Abstract
The proteasome is a multi-catalytic protein degradation enzyme that is regulated by ethanol-induced oxidative stress; such suppression is attributed to CYP2E1-generated metabolites. However, under certain conditions, it appears that in addition to oxidative stress, other mechanisms are also involved in proteasome regulation. This study investigated whether impaired protein methylation that occurs during exposure of liver cells to ethanol, may contribute to suppression of proteasome activity. We measured the chymotrypsin-like proteasome activity in Huh7CYP cells, hepatocytes, liver cytosols and nuclear extracts or purified 20S proteasome under conditions that maintain or prevent protein methylation. Reduction of proteasome activity of hepatoma cell and hepatocytes by ethanol or tubercidin was prevented by simultaneous treatment with S-adenosylmethionine (SAM). Moreover, the tubercidin-induced decline in proteasome activity occurred in both nuclear and cytosolic fractions. In vitro exposure of cell cytosolic fractions or highly purified 20S proteasome to low SAM:S-adenosylhomocysteine (SAH) ratios in the buffer also suppressed proteasome function, indicating that one or more methyltransferase(s) may be associated with proteasomal subunits. Immunoblotting a purified 20S rabbit red cell proteasome preparation using methyl lysine-specific antibodies revealed a 25kDa proteasome subunit that showed positive reactivity with anti-methyl lysine. This reactivity was modified when 20S proteasome was exposed to differential SAM:SAH ratios. We conclude that impaired methylation of proteasome subunits suppressed proteasome activity in liver cells indicating an additional, yet novel mechanism of proteasome activity regulation by ethanol.
Keywords: 20S proteasome, S-adenosylmethionine, S-adenosylhomocysteine, hepatoma cells, hepatocytes, methyl lysine
Introduction
The proteasome is a multi-catalytic enzyme that degrades aged and oxidatively modified proteins, signal transduction factors and peptides. Proteasomal proteolysis contributes to recycling amino acids for de novo protein synthesis and to regulating inter- and intracellular signal communications. The proteasome also generates peptides for MHC class I-restricted antigen presentation. Inhibition of proteasome causes the accumulation of altered proteins in cells and aberrant signal transduction, thereby increasing cell toxicity and apoptotic cell death [1–3].
In the liver, ethanol consumption suppresses proteasome function [4, 5]. Both 26S proteasome and the 20S proteasome are suppressed by ethanol metabolism. These in vivo findings have been confirmed in cultured cells and are related induction of CYP2E1-dependent oxidative stress [6–8]. The proposed mechanism for this suppression appears to be via oxidative modification of proteasomal subunits with consequent reduction in 20S proteasome catalytic core activity [1, 2, 9, 10]. Previously, we demonstrated that hepatoma cells that express the ethanol-metabolizing enzymes, cytochrome P450 2 E1 (CYP2E1) and alcohol dehydrogenase (ADH), exhibit decreased chymotrypsin-like and trypsin-like proteasome activities after ethanol exposure. We further showed that simultaneous treatment with 4-methyl pyrazole (4MP), an inhibitor of ethanol metabolism, prevented the ethanol-elicited decrease in proteasome activity, indicating that ethanol metabolism is required for proteasome inhibition [4, 11]. However, despite oxidative stress being a significant component of ethanol-induced proteasome suppression, treatment with antioxidants does not fully restore proteasome activity suggesting that additional mechanisms are involved in the regulation of proteasome function [12].
We have performed studies with mouse hepatocytes and found that betaine, a methyl group donor that generates SAM and removes SAH to promote methylation reactions, can also alleviate ethanol-elicited proteasome suppression (unpublished observation). Further, ethanol-induced Mallory body formation (the morphological hallmark of alcoholic liver disease, which is induced in liver cells by proteasome inhibition) is also prevented by treatment of hepatocytes with SAM, a universal methylating agent [13, 14]. These observations led us to hypothesize that impaired protein methylation reaction(s) contribute to the suppression of proteasome function.
Ethanol administration causes many defects in the methionine metabolic pathway that result in accumulation of intracellular S-adenosylhomocysteine (SAH) [15]. In turn, this induces a decrease in hepatocellular SAM:SAH ratios that negatively affect the activities of many SAM-specific liver methyltransferases critical to cellular functions [16–18]. However, in addition to compromising methionine metabolism, ethanol generates oxidative stress, which is difficult to dissect from methylation-related events. For this reason, we assessed whether the specific methylation reactions inhibitor, tubercidin, regulates proteasome activity. Tubercidin causes accumulation of S-adenosylhomocysteine (SAH) by blocking S-adenosylhomocysteine hydrolase (SAHH) activity, thereby mimicking ethanol-induced alterations in many crucial methylation reactions. Here, we report that tubercidin suppressed 20S proteasome activity and that this suppression was related to decreased intracellular SAM:SAH ratios. This is a novel mechanism of regulation of 20S proteasome activity that occurs independent of ethanol-induced oxidative stress [1, 2, 9, 10] and decreased methylation of DNA/histones as reported previously [19, 20].
Materials and Methods
High glucose Dulbecco’s minimal essential medium (DMEM), Ham’s F12 Medium, fetal bovine serum (FBS) and blasticidin were purchased from Invitrogen (Carlsbad, CA). Suc-LLVY-AMC fluorogenic substrate and all other analytical grade quality reagents were from Sigma (St. Louis, MO).
Cells and treatments
Huh7 cells were stably transfected with CYP2E1 plasmid as described [21] and were incubated in DMEM supplemented with 10% fetal bovine serum (FBS), 5 μg blasticidin/ml, 100 U penicillin/ml and 100 μg streptomycin/ml in the presence or absence of 10 μM tubercidin for 18h.
Hepatocytes were isolated from livers of C57Bl/6 mice and plated onto collagen-coated 6-well plates. These cells were incubated in William’s E-medium containing 5% FBS in the presence or absence of 2.5 μM tubercidin for 18 hrs. Because hepatocytes are more sensitive than Huh7 cells to tubercidin exposure, they were exposed to the lower dose of this compound. Mouse hepatocytes or prepared cytosols were also exposed to SAM and SAH at ratios of 5 or 2.5, which correspond to the ratios observed in hepatocytes or livers of animals fed control and ethanol diet, respectively [15, 16]. The varying SAM:SAH ratios were achieved by adding increasing amounts of SAH to a fixed amount of SAM (1 mM) in the incubation medium.
Cell viability/death
Cells were stained with propidium iodide (PI) and viability determined by flow cytometry according to kit instructions (BD Biosciences, San Jose, CA).
Reactive oxygen species (ROS)
ROS were measured by 2′7′dichlorodihydrofluorescein diacetate (DCDFA) [22]. Data are expressed as DCFDA units (fluorescence detected at an excitation of 485 nm and an emission of 530 nm).
Cytochrome P450 2E1 (CYP2E1) activity
This activity was measured in whole cell lysates by the formation of 4-nitrocatechol (4NC) as described [11]. CYP2E1 specific activity is expressed as units (nmoles 4NC/hr) per mg protein.
Protein carbonyl levels
These were measured by Western blot as specified in the instructions to an Oxyblot kit (Millipore) after modification of proteins with 2,4, dinitrophenylhydrazine.
Lipid peroxides
These were determined as Thiobarbituric Acid Reactive Substances (TBARS) and measured according to manufacturer’s instructions (Cayman Chemical Company, Ann Arbor, MI).
20S proteasome activity
Proteasome chymotrypsin-like (Cht-L) activity was detected in vitro using the fluorogenic substrate Suc-LLVY-AMC [12].
Subcellular fractions
Cell lysates or liver homogenates were subjected to differential centrifugations to prepare the nuclear and cytosolic extracts [23].
Western Blotting
Purified 20S proteasome (Boston Biochem) was immunoblotted and assessed for methyl arginine and methyl lysine content using appropriate antibodies (Abcam). In addition, the 20 S proteasome was incubated for 15 min in buffers with SAM:SAH ratios of 5 or 2.5 and then immunoblotted and probed with antibody to methyl lysine.
Statistical Analyses
Data are expressed as mean values ± standard deviation from at least 3 experiments/replicates. Multiple comparisons for significance were determined by one-way ANOVA, using a Tukey post-hoc test. Comparison between two groups used Student’s t-test. A probability value of 0.05 or less was considered significant.
Results
To determine whether ethanol treatment affects proteasome activity in a methylation-dependent manner, we exposed mouse hepatocytes to 50 mM ethanol in the presence or absence of 0.1 mM SAM. Proteasome activity was inhibited by ethanol exposure, and this loss was prevented by the inclusion of SAM (Fig. 1A). Ethanol-elicited suppression of proteasome activity was mimicked by exposure of hepatocytes to medium containing SAM and SAH at ratios of 2.5 which correspond to the ratios observed in hepatocytes or livers of ethanol-fed animal [15, 16].
Fig. 1. Effects of impaired methylation/methylation inhibitor on proteasome activity in hepatocytes and Huh7CYP cells.
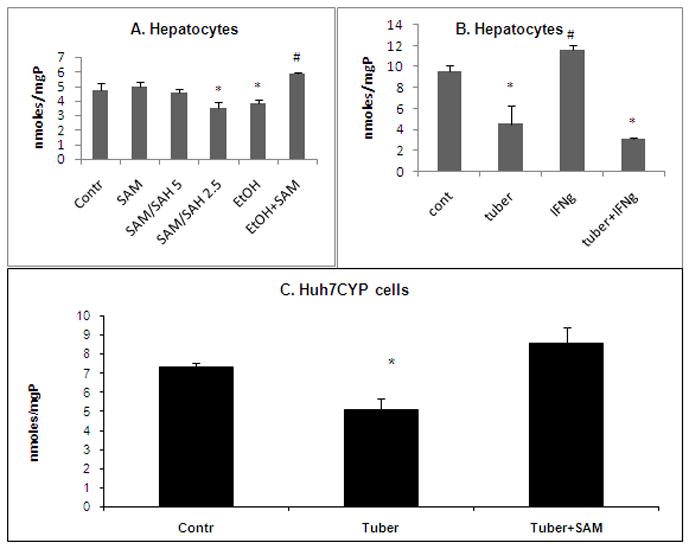
A. Proteasome activity in lysates of hepatocytes following an 18h exposure to 0.1 mM SAM, SAM:SAH ratios 5, 2.5, 50 mM ethanol or 50 mM ethanol+0.1 mM SAM. * is p<0.05 difference in proteasome activity of control vs treated cells; # between ethanol and ethanol+SAM– treated hepatocytes. B. Proteasome activity in lysates of hepatocytes following an 18h exposure to tubercidin (2.5 μM), in the presence or absence of IFNγ (5 nM). * is p<0.05 between proteasome activity in control and tubercidin-treated cells; # is p<0.05 between proteasome activity in IFNγ-treated cells and IFNγ+tubercidin or control cells. C. Proteasome activity in lysates of Huh7CYP cells following exposure to 10 μM tubercidin in the presence or absence of 0.1 mM SAM for 24 hr. * is p<0.05 is difference in proteasome activity between tubercidin-treated cells and other conditions.
Exposure of mouse hepatocytes to tubercidin, a protein methylation reactions inhibitor, suppressed proteasome activity, and this effect was more prominent in IFNγ-pretreated than in untreated cells (53% vs 75% inhibition, respectively, p< 0.05; Fig. 1B). Tubercidin exposure to Huh7CYP cells caused similar inhibition of proteasome activity (Fig. 1C). The doses of tubercidin used for these two cell types produced no toxic effects as revealed from PI staining (not shown).
To determine whether tubercidin affects proteasome activity secondary to induction of oxidative stress, we measured the levels of reactive oxygen species (ROS) production, lipid peroxides, protein carbonyls and total glutathione levels in control and tubercidin-treated HuhCYP cells. Remarkably, while tubercidin treatment elevated CYP2E1 activity two-fold (Fig. 2A), it did not enhance ROS production (Fig. 2B), elevate lipid peroxidation (Fig. 2C), increase protein carbonyls (not shown) or cause a decrease in total glutathione levels (Fig. 2D). Instead, tubercidin exposure suppressed ROS formation (Fig. 2B).
Fig. 2. Effect of tubercidin treatment on indices of oxidative stress.
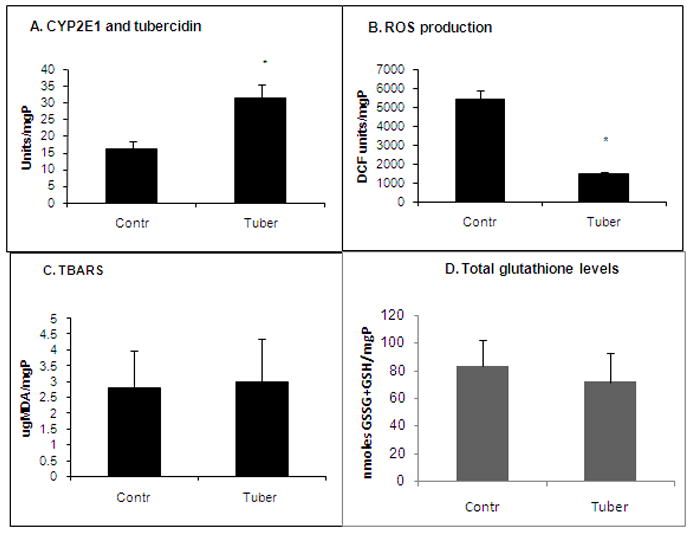
Huh7CYP cells were exposed to tubercidin (10 μM, 24 hr) and then the indices of oxidative stress were measured as indicated in Materials and Methods. * is p<0.05 difference between control and tubercidin treatments.
Both nuclear and cytosolic proteasome activities were lower in 24h tubercidin-treated Huh7CYP cells with a larger reduction in nuclear proteasome (3-fold vs 40%) compared with cytosolic proteasome (Fig. 3A). Furthermore, to exclude the involvement of altered histone or DNA methylation in proteasome suppression by tubercidin, cytosols prepared from livers of chow-fed mice were incubated for 4h in vitro in the presence of varying SAM:SAH ratios (achieved by altering the relative concentration of these two metabolites in the incubation medium). We observed a direct relationship between the SAM:SAH ratios and proteasome activity; i.e. a decrease in the ratio resulted in a corresponding decrease in enzyme activity (Fig. 3B). These results indicated that impaired protein methylation may directly regulate the function of the cytosolic proteasome without the involvement of impaired DNA or histone methylation.
Fig. 3. Effects of tubercidin and SAM:SAH ratios on proteasome activity in cytosol and nuclear fractions.
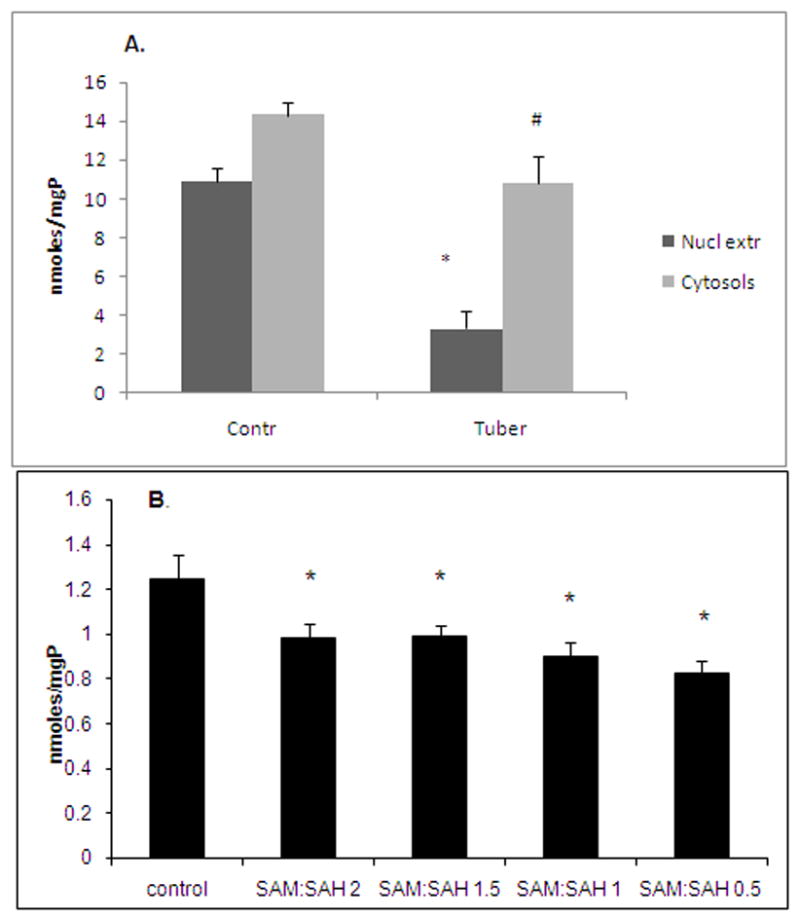
A. Huh7CYP cells were exposed for 24h to 10 μM tubercidin and then proteasome activity was measured in the prepared cytosolic and nuclear fractions. * is p<0.05 in proteasome activity in nuclear extracts, controls vs tubercidin; # is p<0.05 in proteasome activity in cytosols, control vs tubercidin. B. proteasome activity in liver cytosols exposed to low SAM:SAH ratio (2) for 4 hr. * is p<0.05 in proteasome activity between control and treatments.
To determine the interaction of the proteasome with numerous methylation-sensitive cytosolic proteins that, in turn, may regulate proteasome activity, highly purified 20S proteasome (Boston Biochem) was similarly exposed to varying SAM:SAH ratios. Consistent with our findings with the cytosolic proteasome, the purified 20S proteasome also showed a corresponding decrease in activity after exposure to decreasing ratios of SAM:SAH (Fig. 4A). Similarly, a purified immunoproteasome preparation, IPr (Boston Biochem) treated in the same way as the constitutive 20S proteasome, also revealed enzyme inhibition at a SAM:SAH ratio of 2.5 that were even more inhibitory than that observed with 20S proteasome (Fig. 4B). Because in liver cells, the expression of immunoproteasome subunits is high even in the absence of IFNγ stimulation [2], liver proteasome is more sensitive to impaired methylation reactions.
Fig. 4. Effects of various SAM:SAH ratios on proteasome activity in purified preparations of A.
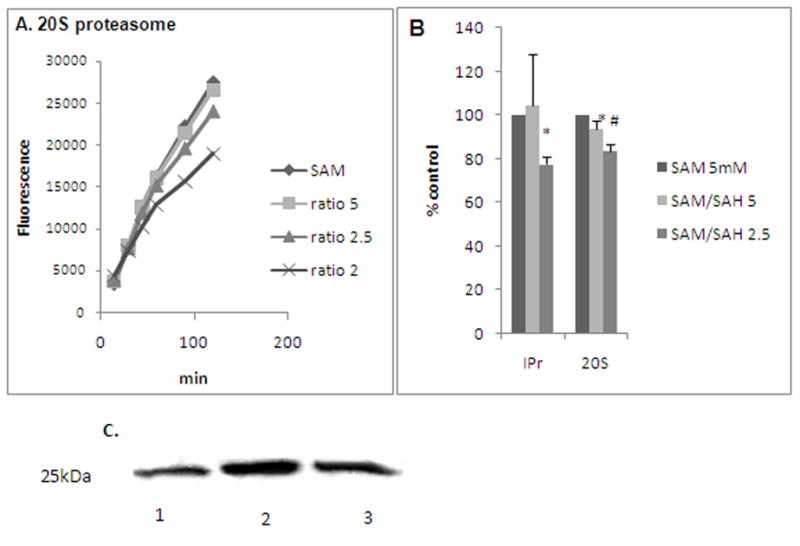
20S proteasome (representative experiment) and B. 20S proteasome vs IPr (immunoproteasome). 20S proteasome or IPr was incubated for 2h in buffers with varying SAM:SAH ratios and then proteasome activity was measured as detailed. * is p<0.05 difference in the activity between 20S proteasome and IPr and # is p<0.05 between control and other treatments (shown as % control, where the activity of both 20S proteasome and IPr in control was considered as 100%). C. Expression of methyl lysine in 20S proteasome under various SAM:SAH ratios. Purified 20S proteasome was incubated for 15 min in buffers with SAM:SAH ratio 5 or 2.5 and then immunoblotted and probed with antibody to methyl lysine. A 25kDa proteasome subunit showed positive reactivity with anti-methyl lysine. 1-no treatment; 2- treated with SAM:SAH ratio 5; 3-treated with SAM:SAH ratio 2.5.
Exposure of purified 20S proteasome to tubercidin alone did not affect chymotrypsin-like activity of the 20S proteasome (data not shown). This finding is important as it indicated that it is the change in SAM:SAH ratios by tubercidin treatment of hepatic cells that affects proteasome function.
To examine whether the purified 20S proteasome is methylated, the commercially-prepared 20S enzyme preparation was subjected to SDS-PAGE, immunoblotted and probed with antibodies to methyl arginine and methyl lysine. A 25kDa proteasome subunit showed positive reactivity with anti-methyl lysine (but not with anti-methyl agrinine). The anti-methyl lysine reactivity was increased when proteasome was incubated at a SAM;SAH ratio of 5; a reduction in reactivity was observed when the ratio was 2.5 (Fig. 4C).
Discussion
Our current understanding of how ethanol regulates proteasome activity is based on studies conducted in ethanol-metabolizing liver cells, which revealed that the ethanol-induced decrease in proteasome activity is due to inhibition by ethanol metabolites, some of which directly forms adducts with proteasomal subunits [5, 24, 25]. In addition, some studies have indicated a link between ethanol-elicited impaired methylation of histones and nuclear proteasome function [26]. However, it was not clear from these studies whether hypomethylation of DNA and/or aberrant histone methylation induces the suppression of proteasome activity or altered methionine metabolism directly regulates proteasome function.
McClain et al postulated a correlation between a decrease in proteasome activity and lower SAM levels in alcoholic liver disease (ALD) [27], but the exact link between these events was not established. Here, we provide evidence that ethanol-induced alteration in methionine metabolism that results in decreased intracellular SAM:SAH ratios directly suppressed 20S proteasome in liver cells. To our knowledge, this is the first indication for a role of impaired methylation reactions as an important factor in the regulation of liver proteasome activity. Here, we report that cultured mouse hepatocytes exposed to a SAM:SAH ratio of 2.5 (as previously seen in hepatocytes and livers of ethanol-fed animals) exhibited decreased proteasome activity compared with cells exposed to a SAM:SAH ratio of 5 (as seen in controls). Further, ethanol-elicited inhibition of proteasome activity was blocked by overnight exposure of hepatocytes to ethanol and 0.1 mM SAM, indicating that ethanol-elicited suppression of proteasome activity can be prevented by this methyl donor. Furthermore, in hepatocyte-like Huh7CYP cells, proteasome was suppressed by exposure to the specific methylation reaction inhibitor, tubercidin, and this effect was also blocked by co-incubation with SAM. Interestingly, proteasome inhibition under hypomethylation conditions seems to be a liver-specific as other investigators reported lower proteasome activity in vascular smooth muscle cells exposed to SAM [28].
The importance of a “normal” SAM:SAH ratio for proteasome function was further validated by experiments, in which we observed an inhibitory effect of tubercidin on proteasome activity in Huh7CYP cells. In addition, it is important to note that because CYP2E1 is a proteasome substrate [29, 30], the tubercidin-mediated increase in CYP2E1 activity, possibly due to CYP2E1 stabilization, could potentially induce oxidative stress. Nevertheless, despite the increase in CYP2E1 activity, tubercidin treatment evidently suppressed oxidative stress indices examined in this study.
Further, we found that tubercidin treatment suppressed proteasome activity in both nuclear and cytosolic fractions of Huh7CYP cells. However, we cannot exclude a possibility that cytosolic proteasome activity was reduced due to lower expression of proteasome components because of impaired gene methylation. To ascertain this, crude cytosolic proteasome from mouse hepatocytes was exposed to varying SAM:SAH ratios. Consistent with the effects observed on proteasome activity in cells exposed to tubercidin, we found that proteasome activity in cytosol was also suppressed when the SAM:SAH ratio was less than 2.5. The latter data suggests that some cytosolic proteins possess SAM-dependent methyltransferase activity, are regulated by altered SAM:SAH ratios and may directly modulate proteasome activity.
The observation that purified 20S proteasome when exposed to SAM:SAH of 2.5 or less possesses significant reduced activity is also noteworthy. In addition, we found that a 25 kDa subunit of purified 20S proteasome preparation exhibited immunoreactivity to antibody against methyl lysine. This is consistent with a recent proteome study that revealed the hepatic 20S proteasome subunits are arginine and lysine methylated [31]. Further, we observed that differential SAM:SAH ratios regulated methylation of lysine residues on the 25 kDa proteasome subunit. This suggests that either the 20S proteasomal subunit(s) and/or other proteins that form a tight complex and are co-purified with commercially obtained 20S proteasome have (i) a SAM-dependent methyltransferase-like activity and; (ii) are sensitive to SAH inhibition.
Although SAM:SAH ratio-induced changes in proteasome activity were modest, this level of proteasome inhibition does result in significant alterations of downstream cell functions including peptide hydrolysis for antigen presentation [32]. Further, the modest decreases in proteasome activity ensure that the viability of treated cells is maintained and that the results are not due to cell death since a profound inhibition of proteasome would likely cause apoptosis/necrosis. Thus, our data indicate that in addition to previously reported methylation-mediated regulation of proteasome activity in cells at the epigenetic level [19, 20], proteasome function is also regulated at the level of SAM:SAH-dependent methylation reactions.
Further, we observed greater inhibition of activity of IPr-enriched proteasome preparation with decreased SAM:SAH compared with 20S constitutive proteasome. This observation is consistent with higher sensitivity of IFNγ-treated hepatocytes to tubercidin compared with untreated cells (Fig. 1B). These findings suggest that there may be an increased association of a lysine methyltransferase (and probably some others) with immunoproteasome subunits. However, establishment of the identity of such methyltransferases that regulate proteasome function is currently under investigation.
Thus, ethanol exposure to liver cells dually regulates proteasome activity: by ethanol-induced oxidative stress [1, 2, 12] and by ethanol-induced defects in protein methylation reactions. The reduction in proteasome activity has serious implications in many important physiologically liver cell functions, such as protein aggregation (Mallory body formation) and antigen presentation. The fact that methylation defects can be corrected by treatment with regulators of methylation, like SAM or betaine, provides another therapeutic target of maintaining proteasome function and protecting against the development of liver injury from alcohol abuse.
We conclude that in liver cells, impaired methylation reactions directly suppress proteasome function, providing a novel mechanism for regulation of 20S proteasome activity.
Acknowledgments
This study is supported by Department of Veterans Affairs National Merit Review grant (KKK), R21AA017296 (KKK) and R21 AA017232 (NAO) from the National Institute of Alcohol Abuse and Alcoholism.
List of abbreviations
- MHC
major histocompatibility complex
- CYP2E1
cytochrome P450 2E1
- ADH
alcohol dehydrogenase
- SAM
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
- TBARS
Thiobarbituric Acid Reactive Substances
- Cht-L
proteasome chymotrypsin-like activity
- ROS
reactive oxygen species
- IFNγ
interferon gamma
- IPr
immunoproteasome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Donohue TM, Jr, Cederbaum AI, French SW, Barve S, Gao B, Osna NA. Role of the proteasome in ethanol-induced liver pathology. Alcohol Clin Exp Res. 2007;31:1446–1459. doi: 10.1111/j.1530-0277.2007.00454.x. [DOI] [PubMed] [Google Scholar]
- 2.Osna NA, Donohue TM., Jr Implication of altered proteasome function in alcoholic liver injury. World J Gastroenterol. 2007;13:4931–4937. doi: 10.3748/wjg.v13.i37.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL, Rock KL. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 4.Osna NA, White RL, Todero S, McVicker BL, Thiele GM, Clemens DL, Tuma DJ, Donohue TM., Jr Ethanol-induced oxidative stress suppresses generation of peptides for antigen presentation by hepatoma cells. Hepatology. 2007;45:53–61. doi: 10.1002/hep.21442. [DOI] [PubMed] [Google Scholar]
- 5.Bardag-Gorce F, Li J, French BA, French SW. The effect of ethanol-induced CYP2E1 on proteasome activity: the role of 4-hydroxynonenal. Exp Mol Pathol. 2005;78:109–115. doi: 10.1016/j.yexmp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Osna N, Clemens D, Donohue TM., Jr Ethanol suppresses interferon gamma-mediated activation of the proteasome by preventing STAT1 phosphorylation in human recombinant Hep G2 cells. Hepatology. 2002;36:334A. (Abstract No. 684). [Google Scholar]
- 7.Bardag-Gorce F, Yuan QX, Li J, French BA, Fang C, Ingelman-Sundberg M, French SW. The effect of ethanol-induced cytochrome p4502E1 on the inhibition of proteasome activity by alcohol. Biochem Biophys Res Commun. 2000;279:23–29. doi: 10.1006/bbrc.2000.3889. [DOI] [PubMed] [Google Scholar]
- 8.Donohue TM, Osna NA, Clemens DL. Recombinant Hep G2 cells that express alcohol dehydrogenase and cytochrome P450 2E1 as a model of ethanol-elicited cytotoxicity. Int J Biochem Cell Biol. 2006;38:92–101. doi: 10.1016/j.biocel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Farout L, Mary J, Vinh J, Szweda LI, Friguet B. Inactivation of the proteasome by 4-hydroxy-2-nonenal is site specific and dependant on 20S proteasome subtypes. Arch Biochem Biophys. 2006;453:135–142. doi: 10.1016/j.abb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Ishii T, Sakurai T, Usami H, Uchida K. Oxidative modification of proteasome: identification of an oxidation-sensitive subunit in 26 S proteasome. Biochemistry. 2005;44:13893–13901. doi: 10.1021/bi051336u. [DOI] [PubMed] [Google Scholar]
- 11.Osna NA, Clemens DL, Donohue TM., Jr Interferon gamma enhances proteasome activity in recombinant Hep G2 cells that express cytochrome P4502E1: modulation by ethanol. Biochem Pharmacol. 2003;66:697–710. doi: 10.1016/s0006-2952(03)00252-1. [DOI] [PubMed] [Google Scholar]
- 12.Osna NA, White RL, Krutik VM, Wang T, Weinman SA, Donohue TM., Jr Proteasome activation by hepatitis C core protein is reversed by ethanol-induced oxidative stress. Gastroenterology. 2008;134:2144–2152. doi: 10.1053/j.gastro.2008.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Bardag-Gorce F, Dedes J, French BA, Amidi F, Oliva J, French SW. S-adenosylmethionine prevents Mallory Denk body formation in drug-primed mice by inhibiting the epigenetic memory. Hepatology. 2008;47:613–624. doi: 10.1002/hep.22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bardag-Gorce F, Dedes J, French BA, Oliva JV, Li J, French SW. Mallory body formation is associated with epigenetic phenotypic change in hepatocytes in vivo. Exp Mol Pathol. 2007;83:160–168. doi: 10.1016/j.yexmp.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barak AJ, Beckenhauer HC, Mailliard ME, Kharbanda KK, Tuma DJ. Betaine lowers elevated s-adenosylhomocysteine levels in hepatocytes from ethanol-fed rats. J Nutr. 2003;133:2845–2848. doi: 10.1093/jn/133.9.2845. [DOI] [PubMed] [Google Scholar]
- 16.Kharbanda KK. Role of transmethylation reactions in alcoholic liver disease. World J Gastroenterol. 2007;13:4947–4954. doi: 10.3748/wjg.v13.i37.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kharbanda KK, Rogers DD, 2nd, Mailliard ME, Siford GL, Barak AJ, Beckenhauer HC, Sorrell MF, Tuma DJ. Role of elevated S-adenosylhomocysteine in rat hepatocyte apoptosis: protection by betaine. Biochem Pharmacol. 2005;70:1883–1890. doi: 10.1016/j.bcp.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 18.Kharbanda KK, Todero SL, Ward BW, Cannella JJ, 3rd, Tuma DJ. Betaine administration corrects ethanol-induced defective VLDL secretion. Mol Cell Biochem. 2009 doi: 10.1007/s11010-009-0044-2. [DOI] [PubMed] [Google Scholar]
- 19.Bardag-Gorce F. Nuclear effects of ethanol-induced proteasome inhibition in liver cells. World J Gastroenterol. 2009;15:1163–1167. doi: 10.3748/wjg.15.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliva J, Dedes J, Li J, French SW, Bardag-Gorce F. Epigenetics of proteasome inhibition in the liver of rats fed ethanol chronically. World J Gastroenterol. 2009;15:705–712. doi: 10.3748/wjg.15.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCartney EM, Semendric L, Helbig KJ, Hinze S, Jones B, Weinman SA, Beard MR. Alcohol Metabolism Increases the Replication of Hepatitis C Virus and Attenuates the Antiviral Action of Interferon. J Infect Dis. 2008;198:1766–1775. doi: 10.1086/593216. [DOI] [PubMed] [Google Scholar]
- 22.Otani K, Korenaga M, Beard MR, Li K, Qian T, Showalter LA, Singh AK, Wang T, Weinman SA. Hepatitis C virus core protein, cytochrome P450 2E1, and alcohol produce combined mitochondrial injury and cytotoxicity in hepatoma cells. Gastroenterology. 2005;128:96–107. doi: 10.1053/j.gastro.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 23.McMullen MR, Pritchard MT, Wang Q, Millward CA, Croniger CM, Nagy LE. Early growth response-1 transcription factor is essential for ethanol-induced fatty liver injury in mice. Gastroenterology. 2005;128:2066–2076. doi: 10.1053/j.gastro.2005.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kessova IG, Cederbaum AI. The effect of CYP2E1-dependent oxidant stress on activity of proteasomes in HepG2 cells. J Pharmacol Exp Ther. 2005;315:304–312. doi: 10.1124/jpet.105.088047. [DOI] [PubMed] [Google Scholar]
- 25.Osna NA, Haorah J, Krutik VM, Donohue TM., Jr Peroxynitrite alters the catalytic activity of rodent liver proteasome in vitro and in vivo. Hepatology. 2004;40:574–582. doi: 10.1002/hep.20352. [DOI] [PubMed] [Google Scholar]
- 26.Bardag-Gorce F, Oliva J, Villegas J, Fraley S, Amidi F, Li J, Dedes J, French B, French SW. Epigenetic mechanisms regulate Mallory Denk body formation in the livers of drug-primed mice. Exp Mol Pathol. 2008;84:113–121. doi: 10.1016/j.yexmp.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClain C, Barve S, Joshi-Barve S, Song Z, Deaciuc I, Chen T, Hill D. Dysregulated cytokine metabolism, altered hepatic methionine metabolism and proteasome dysfunction in alcoholic liver disease. Alcohol Clin Exp Res. 2005;29:180S–188S. doi: 10.1097/01.alc.0000189276.34230.f5. [DOI] [PubMed] [Google Scholar]
- 28.D’Anselmi F, Cucina A, Cavallaro G, Bizzarri M, Cavallaro RA, Fuso A, Cavallaro A, Scarpa S. S-adenosylmethionine inhibits ubiquitin-proteasome system in vitro and on rat vascular smooth muscle cells. Protein Pept Lett. 2008;15:58–62. doi: 10.2174/092986608783330396. [DOI] [PubMed] [Google Scholar]
- 29.Goasduff T, Cederbaum AI. CYP2E1 degradation by in vitro reconstituted systems: role of the molecular chaperone hsp90. Arch Biochem Biophys. 2000;379:321–330. doi: 10.1006/abbi.2000.1870. [DOI] [PubMed] [Google Scholar]
- 30.Goasduff T, Cederbaum AI. NADPH-dependent microsomal electron transfer increases degradation of CYP2E1 by the proteasome complex: role of reactive oxygen species. Arch Biochem Biophys. 1999;370:258–270. doi: 10.1006/abbi.1999.1399. [DOI] [PubMed] [Google Scholar]
- 31.Gomes AV, Young GW, Wang Y, Zong C, Eghbali M, Drews O, Lu H, Stefani E, Ping P. Contrasting proteome biology and functional heterogeneity of the 20 S proteasome complexes in mammalian tissues. Mol Cell Proteomics. 2009;8:302–315. doi: 10.1074/mcp.M800058-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osna NA. Hepatitis C virus and ethanol alter antigen presentation in liver cells. World J Gastroenterol. 2009;15:1201–1208. doi: 10.3748/wjg.15.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]


