terminal residues [7]. Since this derivative lacks the N-terminal domain and the disorder in its membrane orientation could be smaller, its hemolytic as well as antimicrobial activities are considerably reduced. Therefore, there is a clear correlation between the dynamics-induced heterogeneity in the tilt of the C-terminal helix and the barrel-stave mechanism of pardaxin. Therefore, we believe these finding will be useful in the design of potent and highly selective peptides.
Acknowledgments
We thank Dr. Jeffrey Brender for critical reading and help with the preparation of this article. This study was supported by research funds from NIH (AI054515 to A.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Hancock RE, Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends in microbiology. 2000;8:402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 3.Dhople V, Krukemeyer A, Ramamoorthy A. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochimica et biophysica acta. 2006;1758:1499–1512. doi: 10.1016/j.bbamem.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 5.Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochim Biophys Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 6.Matsuzaki K, Sugishita K, Ishibe N, Ueha M, Nakata S, Miyajima K, Epand RM. Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry. 1998;37:11856–11863. doi: 10.1021/bi980539y. [DOI] [PubMed] [Google Scholar]
- 7.Huang HW, Chen FY, Lee MT. Molecular mechanism of Peptide-induced pores in membranes. Physical review letters. 2004;92:198304. doi: 10.1103/PhysRevLett.92.198304. [DOI] [PubMed] [Google Scholar]
- 8.Kandasamy SK, Lee DK, Nanga RP, Xu J, Santos JS, Larson RG, Ramamoorthy A. Solid-state NMR and molecular dynamics simulations reveal the oligomeric ion-channels of TM2-GABA(A) stabilized by intermolecular hydrogen bonding. Biochimica et biophysica acta. 2009;1788:686–695. doi: 10.1016/j.bbamem.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Maloy WL, Kari UP. Structure-activity studies on magainins and other host defense peptides. Biopolymers. 1995;37:105–122. doi: 10.1002/bip.360370206. [DOI] [PubMed] [Google Scholar]
- 10.Gottler LM, de la Salud Bea R, Shelburne CE, Ramamoorthy A, Marsh EN. Using fluorous amino acids to probe the effects of changing hydrophobicity on the physical and biological properties of the beta-hairpin antimicrobial peptide protegrin-1. Biochemistry. 2008;47:9243–9250. doi: 10.1021/bi801045n. [DOI] [PubMed] [Google Scholar]
- 11.Gottler LM, Lee HY, Shelburne CE, Ramamoorthy A, Marsh EN. Using fluorous amino acids to modulate the biological activity of an antimicrobial peptide. Chembiochem. 2008;9:370–373. doi: 10.1002/cbic.200700643. [DOI] [PubMed] [Google Scholar]
- 12.Lipsky BA, Holroyd KJ, Zasloff M. Topical versus systemic antimicrobial therapy for treating mildly infected diabetic foot ulcers: a randomized, controlled, double-blinded, multicenter trial of pexiganan cream. Clin Infect Dis. 2008;47:1537–1545. doi: 10.1086/593185. [DOI] [PubMed] [Google Scholar]
- 13.Giuliani A, Pirri G, Nicoletto SF. Antimicrobial peptides: an overview of a promising class of therapeutics. Central European Journal of Biology. 2007;2:1–33. [Google Scholar]
- 14.Hoskin DW, Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim Biophys Acta. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottler LM, Ramamoorthy A. Structure, membrane orientation, mechanism, and function of pexiganan -- A highly potent antimicrobial peptide designed from magainin. BBA Biomembranes. 2009;1788:1680–1686. doi: 10.1016/j.bbamem.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rapaport D, Peled R, Nir S, Shai Y. Reversible surface aggregation in pore formation by pardaxin. Biophys J. 1996;70:2502–2512. doi: 10.1016/S0006-3495(96)79822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oren Z, Shai Y. A class of highly potent antibacterial peptides derived from pardaxin, a pore-forming peptide isolated from Moses sole fish Pardachirus marmoratus. Eur J Biochem. 1996;237:303–310. doi: 10.1111/j.1432-1033.1996.0303n.x. [DOI] [PubMed] [Google Scholar]
- 18.Shai Y. Pardaxin - Channel Formation by a Shark Repellant Peptide from Fish. Toxicology. 1994;87:109–129. doi: 10.1016/0300-483x(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 19.Rapaport D, Shai Y. Aggregation and Organization of Pardaxin in Phospholipid-Membranes - a Fluorescence Energy-Transfer Study. J Biol Chem. 1992;267:6502–6509. [PubMed] [Google Scholar]
- 20.Shai Y, Hadari YR, Finkels A. pH-dependent pore formation properties of pardaxin analogues. The Journal of biological chemistry. 1991;266:22346–22354. [PubMed] [Google Scholar]
- 21.Shai Y, Bach D, Yanovsky A. Channel formation properties of synthetic pardaxin and analogues. The Journal of biological chemistry. 1990;265:20202–20209. [PubMed] [Google Scholar]
- 22.Zagorski MG, Norman DG, Barrow CJ, Iwashita T, Tachibana K, Patel DJ. Solution structure of pardaxin P-2. Biochemistry. 1991;30:8009–8017. doi: 10.1021/bi00246a019. [DOI] [PubMed] [Google Scholar]
- 23.Porcelli F, Buck B, Lee DK, Hallock KJ, Ramamoorthy A, Veglia G. Structure and orientation of pardaxin determined by NMR experiments in model membranes. The Journal of biological chemistry. 2004;279:45815–45823. doi: 10.1074/jbc.M405454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallock KJ, Lee DK, Omnaas J, Mosberg HI, Ramamoorthy A. Membrane composition determines pardaxin’s mechanism of lipid bilayer disruption. Biophys J. 2002;83:1004–1013. doi: 10.1016/S0006-3495(02)75226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thennarasu S, Nagaraj R. Specific antimicrobial and hemolytic activities of 18-residue peptides derived from the amino terminal region of the toxin pardaxin. Protein engineering. 1996;9:1219–1224. doi: 10.1093/protein/9.12.1219. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto K, Lee DK, Ramamoorthy A. Broadband-PISEMA solid-state NMR spectroscopy. Chemical Physics Letters. 2005;407:289–293. doi: 10.1016/j.cplett.2005.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu CH, Ramamoorthy A, Opella SJ. High-Resolution Heteronuclear Dipolar Solid-State Nmr-Spectroscopy. Journal of Magnetic Resonance Series A. 1994;109:270–272. [Google Scholar]
- 28.Ramamoorthy A, Wei YF, Lee DK. PISEMA solid-state NMR spectroscopy. Annual Reports on NMR Spectroscopy. 2004;52:1–52. [Google Scholar]
- 29.Hallock KJ, Henzler Wildman K, Lee DK, Ramamoorthy A. An innovative procedure using a sublimable solid to align lipid bilayers for solid-state NMR studies. Biophys J. 2002;82:2499–2503. doi: 10.1016/S0006-3495(02)75592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page RC, Kim S, Cross TA. Transmembrane helix uniformity examined by spectral mapping of torsion angles. Structure. 2008;16:787–797. doi: 10.1016/j.str.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page RC, Li C, Hu J, Gao FP, Cross TA. Lipid bilayers: an essential environment for the understanding of membrane proteins. Magnetic Resonance in Chemistry. 2007;45:S2–S11. doi: 10.1002/mrc.2077. [DOI] [PubMed] [Google Scholar]
- 32.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chemical reviews. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. J Magn Reson. 2000;144:150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, Quine JR, Cross TA. Imaging membrane protein helical wheels. J Magn Reson. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 35.Dürr UH, Waskell L, Ramamoorthy A. The cytochromes P450 and b5 and their reductases--promising targets for structural studies by advanced solid-state NMR spectroscopy. Biochim Biophys Acta. 2007;1768:3235–3259. doi: 10.1016/j.bbamem.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Buffy JJ, Traaseth NJ, Mascioni A, Gor’kov PL, Chekmenev EY, Brey WW, Veglia G. Two-dimensional solid-state NMR reveals two topologies of sarcolipin in oriented lipid bilayers. Biochemistry. 2006;45:10939–10946. doi: 10.1021/bi060728d. [DOI] [PubMed] [Google Scholar]
- 37.Dürr UH, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochimica et biophysica acta. 2006;1758:1408–1425. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 38.Afonin S, Grage SL, Ieronimo M, Wadhwani P, Ulrich AS. Temperature-Dependent Transmembrane Insertion of the Amphiphilic Peptide PGLa in Lipid Bilayers Observed by Solid State (19)F NMR Spectroscopy. Journal of the American Chemical Society. 2008;130:16512–16514. doi: 10.1021/ja803156d. [DOI] [PubMed] [Google Scholar]
- 39.Bak M, Bywater RP, Hohwy M, Thomsen JK, Adelhorst K, Jakobsen HJ, Sorensen OW, Nielsen NC. Conformation of alamethicin in oriented phospholipid bilayers determined by (15)N solid-state nuclear magnetic resonance. Biophys J. 2001;81:1684–1698. doi: 10.1016/S0006-3495(01)75822-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aisenbrey C, Bertani P, Henklein P, Bechinger B. Structure, dynamics and topology of membrane polypeptides by oriented 2H solid-state NMR spectroscopy. Eur Biophys J. 2007;36:451–460. doi: 10.1007/s00249-006-0122-2. [DOI] [PubMed] [Google Scholar]
- 41.Brender JR, Durr UH, Heyl D, Budarapu MB, Ramamoorthy A. Membrane fragmentation by an amyloidogenic fragment of human Islet Amyloid Polypeptide detected by solid-state NMR spectroscopy of membrane nanotubes. Biochim Biophys Acta. 2007;1768:2026–2029. doi: 10.1016/j.bbamem.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hallock KJ, Lee DK, Ramamoorthy A. MSI-78, an analogue of the magainin antimicrobial peptides, disrupts lipid bilayer structure via positive curvature strain. Biophys J. 2003;84:3052–3060. doi: 10.1016/S0006-3495(03)70031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wi S, Kim C. Pore structure, thinning effect, and lateral diffusive dynamics of oriented lipid membranes interacting with antimicrobial peptide protegrin-1: 31P and 2H solid-state NMR study. The journal of physical chemistry. 2008;112:11402–11414. doi: 10.1021/jp801825k. [DOI] [PubMed] [Google Scholar]
- 44.Andersson A, Biverstahl H, Nordin J, Danielsson J, Lindahl E, Maler L. The membrane-induced structure of melittin is correlated with the fluidity of the lipids. Biochim Biophys Acta-Biomembr. 2007;1768:115–121. doi: 10.1016/j.bbamem.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 45.Wessman P, Stromstedt AA, Malmsten M, Edwards K. Melittin-Lipid Bilayer Interactions and the Role of Cholesterol. Biophys J. 2008;95:4324–4336. doi: 10.1529/biophysj.108.130559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raghuraman H, Chattopadhyay A. Cholesterol inhibits the lytic activity of melittin in erythrocytes. Chem Phys Lipids. 2005;134:183–189. doi: 10.1016/j.chemphyslip.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 47.Allende D, Simon SA, McIntosh TJ. Melittin-induced bilayer leakage depends on lipid material properties: Evidence for toroidal pores. Biophys J. 2005;88:1828–1837. doi: 10.1529/biophysj.104.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Constantinescu I, Lafleur M. Influence of the lipid composition on the kinetics of concerted insertion and folding of melittin in bilayers. Biochim Biophys Acta-Biomembr. 2004;1667:26–37. doi: 10.1016/j.bbamem.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 49.Raghuraman H, Chattopadhyay A. Interaction of melittin with membrane cholesterol: A fluorescence approach. Biophys J. 2004;87:2419–2432. doi: 10.1529/biophysj.104.043596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benachir T, Monette M, Grenier J, Lafleur M. Melittin-induced leakage from phosphatidylcholine vesicles is modulated by cholesterol: A property used for membrane targeting. Eur Biophys J Biophys Lett. 1997;25:201–210. [Google Scholar]
- 51.Epand RM. Proteins and cholesterol-rich domains. Biochim Biophys Acta-Biomembr. 2008;1778:1576–1582. doi: 10.1016/j.bbamem.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 52.Epand RM. Fusion peptides and the mechanism of viral fusion. Biochim Biophys Acta-Biomembr. 2003;1614:116–121. doi: 10.1016/s0005-2736(03)00169-x. [DOI] [PubMed] [Google Scholar]
- 53.Cecchi C, Baglioni S, Fiorillo C, Pensalfini A, Liguri G, Nosi D, Rigacci S, Bucciantini M, Stefani M. Insights into the molecular basis of the differing susceptibility of varying cell types to the toxicity of amyloid aggregates. Journal of Cell Science. 2005;118:3459–3470. doi: 10.1242/jcs.02473. [DOI] [PubMed] [Google Scholar]
- 54.Kakio A, Nishimoto S, Yanagisawa K, Kozutsumi Y, Matsuzaki K. Cholesterol-dependent formation of GM1 ganglioside-bound amyloid beta-protein, an endogenous seed for Alzheimer amyloid. J Biol Chem. 2001;276:24985–24990. doi: 10.1074/jbc.M100252200. [DOI] [PubMed] [Google Scholar]
- 55.Lau TL, Gehman JD, Wade JD, Masters CL, Barnham KJ, Separovic F. Cholesterol and Clioquinol modulation of A beta(1–42) interaction with phospholipid bilayers and metals. Biochim Biophys Acta-Biomembr. 2007;1768:3135–3144. doi: 10.1016/j.bbamem.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 56.Reid PC, Urano Y, Kodama T, Hamakubo T. Alzheimer’s Disease: cholesterol, membrane rafts, isoprenoids and statins. J Cell Mol Med. 2007;11:383–392. doi: 10.1111/j.1582-4934.2007.00054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sponne I, Fifre A, Kriem B, Koziel V, Bihain B, Oster T, Olivier JL, Pillot T. Membrane cholesterol interferes with neuronal apoptosis induced by soluble oligomers but not fibrils of the amyloid-beta peptide. Faseb J. 2004;18:836-+. doi: 10.1096/fj.03-0372fje. [DOI] [PubMed] [Google Scholar]
- 58.Ramamoorthy A, Thennarasu S, Tan A, Lee DK, Clayberger C, Krensky AM. Cell selectivity correlates with membrane-specific interactions: a case study on the antimicrobial peptide G15 derived from granulysin. Biochim Biophys Acta. 2006;1758:154–163. doi: 10.1016/j.bbamem.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 59.Ramamoorthy A. Beyond NMR spectra of antimicrobial peptides: dynamical images at atomic resolution and functional insights. Solid State NMR Spectrosc. 2009;35:201–207. doi: 10.1016/j.ssnmr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Epand RF, Ramamoorthy A, Epand RM. Membrane lipid composition and the interaction of pardaxin: the role of cholesterol. Protein and peptide letters. 2006;13:1–5. [PubMed] [Google Scholar]