Abstract
Patients with alveolar rhabdomyosarcoma (ARMS) have poorer response to conventional chemotherapy and lower survival rates than those with embryonal RMS (ERMS). To identify compounds that preferentially block the growth of ARMS, we conducted a small-scale screen of 160 kinase inhibitors against the ARMS cell line Rh30 and ERMS cell line RD and identified inhibitors of glycogen synthase kinase 3 (GSK3), including TWS119 as ARMS-selective inhibitors. GSK3 inhibitors inhibited cell proliferation and induced apoptosis more effectively in Rh30 than RD cells. Ectopic expression of fusion protein PAX3-FKHR in RD cells significantly increased their sensitivity to TWS119. Down-regulation of GSK3 by GSK3 inhibitors or siRNA significantly reduced the transcriptional activity of PAX3-FKHR. These results suggest that GSK3 is directly involved in regulating the transcriptional activity of PAX3-FKHR. Also, GSK3 phosphorylated PAX3-FKHR in vitro, suggesting that GSK3 might regulate PAX3-FKHR activity via phosphorylation. These findings support a novel mechanism of PAX3-FKHR regulation by GSK3 and provide a novel strategy to develop GSK inhibitors as anti-ARMS therapies.
Keywords: ARMS, GSK3, Differential cytotoxicity, PAX3-FKHR
Introduction
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma in children. Two subtypes of RMS have been identified on the basis of histopathologic features – embryonal (ERMS) and alveolar (ARMS) – each with distinct clinical and genetic characteristics. Most of the more aggressive ARMSs are associated with either 2;13 or 1;13 chromosomal translocations, which generate PAX3-FKHR and PAX7-FKHR fusion products, respectively. The unique expression, function, and subcellular localization of fusion proteins contribute to their oncogenic behavior by modifying cell growth, differentiation, and apoptosis [1]. Expression of fusion genes is associated with poor prognosis, and patients with ARMS respond more poorly to conventional chemotherapy and radiotherapy and have much lower survival rates than those with ERMS [2, 3]. Target genes of PAX3-FKHR play important roles in ARMS tumorigenesis and are potential therapeutic targets for treating ARMS [4-6]. Directly regulating the transcriptional activity of PAX3-FKHR is also a proposed strategy to treat ARMS [7, 8].
Glycogen synthase kinase 3 (GSK3), having 2 isoforms GSK3α and GSK3β, is a serine/threonine protein kinase known to affect various biological processes such as cell proliferation and apoptosis [9]. Aberrant regulation of GSK3β is implicated in several human diseases, such as diabetes, bipolar disorder, cardiovascular disease, and neurodegenerative diseases [10]. More recently, GSK3β was shown to be up-regulated in gastrointestinal, colon and pancreatic cancers [11, 12]. Inhibition of GSK3β in several cancer cell types reduces expression of anti-apoptotic genes, increases activation of pro-apoptotic genes, and stabilizes the cyclin-dependent kinase inhibitor p27Kip1 to inhibit cell growth and induce apoptosis [13-15]. Hence, GSK3 plays a complex role in cell growth and apoptosis and the biological outcome of GSK3 signaling depends on cell type and tissue [9-15].
Here, we identified GSK3 inhibitors, including TWS119 [16] to be significantly more effective at inhibiting cell growth and inducing apoptosis in ARMS cell line Rh30 than ERMS cell line RD cells. We demonstrate that PAX3-FKHR is responsible for the enhanced cytotoxicity of GSK3 inhibitors, and that GSK3 regulates the activity of PAX3-FKHR. Our findings support a novel mechanism of GSK3-mediated regulation of PAX3-FKHR and provide evidence that GSK3 inhibitors can be developed as anti-ARMS therapeutics.
Materials and methods
Cell culture
Human ERMS cell lines RD and JR1, ARMS cell lines Rh30 and Rh41, RD/PF (RD cells stably transfected with pcDNA3-PAX3-FKHR) and RD/Vector (RD cells stably transfected with pcDNA3 vector) have been described previously [7]. The immortalized human myoblast cell line LHCN-M2 (provided by Dr. Woodring Wright, University of Texas Southwestern Medical Center), was cultured in the growth medium as previously described [17]. The HEK293T cell line [American Type Culture Collection (ATCC), Manassas, VA] was grown in DMEM supplemented with 10% FBS and 2 mM L-glutamine. All cells were cultured in an incubator with a humidified atmosphere maintained at 5% CO2 and 95% air at 37°C. For all luminescence assays, phenol red–free DMEM was used.
Cell proliferation assay and compound screening
The cell proliferation assay was performed by using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI), as previously described [7]. Briefly, cells plated into 384-well plates were treated with compounds for 72 h before the luciferase assay. Data are expressed as percentage of viable cells (%) calculated by the following formula: viable cells (%) = 100%×(compound signal – medium alone signal)/(DMSO control signal – medium alone signal), wherein the DMSO control represents 100% viable cells and the medium alone (no cells) represents 0% viable cells (background). The compounds used for the screen contained 160 kinase inhibitors from EMD Chemicals (San Diego, CA). In all figures, results are expressed as the mean +/− SEM (standard error of mean). Error bars indicate SEM.
Apoptosis assay
Cells were treated with either 0.1% DMSO or 10 μM TWS119 for 48 h. Apoptosis was determined by using the Caspase-Glo® 3/7 Luminescent Assay (Promega), following manufacturer's instructions. Data are expressed as fold-increase in caspases 3 and 7 activity, where DMSO control is set as 1.
Luciferase reporter assay
Rh30 cells were stably transfected with a PAX3-FKHR-responsive firefly luciferase reporter (pGL4.20-6 × PRS9-tk) and maintained in medium containing puromycin (1 μg/ml). 6 × PRS9 contains 6 tandem copies of PAX3-FKHR binding site and has been routinely used to detect the activity of PAX3-FKHR [5, 7]. pGL4.20-6 × PRS9-tk was constructed by subcloning 6 × PRS9-tk from pGL4.14-6 × PRS9-tk [7] into pGL4.20 (Promega). Cells were plated and treated as previously described [7]. Luciferase activity was measured using the SteadyLiteHTS Luciferase Assay System (PerkinElmer), according to manufacturer's instructions. Data are presented as percentage reporter activity (%) of firefly luciferase in the presence of drug compared with a DMSO negative control (set as 100%) and a positive control (10 μM camptothecin as 0%) [7]. In siRNA knockdown experiments, PAX3-FKHR knockdown was used as positive control (0% reporter activity).
Soft agar colony formation assay
The anchorage-independent growth of cells was monitored using CytoSelect™ 96-well Cell Transformation Assay (Cell Biolabs, San Diego, CA). Briefly, the base agar layer was prepared by transferring 50 μL of the mixture of 1.2% agar solution and 2×DMEM/20% FBS medium at equal volumes to each well of a 96-well microplate. To each well, 4 × 103 cells were seeded by transferring 75 μl of the mixture of 1.2% agar solution, 2×RPMI-1640/20% FBS medium, and cell suspension (1.6 × 105 cells/ml medium) at the ratio of 1:1:1. Cells were incubated for 8 days after covering the solidified cell agar layer with 100 μl fresh DMEM containing either vehicle control (0.1% DMSO) or compound. To solubilize the agar, the cultured medium was removed, 50 μL of agar solubilization buffer was added to each well and incubated for 1 h at 37°C. To quantify anchorage-independent growth, 25 μl of 8× lysis buffer was added to each well. Then, 10 μl of cell lysis was mixed with 90 μl of CyQuant GR Dye, and the fluorescence signal (as a readout for anchorage-independent cell growth) was measured using an EnVision plate reader (PerkinElmer) with a 485/535 nm filter set and a FITC optical mirror module.
Western blot analysis
Western blots were performed as previously described [7]. Sources of antibodies were indicated here. Anti-GSK3 (05-412) (Millipore, Chicago, IL); anti-glycogen synthase 3 (3893), anti-phospho-glycogen synthase (S641) (3891), anti-phospho-GSK3(S21/9) (9331), and anti-PARP (9542)(Cell Signaling Technology, Boston, MA); anti-GFP (Santa Cruz Biotechnology); and anti-β-actin (Sigma, St. Louis, MO).
Silencing of PAX3-FKHR with siRNA
A PAX3-FKHR-specific siRNA and a control non-targeting siRNA (NT siRNA) were synthesized as described previously [3, 7]. GSK3α- and GSK3β-specific siRNAs (ON-TARGET plus siRNA) were obtained from Thermo Scientific Genomics (Lafayette, CO). Cells were transfected with 10 nM of siRNAs in a 6-well dish with Lipofectamine™ 2000 reagent, according to manufacturer's instructions. After 6 h of transfection, cells were plated and treated with compounds for cell proliferation and reporter gene assays, as described above. To verify knockdown efficiency, cells treated with siRNAs under the same conditions were analyzed for protein levels by Western blotting.
In vitro kinase assays
HEK293T cells were transfected with either pEGFP-C3 vector (Clontech, Mountain View, CA) or pEGFP-PAX3-FKHR (provided by Dr. David Bouck), in which the coding sequence of PAX3-FKHR from pCD3M8 [5] (provided by Dr. Frederic Barr, University of Pennsylvania) was subcloned into pEGFP-C3 and lysed in RIPA buffer supplemented with protease and phosphatase inhibitors. Anti-GFP agarose beads (Medical and Biological Laboratories, Woburn, MA) were added to the lysates and incubated at 4°C for 3 h. Beads were then washed thrice with prechilled PBS-T (0.1% of Tween). For in vitro phosphorylation of PAX3-FKHR by GSK3, half the beads were incubated with 25μl of the assay buffer containing 15 units of GSK3 (New England Biolabs), 0.5μM cold ATP, and 5μCi [γ-32P] ATP (6000Ci/mmol) at 30°C for 30 min, and the reaction was terminated by adding SDS-PAGE loading buffer prior to electrophoresis. The amount of protein was visualized by SimplyBlue (Invitrogen) stain. The stained gel was desiccated and exposed to the Storage Phosphor Screen (GE Healthcare). Phosphoimages were obtained by using the Storm Scanner (GE Healthcare). To verify the identity of EGFP-PAX3-FKHR, the remaining beads were analyzed by Western blot, using anti-GFP antibody. In vitro GSK3α and GSK3β kinase profiling assays were performed by Ambit Biosciences (San Diego, CA) as previously described [18, 19].
Results
GSK3 inhibitors block cell proliferation more effectively in Rh30 than RD cells
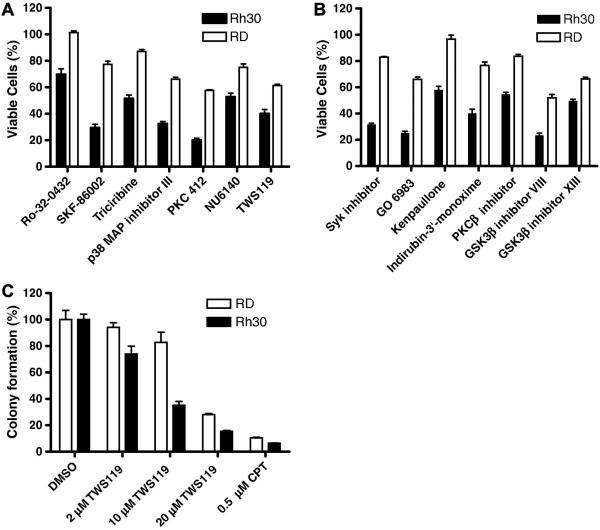
To identify compounds that preferentially block the growth of ARMS, we screened the ERMS cell line RD and the ARMS cell line Rh30 against a collection of 160 kinase inhibitors. In the cell proliferation assay using CellTiter- Glo®, at 10 μM, 7 compounds were significantly more effective (by at least 15%) at inhibiting cell growth of Rh30 than of RD cells (Fig. 1A). At 2 μM, 7 additional compounds were identified as Rh30-selective (Fig. 1B). Consistent with previously published data [8], our screen identified 4 Rh30-selective PKC inhibitors (PKC412, Ro-32 0432, GO 6983, and PKCβ inhibitor). Interestingly, 5 GSK3 inhibitors (TWS119, GSK3 inhibitor XIII, GSK3 inhibitor VIII, indirubin-3′-monoxime, and kenpaullone), two p38 inhibitors (SKF 86002 and p38 MAP kinase inhibitor III), 1 AKT inhibitor (triciribine), 1 CDK inhibitor (NU6140), and 1 Syk inhibitor were also identified as Rh30-selective (Fig. 1A and 1B).
Fig 1.
Identification of compounds that preferentially block the growth of Rh30. Cells were exposed to (A), 10 μM or (B), 2 μM of drugs as indicated for 72 h before determining viable cells (%). (C) Soft agar colony formation assay. Colony formed in vehicle-treated control was set as 100%. Data represent 2 independent experiments, each performed in triplicate.
Our study focused on GSK3 inhibitors, as GSK3 is a promising cancer drug target but its role in RMS remains unknown. To further confirm the effect of GSK3 inhibitors, we showed that other GSK3β inhibitors (SB216763, TDZD-8, and roscovitine) also more effectively inhibited the cell proliferation of Rh30 cells than of RD cells (Fig. S1A).
Anchorage-independent growth is a hallmark of transformed and cancerous cells. In the soft agar colony formation assay, GSK3 inhibitors were more effective at inhibiting anchorage-independent growth in Rh30 cells than in RD cells (Fig. 1C and S1B), confirming the ARMS-selective antitumor potential of GSK3 inhibitors.
TWS119 induces apoptosis more effectively in Rh30 than RD cells
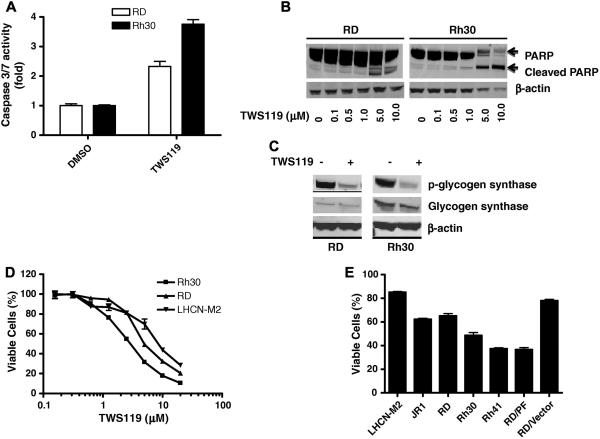
To determine whether GSK3 inhibitor differentially activates caspases 3 and 7 in Rh30 and RD cells, their activities were quantified by the Caspase-Glo® 3/7 Luminescent Assay in cells treated with GSK3 inhibitor. 10 μM of TWS119 [16] was used to inhibit GSK3 in all experiments unless specified otherwise. Basal activities of caspases 3 and 7 were low in both Rh30 and RD cells [7]. Caspase activity increased by approximately 4-fold in Rh30 cells but only by 2.3-fold in RD cells treated with TWS119 (Fig. 2A). Differential induction of apoptosis by TWS119 was confirmed by Poly (ADP ribose) polymerase (PARP) cleavage experiments, in which more than 90% of PARP was cleaved with TWS119 treatment in Rh30 cells but less than 20% was cleaved in RD cells (Fig. 2B). As the basal PARP expression in RD cells was much higher than that in Rh30 cells (Fig. 2B), we are not certain whether differential PARP expression causes the observed difference in TWS119-mediated apoptosis.
Fig. 2.
(A) and (B) TWS119 induces differential activation of caspases 3 and 7 in Rh30 and RD cells. The activity of caspases 3 and 7 (A) was determined as described in Materials and methods. Data represent 3 independent experiments, each performed in triplicate. In the PARP cleavage assay (B), Rh30 and RD cells were exposed to TWS119 for 72 h before Western blot analysis, using anti-PARP antibody and anti β-actin antibody (as loading control). (C) TWS119 inhibits activation of GSK3. Rh30 and RD cells were exposed to DMSO or TWS119 for 24 h and analyzed by Western blot, using indicated specific antibodies. (D) and (E) PAX3-FKHR sensitizes TWS119-mediated inhibition of cell proliferation in RMS cells. Cells were exposed to indicated concentrations (D) or 5 μM (E) of TWS119 for 72 h and viable cells (%) was determined. Data represent 3 independent experiments, each performed in triplicate.
We confirmed the in vivo activity of TWS119 in inhibiting GSK3 by examining the phosphorylation status of glycogen synthase (an endogenous GSK3 substrate) in TWS119-treated RD and Rh30 cells. TWS119 effectively and similarly decreased the phosphorylation of glycogen synthase in both RD and Rh30 cells (Fig. 2C), suggesting that the enhanced sensitivity of Rh30 cells to TWS119 is not due to differential regulation of GSK3 in RD and Rh30 cells.
PAX3-FKHR is at least partially responsible for the enhanced inhibitory effect of TWS119 on growth of ARMS cells
TWS119 differentially inhibited proliferation of RD and Rh30 cells in a dose-dependent manner [IC50 of 2 μM and 6 μM for Rh30 and RD, respectively (Fig. 2D); cells were treated with compounds for 72 h]. Interestingly, TWS119 was significantly less effective at inhibiting cell proliferation of an immortalized human myoblast cell line LHCN-M2 (Fig. 2D). However, the enhanced cytotoxicity of TWS119 was also observed in another ARMS cell line Rh41 but not in the ERMS cell line JR-1 (Fig. 2E). The higher sensitivity of PAX3-FKHR–positive ARMS cell lines Rh30 and Rh41 than of PAX3-FKHR–negative cell lines RD, JR-1, and LHCN-M2 to TWS119 suggests that PAX3-FKHR directly contributes to the enhanced cytotoxicity of TWS119. To test this hypothesis, PAX3-FKHR–negative RD cells stably expressing either ectopic PAX3-FKHR (RD/PF) or vector alone (RD/Vector) were treated with TWS119. Ectopic expression of PAX3-FKHR significantly sensitized RD cells to TWS119-mediated inhibition of cell proliferation (Fig. 2E). Ectopic expression of PAX3-FKHR in the RD/PF stable clone was confirmed as described previously [7]. Taken together, our results suggest that PAX3-FKHR is at least partially responsible for the differential sensitivity of ARMS and ERMS cell lines to inhibition of GSK3.
GSK3 regulates transcriptional activity of PAX3-FKHR
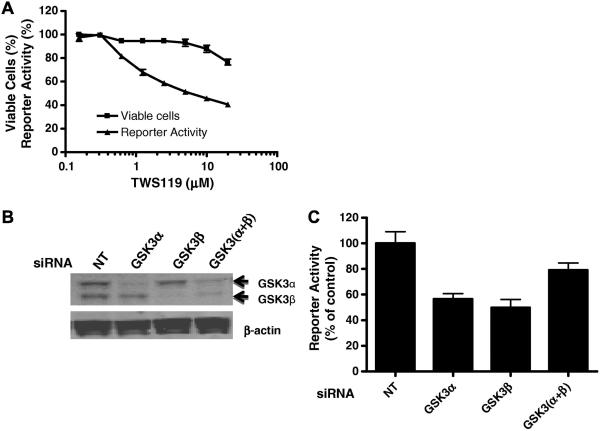
Since PAX3-FKHR is at least partly responsible for enhanced sensitivity of cells to TWS119-mediated growth inhibition, we tested whether TWS119 directly modulates the transcriptional activity of PAX3-FKHR, by measuring the activity of the PAX3-FKHR–regulated luciferase reporter pGL4.20-6 × PRS9-tk stably transfected into Rh30. TWS119 (16 h treatment) inhibited the activity of PAX3-FKHR in a concentration-dependent manner (IC50 ~ 2 μM; Fig. 3A). Treatment of cells with TWS119 for 16 h only slightly decreased the viability of Rh30 cells (Fig. 3A), confirming that the TWS119-mediated decrease in PAX3-FKHR activity was not due to cytotoxicity. Other GSK3 inhibitors also inhibited the transcriptional activity of PAX3-FKHR (Fig. S2). Together, these results (Figs. 3A and S2) suggest that GSK3 plays an important role in regulating the transcriptional activity of PAX3-FKHR.
Fig. 3.
Down-regulation of GSK3 attenuates transcriptional activity of PAX3-FKHR. (A) Inhibition of transcriptional activity of PAX3-FKHR by TWS119. Rh30 cells stably transfected with pGL4.14-6 × PRS9-tk were treated with indicated concentrations of TWS119 for 16 h. Cell viability was determined by the CellTiter-Glo® assay. (B) Verification of GSK3α and GSK3β expression by Western blot analysis with anti-GSK3 antibody. Rh30 cells were treated with NT siRNA (NT) or gene-specific siRNA as indicated. (C) Gene reporter assay. Twenty four hours after transfection with NT siRNA (control) or gene-specific siRNA, luciferase activity was determined. Reporter activity from cells transfected with siRNA against PAX3-FKHR is set as 0% activation and NT siRNA as 100% activation. Data represent 3 independent experiments, each performed in triplicate.
The differential sensitivity of Rh30 and RD cells to TWS119 is not due to differential inhibition of GSK3 in RD and Rh30 (Fig. 2C). GSK3β, but not GSK3α, has been identified as the specific target of TWS119 [16]. To determine whether TWS119 is a GSK3β-selective inhibitor, we performed in vitro kinase profiling assays (see Materials and methods) for GSK3α and GSK3β. TWS119 inhibited the kinase activity of both GSK3α (IC50 = 88 nM) and GSK3β (IC50 = 410 nM), indicating that TWS119 is not isoform selective and is more potent against GSK3α than GSK3β.
Since TWS119 inhibits both GSK3α and GSk3β, we examined the effect of isoform-specific siRNA-mediated GSK3 knockdown on the transcriptional activity of PAX3-FKHR to determine whether GSK3α and GSK3β play different roles in this process. Rh30 cells stably transfected with pGL4.20-6×PRS9-tk were treated with non-targeting (NT), GSK3α-specific, or GSKβ3-specific siRNA before the reporter gene assay. siRNAs decreased levels of GSK3β and GSK3α by more than 50% (Fig. 3B). Reporter gene activity was reduced by approximately 50% by knockdown of either GSK3β or GSK3α, but only by less than 25% in simultaneous knockdown of GSK3β and GSK3α (Fig. 3C), an unexpected observation.
To establish the direct functional relationship among GSK3, PAX3-PKHR, and cell growth, we transiently transfected RD/PF cells, which were more sensitive than RD cells to pharmacological inhibition of GSK3 (Fig. 2E), with specific siRNAs and monitored real-time cell proliferation by the IncuCyte live-cell imaging system. PAX3-FKHR knockdown or GSK3β knockdown significantly decreased proliferation of RD/PF cells (Fig. S3A). However, there was no significant change in cell proliferation with GSK3α-specific siRNA. Interestingly, double knockdown of GSK3α and GSK3β caused only moderate decrease in cell proliferation of RD/PF cells (Fig. S3A), which agrees with our observation of lack of additive effect of GSK3α and GSK3β knockdown on inhibiting transcriptional activity of PAX3-FKHR (Fig. 3C). Western blot analysis showed approximately a 40% reduction in RD/PF cells for knockdown of GSK3α and GSK3β (Fig. S3B). Note that as PAX3-FKHR or GSK3 was down-regulated in only some cells transiently transfected with siRNAs, the growth rate of cells was reduced but cells continued to grow as a mixed population.
Together, our findings support the notion that both GSK3β and GSK3α contribute to the inhibitory effect of TWS119 on the transcriptional activity of PAX3-FKHR, and GSK3β but less likely GSK3α, played an important role in regulating PAX3-FKHR-mediated cell proliferation of PAX3-FKHR-positive ARMS cells.
GSK3 phosphorylates PAX3-FKHR in vitro
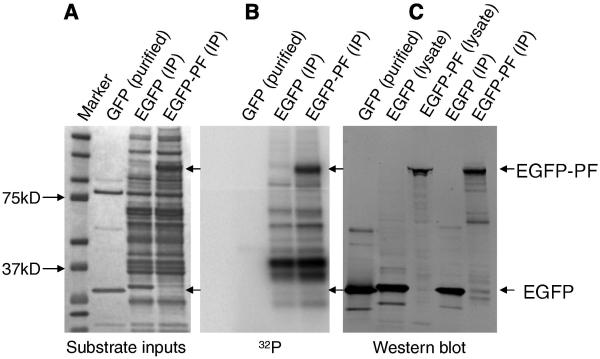
We determined whether GSK3 can directly phosphorylate PAX3-FKHR, as PAX3-FKHR contains multiple putative phosphorylation sites that match the predicted consensus phosphorylation motif for GSK3, including one created by the fusion of PAX3 to FKHR (389SPQNS) (Fig. S4). The consensus motif consists of (S/T)XXX(pS/pT), in which a proline is a preferred residue adjacent to the phosphorylation site. Also, in most cases, a “primed” phosphorylation is required at the +4 position before GSK3-mediated phosphorylation [20]. GSK3 phosphorylated PAX3-FKHR in an in vitro kinase assay (Fig. 4), suggesting that it regulates PAX3-FKHR via phosphorylation.
Fig. 4.
GSK3 phosphorylates PAX3-FKHR in vitro. HEK293T cells were transiently transfected with EGFP-vector or EGFP-PAX3-FKHR plasmids. (A) The amount of EGFP and EGFP-PAX3-FKHR (EGFP-PF) proteins immunoprecipitated by anti-GFP agarose beads were revealed by staining with SimplyBlue. Purified GFP (Millipore) was included as a control. (B) In vitro kinase assays. (C) EGFP and EGFP-PF were revealed by Western blot, using anti-GFP antibody.
Discussion
Camptothecin and PKC inhibitor were previously identified as ARMS-selective inhibitors of cell proliferation [7, 8]. In this study we identified GSK3 inhibitors as ARMS-selective growth inhibitors, and focused on investigating the functional relationship between down-regulation of GSK3 and ARMS growth inhibition.
We down-regulated GSK3 by using either GSK inhibitors, or siRNA specific for GSK3α or GSK3β. Rh30 cells were significantly more sensitive than RD cells to GSK3 inhibitors. TWS119, a novel GSK3 inhibitor [16], induced apoptosis more effectively in Rh30 cells than RD cells, suggesting that increased apoptosis may cause the enhanced sensitivity of Rh30 cells to TWS119. Ectopic expression of PAX3-FKHR sensitized RD cells to TWS119, clearly demonstrating the role of PAX3-FKHR in this process.
Since aberrant transcriptional activity of PAX3-FKHR may increase the proliferation rate and invasiveness of ARMS tumors [21], down-regulation of PAX3-FKHR might inhibit PAX3-FKHR–positive ARMS. Camptothecin inhibits PAX3-FKHR activity by down-regulating its protein levels through an undefined mechanism [7], whereas PKC 412 inhibits the activity of PAX3-FKHR by regulating its phosphorylation [8]. As seen for PKC412, in our study GSK3 inhibitors did not affect the nuclear localization of PAX3-FKHR (data not shown), but effectively inhibited the transcriptional activity of PAX3-FKHR. Inhibition of transcriptional activity of PAX3-FKHR and subsequent growth of ARMS cells by GSK3 inhibition was confirmed by siRNA-mediated down-regulation of GSK3. Our findings underline the importance of GSK3β in positively regulating cell proliferation of PAX3-FKHR–positive ARMS cells, and are consistent with recent findings that GSK3β is a key regulator of cell proliferation of MLL fusion–positive leukemia [15].
However, the role of GSK3 in regulating cell proliferation and apoptosis remains controversial. Early reports suggested that GSK3β is pro-apoptotic in Rat-1 and PC12 cells [22]. In contrast, the discovery that GSK3β-knockout mice are embryonic lethal and fibroblasts derived from these embryos are sensitized to apoptosis suggest that GSK3β is required for cell survival [23]. Recent studies showed that inhibition of GSK3β in glioma [24], multiple myeloma [25], pancreatic cancer [26], colorectal cancer [14] and leukemia cells [15] leads to inhibition of cell proliferation and induction of apoptosis, providing promising evidence that specific GSK3β inhibitors can be developed as anticancer therapy for multiple cancer types. Our findings suggest that GSK3β-specific inhibitors may be developed as novel agents to treat PAX3-FKHR–positive ARMS.
In conclusion, we demonstrate that down-regulation of GSK3 leads to inhibition of ARMS cell proliferation. Our findings reveal a novel role of GSK3 in regulating PAX3-FKHR, and suggest that GSK inhibitors can be used to develop therapeutic agents to treat ARMS. ARMS tumors are more aggressive and associated with higher mortality rates than ERMS tumors and often fail to respond to chemotherapy and radiotherapy, warranting the development of novel, effective treatments.
Supplementary Material
Acknowledgements
We thank Drs. Frederic Barr, Peter Houghton, Woodring Wright, and David Bouck for providing cells or plasmids; Dr. Wenwei Lin and Jing Wu for technical assistance; other members of the Chen group for reagents and their valuable discussions; and Dr. Kip Guy for reviewing and Dr. Vani Shanker for editing the manuscript.
Abbreviations
- GSK3
glycogen synthase kinase 3
- RMS
rhabdomyosarcoma
- ARMS
alveolar RMS
- ERMS
embryonal RMS
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was supported by the American Lebanese Syrian Associated Charities, St. Jude Children's Research Hospital, and National Cancer Institute grant P30CA027165.
References
- 1.Linardic CM. PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer Lett. 2008;270:10–18. doi: 10.1016/j.canlet.2008.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorensen PH, Lynch JC, Qualman SJ, et al. PAX3–FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J. Clin. Oncol. 2002;20:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 3.Kikuchi K, Tsuchiya K, Otabe O, et al. Effects of PAX3–FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem. Biophys. Res. Commun. 2008;365:568–574. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 4.Taulli R, Scuoppo C, Bersani F, et al. Validation of Met as a therapeutic target in alveolar and embryonal rhabdomyosarcoma. Cancer Res. 2006;66:4742–4749. doi: 10.1158/0008-5472.CAN-05-4292. [DOI] [PubMed] [Google Scholar]
- 5.Tomescu O, Xia SJ, Strezlecki D, et al. Inducible short-term and stable long-term cell culture systems reveal that the PAX3-FKHR fusion oncoprotein regulates CXCR4, PAX3, and PAX7 expression. Lab. Invest. 2004;84:1060–1070. doi: 10.1038/labinvest.3700125. [DOI] [PubMed] [Google Scholar]
- 6.Taniguchi E, Nishijo K, McCleish AT, et al. PDGFR-A is a therapeutic target in alveolar rhabdomyosarcoma. Oncogene. 2008;27:6550–6560. doi: 10.1038/onc.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng FY, Cui J, Liu L, Chen T. PAX3-FKHR sensitizes human alveolar rhabdomyosarcoma cells to camptothecin-mediated growth inhibition and apoptosis. Cancer Lett. 2009;284:157–164. doi: 10.1016/j.canlet.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amstutz R, Wachtel M, Troxler H, et al. Phosphorylation regulates transcriptional activity of PAX3/FKHR and reveals novel therapeutic possibilities. Cancer Res. 2008;68:3767–3775. doi: 10.1158/0008-5472.CAN-07-2447. [DOI] [PubMed] [Google Scholar]
- 9.Jope RS, Johnson GV. The glamour and gloom of glycogen synthase-3. Trends Biochem Sci. 2004;29:95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Luo J. Glycogen synthase kinase 3β (GSK3β) in tumorigenesis and cancer chemotherapy. Cancer Lett. 2009;273:194–200. doi: 10.1016/j.canlet.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shakoori A, Ougolkov A, Yu ZW, et al. Deregulated GSK3beta activity in colorectal cancer: its association with tumor cell survival and proliferation. Biochem Biophys Res Commun. 2005;334:1365–1373. doi: 10.1016/j.bbrc.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 12.Ougolkov AV, Fernandez-Zapico ME, Savoy DN, et al. Glycogen synthase 3beta participates in nuclear factor kappaB-mediated gene transcription and cell survival in pancreatic cancer cells. Cancer Res. 2005;65:2076–2081. doi: 10.1158/0008-5472.CAN-04-3642. [DOI] [PubMed] [Google Scholar]
- 13.Ougolkov AV, Bone ND, Fernandez-Zapico ME, et al. Inhibition of glycogen synthase kinase 3 activity leads to epigenetic silencing of nuclear factor kappaB target genes and induction of apoptosis in chronic lymphocytic leukemia B cells. Blood. 2007;110:735–742. doi: 10.1182/blood-2006-12-060947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan J, Zhuang L, Leong HS, et al. Pharmacologic modulation of glycogen synthase 3beta promotes p53-dependent apoptosis through a direct Bax-mediated mitochondrial pathway in colorectal cancer cells. Cancer Res. 2005;65:9012–9020. doi: 10.1158/0008-5472.CAN-05-1226. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Smith KS, Murphy M, et al. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding S, Wu TYH, Brinker A, et al. Synthetic small molecules that control stem cell fate. Proc Natl Acad Sci USA. 2003;100:7632–7637. doi: 10.1073/pnas.0732087100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu CH, Mouly V, Cooper RN, et al. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell. 2007;6:515–523. doi: 10.1111/j.1474-9726.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 18.Fabian MA, Biggs WH, Treiber DK, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 19.Karaman MW, Herrgard S, Treiber DK, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 20.Danek EI, Tcherkezian J, Triki I, et al. Glycogen synthase kinase-3 phosphorylates CdGAP at a consensus ERK 1 regulatory site. J Biol Chem. 2007;282:3624–3631. doi: 10.1074/jbc.M610073200. [DOI] [PubMed] [Google Scholar]
- 21.Davicioni E, Finckenstein FG, Shahbazian V, et al. Identification of a PAX–FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–6946. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 22.Pap M, Cooper GM. Role of glycogen synthase-3 in the phosphatidylinositol 3-kinase/Akt cell survival pathway. J. Biol. Chem. 1998;273:19929–19932. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]
- 23.Hoeflich KP, Luo J, Rubie EA, et al. Requirement for glycogen synthase kinase-3β in cell survival and NF-γB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 24.Kotliarova S, Pastorino S, Kovell LC, et al. Glycogen synthase kinase-3 inhibition induces glioma cell death through c-MYC, nuclear factor-κB, and glucose regulation. Cancer Res. 2008;68:6643–6651. doi: 10.1158/0008-5472.CAN-08-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Uddin S, Zimmerman T, et al. Growth control of multiple myeloma cells through inhibition of glycogen synthase kinase-3. Leukemia & Lymphoma. 2008;49:1945–1953. doi: 10.1080/10428190802304966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaisina IN, Gallier F, Ougolkov AV, et al. From a natural product lead to the identification of potent and selective benzofuran-3-yl-(indol-3-yl)maleimides as glycogen synthase kinase 3beta inhibitors that suppress proliferation and survival of pancreatic cancer cells. J Med Chem. 2009;52:1853–1863. doi: 10.1021/jm801317h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.